Abstract
Bacillus subtilis mutants with high expression of the bacilysin operon ywfBCDEFG were isolated. Comparative genome sequencing analysis revealed that all of these mutants have a mutation in the scoC gene. The disruption of scoC by genetic engineering also resulted in increased expression of ywfBCDEFG. Primer extension and gel mobility shift analyses showed that the ScoC protein binds directly to the promoter region of ywfBCDEFG. Our results indicate that the transition state regulator ScoC, together with CodY and AbrB, negatively regulates bacilysin production in B. subtilis.
Gram-positive model bacterium Bacillus subtilis produces the dipeptide antibiotic bacilysin, which consists of an l-alanine and an unusual amino acid, l-anticapsin (15). We previously reported that a polycistronic operon, ywfBCDEFG, and a monocistronic gene, ywfH, are required for bacilysin production (7). The gene products of ywfB and ywfG are thought to participate in the l-anticapsin biosynthesis pathway, while the ywfE gene product has been assigned as an amino acid ligase involved in alanine-anticapsin ligation (14). The protein encoded by the ywfF gene is necessary for self-protection against bacilysin (13). Thus, the ywfBCDEFG operon has an obligate role in bacilysin production.
We previously showed that a certain rifampin (rifampicin) resistance mutation can activate the B. subtilis dormant secondary metabolism, neotrehalosadiamine (3,3′-diamino-3,3′-dideoxy-α,β-trehalose) synthesis (8). Subsequently, we attempted to activate bacilysin production in the same way. Unexpectedly, we found that the expression of the bacilysin operon ywfBCDEFG was induced by a mechanism independent of the rifampin resistance mutation. Although the expression of the bacilysin operon ywfBCDEFG was previously reported to be negatively regulated by transition state regulators CodY (7) and AbrB (11), the mechanism we found was apparently different from these known mechanisms. Here, we report a novel regulatory mechanism involved in bacilysin production.
Isolation of mutants with marked activation of bacilysin operon promoter.
The B. subtilis strains used in this study are listed in Table 1. To screen the bacilysin-overproducing mutants, a reporter strain TI340 (codY::neo ywfB::pMutinT3), which carries the reporter ywfB′-lacZ, was used. Because this strain does not form blue colonies on L agar plates (7) containing 0.008% 5-bromo-4-chloro-3-indolyl β-d(−)-galactopyranoside (X-Gal), the mutants with enhanced expression of the reporter gene should be distinguishable as blue colonies. A number of spontaneous rifampin-resistant colonies (ca. 10,000 colonies) developed when strain TI340 was plated and incubated on L agar plates containing various concentrations of rifampin (1 to 50 μg/ml) and X-Gal. Among these mutants, eight were found to form a blue colony and were designated RIF1 to RIF8. Backcross transformation analysis, however, showed that all rifampin resistance mutations (rif) were genetically distinct from the mutation leading to the activation of the bacilysin operon promoter (bachy-1 to -8) (data not shown). Because AbrB has been reported to negatively regulate bacilysin production, we sequenced the abrB gene of each chromosomal DNA extracted from RIF1 to RIF8. However, no mutations were found in this gene, indicating that the activation mechanism we found is independent of the known mechanisms mediated by AbrB (and CodY). For further analysis, we constructed strains BAC1 to BAC8 by transforming strain TI336 (codY::neo) with each chromosomal DNA from RIF1 to RIF8 (codY::neo ywfB::pMutinT3 rif bachy-1 to -8) and selecting erythromycin-resistant blue colonies. These transformants showed a significant increase in the promoter activity of the bacilysin operon, indicating that they each contain a bachy mutation (data not shown).
TABLE 1.
Strains used in this study
| Strain | Genotype | Constructiona or source |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| TI94 | trpC2 aspB66 ywfA::neo ywfE179 | 7 |
| TI95 | trpC2 aspB66 codY::neo ywfB::pMutinT3 | 7 |
| TI112 | trpC2 aspB66 ywfH::pMutinT3 | 7 |
| TI294 | trpC2 codY::erm | pCR2.1-codY::erm→168 |
| TI296 | trpC2 abrB::neo | 9 |
| TI298 | trpC2 ΔscoC::spc | pCR2.1-scoC::spc→168 |
| TI302 | trpC2 ΔscoC::spc codY::erm | TI294→TI298 |
| TI303 | trpC2 ΔscoC::spc abrB::neo | TI296→TI298 |
| TI304 | trpC2 ΔscoC::spc abrB::neo codY::erm | TI303→TI302 |
| TI336 | trpC2 codY::neo | TI95→168 |
| TI338 | trpC2 ywfB::pMutinT3 | TI95→168 |
| TI340 | trpC2 codY::neo ywfB::pMutinT3 | TI95→TI336 |
| TI342 | trpC2 ΔscoC::spc ywfB::pMutinT3 | TI298→TI338 |
| TI351 | trpC2 amyE::(PywfB-lacZ cat) | pDL2-PywfB200→168 |
| TI373 | trpC2 ΔscoC::spc amyE::(PywfB-lacZ cat) | TI351→TI298 |
| TI375 | trpC2 ΔscoC::spc abrB::neo amyE::(PywfB-lacZ cat) | TI351→TI303 |
| TI377 | trpC2 ΔscoC::spc codY::erm amyE::(PywfB-lacZ cat) | TI351→TI302 |
| TI381 | trpC2 ΔscoC::spc abrB::neo codY::erm amyE::(PywfB-lacZ cat) | TI351→TI304 |
| TI398 | trpC2 ywfA::neo ywfE179 | TI94→168 |
| TI399 | trpC2 ΔscoC::spc ywfA::neo ywfE179 | TI298→ TI398 |
| TI401 | trpC2 ywfH::pMutinT3 | TI112→168 |
| TI402 | trpC2 ΔscoC::spc ywfH::pMutinT3 | TI298→TI401 |
Arrows indicate transformation of the latter strain by DNA prepared from the former strain.
Identification of bachy mutations.
To identify the bachy mutations, we conducted a comparative genome sequencing analysis (1) using the genomic DNA of the BAC1 mutant and the TI340 strain. As a result, the BAC1 mutant was found to carry an amber mutation at the 38th codon of the scoC gene (Table 2). Strikingly, all other mutants were also found to have a mutation within the same gene, as determined by DNA sequencing. The BAC2, -3, -5, -6, -7, and -8 mutants had a mutation causing an amino acid alteration at Ala26, Lys94, Arg153, Phe164, Asp98, or Val15, respectively. The BAC4 mutant exhibited a nonsense mutation at the 112th codon, producing a C-terminally truncated protein. The ScoC protein belongs to the MarR family of transcriptional regulatory proteins, which contains a “winged-helix” DNA binding motif in the central domain, and forms a tetramer in solution as deposited in the Protein Data Bank (identification number 2fxa). On the basis of its crystal structure, residues Lys94 and Asp98 are located in the DNA binding motif. Therefore, it is likely that the mutations, Lys94 to Gln and Asp98 to Asn, affect the DNA binding activity. Our results also suggest that both N and C termini of this protein are required for its activity. Residues Val15, Ala26, Arg153, and Phe164 may be necessary for its tetramerization.
TABLE 2.
Mutations found in scoC gene
| Strain | Position of mutationa | Amino acid substitution |
|---|---|---|
| TI340 | —b | — |
| BAC1 | G114 → A | Trp38 → Amber |
| BAC2 | C77 → A | Ala26 → Asp |
| BAC3 | A280 → C | Lys94 → Gln |
| BAC4 | G334 →T | Glu112 → Amber |
| BAC5 | C457 → T | Arg153 → Cys |
| BAC6 | T491 → C | Phe164 → Ser |
| BAC7 | G292 → A | Asp98 → Asn |
| BAC8 | G43 → T | Val15 → Phe |
Numbering originated from the start codon of the open reading frame.
—, wild type.
ScoC negatively regulates bacilysin production.
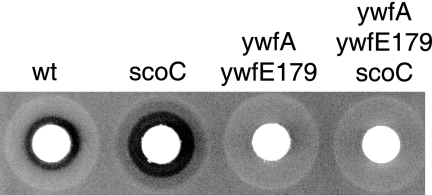
The results described above strongly suggest that ScoC negatively regulates bacilysin production. To confirm this hypothesis, we constructed a scoC disruptant (TI298) by replacing part of the scoC gene with the spectinomycin resistance cassette. A DNA fragment (1,217 bp) containing the complete coding region of the scoC gene and its flanking regions was amplified by PCR using the primer pair ΔscoC-F (5′-CGGCAAAAGAAAGCACGG-3′) and ΔscoC-R (5′-CAGGTACCCCTTCTATGCGC-3′). The amplified DNA was cloned into pCR2.1 (Invitrogen), generating pCR2.1-scoC, and fully sequenced. The spectinomycin resistance cassette was inserted into the region between SspI sites within the scoC gene on pCR2.1-scoC. The resulting plasmid, pCR2.1-ΔscoC::spc, was linearized with BamHI and transformed into B. subtilis 168 with selection for spectinomycin resistance, generating the strain TI298 (ΔscoC::spc). As expected, the disruption of scoC resulted in a significant increase in bacilysin production (Fig. 1). No stimulation of antibiotic production was detected in the genetic background of the ywfE179 strain, which lacks alanine-anticapsin ligase activity, indicating that the scoC disruption was indeed responsible for the observed bacilysin overproduction (Fig. 1).
FIG. 1.
Effect of scoC disruption on antibiotic production. Antibiotic activity was determined by the paper disk-agar diffusion assay, using Staphylococcus aureus 209P as the test organism. B. subtilis strains 168 (wt), TI298 (ΔscoC::spc), TI398 (ywfA::neo ywfE179), and TI399 (ywfA::neo ywfE179 ΔscoC::spc) were grown in S7N medium for 8 h. The culture supernatants (50 μl) obtained after centrifugation were applied onto a paper disk (diameter, 8.0 mm), and the paper disk was placed on a half-strength Mueller-Hinton agar (Difco) plate inoculated with S. aureus 209P.
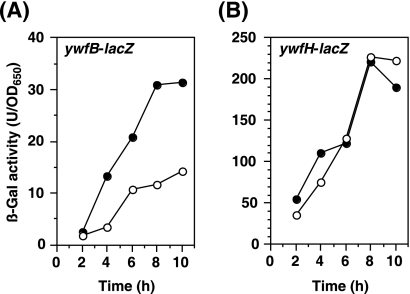
We next examined the effect of scoC disruption on the expression of the reporter genes ywfB-lacZ and ywfH-lacZ. A β-galactosidase (β-Gal) assay was performed as described previously (12). Like in the case of the spontaneous scoC mutations, the disruption of the scoC gene apparently enhanced the expression of ywfB-lacZ (Fig. 2A). In contrast, no significant increase in the expression of ywfH-lacZ was detected in the scoC disruptant, indicating that ScoC is not involved in regulation of ywfH expression (Fig. 2B). To evaluate the effect of double (scoC abrB and scoC codY) or triple (scoC abrB codY) disruption of regulators involved in regulation of bacilysin production, we constructed another reporter strain carrying the ywfB promoter fused to the lacZ gene at the amyE locus. The upstream region of ywfB (192 bp from the translation initiation codon) was amplified by PCR with the primer pair PywfB200-F (5′-GCAAACTTGAGCAGAAGGC-3′) and PywfB-R (5′-GAGCACCAACCAATCTTTTAA-3′). The amplified fragment was cloned into pCR2.1 and fully sequenced to confirm the correct sequence. The EcoRI fragment containing the upstream region of ywfB was inserted into the EcoRI site of pDL2 (4). The resulting plasmid, pDL2-PywfB200, was linearized with PstI and inserted into the amyE locus of the B. subtilis 168 chromosome, selecting for chloramphenicol resistance. The generated strain (amyE::PywfB-lacZ) was designated TI351. Similar to the results shown in Fig. 2A, the activity of ywfB promoter in a scoC disruptant was approximately threefold higher than that in its parent strain TI351 (Table 3). Double (scoC abrB and scoC codY) and triple (scoC abrB codY) mutations resulted in a further increase in the ywfB promoter activity in a stepwise manner. Thus, it is concluded that ScoC, together with CodY and AbrB, is a negative regulator of bacilysin production in B. subtilis.
FIG. 2.
Effect of scoC disruption on the expression of ywfB-lacZ and ywfH-lacZ. (A) Strains TI338 (scoC+ ywfB::pMutinT3) (open circles) and TI342 (ΔscoC::spc ywfB::pMutinT3) (closed circles) were grown in S7N medium. Culture samples were withdrawn at the indicated times, and β-galactosidase (β-Gal) activity was measured. (B) Strains TI401 (scoC+ ywfH::pMutinT3) (open circles) and TI402 (ΔscoC::spc ywfH::pMutinT3) (closed circles) were grown as described for panel A.
TABLE 3.
Effect of disruption of the transition state regulators on PywfB-lacZ expression
| Strain | Relevant genotype | β-Gal activity (U/OD650)a |
|---|---|---|
| TI351 | wtb | 16 ± 1.0 |
| TI373 | scoC | 41 ± 3.8 |
| TI375 | scoC abrB | 52 ± 3.7 |
| TI377 | scoC codY | 61 ± 3.8 |
| TI381 | scoC abrB codY | 87 ± 7.1 |
Cells were grown in S7N medium for 12 h at 37°C. β-Gal activity is expressed in units/cell density at 650 nm.
wt, wild type.
ScoC binds directly to the promoter region of the bacilysin biosynthesis operon.
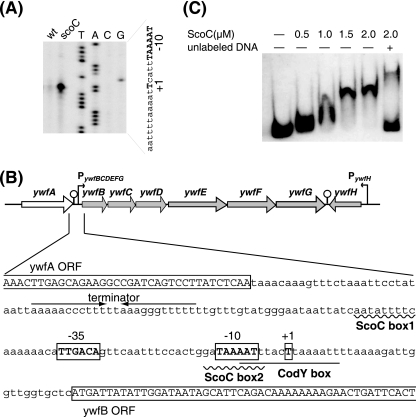
To further investigate the ScoC-mediated regulation, we determined the transcription start site of the bacilysin operon by primer extension analysis. Strains 168 and TI298 (ΔscoC::spc) were grown in S7N medium (7) until the optical density at 650 nm (OD650) reached 6.0. Total cellular RNA was prepared using Isogen reagent (Nippon Gene) as described previously (6). Total RNA (30 μg) was annealed to 1 pmol of an infrared dye-labeled primer IRD800-ywfB (5′-IRD800-GAGCGTGTTGACTGTAATGAG-3′). The primer extension reaction was conducted using SuperScript III reverse transcriptase (Invitrogen). The dideoxy sequencing reactions were also performed with the same primer. The synthesized cDNA and sequencing ladders were subjected to urea-polyacrylamide gel electrophoresis (PAGE). The analysis was performed using the DNA sequencing system LIC-4200L(S)-2 (LI-COR). As a result, we found that transcription of the bacilysin operon was initiated at the T residues 29 bases upstream of the translational start codon (Fig. 3A). It is notable that a much higher level of the ywfB transcript was detected in the scoC disruptant, TI298, than in the wild type, 168. Real-time quantitative PCR analysis confirmed that the level of ywfB transcript in the scoC disruptant is approximately fivefold higher than in its parent strain, 168 (data not shown). The most probable “−35” and “−10” regions (TTGACA and TAAAAT with a 17-bp spacer), which are likely recognized by σA-RNA polymerase (5), were found in the region upstream of the transcriptional start site (Fig. 3B). Inspection of the sequence around the transcription start site revealed the presence of two putative ScoC binding sites located between positions −50 and −42 (ScoC box1, AATATTtTC) and between positions −12 and −4 (ScoC box2, GATAAaATT) with one base mismatch (indicated with a lowercase letter) with the consensus ScoC binding motif (RATANTATY) (3). The CodY binding site, as described by Belitsky and Sonenshein (2), was also found in the region between positions −7 and +8 (CodY box, AATTTaCTtAAAATT) with two base mismatches with the consensus CodY binding motif (AATTTTCWGAAAATT). The putative binding sites, ScoC box2 and CodY box, overlap with the predicted “−10” region of the bacilysin operon promoter (Fig. 3B).
FIG. 3.
ScoC binds directly to the promoter region of the bacilysin operon. (A) Primer extension analysis of the bacilysin operon. Total RNA (30 μg) of strains 168 (wild-type) and TI298 (scoC) was reverse transcribed to generate runoff cDNA. Lanes G, A, T, and C contained the products of the dideoxy sequencing reactions, with the same primer used for reverse transcription. The partial nucleotide sequence of the coding strand corresponding to the ladders is shown, where the “−10” regions and the transcription start sites (+1) are shown by uppercase boldface letters. (B) Organization of the bacilysin operon promoter region. ScoC and CodY binding sites are indicated by a wavy line and underlined letters, respectively. The “−35” and “−10” regions and the transcription start site (+1) of the bacilysin operon promoter are shown by boxed uppercase boldface letters. The transcriptional terminator of the ywfA gene is indicated by the pair of facing arrows. The open reading frames of the ywfA and ywfB genes are labeled and boxed. (C) Gel mobility shift analysis. A DIG-labeled DNA probe containing bacilysin operon promoter (positions −163 to +29) was incubated with His10-tagged ScoC at the indicated concentrations. DNA and DNA-protein complexes were separated on 5% nondenaturing PAGE, transferred onto a membrane, and detected. The position of unshifted DNA is shown in the leftmost lane, which contained no ScoC. The rightmost lane contained 100-fold molar excess of unlabeled DNA.
To determine whether ScoC protein binds directly to the bacilysin operon promoter region, we purified a His10-tagged ScoC protein. The complete coding region of scoC was amplified with the primer pair XhoI-scoCF (5′-CTCGAGATGAATCGAGTGGAACCGC-3′) and BamHI-scoCR (5′-GGATCCTTAACTGTTTACAGGTTCGAGCTC-3′). These primers have the XhoI or BamHI site, as indicated by the underlined letters. The PCR fragment was cloned into pCR2.1 to generate pCR2.1-scoC and fully sequenced to confirm the correct sequence. An XhoI-BamHI fragment containing the entire scoC coding region was inserted into the expression vector pET19b (Novagen), generating pET19b-scoC. Escherichia coli harboring the resultant plasmid can express a His10-tagged ScoC protein, which is extended with Met-Gly-His10-Ser-Ser-Gly-His-Ile-Asp-Asp-Asp-Asp-Lys-His at the N terminus. E. coli BL21(DE3) harboring pET19b-scoC was grown to an OD650 of 1 at 37°C in L medium supplemented with 1% glucose. Isopropyl-β-d-thio-galactopyranoside (IPTG) was added to a final concentration of 2 mM, and the culture was further incubated for 3 h. E. coli cells were harvested by centrifugation and disrupted by sonication. The cell lysate was centrifuged (8,000 × g for 10 min) to remove insoluble material. The crude extract was then fractionated by 50 to 65% saturated ammonium sulfate as described previously (10). The His10-tagged ScoC protein was then purified using a HisTrap HP column (GE Healthcare) as described in the manufacturer's manual. The purified protein (ca. 95% pure) was then stored in storage buffer (20 mM sodium phosphate buffer [pH 7.5], 0.2 M KCl, and 50% glycerol).
Gel mobility shift analysis was performed using a DIG gel shift kit (Roche Diagnostics). To prepare the probe, the promoter region (192 bp) of the bacilysin biosynthesis operon was amplified with the primer pair PywfB200-F and PywfB-R. The resulting PCR product was labeled with digoxigenin (DIG) at the 3′ end as described in the manufacturer's manual. The 100-fold-diluted probe (1 μl) was incubated at 25°C for 15 min with or without His10-tagged ScoC, in 15 μl of binding buffer {20 mM HEPES (pH 7.6), 30 mM KCl, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, 0.2% Tween 20, 6.7 μg/ml poly(Lys), and 67 μg/ml poly[d(I-C)]}. DNA and DNA-protein complexes were separated by 5% nondenaturing PAGE, transferred onto a membrane (Hybond-N+; Amersham), and detected as described in the manufacturer's manual. The result indicated that the His10-tagged ScoC protein bound to the DIG-labeled probe, which contained the ywfB promoter region between positions −163 and +29, with a binding dissociation constant of 1 μM (calculated as a monomer) (Fig. 3C). This association was significantly reduced with a 100-fold molar excess of unlabeled DNA, indicating that the binding to the ywfB promoter region was specific (Fig. 3C). The band which migrates more slowly than the ScoC-probe DNA complex was also detected in the presence of an excess unlabeled probe DNA. Although the reason is still unclear, it is possible that ScoC protein forms a complex with two molecules of the target DNA (probe DNA and unlabeled probe DNA). Our results indicate that ScoC directly binds to the bacilysin operon promoter, eventually leading to repression of its transcription.
Concluding remarks.
Unlike CodY and AbrB, the importance of ScoC in the stationary-phase gene expression of B. subtilis has not been recognized fully, although ScoC has been known to function as a negative regulator of protease production and sporulation (10). Our study is the first to demonstrate unambiguously the important role (negative control) of ScoC in antibiotic production. Owing to the fact that B. subtilis is a model organism of gram-positive bacteria, our results may provide a feasible system for understanding the regulatory circuit of bacterial secondary metabolism.
Acknowledgments
We thank M. Itaya (Keio Univ.) for providing a B. subtilis strain (abrB::neo) and Roche NimbleGen, Inc., Madison, WI, for supporting the mutation search using the comparative genome sequencing technique.
This work was supported by grants from the Effective Promotion of Joint Research of Special Coordination Funds (to K.O.) and from KAKENHI no. 20780064 Grant-in-Aid for Young Scientists (B) (to T.I.).
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., and A. L. Sonenshein. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 5.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inaoka, T., T. Satomura, Y. Fujita, and K. Ochi. 2009. Novel gene regulation mediated by overproduction of secondary metabolite neotrehalosadiamine in Bacillus subtilis. FEMS Microbiol. Lett. 291:151-156. [DOI] [PubMed] [Google Scholar]
- 7.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP cooperatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 8.Inaoka, T., K. Takahashi, H. Yada, M. Yoshida, and K. Ochi. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 279:3885-3892. [DOI] [PubMed] [Google Scholar]
- 9.Itaya, M., and T. Tanaka. 1991. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J. Mol. Biol. 220:631-648. [DOI] [PubMed] [Google Scholar]
- 10.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 11.Karataş, A. Y., S. Cetin, and G. Ozcengiz. 2003. The effects of insertional mutations in comQ, comP, srfA, spo0H, spo0A and abrB genes on bacilysin biosynthesis in Bacillus subtilis. Biochim. Biophys. Acta 1626:51-56. [PubMed] [Google Scholar]
- 12.Kim, J. Y., T. Inaoka, K. Hirooka, H. Matsuoka, M. Murata, R. Ohki, Y. Adachi, Y. Fujita, and K. Ochi. 2009. Identification and characterization of a novel multidrug resistance operon, mdtRP (yusOP), of Bacillus subtilis. J. Bacteriol. 191:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinborn, G., M. R. Hajirezaei, and J. Hofemeister. 2005. bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch. Microbiol. 183:71-79. [DOI] [PubMed] [Google Scholar]
- 14.Tabata, K., H. Ikeda, and S. Hashimoto. 2005. ywfE in Bacillus subtilis codes for a novel enzyme, l-amino acid ligase. J. Bacteriol. 187:5195-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker, J. E., and E. P. Abraham. 1970. The structure of bacilysin and other products of Bacillus subtilis. Biochem. J. 118:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]