Abstract
Type IV secretion systems (T4SS) translocate DNA and protein substrates across prokaryotic cell envelopes generally by a mechanism requiring direct contact with a target cell. Three types of T4SS have been described: (i) conjugation systems, operationally defined as machines that translocate DNA substrates intercellularly by a contact-dependent process; (ii) effector translocator systems, functioning to deliver proteins or other macromolecules to eukaryotic target cells; and (iii) DNA release/uptake systems, which translocate DNA to or from the extracellular milieu. Studies of a few paradigmatic systems, notably the conjugation systems of plasmids F, R388, RP4, and pKM101 and the Agrobacterium tumefaciens VirB/VirD4 system, have supplied important insights into the structure, function, and mechanism of action of type IV secretion machines. Information on these systems is updated, with emphasis on recent exciting structural advances. An underappreciated feature of T4SS, most notably of the conjugation subfamily, is that they are widely distributed among many species of gram-negative and -positive bacteria, wall-less bacteria, and the Archaea. Conjugation-mediated lateral gene transfer has shaped the genomes of most if not all prokaryotes over evolutionary time and also contributed in the short term to the dissemination of antibiotic resistance and other virulence traits among medically important pathogens. How have these machines adapted to function across envelopes of distantly related microorganisms? A survey of T4SS functioning in phylogenetically diverse species highlights the biological complexity of these translocation systems and identifies common mechanistic themes as well as novel adaptations for specialized purposes relating to the modulation of the donor-target cell interaction.
INTRODUCTION
Type IV secretion systems (T4SS) translocate DNA and protein substrates across the cell envelope generally by a mechanism requiring direct contact with a recipient cell. Three types of T4SS have been described: (i) conjugation systems, defined as machines that translocate DNA substrates to recipient cells by a contact-dependent process; (ii) effector translocator systems, functioning to deliver proteins or other effector molecules to eukaryotic target cells; and (iii) DNA release or uptake systems that translocate DNA to or from the extracellular milieu (50). Throughout the past 50 years, investigations of T4SS have focused largely on defining the mechanisms of action of a few model systems of gram-negative bacteria, such as the F (IncF), R388 (IncW), RP4 (IncP), and pKM101 (IncN) plasmid conjugation systems and the Agrobacterium tumefaciens VirB/VirD4 system. One aim of this review is to update the information on these systems with an emphasis on recent exciting structural advances.
A second aim of this review is to broaden the scope of the discussion to include T4SS present in biologically diverse microorganisms. An underappreciated feature of T4SS, notably of the DNA conjugation subfamily, is that they function in many species of gram-negative bacteria, gram-positive bacteria, wall-less bacteria, and even members of the phylum Crenarchaeota of the Archaea. Indeed, T4SS are unique among the known macromolecular translocation systems, now numbering at least seven distinct types in bacteria, in this broad phylogenetic distribution. How these machines induce the formation of and mediate translocation across intercellular junctions is an intriguing area of investigation, especially in view of the striking diversity of prokaryotic cell envelopes. Through this discussion, we hope to convince the reader that, in addition to being intrinsically fascinating machines for structural and mechanistic analyses, the T4SS are excellent subjects for fundamental studies exploring the evolution of biological complexity.
OVERVIEW OF T4SS SUBFAMILIES
Conjugation Systems
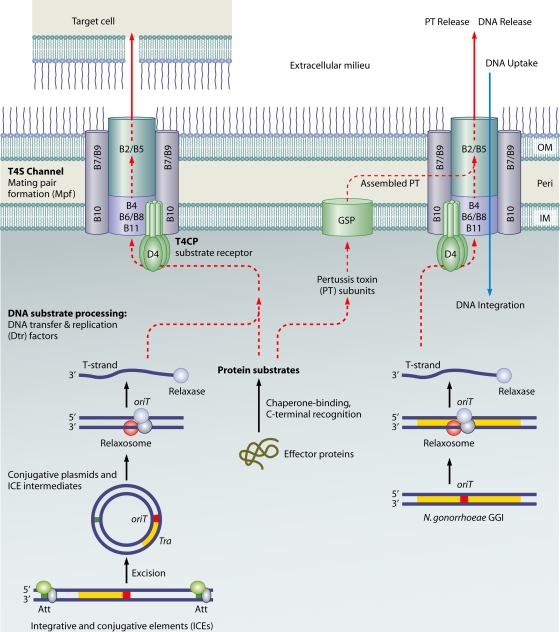
The conjugation systems are the largest and most widely distributed subfamily of T4SS, with systems described for most species of the Bacteria and some members of the Archaea. The overall process of conjugative DNA transfer can be dissected into three biochemical reactions: DNA substrate processing, substrate recruitment, and translocation (Fig. 1) (63, 81, 213, 237). The DNA processing reaction appears to be mechanistically conserved for nearly all conjugation systems. DNA transfer and replication (Dtr) proteins initiate processing by binding a cognate origin-of-transfer (oriT) sequence. The Dtr proteins include a relaxase and one or more accessory factors, and when bound to oriT, the resulting DNA-protein complex is termed the relaxosome. This term originated through the discovery that upon the relaxase-mediated nicking of the DNA strand destined for translocation (hereafter termed the T strand), supercoiled plasmid DNA is converted to the relaxed, open circular form. Accompanying the nicking reaction, relaxase remains bound to the 5′ end of the T strand. The bound relaxase, probably in conjunction with other relaxosome components, confers recognition of the DNA substrate by a cognate T4SS. The relaxase also “pilots” the T strand through the translocation channel. In the recipient cell, the relaxase catalyzes the recircularization of the T strand and may also participate in second-strand synthesis or recombination into the chromosome (52, 84, 105).
FIG. 1.
Mechanisms of T4SS. T4SS mediate the contact-dependent transfer of DNA and protein substrates to target cells, and a subset of systems translocates substrates to or from the extracellular milieu. T4SS are comprised of conjugation, effector translocator, and DNA release/uptake subfamilies. Conjugation systems are comprised of conjugative plasmids and ICEs. For conjugative transfer, DNA substrates are processed by (i) excision from the chromosome by excisionase/integrase enzymes or DDE transposases (for ICEs), (ii) processing of the plasmid or ICE circular transfer intermediate at the origin-of-transfer sequence (oriT) by the Dtr factors (the Dtr-oriT complex is termed a relaxosome), (iii) recruitment of the relaxase-T-strand intermediate to the T4CP, and (iv) translocation through the T4SS channel. Alternatively, protein substrates are maintained in a translocation-competent form and delivered to the T4CP or another receptor or translocation system, e.g., GSP, through the binding of secretion chaperones or other adaptors or spatial-positioning factors. In gram-negative bacteria, T4SS can mediate contact-dependent (left route) or -independent (right route) substrate transfer. The substrate transfer pathway (dashed red lines) through the channel is not clear at this time. DNA uptake by the Helicobacter pylori ComB system (blue line) occurs independently of a T4CP; DNA release by the Neisseria gonorrhoeae GGI-encoded system occurs through a conjugation-like mechanism requiring Dtr factors, a T4CP, and a T4SS channel. In the figure, A. tumefaciens VirD4 is representative of the T4CPs, and the VirB subunits are representative of Mpf channel components; other gram-negative bacterial T4SS are composed of a variable number of VirB homologs. OM, outer membrane; IM, inner membrane; Peri, periplasm.
The self-transmissible plasmids are only one of two major subgroups of conjugative elements. The second set of conjugative elements, originally termed “conjugative transposons” and more recently termed “integrative and conjugative elements” (ICEs), are also present in many bacterial and archaeal species (40, 41, 150, 151, 152, 240, 260). These elements are processed for translocation first by excision from the chromosome through the action of a recombinase/excisionase complex and by the formation of a circular intermediate (Fig. 1). Second, the circularized intermediate is processed at oriT as described above for conjugative plasmids. In the recipient cell, ICEs reintegrate into the chromosome by homologous recombination or through the action of an ICE-encoded integrase.
Conjugative plasmids and ICEs are recruited to the transfer machine through interactions between the relaxosome or processed DNA transfer intermediate and a highly conserved ATPase termed the substrate receptor or type IV coupling protein (T4CP) (Fig. 1). The T4CP physically interacts with the translocation channel, which is comprised of the mating-pair formation (Mpf) proteins (62, 114, 169, 237). Two types of Mpf proteins, an ATPase and a polytopic membrane subunit, are associated with all T4SS, whereas other Mpf proteins are less phylogenetically conserved. In gram-negative bacteria, the Mpf proteins elaborate the secretion channel as well as a pilus or other surface filament to promote attachment to target cells (64, 169). In gram-positive bacteria, surface adhesins rather than conjugative pili mediate attachment (116).
Effector Translocator Systems
A second large subfamily of T4SS, the effector translocators, has gained considerable attention because of its prominent roles in the infection processes of many bacterial pathogens. These systems deliver effector proteins or other macromolecules directly to the cytosols of eukaryotic target cells to aid bacterial colonization and survival within host cells or tissues (18, 50, 97, 195). Described so far only for gram-negative bacteria, these systems lack the Dtr proteins required for the processing of conjugative DNA elements, yet most of them still rely on a T4CP to recruit and bind protein substrates. Some T4SS of medical importance, e.g., the Bordetella pertussis Ptl and Brucella sp. VirB systems, lack T4CPs and instead use another substrate receptor or another mechanism, e.g., the general secretory pathway (GSP), for substrate translocation across the inner membrane (Fig. 1) (39, 51). Effector translocator systems deliver their cargo to eukaryotic target cells through direct cell-to-cell contact, with the exception of the B. pertussis Ptl system, which exports the A/B pertussis toxin (PT) to the extracellular milieu (39).
DNA Uptake and Release Systems
Presently, the third T4SS subfamily is composed of two systems, the Helicobacter pylori ComB system, which acquires DNA from the extracellular milieu, and the Neisseria gonorrhoeae gonococcal genetic island (GGI), which secretes DNA to the extracellular milieu (Fig. 1) (50). The ComB system is ancestrally related to the A. tumefaciens VirB/VirD4 system but lacks the Dtr proteins and T4CP substrate receptor required for DNA export (129, 130, 155). The GGI DNA release system is related to the Escherichia coli F plasmid transfer system and codes for Dtr, T4CP, and Mpf subunits commonly associated with conjugation machines (122, 232). The ComB system is unique among known bacterial competence systems in its phylogenetic relatedness to T4SS. Other competence systems, functioning, for example, in Bacillus subtilis, Streptococcus pneumoniae, N. gonorrhoeae, and Haemophilus influenzae, are ancestrally related to type II secretion systems and type IV pilus assembly systems (see references 54, 55, 56, and 125 for more extensive discussions of these systems).
T4SS Classification Schemes
There have been several attempts to classify T4SS on the basis of phylogenetic relationships. A robust classification nomenclature has been developed for the relaxases of conjugative plasmids and ICEs. In this scheme, the conjugation systems are classified into six groups on the basis of relaxase domain architecture, sequence similarities within the catalytic center, and nic DNA target sequences (104). An earlier scheme classified conjugative elements into incompatibility (Inc) groups, a property referring to the tendency of a resident element, e.g., conjugative plasmid, to inhibit the replication of incoming elements possessing identical or closely related replication systems (53, 169, 204). Accordingly, gram-negative bacterial T4SS resembling the IncF, IncP, and IncI plasmid conjugation systems are designated F-, P-, and I-like systems (169). ICEs are also classified into Tn916/Tn1545, SXT/R391, pKLC102/PAP1, SPI-7, and ICEHin1056 sublineages on the basis of gene sequence similarities and organizations (41, 68, 151). These schemes offer a general way to group ancestrally related T4SS, yet many plasmid and ICE T4SS have undergone extensive modular evolution through recombination, and the resulting mosaicism complicates phylogeny-based classification (157, 209, 244). Our alternative assignment of T4SS on the basis of function as conjugation machines, effector translocators, or DNA release/uptake systems also has its limitations, because conjugation systems also translocate protein substrates independently of DNA, and some effector translocator systems also conjugatively transfer DNA to target cells. Whether T4SS are grouped by phylogeny or function, recent work suggests that all prokaryotic T4SS possess several common mechanistic features, and many have also acquired novel properties for specialized purposes. This review will highlight these mechanistic themes and variations. We refer the reader to several excellent reviews for more detailed discussions of specific T4SS (19, 24, 39, 50, 51, 62-64, 75, 151, 152, 237).
MECHANISM OF T4SS SUBSTRATE PROCESSING
Conjugative Plasmid and ICE Processing Reactions
The enzymes required for conjugative DNA transfer include relaxases acting at oriT sequences and recombinases acting at ICE border sequences (Fig. 1). Relaxases comprise one of two large families of DNA strand transferases; the second is composed of the rolling-circle replicases (Rep) (72, 139). Both families have two signature sequence motifs or domains, an HUH (His-hydrophobic residue-His) or HHH (His-His-His) motif, thought to bind active-site metals (167), and a catalytic pocket with one or two Tyr residues required for the nicking reaction. Relaxases cleave DNA through a transesterification reaction involving nucleophilic attack by the active-site Tyr hydroxyl on the 5′ side of a DNA phosphate. The high-energy phosphodiester bond broken by this reaction is preserved by the formation of a covalent bond between the Tyr residue and the 5′ end of the T strand, and this energy is used for recircularization of the T strand upon translocation to target cells (31, 213, 253).
Relaxases are generally large proteins with two or more domains, one catalyzing the transesterification reaction and others with primase, helicase, or other activities (110). Crystal structures have been presented for DNA strand transferases: (i) a rolling-circle replicase protein from adeno-associated virus (126), (ii) the relaxase domain of TraI from the F plasmid without and with a bound DNA substrate (72, 166, 167, 253), (iii) the relaxase domain of TrwC from plasmid R388 with bound DNA (31, 117), and (iv) the relaxase domain of MobA from IncQ plasmid R1162 (193). (Hereafter, we will identify a specific T4SS or subunit by the system or subunit name followed by a subscript indicating the associated conjugative element or organism, e.g., VirB/VirD4At [for the A. tumefaciens VirB/VirD4 system], TraIF, TrwCR388, and MobAR1162). Despite exhibiting low levels of overall primary-sequence relatedness, all of these structures exhibit similar geometric arrangements in the metal binding and active-site Tyr moieties.
ICEs initiate conjugative transfer by excising from the chromosome and forming a circular double-stranded DNA (dsDNA) transfer intermediate (Fig. 1). Until recently, all ICEs (including elements originally termed “conjugative transposons,” e.g., Enterococcus faecalis Tn916 and Bacteroides fragilis CTnDot) were thought to integrate/excise exclusively by use of Tyr or Ser site-specific recombinases (33, 40, 41, 68). These recombinases are named after the amino acid residue that forms a transient covalent bond with DNA in the reaction intermediate. Recently, a new family of ICEs represented by Streptococcus agalactiae TnGBS2 was shown to use a DDE transposase to catalyze the integration and excision reactions (37). The DDE motif is a catalytic triad of acidic amino acids involved in the DNA cleavage reaction accompanying transposition. In contrast to site-specific recombinases, DDE transposases cleave the ends of insertion sequences or transposases and target-site DNAs without forming a protein-DNA covalent intermediate (124). The TnGBS-like elements thus utilize a bona fide transposition mechanism to generate the conjugative DNA transfer intermediate (37).
Accessory Dtr Processing Factors
Most relaxases require accessory Dtr factors for efficient nicking at oriT (Fig. 1). These factors stimulate the processing reaction by recruiting the relaxase to oriT, and they might also function as molecular wedges to melt dsDNA and facilitate the access of the relaxase to the nic site. Recent studies have identified a family of Dtr accessory factors with a common structural basis for oriT binding and relaxase recruitment. These factors possess a ribbon-helix-helix (RHH) DNA binding motif resembling that described previously for the Arc/CopG/Mnt family of transcriptional regulators (36, 268, 291). Characterized members of the RHH family of Dtr accessory factors include TraYF, TraMF, TrwAR388, NikAR64, TraJRP4, MobCRSF1010, PcfFpCF10, MbeCColE1, and VirC2At (36, 58, 192, 268, 291, 297, 299). X-ray structures are now available for RHH domains of NikA and VirC2, and results of structure-based mutational analyses firmly establish the importance of the RHH domain in oriT binding as well as the recruitment of and nicking by the relaxase (179, 291).
Novel Conjugative DNA Transfer Processing Reactions
Although conjugative plasmids and ICEs generally translocate as elements with defined borders, these elements can also coincidentally cotransfer large chromosomal DNA fragments to target cells. The underlying mechanism is thought to resemble that described for E. coli Hfr strains, in which a relaxase nicks at an oriT sequence associated with an ICE or integrated conjugative plasmid and mediates the unidirectional transfer of a single strand of chromosomal DNA. Interestingly, recent work has shown that relaxases can also initiate DNA transfer from cryptic oriT sequences dispersed in the chromosome and unassociated with mobile elements (38, 189). For example, Dtr factors encoded by plasmid R1162 can recognize cryptic oriT sequences and mediate the transfer of large fragments of the E. coli chromosome. R1162, like other related IncQ plasmids, is capable of transferring to and replicating in a variety of different gram-negative and gram-positive bacterial species. Thus, through the recognition of cryptic oriT sequences in these different hosts, such promiscuous elements likely have exerted profound effects in shaping bacterial genome architectures over evolutionary time (189).
At least two other novel mechanisms are responsible for the conjugative transfer of chromosomal DNA. In the gram-positive Actinomycetes, some conjugative plasmids and ICEs are translocated from mycelial donor to recipient cells as dsDNA transfer intermediates (116, 216, 260). These elements code for an unusual T4CP, which is capable of recognizing and translocating dsDNA substrates independently of other Mpf channel subunits. This protein is phylogenetically related to B. subtilis SpoIIIE and E. coli FtsK, which mediate the translocation of chromosomal DNA across septal membranes during B. subtilis sporulation and E. coli cell division, respectively (30, 187). Not surprisingly, the Actinomycetes ICEs or integrated plasmids can also function as Hfr-like elements and mediate the transfer of large segments of chromosomal DNA across mycelial membranes (116, 260).
A second novel chromosomal transfer system is found in Mycobacterium smegmatis. M. smegmatis lacks genes for classical Dtr, T4CP, or Mpf components, yet this species translocates fragments of its genome to recipient cells. Strikingly, noncontiguous segments of the chromosome are translocated with similar frequencies during mating (278, 279). This contrasts sharply with a classical Hfr transfer mechanism in which chromosomal loci positioned near oriT are transferred at higher frequencies than more distal loci. cis-Acting sequences identified in the chromosome might functionally resemble oriT sequences, but these sequences are larger and more complex than previously described oriT sequences. One model posits that chromosomal transfer initiates at a break in the chromosome and that, upon transfer, the DNA integrates via recombination into the recipient chromosome (278, 279).
Processing for DNA Release and Uptake
The DNA release and uptake systems also process DNA substrates for translocation across membranes. The N. gonorrhoeae GGI DNA release system encodes a relaxase, termed TraI, that binds and nicks the chromosome at a cognate oriT site within the GGI (232). The relaxase domain of TraIGGI is fused to a phosphohydrolase (HD) domain, whose metal-dependent phosphohydrolase activity might also contribute in some way to the processing reaction. TraIGGI also possesses an N-terminal amphipathic helix that mediates binding to the inner membrane (232). Membrane binding, a property shared by other relaxases and accessory Dtr processing factors, might promote the engagement of the relaxase-DNA intermediate with the T4CP receptor at the membrane (see below). Once engaged with the transfer machine, TraIGGI presumptively pilots its chromosomal cargo across the neisserial envelope to the milieu (Fig. 1).
The H. pylori ComB system imports DNA from the extracellular milieu (131, 155). At this time, nothing is known about the requirements for binding and importing exogenous DNA. Two other competence systems functioning in B. subtilis and N. gonorrhoeae are well characterized (54, 55), but as noted above, these systems are phylogenetically unrelated to the H. pylori ComB system. How an ancestral T4SS evolved as a DNA uptake system remains an intriguing question for future study.
SUBSTRATE RECOGNITION SIGNALS, SECRETION CHAPERONES, AND ACCESSORY FACTORS
Perhaps with the sole exception of the H. pylori ComB DNA uptake system, T4SS recognize substrates by virtue of signal sequences carried by relaxase components of DNA transfer intermediates or by effector proteins. Early efforts to define the nature of these peptide signals focused on protein substrates of the A. tumefaciens VirB/VirD4 and Legionella pneumophila Dot/Icm T4SS. The outcome of these investigations led to a general model that secretion signals are positioned near the C termini and consist of clusters of hydrophobic or positively charged residues. More recent findings, however, suggest that substrate recognition is mediated by a combination of C-terminal signals, additional intrinsic motifs, and other cellular factors, e.g., chaperones and accessory proteins (Table 1).
TABLE 1.
Requirements for T4SS substrate-channel docking
| T4SS | Substrate(s)a | Recognition motif(s) (T4CP interaction)b | Accessory protein (function)c | Reference(s) |
|---|---|---|---|---|
| Conjugation systems | ||||
| Gram-negative bacteria | ||||
| A. tumefaciens | TraApTiC58 | C-terminal positive-charge tail sequence | 239 | |
| BID domain | ||||
| P. aeruginosa | MobAR1162 | C-terminal positive charge and overall 2° structure | MobB (binds membrane, stabilizes MobA-T4CP interaction) | 214 |
| NTD recognition signal | ||||
| E. coli | TraIF | ND | TraM (RHH DNA binding protein, tetramer, interacts with C-terminal extension of TraDF T4CP) | 28, 82, 180, 181 |
| TrwCR388 | ND (interacts with TrwBR388 T4CP) | TrwA (RHH DNA binding protein, tetramer, stimulates T4CP ATP hydrolysis) | 176, 192, 257 | |
| TraIRP4 | ND (interacts with TraGRP4 T4CP) | 236 | ||
| MobARSF1010 | CT50aa suffices for translocation of Cre | 269 | ||
| Bordetella bronchiseptica | MobpBHR1 | ND (interacts with TraGRP4) | 254 | |
| B. fragilis | MbpBpLV122 | ND (interacts with the T4CPs TraGRP4 and TraDF deleted of its C terminus) | 262 | |
| Gram-positive bacteria | ||||
| E. faecalis | PcfGpCF10 | ND (interacts with PcfCpCF10 T4CP) | PcfF (interacts with PcfG and PcfC, spatially positions relaxosome or transfer intermediate near the T4CP) | 58, 59 |
| S. agalactiae | TraApIP501 | ND (interacts with the Orf10pIP501 T4CP) | 1 | |
| Effector translocators | ||||
| A. tumefaciens | VirE2VirB | CT50aa sufficient (interacts with the VirD4At T4CP, CT100aaVirE2 sufficient for the T4CP interaction) | VirE1 (chaperone prevents VirE2 aggregation and DNA binding in A. tumefaciens) | 12, 88, 101, 246, 269, 270, 298 |
| VirD2 | ND (VirD2 T strand interacts with VirD4VirB); VirD2 mediates translocation of Cre | VirC1 (spatial positioning), VBP1 (VirD2 binding protein recruits VirD2 to T4SS) | 10, 49, 119, 269 | |
| VirFVirB | C-terminal positive-charge tail, CT19aa sufficient | 269 | ||
| VirD5VirB | C-terminal positive-charge tail, CT50aa sufficient | 269 | ||
| VirE3VirB | C-terminal positive-charge tail, CT50aa sufficient | 270 | ||
| Atu6154TiC58 | C-terminal positive-charge tail, CT50aa sufficient | 269 | ||
| Agrobacterium rhizogenes | GALLSVirB | C-terminal pos. charge tail, CT27aa sufficient | 128 | |
| L. pneumophila | RalFIcm/Dot | C-terminal hydrophobic tail, CT20aa sufficient; Leu-3 required for translocation | Independent of IcmS/IcmW | 23, 194, 203 |
| LepAIcm/Dot, LepBIcm/Dot | ND, presence of the C-terminal half improves but is not required for translocation | Independent of IcmS/IcmW | 57 | |
| AnkBIcm/Dot | CT Val-2 and Leu-3 required for translocation | IcmS/IcmW (chaperones) | 3 | |
| SidGIcm/Dot | C-terminal hydrophobic tail, CT20aa | IcmS/IcmW (chaperones) | 44 | |
| SidCIcm/Dot | C-terminal hydrophobic tail, CT100aa sufficient | IcmS/IcmW (chaperones) | 44, 267 | |
| LegS2Icm/Dot | C-terminal hydrophobic tail, CT30aa sufficient | IcmS/IcmW (chaperones) | 74 | |
| SidAIcm/Dot, SidBIcm/Dot, SidDEIcm/Dot, SidFIcm/Dot, SidHIcm/Dot, PieABCDEFGIcm/Dot, PpeABIcm/DotIcm/Dot, PpgAIcm/Dot, SdeAIcm/Dot, WipAIcm/Dot, WipBIcm/Dot | ND | IcmS/IcmW (chaperones) | 23, 44, 201, 203 | |
| C. burnetii | AnkIIcm/Dot | C-terminal hydrophobic tail, CT82aa sufficient | IcmS (chaperone) | 274 |
| AnkBIcm, AnkHIcm, AnkJIcm, AnkMIcm, AnkNIcm, OIcm | ND | IcmS (chaperone) | 274 | |
| AnkAIcm, AnkFIcm, AnkGIcm, AnkPIcm | ND | Independent of IcmS | 274 | |
| Brucella spp. | VceAVirB and VceCVirB | C-terminal hydrophobic tail, CT20aa required; CT115aa of VceC sufficient for translocation through L. pneumophila Dot/Icm (no T4CP in Brucella VirB T4S) | 76 | |
| Bartonella spp. | BepABCDEFG | C-terminal positive-charge tail sequence BID domain | 239 | |
| H. pylori | CagACag | CT20aa and an intact N terminus | CagF (chaperone) | 70, 132, 215 |
| DNA uptake/release | ||||
| N. gonorrhoeae | TraIGGI | ND | ParA homolog (might function like VirC1At to spatially position substrate near transfer machine) | 122, 232 |
| H. pylori | ComB | 129-131, 155 |
For the conjugation systems, the listed proteins are relaxases that bind a cognate T4CP and are delivered to recipient cells. For the effector translocator systems, the listed proteins are effectors that play a role in the infection processes of the bacterial pathogen. TraIGGI of the N. gonorrhoeae DNA release system is a relaxase required for DNA release, but its translocation to the extracellular milieu has not been shown.
The motifs listed are required for substrate translocation. In some cases, the protein or its C-terminal fragment (CT) is sufficient to mediate translocation to target cells, as shown by fusion to a reporter protein such as Cre recombinase or adenylate cyclase. Amino acids (aa) at positions listed relative to the C-terminal fragment (subscript) are required for translocation, as shown by mutational analysis. ND, not determined. Parentheses indicate that the interaction between a protein substrate and a cognate T4CP has been experimentally shown.
Accessory factors required for T4SS channel docking or translocation. The proposed function in mediating substrate-T4SS channel docking is shown in parentheses.
C-Terminal Secretion Signals
The A. tumefaciens VirB/VirD4 T4SS translocates the VirE2 single-stranded DNA (ssDNA) binding protein to plant cells during infection (81). In the plant, VirE2 binds along the length of a translocated T-DNA substrate, protecting the ssDNA intermediate from nucleases en route to the nucleus. An intact C terminus was shown to be important for VirE2 translocation through the VirB/VirD4 T4SS (246). More recently, an assay termed the Cre reporter assay for translocation confirmed that the C termini of VirE2 as well as other protein substrates of the VirB/VirD4 T4SS carry substrate recognition signals (269). In this assay, Cre recombinase is fused to an intact secretion substrate or peptide fragments carrying suspected secretion signals, and the resulting fusion protein is assayed for translocation to eukaryotic cells, as monitored by Cre recombination at lox sites engineered into the target cells (269). In L. pneumophila, a similar assay using adenylate cyclase (CyaA) as a reporter for translocation established that the C terminus of the secretion substrate RalF mediates transfer through the Dot/Icm T4SS (194). The C termini of VirE2 and other VirB/VirD4 substrates carry clusters of positively charged residues, whereas the C terminus of RalF carries hydrophobic residues. Mutational analyses of VirE2 and RalF confirmed the importance of these charged and hydrophobic residues for translocation (Table 1) (194, 269).
The C termini of both RalF and VirE2 are disordered and solvent exposed, as first indicated by secondary structure prediction algorithms and more recently by X-ray crystallography (5, 88). These C-terminal domains (CTDs) are thus likely accessible to bind cognate T4SS receptors. VirE2 was crystallized as a complex with its secretion chaperone, VirE1 (88). This structure establishes the importance of substrate-chaperone complex formation for VirE2 translocation. VirE2 possesses two structurally similar N-terminal domains (NTDs) and CTDs that clamp tightly around a single α-helix of VirE1 (88). This substrate-chaperone complex is competent for translocation by virtue of its solubility and C-terminal tail accessibility. In the absence of a chaperone, VirE2 aggregates and forms solenoid filaments, and it also binds prematurely to ssDNA substrates (88, 101, 298).
In Brucella spp., a VirB T4SS related to that of A. tumefaciens contributes to virulence, although the identification of translocated effector proteins has proven challenging (34, 78). Recently, two substrates, VceA and VceC, were identified through the use of CyaA or TEM1 β-lactamase fusion assays (Table 1) (76). Consistent with the above-described findings, both VceA and VceC require intact C termini for translocation through the Brucella VirB system. A VceC-CyaA fusion protein also was shown to translocate through the heterologous L. pneumophila Dot/Icm system, and moreover, the C-terminal 115 residues of VceA were sufficient for CyaA translocation. The latter finding is quite surprising because the L. pneumophila Dot/Icm system possesses a T4CP substrate receptor, but to date, no T4CP has been identified for the Brucella sp. VirB T4SS. How VceC is recognized as a substrate by these two quite distinct T4SS remains an interesting question for future investigation.
The importance of C-terminal recognition signals is not confined to protein substrates of T4SS. Where characterized, relaxases covalently bound to the 5′ end of T strands carry C-terminal signals specifying DNA substrate-channel interactions. Moreover, several relaxases including MobARSF1010, VirD2At, TraApATC58, and TrwCR388 are bona fide substrates of T4SS even without any associated DNA, as demonstrated by the Cre reporter assay for translocation (84, 214, 239, 269). With this assay, it was also shown that the C-terminal 50 residues of MobARSF1010 are sufficient to mediate Cre translocation (269).
Other Intrinsic Secretion Signals
While important, C-terminal signals are probably not sufficient for mediating the translocation of most native substrates (Table 1). In the Brucella VirB system described above, C-terminal deletions of VceA and VceC diminished but did not abolish substrate translocation, which is suggestive of an alternative or additional recognition signal(s) within these proteins (76). In Bartonella spp., the Bartonella-translocated effector proteins (Bep proteins) are translocated through a VirB/VirD4 T4SS to human cells. Bep proteins display a modular architecture with a bipartite secretion signal composed of a positively charged C terminus and at least one internal domain, termed a Bep intracellular delivery (BID) domain (239). BID domains are also present in a family of relaxases associated with some conjugative plasmids in alphaproteobacteria, and both the C terminus and BID domains of one such relaxase, TraApATC58, were required for Cre translocation through the Bartonella henselae VirB/VirD4 system.
In H. pylori, a positively charged C-terminal tail is important for the translocation of the CagA substrate, yet this sequence can be replaced with C-terminal sequences of heterologous substrates including VirE3At and MobARSF1010 (132). However, unlike most other T4SS substrates analyzed so far, CagA tolerates the addition of an epitope tag to its C terminus but not N-terminal deletion mutations (132). The N terminus of CagA thus might also contribute to substrate transfer, perhaps supplying a second recognition sequence or fold important for docking with the translocation channel. Finally, as noted above, MobA relaxases of IncQ plasmids can translocate through a type IV channel independently of DNA, as monitored with the Cre fusion assay. MobAR1162 is composed of two domains, an N-terminal relaxase domain, and a C-terminal primase domain, and recent work has shown that each domain can separately mediate Cre translocation. Furthermore, mutations in the relaxase and primase catalytic sites abolish translocation, suggesting that secondary structures of both domains might be important for translocation (214).
As discussed further below, T4CPs probably function as receptors through the binding of one or more substrate signals. In A. tumefaciens, however, other Mpf channel subunits also specify substrate recognition, as shown by the isolation of channel mutations conferring the selective transfer of one secretion substrate but not others (142, 145, 230). Most intriguingly, some specificity mutations map within the outer membrane channel subunit VirB9. These mutations can block the transfer of VirE2 but not DNA substrates or the transfer of plasmid RSF1010 but not T-DNA or vice versa (142). VirB9 thus appears to function as a substrate specificity checkpoint at the distal portion of the secretion channel. Such checkpoints along the translocation pathway might serve to optimize the delivery of multiple substrates in space and time.
Contributions of Accessory Proteins to Substrate Recognition
Chaperones.
Certain T4SS substrates require secretion chaperones for translocation (Table 1). These chaperones often possess physical properties (small size of ∼15 kDa, acidic pI, and amphipathic helices) resembling those of chaperones associated with the type III secretion systems, a family of macromolecular translocation systems ancestrally related to bacterial flagella (211). As mentioned above, an X-ray structure of the VirE1 chaperone/VirE2 effector complex showed the unusual structural arrangement of two VirE2 domains wrapped around the VirE1 chaperone (88). VirE1 does not participate directly in VirE2 docking with the VirB/VirD4 channel (12, 270); rather, complex formation blocks VirE2 aggregation and exposes the C-terminal tail sequence for productive contacts with the substrate receptor (77, 81, 88).
In L. pneumophila, the Dot/Icm T4SS translocates more than 50 effectors to mammalian cells during the course of infection (202). At least three T4SS chaperones, IcmS, IcmW, and LvgA, are required for the translocation of effector proteins (44, 203, 271). Interestingly, IcmS forms heterodimeric interactions with IcmW or LvgA, and the resulting chaperone complexes interact with and mediate the translocation of many effectors through the Dot/Icm T4SS. Both the IcmS/IcmW and IcmS/LvgA complexes functionally resemble VirE1 by inducing a conformation necessary for substrate docking with the T4SS apparatus. IcmS/IcmW chaperone binding, for example, induces a conformational change in the effector SidG required for the exposure of the C-terminal recognition sequence (44).
In H. pylori, the CagF chaperone is required for CagA translocation through the Cag T4SS. CagF resembles the above-described chaperones in physical properties (acidic pI and α-helical) but is much larger (∼35 kDa) and also localizes in both the cytosol and inner membrane (215). The CagF-CagA complex associates predominantly with the membrane, although CagF and CagA also bind the membrane independently of each other. There is some evidence that CagF binds the membrane at or near the Cag T4SS, which is suggestive of a possible role in the spatial coordination of CagA docking with the translocation channel (215).
Accessory factors as spatial adaptors.
There is additional evidence that chaperones or other accessory factors function to position secretion substrates near or at the T4SS channel entrance (Table 1). In the E. faecalis pCF10 transfer system, the relaxase PcfG requires PcfF to nick at the pCF10 oriT sequence (58). PcfF and PcfG interact with each other, and both Dtr factors also bind the PcfC T4CP (59). Furthermore, all three proteins colocalize at discrete sites at the cell membrane (59). Thus, a working model proposes that PcfF recruits PcfG to oriT, and through interactions with unknown membrane constituents, both factors then mediate the binding of the relaxosome (or the pCF10 transfer intermediate) at the membrane near the T4SS channel (59). A similar mechanism was postulated for MobBR1162, an accessory factor that stimulates R1162 transfer. Interestingly, MobB stimulates the translocation of both relaxase and primase domains when separately produced (214). R1162 lacks genes for its own Mpf channel but promiscuously translocates through other plasmid- or ICE-encoded channels. The promiscuity of this and related IncQ plasmids could be explained by a combination of MobB interactions, with relaxase, the membrane, and a conserved domain(s) carried by T4CPs, together serving to tether the R1162 relaxosome or transfer intermediate near various T4SS channels (214).
A. tumefaciens VirC1 provides another example of a Dtr accessory factor functioning as a spatial determinant. VirC1 and its binding partner, VirC2 (see above), stimulate processing at oriT-like T-DNA border sequences by binding an adjacent sequence termed overdrive (265). The formation of the VirC1/VirC2/overdrive complex probably recruits the VirD2 relaxase to the border sequence and might also melt dsDNA to allow relaxase access to the nic site (265). VirC1 is a member of the ParA/Soj/MinD family of ATPases, and like other family members, VirC1 localizes at specific sites in the cell (10). Specifically, VirC1 localizes at cell poles, which are also the sites of VirB/VirD4 channel assembly (10, 148, 162). Interestingly, VirC1 was also found to recruit the VirD2 relaxase as well as the processed T strand to the polar membrane (10). Through a demonstrated interaction with the polar-membrane-localized VirD4 T4CP, VirC1 thus stimulates the binding of the T-DNA substrate with the T4CP receptor (10). Another family of proteins, termed VirD2 binding proteins, also function to recruit the VirD2-T-strand complex to the VirB/VirD4 machine; however, VirD2 binding proteins appear to function nonspecifically because they also recruit other conjugative plasmid intermediates to their cognate channels (119).
There is growing evidence that other ParA/Soj/MinD ATPases, or other cytoskeletal proteins, function as spatial determinants to promote the docking of conjugative DNA elements with cognate T4SS channels. For example, many ICEs encode ParA-like proteins. Such proteins probably play no role in the maintenance of ICEs, because these elements replicate and segregate with chromosomes during cell division. Instead, like VirC1, ICE-encoded ParA proteins might coordinate the docking of the excised ICE intermediate with the cognate translocation channel. In support of this proposal, the N. gonorrhoeae GGI codes for a ParA-like protein, and mutational studies have confirmed its importance for DNA release (122). Genes for other cytoskeletal proteins are also linked to T4SS loci; for example, a T4SS gene cluster in Spiroplasma carries a gene for an mreB homolog (20). Whether MreB or other cytoskeletal factors spatially coordinate type IV secretion awaits further study.
THE T4CP: A SUBSTRATE RECEPTOR AND POSSIBLE DNA TRANSLOCASE
The T4CPs are a fascinating family of ATPases associated with nearly all prokaryotic conjugation systems. T4CPs are also associated with most effector translocator systems, and a T4CP is required for DNA release by the N. gonorrhoeae GGI. T4CPs have been the subject of several excellent reviews (63, 114, 169, 237); therefore, only a brief update on T4CP biochemistry and subunit interactions is warranted. We will, however, highlight results of our analyses of T4CPs from phylogenetically distant organisms indicating that these proteins display extensive sequence heterogeneity and distinct domain architectures. In this context, we raise the question of whether T4CP functions proposed on the basis of studies of a few “paradigmatic” gram-negative systems are universally applicable.
Biochemical and Structural Properties of Paradigmatic T4CPs
T4CPs of gram-negative conjugation machines, including TrwBR388, TraGRP4, TraDF, and VirD4At, have been characterized in considerable biochemical and structural detail (112, 114, 236, 238). Recently, biochemical studies were expanded to include T4CPs of gram-positive conjugation machines, including E. faecalis PcfCpCF10 and Clostridium perfringens TcpApCW3 (59, 251). In general, T4CPs possess three domains, an N-terminal transmembrane (TM) domain, a nucleotide binding domain (NBD), and an all-α-domain (AAD). A soluble fragment of TrwBR388, comprised of the NBD and AAD, crystallized as a homohexameric sphere with dimensions of 110 Å in diameter and 90 Å in height and a central channel of 20 Å in diameter. The NBD is structurally similar to RecA and DNA ring helicases, and the AAD is structurally similar to an NTD of the site-specific recombinase XerD of the λ integrase family (111, 113). The TM domain, modeled on the basis of electron microscopy, is depicted as projecting across the cytoplasmic membrane, giving rise to an F1Fo-like, ball-stem structure for the full-length protein (Fig. 1) (112). TrwBR388 and other T4CPs carry conserved Walker nucleoside triphosphate (NTP) binding motifs required for function, and the soluble fragment of TrwBR388 displays a DNA-dependent ATPase activity in vitro. Several T4CPs bind ssDNA and dsDNA substrates nonspecifically in vitro, and TrwBR388 also oligomerizes upon DNA binding in vitro (59, 257). Finally, the TM domain of TrwBR388 contributes to hexamer formation and influences nucleotide binding properties (134, 135).
T4CPs are phylogenetically and structurally related to the FtsK and SpoIIIE ATPases (Fig. 2) (30, 187). The latter proteins are dsDNA translocases, and in the crystal structure, duplex DNA was detected in the annulus of the FtsK hexamer (187). Correspondingly, T4CP hexameric complexes were proposed to function as translocases by encircling ssDNA substrates and energizing DNA transfer across the cytoplasmic membrane through the lumen of the TM domain. The AAD, which occupies the cytoplasmic entrance to the opening of the hexamer, could bind DNA substrates or be involved with the processing of protein or DNA substrates prior to transfer. Consistent with the above-described proposal, the TM domain is indispensable for T4CP function among gram-negative systems characterized to date. However, as discussed further below, T4CPs associated with several gram-negative and -positive systems lack discernible N-terminal TM domains, and there is even an example of a T4CP that retains function when deleted of its TM domain.
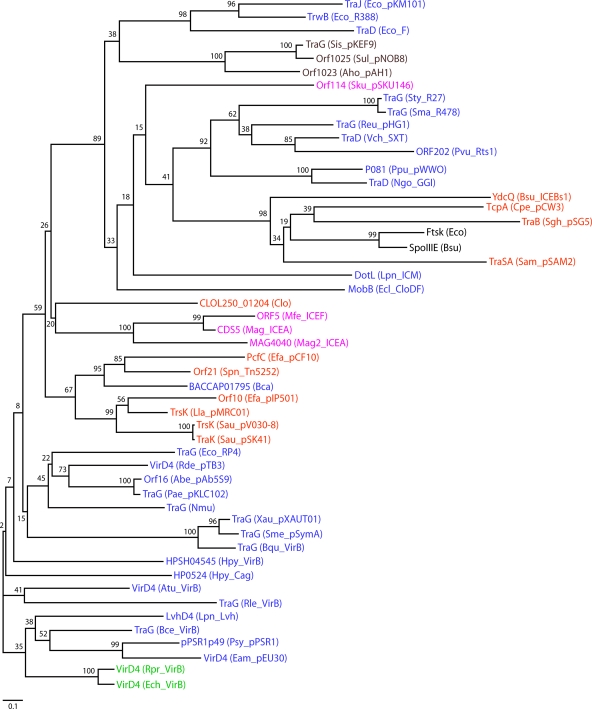
FIG. 2.
Phylogenetic tree of T4CP family members. Sequence alignment and phylogeny estimation were performed using the program MAFFT, version 6.0 (available at http://align.bmr.kyushu-u.ac.jp/mafft/software). The sequence alignment was performed by using the E-INS-i method and default parameters of the program. The tree was constructed using the neighbor-joining method and the following parameters: all ungapped sites from the alignment, JTT amino acid substitution model, and ignore heterogeneity among sites. Bootstrap values for 500 replicates are indicated. The different groups of bacteria are indicated by the following color scheme: blue for gram-negative bacteria, green for gram-negative obligate intracellular pathogens, orange for gram-positive bacteria, pink for cell wall-less bacteria, dark red for Archaea, and black for the related DNA translocases FtsK and SpoIIIE. T4CP designations include the following protein names followed by the species name and plasmid, ICE, or T4SS in parentheses, according to the GenBank database. Accession numbers of T4CPs are AAB58711.1 for Eco_pKM101_TraJ, CAA44852.1 for Eco_R388_TrwB, BAA97972 for Eco_F_TraD, YP_138373.1 for Sis_pKEF9_TraG, CAA09120.1 for Sul_pNOB8_Orf1025, ACI15704.1 for Aho_pAH1_Orf1023, AAS59568.1 for Sku_pSKU146_Orf14, NP_058332.1 for Sty_R27_TraG, NP_941281.1 for Sma_R478_TraG, NP_943001.1 for Reu_pHG1_TraG, AAL59680.1 for Vch_SXT_TraD, NP_640162.1 for Pvu_Rts1_ORF202, NP_542873.1 for Ppu_pWWO_p081, AAW83057.1 for Ngo_GGI_TraD, CAB12293.1 for Bsu_ICEBs1_YdcQ, ABF47325.1 for Cpe_pCW3_TcpA, CAA56759.1 for Sgh_pSG5_TraB, CAA90178.1 for Eco_FtsK, NP_389562.1 for Bsu_SpoIIIE, CAA06449.1 for Sam_pSAM2_TraSA, O54524 for Lpn_ICM_DotL, CAB62409.1 for Ecl_CloDF_MobB, ZP_02074434.1 for Clo_CLOL250_01204, AAN85238.1 for Mfe_ICEF_Orf5, CAJ32610.1 for Mag_ICEA_CDS5, CAL59102.1 for Mag2_ICEA_MAG4040, YP_195789.1 for Efa_pCF10_PcfC, AAG38037.1 for Spn_Tn5252_Orf21, ZP_02036195.1 for Bca_BACCAP01795, CAD44390.1 for Efa_pIP501_Orf10, NP_047302.1 for Lla_pMRC01_TrsK, YP_001653098.1 for Sau_pV030-8_TrsK, NP_863634.1 for Sau_pSK41_TraK, CAA38334.1 for Eco_RP4_TraG, YP_771875.1 for Rde_pTB3_VirD4, YP_001220615.1 for Abe_pAb5S9_Orf16, AAP22624.1 for Pae_pKLC102_TraG, YP_413489.1 for Nmu_TraG, YP_001409435.1 for Xau_pXAUT01_TraG, NP_435748.1 for Sme_pSymA_TraG, YP_032630.1 for Bqu_VirB_TraG, YP_001910374.1 for Hpy_VirB_HPSH04545, NP_207320.1 for Hpy_Cag_HP0524, AAF77174.1 for Atu_VirB_VirD4, ZP_02859218.1 for Rle_VirB_TraG, CAB60062.1 for Lpn_Lvh_LvhD4, ABF79722.1 for Bce_VirB_TraG, NP_940734.1 for Psy_pPSR1_pPSR1p49, NP_943287.1 for Eam_pEU30_VirD4, H71684 for Rpr_VirB_VirD4, and Q8RPL9 for Ech_VirB_VirD4. Species name abbreviations are as follows: Eco, E. coli; Sis, Sulfolobus islandicus; Sul, Sulfolobus sp. strain NOB8H2; Aho, “Acidianus hospitalis”; Sku, S. kunkelii; Sty, S. enterica serovar Typhi; Sma, Serratia marcescens; Reu, Ralstonia eutropha; Vch, V. cholerae; Pvu, Proteus vulgaris; Ppu, Pseudomonas putida; Ngo, N. gonorrhoeae; Bsu, B. subtilis; Cpe, C. perfringens; Sgh, Streptomyces ghanaensis; Sam, Streptomyces ambofaciens; Lpn, L. pneumophila; Eclo, Enterobacter cloacae; Clo, Clostridium sp. strain L2-50; Mfe, M. fermentans; Mag, M. agalactiae strain 5632; Mag2, M. agalactiae PG2; Efa, E. faecalis; Spn, S. pneumoniae; Bca, B. capillosus; Lla, L. lactis; Sau, S. aureus; Rde, Roseobacter denitrificans; Abe, Aeromonas bestiarum; Pae, Pseudomonas aeruginosa; Nmu, Nitrosospira multiformis; Xau, Xanthobacter autotrophicus; Sme, Sinorhizobium meliloti; Bqu, Bartonella quintana; Hpy, H. pylori; Atu, A. tumefaciens; Rle, Rhizobium leguminosarum; Bce, B. cenocepacia; Psy, P. syringae; Eam, Erwinia amylovora; Rpr, R. prowazekii; Ech, E. chaffeensis.
Substrate Receptor Activity
A combination of genetic and biochemical data further indicate that T4CPs function as docking sites for T4SS substrates (Table 1). Some T4CPs, for example, functionally substitute for others in heterologous T4SS. TraGRP4 and TrwBR388 substitute for each other in the heterologous plasmid R388 and RP4 transfer systems, as monitored by the capacity of the chimeric systems to translocate promiscuous plasmid RSF1010. VirD4At can also replace TraGpTiC58 in mediating RSF1010 transfer through the pTiC58 channel (42, 121). T4CPs have also been shown to interact with relaxosome components or protein substrates in vitro. Although the well-characterized gram-negative plasmid conjugation systems dominate this list of interactions (Table 1), T4CPs of gram-positive systems also bind relaxases; e.g., PcfCpCF10 binds PcfG relaxase, and Orf10pIP501 binds TraA relaxase (1, 59).
The physiological relevance of T4CP-relaxase interactions has been confirmed by use of a chromatin immunoprecipitation assay termed transfer DNA immunoprecipitation (TrIP), which was developed for the detection of DNA substrate contacts with type IV machine components (49). By use of TrIP, it was shown that the VirD4At T4CP interacts with the T-DNA substrate and, furthermore, that substrate binding requires catalytically active VirD2 relaxase. These findings indicate that the T4CP binds only the processed form of the T-DNA substrate in vivo (49). This interaction was detected in A. tumefaciens mutants lacking VirB channel components, confirming that the T4CP functions as a substrate receptor even independently of the channel components (49). Similar lines of investigation have now established that B. fragilis pLV22a (which can translocate through the plasmid RP4 T4SS) binds the TraGRP4 T4CP (262) and that the E. faecalis pCF10 transfer intermediate binds the PcfCpCF10 T4CP (59).
Recent structural studies have shed light on a possible mechanism for the T4CP-DNA substrate interaction. The TraDF T4CP possesses a C-terminal extension that is required for efficient F transfer but is inhibitory for RSF1010 transfer (28, 82, 180). This region of TraD binds TraM, an accessory factor with an RHH DNA binding domain that functions as a tetramer. An X-ray structure of the TraD C-terminal tail bound to TraM shows that the TraD tail forms extensive contacts with the TraM monomer and, furthermore, that as many as four TraD C-terminal tails can bind a single TraM tetramer. These findings imply that a TraD hexamer establishes extensive contacts with TraM in vivo, thus forming the basis of a highly specific relaxosome-T4CP interaction (181). In this system, the T4CP carries the unstructured C-terminal tail responsible for F plasmid recognition and binding. In other systems, C-terminal tails carried not by the T4CP but instead by the substrate, e.g., relaxase, VirE2, and RalF, could mediate specific binding with the cognate T4CP. The nature of these or other types of substrate-T4CP interactions awaits further study.
T4CP Heterogeneity
We have analyzed the phylogenies and predicted physical characteristics of over 50 T4CPs associated with T4SS from gram-negative and -positive bacteria, wall-less bacteria, and archaea (Fig. 2). Although sequence conservation among motifs implicated in nucleotide binding defines this family of ATPases, in fact, considerable sequence variation exists along the lengths of these proteins (Fig. 2). In general, T4CPs from closely related species display higher sequence similarities, but interesting exceptions exist. For example, BACCAP01795 from gram-negative Bacteroides capillosus clusters with gram-positive T4CPs. While most T4CPs from wall-less species cluster with those from phylogenetically closely related gram-positive bacteria, Sulfolobus kunkelii Orf14 clusters with gram-negative and archaeal T4CPs. Several gram-positive T4CPs cluster with E. coli FtsK and B. subtilis SpoIIIE, but several cluster with gram-negative T4CPs. Overall, the most extensive sequence heterogeneity among T4CPs exists in the N-terminal regions that often carry membrane-spanning domains (Fig. 3). A number of T4CPs also possess distinct C-terminal extensions of 50 or more residues. T4CPs thus appear to possess conserved NBDs, but many have also acquired novel N- or C-terminal structural motifs or domains of likely functional importance. This heterogeneous domain architecture establishes a basis for grouping T4CPs into distinct subfamilies (Fig. 3).
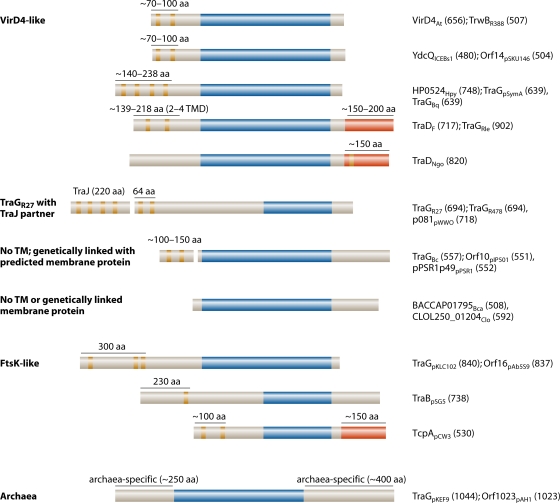
FIG. 3.
Distinct molecular architectures of T4CP family members. T4CPs possess recognizable P-loop NTP binding domains but display considerable variation in NTDs and CTDs. The different types of T4CPs are listed at the left, with representatives of each type at the right. T4CPs are designated with the protein name or accession number, with a subscript identifying the species or plasmid origin, followed by the length in amino acid (aa) residues in parentheses. The domains are indicated with the following color scheme: tan, predicted TM helices; dark blue, P-loop domains; red, C-terminal extensions that are ancestrally unrelated to C termini of other T4CPs. Protein sequence analyses were performed by using the programs TMHMM (TM helix prediction), BLASTP (local alignment), and the NCBI conserved-domain search tool. Protein sequences were obtained from the GenBank database (NCBI), and accession numbers are presented in the legend of Fig. 2. TMD, TM domain.
The well-characterized T4CPs from gram-negative T4SS, e.g., TraDF, TraGRP4, TrwBR388, and VirD4At, typically range in molecular sizes from ∼600 to 750 residues and possess a minimum of two predicted N-terminal TM domains with an intervening periplasmic loop of ∼30 to 50 residues. These T4CPs, here designated VirD4-like T4CPs (Fig. 3), generally display low overall sequence identities (15 to 20%), yet as mentioned above, some of these T4CPs functionally substitute for one another in mobilizing the transfer of promiscuous IncQ plasmids through chimeric T4CP/Mpf systems. Many T4CPs from gram-positive bacteria, wall-less bacteria, and archaeal species are also classified as being VirD4-like on the basis of sequence similarities and conserved domain architectures (Fig. 3).
A distinct clade of T4CPs was previously recognized (Fig. 2 and 3) (118). These T4CPs resemble TraGRP4, TrwBR388, and VirD4At in overall size and predicted N-terminal TM domains, but they typically possess much smaller (∼4 residues) periplasmic domains. TraG of Salmonella enterica serovar Typhi plasmid R27 (IncH) is the archetype for this T4CP subfamily, which consists of about 40 members from gamma- and betaproteobacteria. These T4CPs, designated TraGR27-like, are considerably more related to each other than to TraDF, TraGRP4, TrwBR388, and VirD4At. Not unexpectedly, TraGR27 homologs (but not TraGRP4, TraDF, and TrwBR388) can functionally substitute for TraGR27 in mating assays (118). Also of importance, TraGR27 and its homologs function together with another protein termed TraJ. TraJ is often synthesized from an open reading frame (ORF) located immediately upstream or within a few hundred base pairs of the cognate T4CP gene (Fig. 3). TraJ proteins are typically ∼200 residues in length and are predicted to span the membrane up to five times. TraGR27-like proteins (but not TraGRP4, TraDF, or TrwBR388) bind TraJR27 in vitro. The close juxtaposition of genes for TraJ and TraG, the multimembrane-spanning character of TraJ proteins, and a sequence identity of 21.4% between TraJR27 and the N terminus of the FtsK translocase support a proposal that TraJ and TraG cumulatively represent the domain architecture of the larger FtsK/SpoIIIE DNA translocases (118).
In contrast to the above-described subfamilies, a subset of T4CPs associated with gram-negative and -positive systems lacks discernible TM domains (Fig. 3). Sequence alignments show that these T4CPs, generally shorter than other T4CPs, possess NBDs but lack predicted N-terminal membrane-spanning regions. Notable members of this T4CP subfamily include Orf10 of the S. agalactiae pIP501 transfer system and several T4CPs associated with Staphylococcus aureus and Lactococcus lactis conjugation systems. In gram-negative bacteria, examples include T4CPs associated with putative effector translocator systems encoded on the Burkholderia cenocepacia AU1054 chromosome and Pseudomonas syringae conjugative plasmid pSR1 (Fig. 3). In some cases, ORFs coding for small proteins (∼150 to 200 residues) with two to four predicted TM domains reside upstream of the T4CP gene. The predicted membrane proteins are not related to TraJR27; however, they might function like TraJ in partnering with the cognate T4CP. Of further interest, we found that the introduction of a reading frameshift near the ends of upstream ORFs places these ORFs in frame with the B. cenocepacia AU1054 virD4 and S. agalactiae pIP501 orf10 T4CP genes. Accordingly, the predicted proteins of ∼650 residues display sequence similarities with VirD4-like T4CPs across their entire lengths, and they also possess the characteristic domain architecture of the VirD4-like T4CPs (our unpublished observations). A single frameshift mutation thus might have resulted in a novel subfamily of T4CPs in which N-terminal TM and NBDs are synthesized as separate polypeptides but interact to form a functional complex. Whether such two-partner T4CPs indeed exist remains to be experimentally shown, as it is also possible that in these cases a programmed translational frameshift results in the production of a classical VirD4-like T4CP.
Among this group of TM-less T4CPs are a few examples that appear to lack a membrane partner entirely insofar as no gene for a membrane protein resides in the vicinity of the T4CP gene. Examples include T4CPs from Clostridium sp. strain L2-50 and Bacteroides capillosus ATCC 29799 (Fig. 2 and 3). Whether these T4CPs in fact function in type IV secretion remains to be determined. However, in this context, it is intriguing that while TM domains of well-characterized T4CPs (TraDF, TrwBR388, and VirD4At) clearly are essential for T4CP function, the deletion of the TM domain from the C. perfringens TcpApCW3 T4CP reduces but does not abolish the conjugative transfer of plasmid pCW3 (251). The identification of possible TM-less T4CPs, coupled with a demonstration that the TM domain of TcpApCW3 is dispensable for function, calls into question the model that TM domains of T4CPs obligatorily serve as channels for substrate passage across the membrane.
Another distinct clade of T4CP-like proteins is comprised of FtsK, SpoIIIE, and Streptomyces Tra proteins (Fig. 3). These translocases, which function independently of other T4SS subunits, are often large proteins (>750 residues) with multiple predicted N-terminal TM domains separated from the NBDs by large linker sequences. As mentioned above, the Streptomyces Tra proteins resemble FtsK and SpoIIIE in translocating dsDNA forms of conjugative plasmids, ICEs, or chromosomal DNA across the mycelial membrane (116). While Tra-mediated dsDNA translocation could be considered the simplest T4SS, perhaps intermycelial DNA transfer is more appropriately viewed as an evolutionary adaptation of an ancestral FtsK translocase activity.
Finally, as noted above, a few T4CPs carry C-terminal extensions (Fig. 3). Besides TraDF, other T4CPs with C-terminal extensions include C. perfringens TcpApCW3 and TraD encoded by the N. gonorrhoeae GGI DNA release system. As shown for TraDF, the C-terminal extension might generally contribute to substrate specificity. The C terminus of TraDGGI has not been characterized, but that of TcpApCW3 enhances pCW3 conjugation transfer frequencies by 3 orders of magnitude and also mediates the interaction of TcpA with the TcpC channel subunit (see below) (251).
T4SS Lacking a Cognate T4CP
A few T4SS lack cognate T4CPs and thus rely on another mechanism for substrate recognition or translocation across the cytoplasmic membrane. For example, as mentioned above, subunits of the pentameric PT are translocated across the B. pertussis cytoplasmic membrane via the GSP (Fig. 1) (39, 287). In the periplasm, PT assembles and then engages in an unknown way with the Ptl T4SS for delivery across the outer membrane. Other T4SS lacking a T4CP might use a similar two-step translocation mechanism for the delivery of effector proteins across the cell envelope. However, the recent discovery that the Brucella effectors VceA and VceC translocate through both the Brucella suis VirB (VirBBs) system and the L. pneumophila Dot/Icm system (which depends on the DotL T4CP) suggests that an unidentified VirB (or other) membrane protein supplies a T4CP-like receptor activity to mediate transfer through the VirBBs channel (76).
The Bartonella sp. Trw system also lacks a T4CP protein. This system elaborates antigenically variant pili that are important for the infection of erythrocytes (see below). To date, no Trw secretion substrates have been identified, and thus, it is possible that this system functions exclusively to elaborate adhesive pili. However, this system could rely on another T4CP, e.g., from a coresident T4SS, or another uncharacterized receptor for substrate translocation (75).
There are a couple of reports documenting DNA transfer by a conjugation-like mechanism in the absence of a discernible T4CP homolog. As mentioned above, M. smegmatis translocates distinct fragments of chromosomal DNA to recipient cells by a conjugation-like mechanism in the apparent absence of a T4CP or Mpf channel (69). Another intriguing example involves the transfer of an erythromycin resistance gene from Borrelia burgdorferi to gram-positive bacteria including B. subtilis and E. faecalis (141). The B. burgdorferi genome also lacks obvious T4CP/Mpf genes, which again is suggestive of a transfer mechanism distinct from other known systems.
T4SS ARCHITECTURES: THE GRAM-NEGATIVE PARADIGM
Although T4SS vary extensively in subunit number and composition, most gram-negative systems are composed of Mpf channel subunits related to a core set of the A. tumefaciens VirB subunits. In the next sections, we will summarize features of the VirBAt subunits and the VirB/VirD4 channel to define fundamental requirements for type IV secretion across the gram-negative cell envelope. We will then draw on this information to explore features of T4SS functioning in other prokaryotes.
Energetic Subunits: VirB4 and VirB11
Gram-negative systems employ one or two ATPases besides the T4CP to energize early steps of machine biogenesis or substrate transfer. These are homologs of the A. tumefaciens VirB4 and VirB11 ATPases (Fig. 4).
FIG. 4.
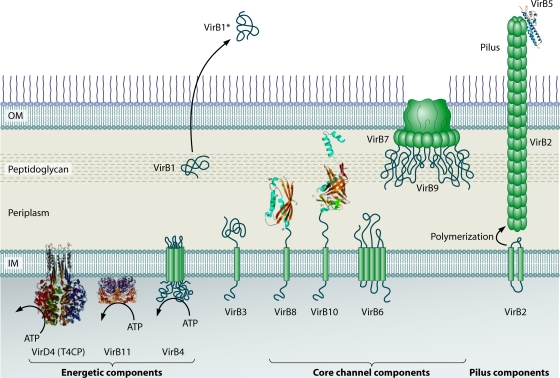
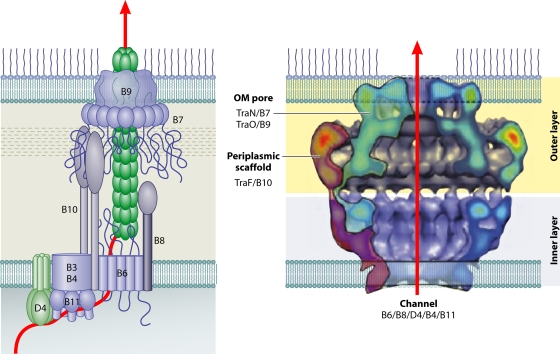
Localization of the A. tumefaciens VirB/VirD4 subunits. The coupling protein VirD4 and the Mpf components (VirB1 to VirB11) are represented according to their proposed functions: energetic, channel, or pilus components. IM, inner membrane; OM, outer membrane. VirB1 is processed to form VirB1*, which is exported across the outer membrane. VirB2 undergoes a novel head-to-tail cyclization reaction and polymerizes to form the T pilus. A VirB7 lipoprotein-VirB9-VirB10 complex forms a multimeric channel across the outer membrane. Crystal structures are shown for homologs of VirD4 (TrwBR388 soluble domain with an N-terminal TM domain modeled from electron microscopy images) (adapted from reference 113 by permission of Macmillan Publishers Ltd., copyright 2001), VirB11 (HP0525Hp) (adapted from reference 289 with permission of Elsevier), VirB5 (TraCpKM101) (adapted from reference 290 with permission of the publisher; copyright 2005 National Academy of Sciences, U.S.A.), VirB8 (soluble domain of VirB8Bs), and VirB10 (soluble domain of ComB10Hp) (VirB8 and VirB10 adapted from reference 261 with permission of the publisher. Copyright 2003 National Academy of Sciences, U.S.A.). VirB7, VirB9, and VirB10 assemble to form a transenvelope “core” complex (Fig. 5).
VirB4.
VirB4 proteins are large (>70-kDa) proteins with consensus Walker A and B NTP binding domains and additional sequence similarities among domains distributed along the entire polypeptide (219). C-terminal residues 426 to 787 of VirB4 resemble those of the TrwBR388 T4CP (190). This finding prompted speculation that these subunits might assemble as higher-order homohexamers and serve as docking sites for substrates (190), but at this time, there is no experimental support for this proposal. VirB4 subunits reside at the cytoplasmic face of the inner membrane, and there is evidence that one or two domains of VirB4At embed into or protrude across the inner membrane (71). An integral membrane association might not be obligatory, however, because VirB4-like TrwKR388 cofractionates with the membrane and the cytosol, and soluble TrwKR388 exhibits ATPase activity in vitro (9). Furthermore, TraCF and TrhCR27 associate with the inner membrane but probably peripherally through interactions with other T4SS subunits (109, 234).
VirB4 subunits are associated with every T4SS described to date. All homologs display similar physical properties such as a large molecular size and the presence of conserved NTP binding motif domains, although the number and location of predicted TM domains vary among family members. In scanning the virB4 loci of diverse prokaryotic species, we noticed a general feature that genes predicted to encode small integral membrane proteins typically reside upstream of and overlap with a virB4 gene. In gram-negative systems, this gene arrangement often consists of genes coding for homologs or orthologs of VirB2At pilin, VirB3At, and VirB4At. Indeed, among at least eight gram-negative T4SS, the virB3- and virB4-like genes are joined in frame and encode a VirB3/VirB4 fusion protein (63). Thus, it is reasonable to predict that complex formation between small membrane proteins and VirB4-like subunits is a general feature of diverse T4SS.
In the F plasmid transfer system, VirB4-like TraC is the only ATPase required for F pilus production. This is of interest because F pili are the only conjugative pili thus far shown to undergo dynamic extension and retraction (66). This strongly implicates TraC in energizing pilus polymerization and/or depolymerization reactions. However, the fact that VirB4-like subunits are signature components of T4SS regardless of whether the system produces pili suggests that these ATPases also energize the assembly or activity of the secretion channel (125, 169).
VirB11.
VirB11 subunits are structurally related to a large family of ATPases commonly associated with macromolecular trafficking systems, including type II secretion systems, type IV pilus systems, and archaeal flagellar biogenesis systems (289). Several VirB11 homologs, including VirB11At, TrbBRP4, TrwCR388, and H. pylori HP0525Cag, have been biochemically and structurally characterized. These proteins hydrolyze ATP, and this activity is stimulated by lipid in vitro, consistent with the affinity of these ATPases for membranes in vivo (159, 160, 224). TrbBRP4, TrwCR388, and H. pylori HP0525Cag assemble as homohexameric rings as shown by electron microscopy or X-ray crystallography (Fig. 4) (159, 160). In the HP0525Cag crystal structure (289), the monomer presents as two domains corresponding to the N-terminal (NTD) and C-terminal (CTD) halves of the protein. In the hexamer, the NTDs and CTDs form two separate rings, defining a chamber of ∼50 Å in diameter, which is open on the NTD side and closed on the CTD side. The CTD adopts a RecA fold, whereas the NTD is unique to HP0525Cag. The overall HP0525Cag structure appears to be highly conserved, even among the more distantly related ATPases associated with other transport or fimbrial biogenesis systems (289). More recently, an X-ray structure of Brucella suis VirB11 (VirB11Bs) was determined (123). The VirB11Bs monomer differs dramatically from that of HP0525 by a large domain swap caused by the insertion of additional sequences into the linker between the NTD and the CTD. The overall assembly of the VirB11 hexamer remains intact compared to HP5025Cag, but the domain organization modifies the nucleotide binding site and the interface between subunits. Based on sequence comparisons, most VirB11 subunits probably display a VirB11Bs-like architecture (123). VirB11At associates tightly with the cytoplasmic membrane, although other homologs, e.g., TrbBRP4, are predominantly cytosolic.
VirB11 homologs are widely distributed among gram-negative conjugation systems and effector translocator systems, and in these systems, these ATPases are essential for the assembly of secretion channels as well as pili. Interestingly, these ATPases are not associated with the H. pylori ComB DNA uptake system, the F plasmid transfer system (63, 155), or, with a few notable exceptions, gram-positive bacterial or archaeal systems (see below).
Inner Membrane Channel/Scaffold Subunits: VirB3, VirB6, VirB8, and VirB10
Besides the ATPases, the A. tumefaciens VirB/VirD4 system is composed of four inner membrane proteins, VirB3, VirB6, VirB8, and VirB10, that contribute in various ways to channel formation and activity (Fig. 4).
VirB3.
VirB3 was originally reported to associate with the A. tumefaciens outer membrane (146), but this location is inconsistent with hydropathy analyses predicting one or two α-helical TM domains for insertion into the cytoplasmic membrane. An inner membrane topology also fits with the finding that some T4SS are comprised of VirB3/VirB4 fusion proteins and observations that VirB4At stabilizes VirB3At (146, 292). VirB3At has been linked with a pilus assembly pathway (292), but it is also essential for substrate translocation (29). The available data suggest that VirB3 interacts with VirB2 and VirB4 at the inner membrane, but its precise contribution to machine function is presently unknown.
VirB6.
VirB6At is a polytopic membrane protein with a periplasmic N terminus, five TM domains, and a cytoplasmic C terminus (144, 145, 149). Polytopic subunits are features of all T4SS and most other bacterial secretion systems. As exemplified with the GSP subunit SecY, polytopic proteins generally assemble as dimeric or higher-order multimeric channels (85, 99). Consistent with such a channel activity, VirB6At forms formaldehyde-cross-linkable contacts with the T-DNA substrate during translocation, as shown with the TrIP assay, and VirB6 functionally interacts with other putative channel subunits, including VirB8 and a VirB7-VirB9-VirB10 core complex (see below) (49, 144). VirB6At possesses a large central periplasmic domain that plays an important role in mediating DNA substrate transfer through the distal portion of the secretion channel (144).
Although all described T4SS invariably possess one highly hydrophobic membrane protein with five or more predicted TM domains, the VirB6 family members display low overall sequence similarities. Most VirB6 subunits have a molecular mass of ∼30 to 35 kDa, but considerably larger polytopic subunits also exist. Here, we will term these proteins “extended VirB6” because they typically are composed of VirB6-like N-terminal regions bearing multiple TM domains joined to a large C-terminal hydrophilic domain. Such proteins often exceed 60 kDa, more than twice the molecular mass of VirB6At. This subfamily includes TraG subunits of the E. coli F plasmid and Vibrio cholerae SXT ICE (15, 185, 186). Like VirB6At, the N-terminal regions of extended-VirB6 subunits probably comprise part of the cytoplasmic membrane channel. However, recent studies indicate that the C-terminal extensions of these proteins display a range of biologically important extracytoplasmic functions. For example, some extended-VirB6 subunits participate in mating-pair stabilization and/or entry exclusion. The latter property, common among conjugative plasmids and some ICEs, prevents the redundant transfer of conjugative elements to donor cells. For two extended-VirB6 subunits, TraGF and TraGSXT, residues important for entry exclusion were mapped to the CTDs. Furthermore, these residues were shown to mediate specific interactions with entry exclusion proteins (Eex proteins) located in the inner membrane proteins of other donor cells (15, 185, 186). Two mechanisms were envisioned for the establishment of these distal contacts: the C-terminal domains might simply protrude through the T4SS and into the target cell, or they might be proteolytically released from the N-terminal domain and then translocate via the T4SS into the target cell (15, 185, 186).
Although these are the best-characterized examples of extended-VirB6 CTDs with extracytoplasmic functions, there is evidence for a surface exposure of extended-VirB6 subunits among rickettsial species (see below). Furthermore, it is interesting that ComEC, a channel subunit required for DNA uptake by the B. subtilis competence system, has extended-VirB6 features, although the disposition of the ComEC CTD at the B. subtilis cell envelope has not been rigorously examined (55).
VirB8 and VirB10.
The A. tumefaciens VirB/VirD4 system encodes two bitopic subunits, VirB8 and VirB10 (Fig. 4). VirB8 subunits display sequence similarities mainly in two regions corresponding to residues 100 to 143 and 190 to 235 of VirB8At. X-ray structures for the periplasmic fragments of B. suis and A. tumefaciens VirB8 subunits each present as a large extended β-sheet with five α-helices, giving rise to an overall globular fold (21, 261). The VirB8 subunits pack as dimers in the crystal structures, and results of mutational analyses suggest that dimerization is physiologically relevant. VirB10 subunits are bitopic inner membrane proteins typically with a short cytoplasmic domain, a TM domain, a proline-rich or coiled-coiled domain, and a large globular CTD (143). An X-ray structure for the CTD of H. pylori ComB10 presents as an extensively modified β-barrel with an α-helix projecting off one side and a second, flexible helix-loop-helix of 70 Å in length projecting off the top (Fig. 4) (261). ComB10 crystallized as a head-to-tail dimer; recent studies indicate that this arrangement is probably a packing artifact, although the VirB10 subunits do assemble as homomultimers (see below; 143).
Various domains of VirB8 and VirB10 contribute to self-association and interactions with other channel subunits. VirB8At interacts with multiple partners, including VirB1, VirB4, VirB5, VirB8, VirB9, VirB10, and VirB11, and there is also indirect evidence that VirB8At interacts with polytopic VirB6 (144, 161, 281, 292). Consistent with its multiple contacts, VirB8 is important for the spatial positioning of VirB proteins at the A. tumefaciens cell pole, leading to a proposal that VirB8 subunits function generally as nucleation factors during the assembly of T4SS (148, 162). In this context, it is noteworthy that VirB8At binds VirB1At, a transglycosylase that catalyzes the degradation of the peptidoglycan (see below) (281). The positioning of VirB8 at the cell envelope might determine the site of machine assembly through a combination of VirB partner contacts and the recruitment of a transglycosylase for localized murein degradation. Underscoring the functional importance of the VirB8At/VirB1At interaction, the B. pertussis Ptl system lacks a VirB1 homolog, but VirB8-like PtlE is fused to a transglycosylase domain (222).
VirB10 subunits are conserved among gram-negative bacterial T4SS and, like VirB8, form multiple subunit contacts with other Mpf subunits. VirB10At also functions dynamically by undergoing a conformational change in response to ATP energy use at the inner membrane (48). This structural transition, induced by the VirD4At T4CP and VirB11At ATP binding or hydrolysis, is necessary for the formation of a stable VirB7-VirB9-VirB10 channel complex and for DNA substrate passage through the distal portion of the translocation channel (48). Energized VirB10 is thus postulated to physically bridge inner and outer membrane subassemblies of gram-negative T4SS. Recent ultrastructural findings described below now firmly support the notion that VirB10 subunits function as structural scaffolds across the entire cell envelope.
Although most VirB10 homologs share similar topological and domain architectures, some subunits designated VirB10 in fact bear little similarity to other VirB10 subunits. Most notably, in H. pylori, proteins designated VirB10-like, e.g., HP0527, are associated with different Cag pathogenicity islands. This protein family ranges considerably in molecular mass from ∼200 to >1,000 kDa, and only the extreme C-terminal regions display sequence similarities with other VirB10 subunits. Most of these proteins possess centrally located, multiple-repeat sequences. These regions are thought to comprise hot spots for intragenic recombination that, over evolutionary time, resulted in the observed size variability. Unlike other VirB10 subunits, the H. pylori counterparts form part of a sheathed filament that extends from the H. pylori cell surface (227).
Periplasmic/Outer Membrane Channel Subunits: VirB1, VirB2, VirB5, VirB7, and VirB9
Besides the periplasmic domains of VirB6, VirB8, and VirB10, several other VirB subunits contribute to the assembly of the A. tumefaciens VirB/VirD4 secretion channel across the periplasm and outer membrane (Fig. 4).
VirB1 transglycosylases.
VirB1 homologs or other putative muramidases are associated with many T4SS of gram-negative and -positive bacteria. These subunits are delivered across the cytoplasmic membrane by the GSP and, as noted above, are thought to facilitate the assembly of the T4SS channel complex through the localized degradation of the peptidoglycan (294). In the A. tumefaciens VirB/VirD4 system, VirB1 is actually dispensable for channel assembly but still required for the biogenesis of the T pilus (29, 133). VirB1At is also proteolytically cleaved to liberate a C-terminal peptide, VirB1*; this fragment is delivered to the cell exterior, but its extracellular function is unknown (177, 300).
Outer membrane-associated subunits VirB7 and VirB9.
VirB7 subunits are small lipoproteins found in only a subset of gram-negative systems. In A. tumefaciens, VirB7 stabilizes VirB9, in part through the formation of a disulfide cross-link (Fig. 4) (6, 249). VirB7At localizes predominantly at the outer membrane, but cytoplasmic membrane, pilus-associated, and other extracellular forms also exist (96). In B. pertussis and Brucella abortus, VirB7 homologs also stabilize VirB9-like partners as well as other VirB subunits (78, 93). In B. abortus, mature VirB7 lacks an N-terminal Cys residue and thus is not lipid modified. B. abortus VirB7 is also not required for the formation of a functional secretion system (78). Many gram-negative T4SS encode lipoproteins with little sequence similarity to VirB7At that could supply a VirB7-like function or another novel activity. The H. pylori HP0532 lipoprotein, for example, is considerably larger than VirB7At and localizes extracellularly as a component of a large sheathed filament produced by the Cag T4SS (227).
VirB9 subunits are hydrophilic and localize in the periplasm and the outer membrane of gram-negative bacteria. VirB9At is composed of two conserved domains with an intervening nonconserved linker (142). As shown by the nuclear magnetic resonance structure, the C-terminal one-third of VirB9-like TraNpKM101 adopts a β-sandwich fold around which VirB7-like TraOpKM101 winds (26). The C-terminal region of VirB9At is part of the outer membrane channel, as shown by the surface accessibility of cysteine residues and epitopes introduced into this region (26). As described in more detail below, recent studies have shown that VirB7 and VirB9 form a central channel that, together with VirB10, spans the entire gram-negative bacterial cell envelope.
Pilus subunits VirB2 and VirB5.
VirB2 pilin proteins assemble as components of the secretion channel as well as the conjugative pilus. Typically, these are small (∼5- to 10-kDa) hydrophobic proteins with low overall levels of sequence relatedness. Upon cleavage of the unusually long signal sequence, the proproteins are further processed into the mature pilin. For example, the N terminus of the TraAF pilin is N acetylated (169), and TrbBRP4 and VirB2At undergo novel cyclization reactions resulting in the covalent linkage of their N and C termini (Fig. 4) (90, 153). Mature pilin integrates into the cytoplasmic membrane via two hydrophobic α-helices, presumably forming a pool for subsequent use in building the channel and pilus (169, 245).
Gram-positive bacterial and archaeal T4SS lack VirB2 homologs, but as noted above, genes encoding small proteins with pilin-like hydropathy profiles are often present and colinear with genes encoding VirB4-like subunits. In some systems, such components might in fact assemble as conjugative pili extending from the cell surface or pseudopili extending across the cell wall. Although no such pili have been detected on the surfaces of gram-positive bacteria, in the crenarchaeote Sulfolobus, strains carrying conjugative plasmids, e.g., pKEF9, elaborate long pili. A pKEF9-encoded subunit, P05, bears pilin-like features and might correspond to the pilin subunit (217, 235).
VirB5 subunits are exported to the periplasm, where they contribute to substrate transfer and pilus assembly. In an early study, it was shown that the conjugative donor activity of E. coli cells carrying a pKM101 derivative lacking the gene for VirB5-like TraC was restored upon mixture with TraC-producing cells (288). TraC mutations were also shown to disrupt bacteriophage attachment to pilus receptors, leading to a proposal that VirB5-like subunits are pilus constituents that mediate contacts between donor and recipient cells (288). Consistent with this proposal, VirB5At localizes at the tip of the A. tumefaciens T pilus, as shown by immunoelectron microscopy (4). As discussed further below, there is increasing evidence that extracellular forms of VirB2 and VirB5 homologs function generally in target cell attachment and host immune evasion in pathogens.
A T4SS CORE COMPLEX AND ROUTE OF SUBSTRATE TRANSLOCATION ACROSS THE GRAM-NEGATIVE BACTERIAL CELL ENVELOPE
Exciting structural and functional advancements are unveiling how T4SS machines are architecturally configured and how substrates are delivered across the gram-negative cell envelope (Fig. 5). VirB7, VirB9, and VirB10 homologs from the pKM101 T4SS were purified and shown by cryo-electron microscopy (CryoEM) to assemble as a 1-MDa channel with the potential of spanning the entire gram-negative bacterial cell envelope (102). The structure, determined at a 15-Å resolution, consists of 14 copies of each homolog. It is configured as a double-walled ring-like structure with an inner layer (I layer) composed of the N-terminal domains of the VirB9 and VirB10 homologs anchored in the inner membrane and opened at the base by a 55-Å-diameter hole. The outer layer (O layer), composed of the VirB7 homolog and the C-terminal domains of the VirB9 and VirB10 homologs, forms the main body and a narrow cap with a hole of 10 Å in diameter. The cap presumptively forms the channel across the outer membrane, but due to the small size of the opening, there are probably significant structural rearrangements during substrate translocation or pilus assembly (102).
FIG. 5.
Substrate translocation pathway and architecture of a T4SS core complex. The substrate translocation pathway depicted at the left (red arrow) was developed for the A. tumefaciens VirB/VirD4 system on the basis of DNA-channel subunit contacts identified by formaldehyde cross-linking (49). The DNA substrate cross-links with the VirD4 T4CP, VirB11 ATPase, polytopic VirB6, bitopic VirB8, VirB2 pilin, and outer membrane (OM) VirB9. These subunits are postulated to comprise the secretion channel, whereas other components, including VirB3, VirB4, and VirB10, promote channel assembly as protein scaffolds or through ATP-mediated conformational changes. A CryoEM structure of a core complex composed of pKM101 VirB7-like TraN, VirB9-like TraO, and VirB10-like TraF is presented at right (102). Fourteen copies of each subunit assemble to form a large double-walled chamber that presumptively spans the entire gram-negative cell envelope. Additional subunits listed at the entrance are postulated to position within the core chamber. (The cryo-electron microscopy structure is reprinted from reference 102 with permission from AAAS.)
A crystal structure of the C-terminal domain of a VirB10 homolog (H. pylori ComB10 [ComB10Hp]) was fitted into the electron density of the external wall of the O layer, with its N terminus directed toward the I layer (102, 261). In this location, VirB10 forms a scaffold linking the inner and outer membrane channel subunits and also energizes substrate translocation across the outer membrane, through ATP-mediated conformational changes. The I layer of the core complex is inserted into the inner membrane via the VirB10 TM domains. Other subunits required for translocation, including VirB6, VirB8, and the VirD4, VirB4, and VirB11 ATPases, are postulated to assemble as the inner membrane translocase within the 55-Å hole formed by the VirB10 scaffold. Pilin subunits, e.g., VirB2 and VirB5, could also be located within the VirB9/VirB10 chamber, forming a conduit for substrate transfer through the periplasm and across the outer membrane (Fig. 5) (102).
Complementing this VirB7/VirB9/VirB10 structure, as mentioned above, a formaldehyde-cross-linking assay termed TrIP was developed to identify contacts between translocating DNA substrates and the Mpf subunits of the A. tumefaciens VirB/VirD4 system (Fig. 5) (49, 64). Substrate contacts were detected with the VirD4 T4CP, VirB11 ATPase, VirB6 and VirB8, and VirB2 and VirB9. Further TrIP studies with nonpolar virB and virD4 null mutants identified the subunit requirements for channel-substrate contacts and enabled the definition of the substrate translocation pathway across the cell envelope. Accordingly, the VirD4 T4CP receptor first binds the substrate and delivers it to the VirB11 ATPase. VirB11 then coordinates with the VirD4 and VirB4 ATPases to transfer the substrate to channel the subunits VirB6 and VirB8 for passage across the inner membrane. Finally, the substrate is delivered through the portion of the channel composed of VirB2 and VirB9 for translocation across the periplasm and outer membrane (11, 49, 142, 144).
Taken together, the definition of the VirB7/VirB9/VirB10 core structure by CryoEM and the route of substrate translocation by TrIP offer a cohesive view of how type IV secretion substrates are delivered across the gram-negative cell envelope (Fig. 5). This general machine architecture and translocation mechanism are probably similar at least among dedicated DNA transfer systems. Whether the same channel structure mediates the translocation of protein substrates is not yet known. Particularly noteworthy in this context is the B. pertussis Ptl system, wherein the protein substrate is a multisubunit complex (pentameric PT) that assembles in the periplasm and engages with the T4SS specifically for transit across the outer membrane (39). It is difficult to envision how PT could enter a VirB7/VirB9/VirB10-like chamber for translocation across the outer membrane without significant conformational rearrangements. Also, as discussed in further detail below, the A. tumefaciens VirB/VirD4 T4SS and other gram-negative systems elaborate two surface structures, a secretion channel and a conjugative pilus or other surface filament. It is unclear whether the VirB7/VirB9/VirB10 channel complex visualized by CryoEM represents a scaffold for the secretion channel, the surface filament, or both organelles.
SURFACE STRUCTURES OF GRAM-NEGATIVE SYSTEMS
Surface structures contribute in a number of ways to biological functions of the T4SS. They establish contacts with target cells and promote attachment to abiotic surfaces to facilitate biofilm formation, and there is increasing evidence that they can contribute to the evasion of host immune defenses in pathogenic settings. The pili of gram-negative bacterial conjugation systems are the best characterized of the T4SS surface structures. The conjugative pili are composed of a pilin subunit and often a minor pilin subunit attached at the pilus tip and/or base. F plasmid-encoded pili are long (2 to 20 μm) and flexible, with a diameter of 8 nm and a central lumen of 2 nm, whereas the P pili elaborated by IncP plasmid RP4 are short (<1 μm) and rigid, with a diameter of 8 to 12 nm (154, 169, 245). F-like pili enable E. coli donor cells to transfer DNA in liquid media, whereas P-like pili support efficient transfer only on solid surfaces. F-like pili can be detected on the surfaces of F plasmid-carrying cells, and they can undergo cycles of extension and retraction to promote the formation of conjugative junctions (66). P-like pili are rarely detected on donor cell surfaces and probably do not retract; rather, these hydrophobic filaments are sloughed from the cell surface, where their adhesive properties are thought to mediate the aggregation of donor and recipient cells (233, 237).
Recent microscopy studies of the F pili have supplied both high-resolution structural information and insights into the dynamics and biological importance of pilus extension and retraction (66, 280). The structure of the F pilus was examined using CryoEM and single-particle methods. The tubular structure has a diameter of ∼8.5 nm, with two different subunit packing arrangements. One is a stack of pilin rings of C4 symmetry, and the second is a one-start helical symmetry with an axial rise of ∼3.5 Å per subunit and a pitch of ∼12.2 Å. These two packing arrangements seem to coexist within the pilus structure. The central lumen of the pilus is estimated to be ∼30 Å, large enough to accommodate ssDNA and an unfolded protein(s), although the question of whether the conjugative pilus serves as a conduit for substrate transfer remains a subject of debate (280).
F pilus extension and retraction has now been visualized with living cells by laser scanning confocal microscopy (66). Intriguingly, the images suggest that E. coli donor cells undergo cycles of pilus extension and retraction to sample the immediate surroundings. Extension involves the addition of pilin subunits to the cell-proximal base of the pilus. If the extended pilus establishes contact with a recipient cell, retraction generates a force sufficient to bring the cells together.
The transfer of F plasmid DNA to recipient cells has also been monitored in real time (16). In that study, an assay based on the binding of SeqA to hemimethylated DNA enabled the visualization of newly acquired DNA during conjugation. Recipient cells deficient in guanosine methylation at GATC sequences are engineered to synthesize SeqA fused to yellow fluorescent protein. When Dam+ donor cells are mated with Dam− recipient cells, the ssDNA substrate from the donor becomes hemimethylated upon second-strand synthesis in the recipient. SeqA-yellow fluorescent protein binds the hemimethylated DNA, enabling the visualization of newly acquired DNA by fluorescence microscopy. By use of this assay, newly acquired DNA could be localized within the recipient cell. Interestingly, DNA acquisition could be detected in the absence of direct cell-to-cell contact albeit at frequencies several orders of magnitude lower than previously recorded frequencies of F plasmid transfer (16). In these instances, DNA might translocate through the extended pilus, although other mechanisms such as transformation or trafficking through membrane blebs were not ruled out.
Although the F pilus might function as a conduit for DNA transfer, a large amount of genetic and biochemical data for the F plasmid transfer system strongly indicates that efficient transfer requires direct cell-to-cell contact followed by the stabilization of mating junctions (87, 233). Similarly, for systems in which conjugative pili are sloughed from the cell surface, e.g., plasmid RP4 and VirBAt/VirD4At systems, the isolation of “uncoupling” mutations that block detectable pilus production while permitting efficient DNA transfer establishes that conjugative pili are dispensable for substrate translocation. Such mutations have been isolated in pilin subunits and several other Mpf channel subunits (90, 142, 145, 230). The notion that pili are not required for T4SS has gained further support from studies showing that the B. pertussis Ptl system lacks a detectable pilus yet translocates PT into the milieu by a mechanism that is still dependent on the production of VirB2-like PtlA (287).
T4SS of gram-negative bacteria also elaborate a variety of other surface structures that are thought to function primarily or exclusively in attachment to target cells or abiotic surfaces. In H. pylori, the Cag pathogenicity island mediates the formation of sheathed appendages of 100 to 200 nm in length (227). The needle portion of this structure has a diameter of 40 nm, and the sheathed structure has a diameter of 70 nm. Subunits associated with this sheathed structure include HP0527 (VirB10-like domain), HP0532 (VirB7-like lipoprotein), HP0528 (VirB9-like), and, at its tip, the CagA effector protein (227, 256). Of further interest, HP0525 (VirB11-like ATPase) is dispensable for HP0532 surface localization and associations with extracellular filaments (256). HP0525 thus might contribute to morphogenesis or the function of the Cag T4SS in ways that are distinct from those of other VirB11 ATPases in mediating pilus assembly. H. pylori appears to elaborate another type of Cag-dependent pilus composed of HP0546 (VirB2At-like) and CagL (VirB5At-like) (17, 19, 164). Interestingly, CagL is a specialized adhesin that binds to and activates integrin receptors on mammalian epithelial cells through an Arg-Gly-Asp (RGD) motif (see below; 164). The relationship, if any, between the sheathed structure and the VirB2/VirB5-like pilus is not clear at this time. In L. pneumophila, the Dot/Icm system elaborates a different type of surface structure, a fibrous mesh that covers the cell surface and is composed of DotO and DotH subunits (283). This fibrous mesh is thought to facilitate specific stages of the L. pneumophila infection cycle. Below, we discuss other bacterial and archaeal surface appendages or adhesins associated with T4SS.
BIOLOGICAL DIVERSITY OF T4SS: MECHANISTIC THEMES AND VARIATIONS
In the next sections, we will summarize information available for T4SS functioning in phylogenetically diverse prokaryotes. The discussion will focus mainly on recently described systems in gram-negative obligate intracellular bacteria, gram-positive bacteria, wall-less bacteria, and members of the Archaea. Sometimes, information is limited to the identification of T4SS genes through genome sequencing, yet even in silico analyses can offer valuable insights regarding possible machine architectures and functions.
More Gram-Negative Systems
Gram-negative obligate intracellular bacteria.
Rickettsia spp. are a group of gram-negative obligate intracellular alphaproteobacteria responsible for a variety of human diseases (275). Members of the families Anaplasmataceae and Rickettsiaceae of Rickettsia are responsible for various arthropod-borne diseases of mammalian hosts. The genomes of these intracellular pathogens are generally small (∼1 to 1.5 Mb) and honed for survival in the specialized niches of arthropod and mammalian hosts. In this context, it is intriguing that these pathogens often possess T4SS gene clusters bearing duplications of certain virB-like genes. Homologs of most of the VirB/VirD4 genes were identified in all species of the Anaplasmataceae (Ehrlichia chaffeensis, Ehrlichia ruminantium, Anaplasma phagocytophilum, Anaplasma marginale, Wolbachia spp., Neorickettsia sennetsu) as well as the Rickettsiaceae (Rickettsia typhi, Rickettsia prowazekii, and Rickettsia conorii) (7, 61, 106, 108, 136, 140, 188, 205, 206).
As depicted in Fig. 6, the T4SS genes generally cluster into two or three groups (95, 106, 136, 207). One group codes for proteins resembling A. tumefaciens VirB3, VirB4, and VirB6. Particularly noteworthy is that these gene clusters code for three to four copies of VirB6 paralogs that we classify as extended VirB6 due to their considerably larger sizes (∼850 to >2,350 kDa) than VirB6At. These are polytopic membrane proteins with large hydrophilic domains located N terminally, centrally, or C terminally. VirB6-2 of E. chaffeensis undergoes proteolysis, resulting in the release of an 80-kDa fragment that accumulates in E. chaffeensis-containing vacuoles (22). Evidence was also presented for pairwise interactions among the E. chaffeensis VirB6 paralogs and for interactions between the 80-kDa proteolytic fragment and two VirB6 paralogs (22). In addition, a VirB6 subunit appears to localize at the cell surface of Wolbachia in infected insect cultured cell lines (223). The presence of multiple variant forms of VirB6-like subunits in obligate intracellular bacteria suggests the intriguing possibility that these proteins promote survival in the host through the binding of host cell receptors or evasion of host immune defenses.
FIG. 6.
T4SS genes in the genomes of gram-negative obligate intracellular pathogens are organized in distinct clusters. (A) Organizations of T4SS genes in some completed genomes of intracellular pathogens; noncontiguous gene clusters are separated by double cross-hatched lines (106, 136, 199). All genomes examined possess multiple copies of virB6-like genes. (B) Anaplasmataceae genomes also possess several copies of virB2-like genes often next to virB4-like genes. GenBank accession numbers are CP000235 for A. phagocytophilum, CP000236 for E. chaffeensis, CP000237 for N. sennetsu, and AE017196 for Wolbachia pipientis ωMel. Orthologous and paralogous genes are shown in the same color for all the genomes; gene sizes are not depicted to scale.
A second gene cluster codes for homologs of A. tumefaciens VirB8, VirB9, VirB10, VirB11, and VirD4 (Fig. 6). There is evidence for the transcription of this gene cluster and also for the surface exposure of the VirB9 subunits of E. chaffeensis and A. phagocytophilum (22, 94, 95, 178). Additionally, a VirD4-like T4CP is required for the translocation of the ankyrin repeat protein AnkA into the mammalian host cytoplasm (173). Thus, the data indicate that the virB-virD4 genes are expressed and that a functional VirB/VirD4 system assembles in these obligate intracellular pathogens.
A third gene cluster recently identified in species including A. phagocytophilum, A. marginale, Ehrlichia canis, E. chaffeensis, and Wolbachia spp. codes for several copies of VirB2 orthologs and VirB4 (Fig. 6) (106, 199). In view of the proposed role for VirB4 in mediating the assembly of conjugative pili, the preservation of the virB2-virB4 gene arrangement suggests that these proteins might assemble as an antigenically variable surface organelle in intracellular bacteria either independently of or in conjunction with the other VirB subunits. An interesting parallel exists in the related alphaproteobacterium Bartonella tribocorum, in which the Trw T4SS carries multiple tandem copies of virB2- and virB5-like genes (75). The Trw system also encodes tandemly duplicated copies of VirB6-like TrwI, although the variant TrwI proteins closely resemble VirB6At family members in size and hydropathy. These findings suggest that the obligate intracellular pathogens might have adopted a strategy similar to that of Bartonella spp. in synthesizing variant pili to promote survival in the host cell (see below).
Flanking the virB-virD4 loci of various intracellular pathogens are several additional genes of interest. For example, in A. phagocytophilum and E. chaffeensis (but not Rickettsia spp.), sodB, encoding an iron superoxide dismutase, is cotranscribed with the virB3-virB6 genes (60, 207). A protective oxidative stress response might thus be coupled with T4SS machine assembly in environments, e.g., the eukaryotic cytoplasm or blood-feeding ticks, where both activities are needed for cell survival or proliferation. In addition, flanking virB2-virB4 gene clusters of A. phagocytophilum and E. canis are genes encoding P44 outer membrane proteins or surface antigens, as well as membrane proteases. In A. phagocytophilum, P44 outer membrane protein genes also flank the virB8-virD4 gene cluster, and in E. chaffeensis, genes encoding a 120-kDa immunodominant surface antigen and a tetricopeptide repeat protein flank the corresponding virB-virD4 gene cluster (136). These gene relationships might reflect a coordination of expression and function among T4SS and various surface constituents. Finally, besides the virB-virD4 gene clusters described above, orphan virB genes are distributed throughout the genomes of Rickettsia species. These are generally virB2 or virB9 orthologs, but further studies are needed to assess their biological importance.
Although Rickettsia felis and other Rickettsia species have plasmids carrying a subset of F plasmid-like tra genes (107), a gene cluster on the Rickettsia bellii chromosome coding for 12 of the 17 F tra genes represents the most intact Tra system identified to date for Rickettsia organisms (107, 205). This gene cluster might elaborate a functional T4SS, as further suggested by electron microscopy images showing the presence of F-like sex pili extending from Rickettsia bellii cell surfaces and forming cell-to-cell contacts (205).
Genomes of all obligate intracellular Rickettsia spp. carry multiple T4SS gene clusters; however, the intracellular pathogen Orientia tsutsugamushi displays the most dramatic example of T4SS gene fragmentation described to date (61). The O. tsutsugamushi genome carries T4SS genes closely related to those of the F plasmid tra region arranged into 24 fragmented repeat clusters with eight or more genes per cluster as well as additional shorter gene clusters. Most of the gene clusters are flanked by tRNA or integrase genes, suggesting that they are remnants of ancestral genomic islands. Mutations and gene rearrangements generate a mosaic of intact genes and pseudogenes among all T4SS gene clusters (103). Nevertheless, the T4SS-mediated translocation of effector proteins might be important for O. tsutsugamushi intracellular survival and host switching, as suggested by the findings that the genome also carries genes for over 40 ankyrin repeat (Ank) proteins and that many of these are flanked on one or both sides by T4SS genes. Deciphering how T4SS gene fragmentation arose and how the putative T4SS gene products contribute to the intracellular life-style of O. tsutsugamushi are fascinating areas for future study.
Studies with chimeric T4SS and surrogate hosts.
Mechanistic studies of T4SS in obligate intracellular bacteria and other gram-negative pathogens are often constrained by the failure of these organisms to grow in laboratory culture. Efforts to reconstitute entire T4SS in heterologous hosts have met with limited success, but the use of chimeric systems or surrogate hosts has been informative. In Bartonella tribocorum, for example, the Trw system is highly related to the TrwR388 DNA conjugation system (75, 79, 241). Homologs of the two systems share 20 to 80% sequence identities, and the VirB7-, VirB9-, and VirB10-like core subunits exhibit >50% identities. In contrast to the TrwR388 system, the B. tribocorum Trw system lacks a cognate T4CP, fails to translocate DNA, and instead synthesizes an antigenically variable pilus thought to be important for the establishment of persistent erythrocyte infections in mammals (see below). Remarkably, B. tribocorum Trw genes encoding VirB5-like TrwH, VirB10-like TrwE, and VirB11-like TrwD substitute for their homologs in the TrwR388 system (79, 241). This finding underscores the conservation of machine architecture between two T4SS that appear to have evolved for completely different functions.
Studies of the intracellular Brucella sp. VirB systems are limited in part because of biosafety concerns. Brucella is phylogenetically closely related to Agrobacterium, and although the Brucella VirB system does not mediate substrate secretion when produced in A. tumefaciens, it assembles at least partly in A. tumefaciens. Years ago, it was shown that the synthesis of the A. tumefaciens VirB proteins in agrobacterial recipient cells greatly stimulates the acquisition of plasmid DNA during matings with agrobacterial donor cells (32, 175, 282). Intriguingly, the VirBBs proteins also stimulate DNA uptake when synthesized in agrobacterial recipient cells. Although the underlying mechanism for enhanced DNA uptake is unknown, this assay enables further structure-function studies of the Brucella VirB machinery in the A. tumefaciens surrogate host (45). Additionally, although most Brucella subunits are not interchangeable with homologs of other T4SS, VirB1Bs functionally substitutes for VirB1At (133). The periplasmic domain of VirB8Bs can also be swapped with the corresponding domain of a homolog encoded by the pSB102 plasmid transfer system (35). These findings support predictions from crystallographic studies that the CTDs of VirB8-like subunits are structurally conserved.
The use of a surrogate host also enabled the identification of secretion substrates of a T4SS functioning in the obligate intracellular pathogen Coxiella burnetii (273). C. burnetii, the causative agent of Q fever, is phylogenetically closely related to L. pneumophila and carries a Dot/Icm system resembling that of L. pneumophila. Several Coxiella dot/icm genes (dotB, icmS, icmW, and icmT) complement corresponding gene mutations in L. pneumophila, whereas a few do not (icmX, icmQ, dotM, dotL, dotN, and dotO) (296). The L. pneumophila Dot/Icm system delivers over 70 effector proteins during infection (202), but the Coxiella genome lacks genes for most or all of these effectors. Bioinformatic screens have, however, identified proteins with eukaryote-like domains including Ank repeats, tetratricopeptide repeats, coiled-coil domains, leucine-rich repeats, GTPase domains, ubiquitination-related motifs, and eukaryote-like kinases and phosphatases (273, 274). To test whether the Ank proteins might be T4SS substrates, C. burnetii genes were expressed in L. pneumophila cells and assayed for translocation through the L. pneumophila Dot/Icm system. Eleven C. burnetii Ank proteins were found to translocate through this heterologous system. These Ank proteins possess C-terminal regions necessary for translocation, and some Ank proteins also require the chaperone IcmS for secretion (Table 1) (273, 274). The use of L. pneumophila as a surrogate host has been valuable because until recently, C. burnetii cells could not be grown in laboratory culture or genetically manipulated. However, in two recent reports, both a medium supporting the axenic growth of C. burnetii cultures and the application of mariner-based Himar transposition for mutant constructions were described (27, 208). These significant advancements promise to accelerate genetic and biochemical studies elucidating the contribution of the C. burnetii Dot/Icm system to infection.
Gram-Positive T4SS
Conjugation among the gram-positive bacteria is a principal mechanism for the dissemination of multiple-antibiotic resistance and other virulence traits in clinical settings. An excellent review on conjugation in gram-positive bacteria appeared in 2003 (116). Here, we will update information on gram-positive elements by focusing on a few representative conjugation systems, mainly to draw comparisons with gram-negative systems and identify fundamental requirements for DNA translocation across the gram-positive envelope.
Conjugative DNA processing.
Relaxases associated with gram-positive T4SS have been classified into three phylogenetic groups (100, 116), but the characterized relaxases catalyze nicking by the same transesterase mechanism as that described above for gram-negative enzymes. Characterized relaxases include MobA (staphylococcal pC221) (46, 47), PcfG (E. faecalis plasmid pCF10) (58, 59), MobM (streptococcal plasmid pMV158) (92, 120), and TraA (S. agalactiae plasmid pIP501) (158, 163, 276). In vitro studies of the staphylococcal pC221 and E. faecalis pCF10 processing reactions established the importance of the accessory factors MobC and PcfF in coordinating the binding and catalytic activities of the relaxases MobA and PcfG at their respective oriT target sites (46, 47, 58). Studies of both systems support a model in which the accessory factors recruit MobA to the nic site and also facilitate nicking through strand opening. Furthermore, by examining the efficiency with which the pC221 and pCF10 Dtr factors catalyze nicking at oriT sequences of cognate and closely related plasmids, data were presented showing that minor nucleotide variations, specifically within the relaxase binding site, form the basis for the specificity of the nicking reaction (47, 58).
Like the gram-negative systems, some gram-positive relaxases, e.g., MobMpMV158 and TraApIP501, mediate plasmid transfer independently of other Dtr processing factors (92, 158). For both pMV158 and pIP501, the oriT regions are compactly organized, and the promoter for the relaxase gene overlaps the nic site (92, 163). Correspondingly, in the pIP501 system, relaxase binding to oriT autoregulates the expression of the relaxase and downstream transfer genes (163). Gram-negative plasmid pBBR1 has a nic region identical to that of pMV158, and reminiscent of pIP501, the binding of pBBR1 relaxase at oriT autoregulates downstream tra gene expression (255). It is postulated that this mode of regulation evolved for the feedback control of plasmid transfer rates to minimize the metabolic burden associated with conjugative DNA transfer (163).
Gram-positive ICEs are also processed for transfer with or without accessory factors. One particularly interesting ICE recently identified in B. subtilis, ICEBs1, efficiently transfers between cells in response to global DNA damage, and transfer among donor cell populations is also inhibited by a quorum-sensing immunity system (13, 14). Once the element excises from the chromosome, NicK relaxase catalyzes nicking at the oriT sequence in the absence of other factors (170, 171). In contrast, for the well-characterized ICE Tn916, the Orf20 relaxase catalyzes efficient and site-specific nicking at oriT only in the presence of integrase. In the absence of integrase, Orf20 functions not as a nickase but as an endonuclease that cleaves both strands of the oriT region at distinct sites enriched in GT dinucleotides (226).
T4CPs.
The T4CPs of gram-positive systems also resemble those of their gram-negative counterparts, with conserved NBDs and variable NTDs and CTDs enabling subclassification: PcfCpCF10 and ICEBs1 (VirD4At-like), Orf10pIP501 (short and hydrophilic with an associated membrane protein), TcpApCW3 (divergent N- and C-terminal regions), and Streptomyces FtsK/SpoIIIE-like T4CPs (dsDNA transfer across mycelial membranes) (Fig. 3). A few of these T4CPs have been characterized biochemically. The soluble form of PcfCpCF10 (deleted of its NTD) binds ssDNA and dsDNA substrates in vitro, and the native protein binds the pCF10 transfer intermediate in vivo (59). This form of PcfC binds ATP but does not display detectable levels of ATPase activity and exists in solution predominantly as a monomer (59). Some evidence was presented for the assembly of the Orf10pIP501 and TcpApCW3 T4CPs as dimers and higher-order multimers (1, 251), but presently, there is no firm evidence for hexamer formation by these or any other gram-positive T4CP. As mentioned above, TcpApCW3 deleted of its N-terminal putative TM domain still mediates plasmid transfer, in striking contrast to similar N-terminal mutant forms of T4CPs from gram-negative systems (259). At the least, this finding highlights a fundamental difference between TcpA and other characterized T4CPs. More significantly, it might raise questions about the T4CP channel model.
Further studies have identified several T4CP partner interactions. PcfCpCF10 and Orf10pIP501 interact with the cognate relaxases PcfG and TraA, and PcfC also interacts with the PcfF accessory factor in vitro (1, 59). PcfC also interacts with the processed form of pCF10 in vivo, as shown with the TrIP assay (59). Among the Mpf channel subunits, PcfC interacts with VirB6-like PrgH and VirB4-like PrgJ (C. E. Alvarez-Martinez, Y. Chen, and P. J. Christie, unpublished data), with the latter interaction being reminiscent of that between the VirD4At and VirB4At ATPases (11). TcpApCW3 and Orf10pIP501 also bind several putative channel subunits. For example, TcpApCW3 binds VirB6-like TcpH and a ∼350-kDa bitopic subunit, TcpCpCW3 (251), whereas Orf10pIP501 interacts with Orf6pIP501, a ∼450-kDa protein with a predicted central TM domain (1). Interestingly, these T4CPs also bind the putative peptidoglycan hydrolases TcpGpCW3 and Orf7pIP501, respectively (1, 251). This type of interaction has not been shown for gram-negative T4CPs, and the findings may indicate that gram-positive T4CPs supply a VirB8At-like function by recruiting cell wall-degrading enzymes to sites of machine assembly.
Type IV secretion channels.
Little is known about the architectures or mechanisms of action of type IV secretion channels in gram-positive bacteria. To identify possible mechanistic themes, here we summarize recent findings for five systems currently under study, three conjugative plasmids (E. faecalis pCF10, S. agalactiae pIP501, and C. perfringens pCW3) and two ICEs (enterococcal Tn916 and B. subtilis ICEBs1). The putative channel components of each T4SS and their predicted locations at the gram-positive cell envelope are depicted in Fig. 7.
FIG. 7.
Predicted subunits of five representative T4SS functioning in gram-positive bacteria. Dispositions of the T4SS subunits at the cell envelope are depicted schematically; topology predictions were derived from the TMHMM algorithm and have not been experimentally confirmed. ORFs 9 and 10 from pIP501 might comprise a novel two-partner T4CP, and the VirB4-like proteins might form a complex with an adjacent gene product, as indicated with double arrows and “?.” The VirB6-like subunits are subgrouped as VirB6 (PrgH and Orf12) or extended VirB6 (YddG, TcpH, and Orf15) (see the text for details). Proteins with topologies and/or domains indicative of a role as a T4SS component are depicted, with the exception of TcpIpCW3, a possible membrane-bound metal hydrolase important for pCW3 transfer. Proteins of similar sizes and predicted membrane topologies (TMHMM algorithm) are listed with the same color, and proteins exhibiting sequence relatedness in local alignments using the BLASTP or Psi-BLAST algorithm are listed with the same color and an asterisk. A minus sign indicates that no corresponding subunit is produced by the transfer system. CW, cell wall; CM, cytoplasmic membrane. GenBank accession numbers are NC_006827 for E. faecalis pCF10, L39769.1 and AJ505823.1 for S. agalactiae pIP501, AB001488.1 for B. subtilis ICEBs1, NC_010937.1 for C. perfringens pCW3, and NC_006372.1 for E. faecalis Tn916.
All gram-positive T4SS encode a VirB4-like putative ATPase of >750 residues. As mentioned above, a characteristic feature of many T4SS is that genes encoding small proteins with two or three predicted TM domains reside upstream of and often overlap with the virB4-like genes. In the pCF10 system, prgI overlaps prgJ, and we postulate that the two gene products functionally interact (Fig. 7). The pCF10 and pIP501 systems encode the polytopic subunits PrgH and Orf12, resembling VirB6At in size and number of predicted TM domains. The pCW3, Tn916, and ICEBs1 systems encode extended-VirB6 forms, each with large C-terminal extensions. These extensions of Orf15Tn916 and TcpHpCW3 carry coiled-coil domains, and Orf15Tn916 also has a putative eukaryotic microtubule binding protein MIPP-T3 domain (see below). We depict that these domains extend to the cell surface, where they might bind target cell receptors (Fig. 7); however, further studies are needed to experimentally test this prediction. The deletion of the C-terminal 251 or 318 residues of TcpHpCW3, currently the only characterized VirB6-like subunit of a gram-positive T4SS, diminishes or abolishes plasmid transfer, establishing the importance of this domain for protein function. A central periplasmic domain of TcpH carries a sequence motif, VQQPW, that is also found in Orf15Tn916, and a mutation of this motif abolishes TcpH function (251). As mentioned above, TcpH also self-associates and interacts with TcpC, a bitopic protein related to Orf13Tn916. Interestingly, both TcpH and VirB4-like TcpF localize at the cell poles of C. perfringens, raising the possibility that the Tcp channel assembles at the polar membrane, as shown for the A. tumefaciens VirB/VirD4 system (259).
Each of the five transfer systems also has one or more bitopic membrane proteins of ∼300 to 350 residues with N-proximal or central TM domains (Fig. 7). Bitopic TcpCpCW3 is related to Orf13Tn916 and YddBICEBs1, whereas PrgDpCF10 is related to Orf13pIP501. These subunits might provide VirB8- or VirB10-like scaffold functions for the portion of the secretion channel extending distally from the membrane. Another common feature is the presence of one or more small membrane proteins with two or three predicted TM domains that also might assemble as structural subunits of the membrane translocase. Another intriguing possibility warranting investigation is that one or more of these small hydrophobic subunits polymerize to form fibers extending across the thick murein cell wall, along or through which DNA substrates are translocated, reminiscent of the pseudopili produced in the B. subtilis competence system (56).
Each system also encodes a putative murein hydrolase (Fig. 7). Although these hydrolases fall into different classes on the basis of the predicted peptidoglycan linkages targeted for degradation, they all probably punch holes in the thick murein wall to enable channel assembly. Hydrolases are dispensable for channel formation across gram-negative envelopes (29, 165), but these enzymes might be essential for building type IV structures across the thick murein wall of gram-positive envelopes.
Finally, it is interesting that the five systems lack a VirB11 ATPase homolog. In fact, a few gram-positive systems do carry a VirB11 homolog (116), but these systems are confined to a group of Bacillus sp. plasmids that are closely related to Bacillus anthracis pX02. Plasmids of this group include pX02, pAW63, and pBT9727, and a recent survey also identified pX02-like plasmids encoding VirB11 homologs in Bacillus isolates from a variety of environmental niches (137). Many of these plasmids encode functional T4SS, as shown by their capacity to transfer small mobilizable plasmids (137). Why VirB11-like ATPases are common among gram-negative systems but restricted to a subset of Bacillus sp. systems is an intriguing question for further study.
Surface adhesins/factors.
Among the five systems under discussion, only pCF10 and pIP501 encode proteins with features (signal sequences and anchor domains) suggestive of surface display (Fig. 7). The pCF10 tra region codes for three surface proteins, PrgA, PrgB, and PrgC (Fig. 7) (127). PrgA functions in surface exclusion, and PrgB (also called aggregation substance [AS] or Asc10) mediates the attachment of E. faecalis cells to other bacterial and eukaryotic cells (65, 67, 86). PrgB is a large ∼137-kDa protein with homologs identified primarily in related pheromone-inducible conjugative plasmids in enterococci. In addition to an N-terminal signal sequence and C-terminal LPXTG cell wall anchor motif, PrgB contains internal RGD motifs and aggregation domains shown to be important for the binding of lipoteichoic acid, adherence to eukaryotic cells, and bacterial internalization by epithelial cells (see below) (65, 67, 221, 266, 284, 285, 286). prgB mutants show reductions in pCF10 transfer efficiencies by several orders of magnitude, although transfer still occurs on solid surfaces.
PrgCpCF10 is related to Orf15pIP50, and both proteins carry C-terminal cell wall anchor-like motifs (264). Both proteins also contain highly repetitive sequence motifs of a three-residue periodicity comprised of Pro-uncharged Glu/Asp. Orf15pIP50 possesses 47 of these tandem repeats along most of the protein (see below). Repeat regions enriched with Pro and negatively charged residues are features of other gram-positive surface proteins, including Streptococcus pyogenes Sfb, Iga binding proteins of S. agalactiae, and S. aureus and streptococcal fibronectin binding proteins (138, 198). Such Pro-rich regions characteristically adopt extended structures and also comprise protein-protein interaction surfaces (156). PrgC and Orf15 might thus function as structural scaffolds for the portion of the type IV channel that extends across the cell wall, or they might mediate specific contacts with target cells.
Although the pCW3, Tn916, and ICEBs1 systems apparently lack cell wall-anchored proteins, these systems encode extended-VirB6 subunits that potentially protrude to the cell surface (Fig. 7). It is intriguing to speculate that these systems have adapted extended-VirB6 subunits to provide functions analogous to those of the pCF10 and pIP501 cell wall-anchored proteins.
Cell Wall-Less Systems
Bacteria lacking cell walls are phylogenetically most closely related to gram-positive bacteria, and at least two genera, Mycoplasma spp. and Spiroplasma spp., have been shown to conjugatively transfer DNA (25, 73, 182, 183, 225, 258, 272). Mycoplasma pulmonis and Spiroplasma citri can transfer chromosomal DNA markers, and some early evidence suggested that bidirectional chromosomal transfer can occur during conjugation (25, 182). It was proposed that gene transfer might arise through the fusion of donor and recipient membranes, reminiscent of DNA exchange resulting from protoplast fusion in gram-positive bacteria. More recent studies have confirmed that the genomes of S. citri, Spiroplasma kunkelii, and Mycoplasma agalactiae carry T4SS genes (147, 183, 231). Thus far, discernible homologs are limited to T4CPs, VirB4-like ATPases, and polytopic VirB6-like proteins, but a minimized T4SS apparatus might suffice for substrate transfer across the simplified cell envelopes of wall-less bacteria.
S. citri plasmids and ICEs in Mycoplasma fermentans and M. agalactiae carry T4SS gene clusters (20, 73, 183, 231, 247). Interestingly, some M. fermentans ICEs lack homologs of known integrases, transposases, or recombinases, suggesting that a novel enzyme might be involved in the excision of these elements (43). The S. kunkelii genome also carries four virB4 genes, two on the chromosome and two presumptively on plasmids; virB4 transcripts have been detected in infected insects and plants, providing some evidence for biological function (20). Pili were also detected on the S. kunkelii cell surface, sometimes connecting two cells (210). If cells with simplified envelopes in fact elaborate conjugative pili, it will be exciting to define factors contributing to their spatial organization and biogenesis.
Another feature of interest is the association of genes for Spiroplasma adhesion-related proteins with T4SS loci (20, 73, 231). These proteins possess a repeated amino acid domain, localize extracellularly, and might mediate attachment to host target cells (see below). Other commonly associated genes code for ParA or Soj proteins or, intriguingly, in the case of S. kunkelii, MreB, a cell shape-determining protein postulated to be important for the helical morphology of spiroplasmas (20, 183, 231). These cytoskeletal proteins might function as spatial determinants for the assembly of the T4SS, reminiscent of the ParA-like proteins of gram-negative systems.
Mycobacterial Conjugation
Mycobacteria possess cell walls with complex branched hydrocarbons known as mycolic acids. These mycolic acid layers are exceptionally thick and form a highly ordered lipid bilayer membrane that is covalently attached to layers of complex sugars and the murein cell wall. In view of this novel cell envelope composition, it is perhaps not surprising that M. smegmatis conjugatively transfers DNA independently of classical T4SS components (98, 277). Initial screens for mutations affecting DNA transfer initially failed to identify any transfer-defective mutations but did result in the isolation of a number of hyperconjugative mutations. Intriguingly, these mutations mapped to a locus that codes for the ESX-1 secretion apparatus; this apparatus has been shown to secrete EsxA and EsxB as well as other proteins in M. smegmatis as well as Mycobacterium tuberculosis. The M. smegmatis esx-1 mutations conferred elevated transfer frequencies of ∼20- to 7,000-fold, leading to a proposal that the Esx-1 system negatively regulates DNA transfer through the release of signaling proteins or other molecules (278).
Also intriguingly, a more recent screen for mutations affecting DNA uptake by M. smegmatis recipient cells resulted in the isolation of mutations mapping not only to the esx-1 locus but also to the tad locus (69). The Tad system, originally identified in Aggregatibacter actinomycetemcomitans, mediates the assembly of Flp pili in that species (263). Further studies showed that tad mutations also conferred a hyperconjugative transfer phenotype in M. smegmatis donor cells (69). The Esx-1 and Tad secretion systems thus negatively regulate donor transfer and are also required for DNA acquisition by recipient cells. A working model posits that proteins secreted by these systems regulate specific donor/recipient cell contacts necessary for DNA transfer. In M. tuberculosis, the Esx-1 system does not modulate DNA transfer but instead secretes virulence factors that target mammalian cell receptors to aid in intracellular survival and the suppression of host responses (2, 80). Therefore, in M. smegmatis, the Esx system appears to have evolved for interbacterial communication at least in part to modulate DNA transfer, whereas in M. tuberculosis, this system was adapted to secrete factors to modulate the pathogen-host interaction (69).
In a recent update, a lipoprotein-metalloprotease, LpqM, was shown to be important for M. smegmatis DNA donor activity. LpqM probably does not contribute directly to DNA transfer but instead might generate extracellular signal molecules through the proteolysis of proteins released by the Esx-1 or Tad systems (200). These peptide signals could in turn induce the expression of the DNA processing and transfer genes, reminiscent of the pheromone-inducible plasmid transfer systems of gram-positive bacteria.
Archaeal Systems
Archaeal cell envelopes can also possess novel constituents, including mono- or bilayer lipid membranes, rigid cell walls made up of polysaccharides or pseudomurein, no cell walls, or glycoprotein S layers. Only a few archaeal conjugation systems have been described, so far only for crenarchaeal species (91, 174, 217, 243, 250). An early study reported that Haloferax volcanii bidirectionally translocates chromosomal DNA transfer during conjugation, and large structures (2 mm long and 0.1 mm wide) bridging cells were postulated to mediate transfer (228). Recent genome-sequencing studies have identified at least two Haloferax plasmids with genes coding for T4CP and VirB4 homologs, although transmissibility of these plasmids has not been demonstrated (248).
Thermophilic crenarchaeal Sulfolobus spp. carry a number of conjugative plasmids grouped as pKEF or pARN plasmids (115, 174). Both plasmid groups encode T4CP- and VirB4-like proteins, several other predicted membrane proteins, and possible pilin subunits. The Sulfolobus T4CPs are considerably larger than VirD4At or TrwBR388, and their N- and C-terminal regions are unrelated to other T4CPs (Fig. 3) (115). Neither the N-terminal region nor the upstream ORFs carry predicted TM domains, although possible TM domains might be located between the Walker A and B NTP binding motifs. The VirB4-like proteins are slightly smaller than their bacterial counterparts, their N termini are unrelated to other family members, and they lack predicted TM domains (174). The polytopic proteins fall into the subfamily of extended-VirB6 subunits with C-terminal hydrophilic domains. As noted above, hydrophobic pilin-like proteins, e.g., pKEF9 p05, might assemble to form the pili detected on Sulfolobus solfataricus donor cells (235).
THE EVOLUTION OF T4SS
T4SS have evolved to perform a wide variety of biological activities profoundly influencing genome architectures and infection processes. The biological consequences of T4SS are subjects of several excellent reviews (18, 19, 24, 63, 89, 202, 212, 218). Here, we wish to explore two ideas relating to the evolution and biological complexity of T4SS. The first is that T4SS, particularly of the conjugation subfamily, have acquired many regulatory mechanisms to ensure their maintenance while at the same time mitigating the metabolic burden that they impose on their prokaryotic hosts. The second is that T4SS, particularly of the effector translocator subfamily, have acquired novel surface features as a result of an ongoing dialogue between the bacterial pathogen and eukaryotic target.
Modulation of Lateral Gene Transfer
Conjugative elements clearly can benefit their prokaryotic hosts because cargo genes often encode antibiotic resistance, virulence traits, or other metabolic functions that enhance cell survival in specific environmental niches. However, mobile elements in general also impose a metabolic burden, and to minimize the risk of killing the bacterial host, various mechanisms have evolved, by both the element and the prokaryotic host, to regulate propagation. Mechanisms controlling T4SS gene expression range from simple feedback systems monitoring intracellular levels of Dtr factors, e.g., relaxases, to more complex regulatory systems linking the cell envelope stress response or various environmental signals to T4SS gene expression (168, 220, 293, 295). Other mechanisms that have evolved to control propagation operate at the level of replication (copy number control and incompatibility), partitioning, or entry exclusion (15, 185, 186, 204). While several of these are general plasmid maintenance mechanisms with which T4SS have become genetically linked, the intriguing example of entry exclusion discussed above, in which the F plasmid and V. cholerae SXT ICE prevent the redundant acquisition of related elements through TraG-Eex protein interactions, underscores the specificity with which T4SS have evolved to distinguish self from nonself.
The above-described plasmid/ICE-encoded regulatory mechanisms are well known, but recent work indicates that prokaryotic hosts also have evolved at least two immunity systems for controlling “infection” by mobile elements. One involves the acquisition of elements in the genome of clustered regularly interspaced short palindromic repeats (CRISPR) and associated CRISPR-associated sequence proteins (184, 191, 242). These elements, present in about 40% of bacterial species and almost all archaeal species, confer sequence-directed immunity against incoming plasmids and phages (172). The exact mechanism of action is not yet clear but appears to involve an interference machinery that directly targets the incoming DNA through the recognition of homologous sequences (191, 242). An interesting example of CRISPR-mediated inhibition of conjugative DNA transfer occurs in Staphylococcus epidermidis, wherein a CRISPR sequence matching a relaxase gene that is present in nearly all conjugative plasmids of staphylococci prevents plasmid acquisition during conjugation (184).
The second immunity mechanism involves the recognition of foreign DNA through HN-S silencing. HN-S is a DNA binding protein that is well known for its role in transcription silencing through the blockage of RNA polymerase access to or progression along DNA (196, 197). HN-S preferentially binds A+T-rich DNA and thus is thought to recognize and silence A+T-rich foreign DNA. Interestingly, through the ongoing dialogue between mobile elements and prokaryotic hosts, several mechanisms have also evolved to counteract HN-S silencing of foreign DNA. Such counteractive tactics are encoded by either the bacterial host or an incoming mobile element (83, 252). If encoded by the bacterial host, antisilencing might allow the sampling of incoming DNAs for potentially beneficial traits. If encoded by an incoming element, antisilencing likely evolved for means of selfish propagation.
Evolution of T4SS-Associated Surface Structures
There is also accumulating evidence that T4SS have adapted or acquired a variety of surface organelles or adhesins over evolutionary time (Fig. 8). Clearly, one function of T4SS-associated surface factors, e.g., conjugative pili, is to mediate attachment to target cells specifically to promote efficient substrate transfer. However, recent evidence suggests that T4SS surface constituents can also mediate attachment to a variety of biotic or abiotic surfaces to promote biofilm formation, expand the repertoire of susceptible target cells, or modulate interactions with host cells in other ways. In considering the spectrum of surface-variable components associated with T4SS, many of which have been discussed in this review, we can identify three main mechanisms or origins.
FIG. 8.
Surface proteins shown or predicted to be associated with T4SS. (Left) Proteins are grouped according to their sequence relatedness to A. tumefaciens VirB subunits. (Center) Schematics of novel features and domains. SP, signal peptide; lipo box, lipoprotein motif; c-coil, coiled coil; anchor, cell wall anchor motif; aa, amino acids. Gene (arrows, top) and protein (rectangles) sizes are not to scale. (Right) Description of surface variation and possible function. Studies describing demonstrated or postulated functions in the environmental response and/or adaptation are cited. PMN, polymorphonuclear leukocytes; LTA, lipoteichoic acid.
One mechanism, illustrated by the Bartonella Trw system, possibly the rickettsial systems, the H. pylori Cag system, and others is the acquisition of antigenically variable pilin or pilus-associated subunits (Fig. 8). For example, the Bartonella Trw system possesses tandem copies of VirB2- and VirB5-like genes and is thought to be specifically adapted for the production of antigenically variable pili through intergenic recombination (75). Among Rickettsia species, the presence of tandem virB2-like genes often adjacent to a virB4-like gene suggests that these organisms similarly display surface-variable pilin or pili as a result of recombination or differential gene expression (106). H. pylori does not encode multiple copies of pilin subunits but rather evolved a novel VirB5-like CagL subunit with an RGD motif (Fig. 8). As shown for VirB5At, CagL is thought to localize at the tip of a pilus-like structure where its RGD mediates binding to host cell integrin (17). For these systems, the presentation of variable pilin or pilus structures at the cell surface presumably enables bacteria to attach to distinct cell types or evade the host immune system for enhanced survival within the eukaryotic host.
A second mechanism by which T4SS have acquired novel surface functions is through the adaptation of other Mpf subunits (Fig. 8). In the Bartonella Trw system, clusters of trwJIH genes, encoding homologs of VirB5, VirB6, and VirB7, occur five times in tandem, potentially representing a source of structural and functional variation among secretion channels as a result of intergenic recombination or differential gene expression. Several other T4SS have also evolved distinct forms of VirB6 subunits. As discussed above, the C termini of least two “extended-VirB6” subunits, TraG associated with the F plasmid and SXT ICE systems, mediate specific interactions with proteins in the recipient cell either through protrusion or through the proteolytic release/active translocation mechanisms. Obligate intracellular Rickettsia spp. also code for multiple VirB6 variants, and a processed form of one such protein is released into Ehrlichia-containing vacuoles (22). Several gram-positive T4SS also encode extended-VirB6 subunits, and some of these carry protein-protein interaction motifs of possible importance for host cell interactions (Fig. 8).
In the H. pylori Cag system, a number of Mpf subunits designated “VirB-like” in fact bear little resemblance to the VirBAt proteins (17, 164, 227, 256) (Fig. 8). Most interesting are CagT and CagY, two subunits reported to be similar to the A. tumefaciens core subunits VirB7 and VirB10, respectively. However, CagT and CagY are considerably larger than their VirB counterparts, both localize extracellularly as a component of a large sheathed filament produced by the Cag T4SS, and both possess variable- or multiple-repeat regions (8, 227). Among Cag systems of different H. pylori isolates, repeat regions of these subunits display considerable variation in size and composition. Surface variation resulting from intragenic recombination in cagT or cagY might play a role in the modulation of H. pylori-host cell interactions or immune evasion.
A third mechanism with which T4SS have acquired surface variability is through genetic linkage and the coordinated expression of genes encoding novel surface proteins (Fig. 8). This mechanism seems to predominate among gram-positive and wall-less bacteria. For example, in the gram-positive pCF10 and pIP501 systems, genes encoding cell wall-anchored surface proteins are coexpressed with T4SS subunit genes (65, 67, 86). In pCF10, at least two cell wall-anchored proteins, termed AS (PrgB) and surface exclusion protein (PrgA), are coproduced with the channel subunits. Both proteins modulate DNA transfer efficiencies; however, both are dispensable for DNA transfer and thus are not structural subunits of the secretion channel (59, 65, 67, 86). AS does, however, possess variable domains and RGDs that are implicated as contributing factors in binding, internalization, and the intracellular survival of E. faecalis cells during infection of human hosts (285) (Fig. 8). pCF10 and pIP501 also encode the putative surface factors PrgCpCF10 and Orf15pIP501, both of which carry Pro-X-Glu/Asp repeat triads reminiscent of repeats carried by other gram-positive surface proteins (Fig. 8). Although little is known about these proteins, they also probably are dispensable for DNA transfer and instead benefit the bacterial host in another unspecified way(s). Finally, in wall-less bacteria, genes encoding outer membrane proteins or surface adhesins are commonly linked to T4SS gene clusters (20, 73). Among these, the Spiroplasma adhesion-related proteins contain repeat domains as well as variable regions that are thought to play a role in mediating adhesion to insect or other cells (Fig. 8) (231).
SUMMARY AND FUTURE DIRECTIONS
Studies of a few “paradigmatic” T4SS in gram-negative bacteria have shaped our views of how these machines move macromolecules across the cell envelope. Among the three broad biochemical processes required for substrate transfer (processing, recruitment, and translocation), DNA processing by Dtr factors is the best understood at a molecular level. This is largely because the catalytic Dtr proteins or domains have proven amenable to purification and in vitro biochemical and structural characterizations. Despite these advances, however, we still do not know the details of how DNA or protein substrates are recognized and recruited to the T4SS channel. Relaxases carry the information necessary for the translocation of specific DNA substrates, and there is evidence that at least some relaxases carry C-terminal recognition signals. However, the importance of C-terminal substrate signals is better established with protein substrates of effector translocator systems. Where characterized, these signals consist of unstructured termini with clusters of positively charged or hydrophobic residues (Fig. 1 and Table 1). How these tail sequences are recognized or interact with the T4SS channel remains to be determined, but the X-ray structure of the TraDF T4CP C-terminal extension with the accessory factor TraM might form the basis for a general docking model. How other signals or motifs, e.g., BID domains, coordinate substrate-channel docking or substrate translocation through the T4SS channel also remains to be elucidated.
There is increasing evidence for contributions of accessory Dtr or other adaptor proteins to substrate recruitment (Fig. 1). Proteins such as ParA/Soj-like VirC1At, E. faecalis PcfFpCF10, and MobBR6112 appear to mediate a complex network of interactions between the substrate, the membrane, and the T4CP receptor, presumably to enhance the efficiency of the docking reaction. At this time, none of these factors has been shown to move dynamically in the cell, as might be expected of a factor engaged in substrate recruitment to T4SS channels localized at specific sites on the cell envelope. However, such a scanning mechanism cannot be excluded, nor can one involving the directed movement of substrates along an actin- or tubulin-like cytoskeleton. The association of ParA- or MreB-like proteins with ICEs (as well as many conjugative plasmids) raises the intriguing possibility that cytoskeletal factors contribute generally to the positioning of T4SS substrates at or near the secretion channel entrance.
Genetic, biochemical, and structural data support the notion that T4CPs function as substrate receptors; however, the molecular details underlying the substrate-T4CP interaction or the contribution of T4CPs to translocation remain undefined. In this review, we have highlighted the phylogenetic diversity and domain heterogeneity of T4CPs (Fig. 2 and 3). Among the T4CPs warranting further structure-function studies, clearly, analyses of those lacking predicted TM domains, e.g., Orf10pIP501 and VirD4-like TcpHpCW3, will permit a rigorous test of the working model suggested by the TrwB structure that these ATPases deliver substrates across the cytoplasmic membrane through a lumen formed by the hexameric NBD and TM stem (Fig. 4).
Only a few subunits are widely conserved among T4SS channels: (i) a VirB4-like ATPase (all systems), (ii) a VirB6-like polytopic subunit (all systems), (iii) a VirB1-like transglycosylase (many systems), (iv) VirB7-VirB10 core subunits (most or all gram-negative systems), and (v) a VirB11-like ATPase (most gram-negative and a few gram-positive systems). On the basis of the DNA translocation pathway defined by TrIP for the A. tumefaciens VirB/VirD4 system and the CryoEM structure of the pKM101 homologs of VirB7, VirB9, and VirB10 (Fig. 5), we envision the following general architecture for T4SS channels of gram-negative bacteria. The cytoplasmic membrane translocase, composed of a ternary complex of two or three ATPases (T4CP, VirB4, and VirB11), polytopic VirB6, and bitopic VirB8 assembles within the base of the core chamber complex comprised of VirB7, VirB9, and VirB10 (Fig. 5). In the more distal portion of the chamber, the periplasmic domain of VirB8 assembles with VirB2, VirB9, and VirB10 to form the channel extending through the periplasm and outer membrane. Accordingly, the chamber complex serves as a scaffold linking the cytoplasmic membrane translocase to the outer membrane pore.
While this proposed architecture is conceptually gratifying, many fundamental questions remain. First, is it correct? Both the TrIP and CryoEM data should be interpreted cautiously, the former because of the possibility of recovering indirect or nonspecific DNA substrate-subunit contacts by formaldehyde cross-linking and the latter because only 3 of the more than 10 required Mpf subunits are represented in the structure. If the channel structure is architecturally configured as we suggest, does it function to translocate substrates, assemble extracellular pili, or both? At this time, contrary to depictions by many science textbooks and reviews on conjugation, there is no firm evidence that the gram-negative T4SS is composed of a channel attached to a pilus or that the pilus serves as a conduit for substrate transfer. In fact, compelling evidence exists to the contrary, e.g., mutations that “uncouple” substrate transfer from pilus formation and electron microscopy images of tight mating junctions without a detectable pilus. These findings are at least consistent with the notion that the Mpf subunits can alternatively assemble as a translocation channel or pilus. If so, how do the channel versus pilus basal structures differ, and what signals regulate channel versus pilus assembly? With respect to the energetics of translocation, what are the contributions of the ATPases to the biogenesis of the translocation channel or pilus or to substrate translocation? How do the ATPases coordinate their activities to energize these processes? Does the channel assemble dynamically in response to substrate engagement and/or ATP hydrolysis? Future studies are needed to address all of these questions.
Can we extrapolate from data obtained for a few paradigmatic T4SS to other gram-negative systems or systems functioning in other prokaryotes? At this time, it seems reasonable to predict that all T4SS employ a common mechanism for delivering substrates across the cytoplasmic membrane. This membrane translocase consists minimally of a T4CP ATPase, a VirB4-like ATPase, and a polytopic membrane protein. We suggest that the main force driving the evolution of genetic complexity among T4SS was the requirement that these machines span anatomically diverse envelopes to convey substrates from the cytoplasmic membrane to the cell surface. T4SS of gram-negative bacteria appear to have accomplished this through the assembly of a core structural scaffold comprised of VirB10- and VirB9-like subunits for housing the transenvelope channel. We predict that another type of common scaffold might have evolved for T4SS of gram-positive bacteria, possibly a structure resembling the B. subtilis competence pseudopilus along or through which substrates pass.
Finally, although the ancestral T4SS-associated surface organelles or adhesins likely functioned exclusively to promote the formation of stable mating junctions, it is now evident that many T4SS have adapted a variety of surface components for novel purposes relating to the ongoing dialogue between donor and target cells. Many gram-negative systems express variant forms of pilus subunits, e.g., VirB2 and VirB5, or channel subunits, e.g., VirB6, VirB7, and VirB10, whereas systems functioning in gram-positive and wall-less bacteria have acquired a variety of different surface factors. These are not Mpf channel components per se and are thus not essential for substrate transfer. In pathogenic settings, these surface factors might promote adherence or, through surface variability, the colonization of different cell types or evasion of host immune defenses. Indeed, some T4SS, e.g., the Bartonella Trw system, apparently evolved from an ancestral conjugation system so that it now functions exclusively to modulate the bacterium-eukaryotic host cell interaction. A large number of T4SS-associated surface organelles or proteins have been identified, yet defining how they contribute to substrate transfer or other aspects of T4SS biology awaits further study.
In sum, these are exciting times for the field of T4SS. Our overarching premise is that we will come to understand the full biological complexity of T4SS only through a variety of experimental approaches directed toward the characterization of many different prokaryotic systems. We hope that this review stimulates research exploring these richly diverse organelles.
ADDENDUM IN PROOF
A recently published manuscript (V. Chandran et al., Nature doi:10.1038/nature08588, 2009) presents an X-ray structure of the O-layer of the Escherichia coli pKM101 core complex (see Fig. 5). The structure reveals a strikingly different outer membrane architecture of gram-negative T4SS than previously depicted (see Fig. 4 and 5). Most notably, the antenna projection of VirB10-like subunits (see Fig. 4) extends across the outer membrane and forms a pore comprised of alpha-helices. This is the first crystal structure of a heteromultimeric outer membrane channel complex. Furthermore, this is the second-described example of an alpha-helical outer membrane channel (the other is formed by the Wza octomer). Finally, taken together with previous findings (E. Cascales and P. J. Christie, Proc. Natl. Acad. Sci. USA 101:17228-17233, 2004; S. J. Jakubowski et al., Mol. Microbiol. 71:779-794, 2009), these results establish that VirB10-like subunits are the first-described proteins to span the entire gram-negative cell envelope. As mentioned in the text, VirB10-like subunits are postulated to function both as scaffolds for machine assembly and also as sensors of ATP energy use by the VirD4/VirB ATPases. These new findings suggest that VirB10 orchestrates the conversion of chemical energy derived from ATP hydrolysis at the inner membrane to a structural change in the alpha-helical pore to enable substrate passage and/or pilus polymerization across the outer membrane.
Acknowledgments
We thank laboratory members for insightful comments and discussions.
Work in this laboratory is supported by the National Institutes of Health (grant GM48746). C.E.A.-M. was supported in part from a Brazilian postdoctoral fellowship from CAPES-Brazil.
REFERENCES
- 1.Abajy, M. Y., J. Kopec, K. Schiwon, M. Burzynski, M. Doring, C. Bohn, and E. Grohmann. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J. Bacteriol. 189:2487-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdallah, A. M., N. C. G. van Pittius, P. A. Champion, J. Cox, J. Luirink, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, and W. Bitter. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883-891. [DOI] [PubMed] [Google Scholar]
- 3.Al-Khodor, S., C. T. Price, F. Habyarimana, A. Kalia, and Y. Abu Kwaik. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70:908-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aly, K. A., and C. Baron. 2007. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153:3766-3775. [DOI] [PubMed] [Google Scholar]
- 5.Amor, J. C., J. Swails, X. Zhu, C. R. Roy, H. Nagai, A. Ingmundson, X. Cheng, and R. A. Kahn. 2005. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J. Biol. Chem. 280:1392-1400. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 8.Aras, R. A., W. Fischer, G. I. Perez-Perez, M. Crosatti, T. Ando, R. Haas, and M. J. Blaser. 2003. Plasticity of repetitive DNA sequences within a bacterial (type IV) secretion system component. J. Exp. Med. 198:1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arechaga, I., A. Pena, S. Zunzunegui, M. del Carmen Fernandez-Alonso, G. Rivas, and F. de la Cruz. 2008. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J. Bacteriol. 190:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atmakuri, K., E. Cascales, O. T. Burton, L. M. Banta, and P. J. Christie. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 26:2540-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atmakuri, K., Z. Ding, and P. J. Christie. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auchtung, J. M., C. A. Lee, K. L. Garrison, and A. D. Grossman. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 64:1515-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auchtung, J. M., C. A. Lee, R. E. Monson, A. P. Lehman, and A. D. Grossman. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA 102:12554-12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Audette, G. F., J. Manchak, P. Beatty, W. A. Klimke, and L. S. Frost. 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153:442-451. [DOI] [PubMed] [Google Scholar]
- 16.Babic, A., A. B. Lindner, M. Vulic, E. J. Stewart, and M. Radman. 2008. Direct visualization of horizontal gene transfer. Science 319:1533-1536. [DOI] [PubMed] [Google Scholar]
- 17.Backert, S., R. Fronzes, and G. Waksman. 2008. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 16:409-413. [DOI] [PubMed] [Google Scholar]
- 18.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 19.Backert, S., and M. Selbach. 2008. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell. Microbiol. 10:1573-1581. [DOI] [PubMed] [Google Scholar]
- 20.Bai, X., T. Fazzolari, and S. A. Hogenhout. 2004. Identification and characterization of traE genes of Spiroplasma kunkelii. Gene 336:81-91. [DOI] [PubMed] [Google Scholar]
- 21.Bailey, S., D. Ward, R. Middleton, J. G. Grossmann, and P. C. Zambryski. 2006. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc. Natl. Acad. Sci. USA 103:2582-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao, W., Y. Kumagai, H. Niu, M. Yamaguchi, K. Miura, and Y. Rikihisa. 2009. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J. Bacteriol. 191:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardill, J. P., J. L. Miller, and J. P. Vogel. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56:90-103. [DOI] [PubMed] [Google Scholar]
- 24.Baron, C., and B. Coombes. 2007. Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect. Disord. Drug Targets 7:19-27. [DOI] [PubMed] [Google Scholar]
- 25.Barroso, G., and J. Labarere. 1988. Chromosomal gene transfer in Spiroplasma citri. Science 241:959-961. [DOI] [PubMed] [Google Scholar]
- 26.Bayliss, R., R. Harris, L. Coutte, A. Monier, R. Fronzes, P. J. Christie, P. C. Driscoll, and G. Waksman. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl. Acad. Sci. USA 104:1673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beare, P. A., D. Howe, D. C. Cockrell, A. Omsland, B. Hansen, and R. A. Heinzen. 2009. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J. Bacteriol. 191:1369-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bigot, S., V. Sivanathan, C. Possoz, F. X. Barre, and F. Cornet. 2007. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 64:1434-1441. [DOI] [PubMed] [Google Scholar]
- 31.Boer, R., S. Russi, A. Guasch, M. Lucas, A. G. Blanco, R. Perez-Luque, M. Coll, and F. de la Cruz. 2006. Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J. Mol. Biol. 358:857-869. [DOI] [PubMed] [Google Scholar]
- 32.Bohne, J., A. Yim, and A. N. Binns. 1998. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc. Natl. Acad. Sci. USA 95:7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41-51. [DOI] [PubMed] [Google Scholar]
- 34.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Lavigne, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. Type IV secretion and Brucella virulence. Vet. Microbiol. 90:341-348. [DOI] [PubMed] [Google Scholar]
- 35.Bourg, G., R. Sube, D. O'Callaghan, and G. Patey. 2009. Interactions between Brucella suis VirB8 and its homolog TraJ from the plasmid pSB102 underline the dynamic nature of type IV secretion systems. J. Bacteriol. 191:2985-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowie, J. U., and R. T. Sauer. 1990. TraY proteins of F and related episomes are members of the Arc and Mnt repressor family. J. Mol. Biol. 211:5-6. [DOI] [PubMed] [Google Scholar]
- 37.Brochet, M., V. Da Cunha, E. Couve, C. Rusniok, P. Trieu-Cuot, and P. Glaser. 2009. Atypical association of DDE transposition with conjugation specifies a new family of mobile elements. Mol. Microbiol. 71:948-959. [DOI] [PubMed] [Google Scholar]
- 38.Brochet, M., C. Rusniok, E. Couve, S. Dramsi, C. Poyart, P. Trieu-Cuot, F. Kunst, and P. Glaser. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 105:15961-15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 40.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 41.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 42.Cabezon, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 43.Calcutt, M. J., M. S. Lewis, and K. S. Wise. 2002. Molecular genetic analysis of ICEF, an integrative conjugal element that is present as a repetitive sequence in the chromosome of Mycoplasma fermentans PG18. J. Bacteriol. 184:6929-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cambronne, E. D., and C. R. Roy. 2007. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carle, A., C. Hoppner, K. A. Aly, Q. Yuan, A. den Dulk-Ras, A. Vergunst, D. O'Callaghan, and C. Baron. 2006. The Brucella suis type IV secretion system assembles in the cell envelope of the heterologous host Agrobacterium tumefaciens and increases IncQ plasmid pLS1 recipient competence. Infect. Immun. 74:108-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caryl, J. A., M. C. Smith, and C. D. Thomas. 2004. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J. Bacteriol. 186:3374-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caryl, J. A., and C. D. Thomas. 2006. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol. Microbiol. 60:1302-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cascales, E., and P. J. Christie. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA 101:17228-17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Celli, J., and J. P. Gorvel. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 52.Cesar, C. E., C. Machon, F. de la Cruz, and M. Llosa. 2006. A new domain of conjugative relaxase TrwC responsible for efficient oriT-specific recombination on minimal target sequences. Mol. Microbiol. 62:984-996. [DOI] [PubMed] [Google Scholar]
- 53.Cevallos, M. A., R. Cervantes-Rivera, and R. M. Gutierrez-Rios. 2008. The repABC plasmid family. Plasmid 60:19-37. [DOI] [PubMed] [Google Scholar]
- 54.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen, I., and D. Dubnau. 2003. DNA transport during transformation. Front. Biosci. 8:S544-S556. [DOI] [PubMed] [Google Scholar]
- 56.Chen, I., R. Provvedi, and D. Dubnau. 2006. A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J. Biol. Chem. 281:21720-21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, J., M. Reyes, M. Clarke, and H. A. Shuman. 2007. Host cell-dependent secretion and translocation of the LepA and LepB effectors of Legionella pneumophila. Cell. Microbiol. 9:1660-1671. [DOI] [PubMed] [Google Scholar]
- 58.Chen, Y., J. H. Staddon, and G. M. Dunny. 2007. Specificity determinants of conjugative DNA processing in the Enterococcus faecalis plasmid pCF10 and the Lactococcus lactis plasmid pRS01. Mol. Microbiol. 63:1549-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen, Y., X. Zhang, D. Manias, H. J. Yeo, G. M. Dunny, and P. J. Christie. 2008. Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J. Bacteriol. 190:3632-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng, Z., X. Wang, and Y. Rikihisa. 2008. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J. Bacteriol. 190:2096-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho, N. H., H. R. Kim, J. H. Lee, S. Y. Kim, J. Kim, S. Cha, S. Y. Kim, A. C. Darby, H. H. Fuxelius, J. Yin, J. H. Kim, J. Kim, S. J. Lee, Y. S. Koh, W. J. Jang, K. H. Park, S. G. Andersson, M. S. Choi, and I. S. Kim. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. USA 104:7981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christie, P. J. 2004. Bacterial type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christie, P. J., and E. Cascales. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol. Membr. Biol. 22:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chuang, O. N., P. M. Schlievert, C. L. Wells, D. A. Manias, T. J. Tripp, and G. M. Dunny. 2009. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect. Immun. 77:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clarke, M., L. Maddera, R. L. Harris, and P. M. Silverman. 2008. F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. USA 105:17978-17981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clewell, D. B. 2007. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid 58:205-227. [DOI] [PubMed] [Google Scholar]
- 68.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 69.Coros, A., B. Callahan, E. Battaglioli, and K. M. Derbyshire. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 69:794-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Couturier, M. R., E. Tasca, C. Montecucco, and M. Stein. 2006. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect. Immun. 74:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dang, T. A., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Datta, S., C. Larkin, and J. F. Schildbach. 2003. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure 11:1369-1379. [DOI] [PubMed] [Google Scholar]
- 73.Davis, R. E., E. L. Dally, R. Jomantiene, Y. Zhao, B. Roe, S. Lin, and J. Shao. 2005. Cryptic plasmid pSKU146 from the wall-less plant pathogen Spiroplasma kunkelii encodes an adhesin and components of a type IV translocation-related conjugation system. Plasmid 53:179-190. [DOI] [PubMed] [Google Scholar]
- 74.Degtyar, E., T. Zusman, M. Ehrlich, and G. Segal. 2009. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell. Microbiol. 11:1219-1235. [DOI] [PubMed] [Google Scholar]
- 75.Dehio, C. 2008. Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell. Microbiol. 10:1591-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Jong, M. F., Y. H. Sun, A. B. den Hartigh, J. M. van Dijl, and R. M. Tsolis. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 70:1378-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng, W., L. Chen, W. T. Peng, X. Liang, S. Sekiguchi, M. P. Gordon, L. Comai, and E. W. Nester. 1999. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 31:1795-1807. [DOI] [PubMed] [Google Scholar]
- 78.den Hartigh, A. B., H. G. Rolan, M. F. de Jong, and R. M. Tsolis. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J. Bacteriol. 190:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Paz, H. D., F. J. Sangari, S. Bolland, J. M. Garcia-Lobo, C. Dehio, F. de la Cruz, and M. Llosa. 2005. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiology 151:3505-3516. [DOI] [PubMed] [Google Scholar]
- 80.DiGiuseppe Champion, P. A., and J. S. Cox. 2007. Protein secretion systems in mycobacteria. Cell. Microbiol. 9:1376-1384. [DOI] [PubMed] [Google Scholar]
- 81.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Disque-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dorman, C. J., and K. A. Kane. 2009. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol. Rev. 33:587-592. [DOI] [PubMed] [Google Scholar]
- 84.Draper, O., C. E. Cesar, C. Machon, F. de la Cruz, and M. Llosa. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. USA 102:16385-16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Driessen, A. J., and N. Nouwen. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643-667. [DOI] [PubMed] [Google Scholar]
- 86.Dunny, G. M. 2007. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durrenberger, M. B., W. Villiger, and T. Bachi. 1991. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J. Struct. Biol. 107:146-156. [DOI] [PubMed] [Google Scholar]
- 88.Dym, O., S. Albeck, T. Unger, J. Jacobovitch, A. Branzburg, Y. Michael, D. Frenkiel-Krispin, S. G. Wolf, and M. Elbaum. 2008. Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners. Proc. Natl. Acad. Sci. USA 105:11170-11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Economou, A., P. J. Christie, R. C. Fernandez, T. Palmer, G. V. Plano, and A. P. Pugsley. 2006. Secretion by numbers: protein traffic in prokaryotes. Mol. Microbiol. 62:308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eisenbrandt, R., M. Kalkum, R. Lurz, and E. Lanka. 2000. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erauso, G., K. M. Stedman, H. J. van de Werken, W. Zillig, and J. van der Oost. 2006. Two novel conjugative plasmids from a single strain of Sulfolobus. Microbiology 152:1951-1968. [DOI] [PubMed] [Google Scholar]
- 92.Farias, M. E., E. Grohmann, and M. Espinosa. 1999. Expression of the mobM gene of the streptococcal plasmid pMV158 in Lactococcus lactis subsp. lactis. FEMS Microbiol. Lett. 176:403-410. [DOI] [PubMed] [Google Scholar]
- 93.Farizo, K. M., T. G. Cafarella, and D. L. Burns. 1996. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 271:31643-31649. [DOI] [PubMed] [Google Scholar]
- 94.Felek, S., H. Huang, and Y. Rikihisa. 2003. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect. Immun. 71:6063-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Felix, C., S. Pichon, C. Braquart-Varnier, H. Braig, L. Chen, R. A. Garrett, G. Martin, and P. Greve. 2008. Characterization and transcriptional analysis of two gene clusters for type IV secretion machinery in Wolbachia of Armadillidium vulgare. Res. Microbiol. 159:481-485. [DOI] [PubMed] [Google Scholar]
- 96.Fernandez, D., T. A. Dang, G. M. Spudich, X. R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fischer, W., R. Haas, and S. Odenbreit. 2002. Type IV secretion systems in pathogenic bacteria. Int. J. Med. Microbiol. 292:159-168. [DOI] [PubMed] [Google Scholar]
- 98.Flint, J. L., J. C. Kowalski, P. K. Karnati, and K. M. Derbyshire. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 101:12598-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flower, A. M. 2007. The SecY translocation complex: convergence of genetics and structure. Trends Microbiol. 15:203-210. [DOI] [PubMed] [Google Scholar]
- 100.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 101.Frenkiel-Krispin, D., S. G. Wolf, S. Albeck, T. Unger, Y. Peleg, J. Jacobovitch, Y. Michael, S. Daube, M. Sharon, C. V. Robinson, D. I. Svergun, D. Fass, T. Tzfira, and M. Elbaum. 2007. Plant transformation by Agrobacterium tumefaciens: modulation of single-stranded DNA-VirE2 complex assembly by VirE1. J. Biol. Chem. 282:3458-3464. [DOI] [PubMed] [Google Scholar]
- 102.Fronzes, R., E. Schafer, L. Wang, H. R. Saibil, E. V. Orlova, and G. Waksman. 2009. Structure of a type IV secretion system core complex. Science 323:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fuxelius, H. H., A. Darby, C. K. Min, N. H. Cho, and S. G. Andersson. 2007. The genomic and metabolic diversity of Rickettsia. Res. Microbiol. 158:745-753. [DOI] [PubMed] [Google Scholar]
- 104.Garcillan-Barcia, M. P., M. V. Francia, and F. de la Cruz. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657-687. [DOI] [PubMed] [Google Scholar]
- 105.Garcillán-Barcia, M. P., P. Jurado, B. Gonzalez-Pérez, G. Moncalián, L. A. Fernández, and F. de la Cruz. 2007. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 63:404-416. [DOI] [PubMed] [Google Scholar]
- 106.Gillespie, J. J., N. C. Ammerman, S. M. Dreher-Lesnick, M. S. Rahman, M. J. Worley, J. C. Setubal, B. S. Sobral, and A. F. Azad. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE 4:e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gillespie, J. J., M. S. Beier, M. S. Rahman, N. C. Ammerman, J. M. Shallom, A. Purkayastha, B. S. Sobral, and A. F. Azad. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE 2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gillespie, J. J., K. Williams, M. Shukla, E. E. Snyder, E. K. Nordberg, S. M. Ceraul, C. Dharmanolla, D. Rainey, J. Soneja, J. M. Shallom, N. D. Vishnubhat, R. Wattam, A. Purkayastha, M. Czar, O. Crasta, J. C. Setubal, A. F. Azad, and B. S. Sobral. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE 3:e2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gilmour, M. W., T. D. Lawley, M. M. Rooker, P. J. Newnham, and D. E. Taylor. 2001. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol. Microbiol. 42:705-715. [DOI] [PubMed] [Google Scholar]
- 110.Gomis-Ruth, F. X., and M. Coll. 2006. Cut and move: protein machinery for DNA processing in bacterial conjugation. Curr. Opin. Struct. Biol. 16:744-752. [DOI] [PubMed] [Google Scholar]
- 111.Gomis-Ruth, F. X., and M. Coll. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33:839-843. [DOI] [PubMed] [Google Scholar]
- 112.Gomis-Ruth, F. X., F. de la Cruz, and M. Coll. 2002. Structure and role of coupling proteins in conjugal DNA transfer. Res. Microbiol. 153:199-204. [DOI] [PubMed] [Google Scholar]
- 113.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 114.Gomis-Ruth, F. X., M. Sola, F. de la Cruz, and M. Coll. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 10:1551-1565. [DOI] [PubMed] [Google Scholar]
- 115.Greve, B., S. Jensen, K. Brugger, W. Zillig, and R. A. Garrett. 2004. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea 1:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guasch, A., M. Lucas, G. Moncalian, M. Cabezas, R. Perez-Luque, F. X. Gomis-Ruth, F. de la Cruz, and M. Coll. 2003. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 10:1002-1010. [DOI] [PubMed] [Google Scholar]
- 118.Gunton, J. E., M. W. Gilmour, K. P. Baptista, T. D. Lawley, and D. E. Taylor. 2007. Interaction between the co-inherited TraG coupling protein and the TraJ membrane-associated protein of the H-plasmid conjugative DNA transfer system resembles chromosomal DNA translocases. Microbiology 153:428-441. [DOI] [PubMed] [Google Scholar]
- 119.Guo, M., S. Jin, D. Sun, C. L. Hew, and S. Q. Pan. 2007. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc. Natl. Acad. Sci. USA 104:20019-20024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guzman, L. M., and M. Espinosa. 1997. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J. Mol. Biol. 266:688-702. [DOI] [PubMed] [Google Scholar]
- 121.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hamilton, H. L., N. M. Dominguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 123.Hare, S., R. Bayliss, C. Baron, and G. Waksman. 2006. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J. Mol. Biol. 360:56-66. [DOI] [PubMed] [Google Scholar]
- 124.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 125.Hazes, B., and L. Frost. 2008. Towards a systems biology approach to study type II/IV secretion systems. Biochim. Biophys. Acta 1778:1839-1850. [DOI] [PubMed] [Google Scholar]
- 126.Hickman, A. B., D. R. Ronning, Z. N. Perez, R. M. Kotin, and F. Dyda. 2004. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell 13:403-414. [DOI] [PubMed] [Google Scholar]
- 127.Hirt, H., D. A. Manias, E. M. Bryan, J. R. Klein, J. K. Marklund, J. H. Staddon, M. L. Paustian, V. Kapur, and G. M. Dunny. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J. Bacteriol. 187:1044-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hodges, L. D., A. C. Vergunst, J. Neal-McKinney, A. den Dulk-Ras, D. M. Moyer, P. J. Hooykaas, and W. Ream. 2006. Agrobacterium rhizogenes GALLS protein contains domains for ATP binding, nuclear localization, and type IV secretion. J. Bacteriol. 188:8222-8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hofreuter, D., A. Karnholz, and R. Haas. 2003. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int. J. Med. Microbiol. 293:153-165. [DOI] [PubMed] [Google Scholar]
- 130.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 131.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 132.Hohlfeld, S., I. Pattis, J. Puls, G. V. Plano, R. Haas, and W. Fischer. 2006. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol. Microbiol. 59:1624-1637. [DOI] [PubMed] [Google Scholar]
- 133.Hoppner, C., Z. Liu, N. Domke, A. N. Binns, and C. Baron. 2004. VirB1 orthologs from Brucella suis and pKM101 complement defects of the lytic transglycosylase required for efficient type IV secretion from Agrobacterium tumefaciens. J. Bacteriol. 186:1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hormaeche, I., I. Iloro, J. L. Arrondo, F. M. Goni, F. de la Cruz, and I. Alkorta. 2004. Role of the transmembrane domain in the stability of TrwB, an integral protein involved in bacterial conjugation. J. Biol. Chem. 279:10955-10961. [DOI] [PubMed] [Google Scholar]
- 135.Hormaeche, I., R. L. Segura, A. J. Vecino, F. M. Goni, F. de la Cruz, and I. Alkorta. 2006. The transmembrane domain provides nucleotide binding specificity to the bacterial conjugation protein TrwB. FEBS Lett. 580:3075-3082. [DOI] [PubMed] [Google Scholar]
- 136.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. A. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hu, X., G. Van der Auwera, S. Timmery, L. Zhu, and J. Mahillon. 2009. Distribution, diversity, and potential mobility of extrachromosomal elements related to the Bacillus anthracis pXO1 and pXO2 virulence plasmids. Appl. Environ. Microbiol. 75:3016-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iannelli, F., M. R. Oggioni, and G. Pozzi. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63-71. [DOI] [PubMed] [Google Scholar]
- 139.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ishmael, N., J. C. D. Hotopp, P. Ioannidis, S. Biber, J. Sakamoto, S. Siozios, V. Nene, J. Werren, K. Bourtzis, S. R. Bordenstein, and H. Tettelin. 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 9:1252-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jackson, C. R., J. A. Boylan, J. G. Frye, and F. C. Gherardini. 2007. Evidence of a conjugal erythromycin resistance element in the Lyme disease spirochete Borrelia burgdorferi. Int. J. Antimicrob. Agents 30:496-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jakubowski, S. J., E. Cascales, V. Krishnamoorthy, and P. J. Christie. 2005. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol. 187:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jakubowski, S. J., J. E. Kerr, I. Garza, V. Krishnamoorthy, R. Bayliss, G. Waksman, and P. J. Christie. 2009. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 71:779-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jakubowski, S. J., V. Krishnamoorthy, E. Cascales, and P. J. Christie. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341:961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jones, A. L., K. Shirasu, and C. I. Kado. 1994. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 176:5255-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Joshi, B. D., M. Berg, J. Rogers, J. Fletcher, and U. Melcher. 2005. Sequence comparisons of plasmids pBJS-O of Spiroplasma citri and pSKU146 of S. kunkelii: implications for plasmid evolution. BMC Genomics 6:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Judd, P. K., R. B. Kumar, and A. Das. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. USA 102:11498-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Judd, P. K., R. B. Kumar, and A. Das. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55:115-124. [DOI] [PubMed] [Google Scholar]
- 150.Juhas, M., D. W. Crook, I. D. Dimopoulou, G. Lunter, R. M. Harding, D. J. Ferguson, and D. W. Hood. 2007. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Juhas, M., D. W. Crook, and D. W. Hood. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10:2377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Juhas, M., J. R. van der Meer, M. Gaillard, R. M. Harding, D. W. Hood, and D. W. Crook. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33:376-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kalkum, M., R. Eisenbrandt, and E. Lanka. 2004. Protein circlets as sex pilus subunits. Curr. Protein Pept. Sci. 5:417-424. [DOI] [PubMed] [Google Scholar]
- 154.Kalkum, M., R. Eisenbrandt, R. Lurz, and E. Lanka. 2002. Tying rings for sex. Trends Microbiol. 10:382-387. [DOI] [PubMed] [Google Scholar]
- 155.Karnholz, A., C. Hoefler, S. Odenbreit, W. Fischer, D. Hofreuter, and R. Haas. 2006. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J. Bacteriol. 188:882-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 157.Koonin, E. V., and Y. I. Wolf. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kopec, J., A. Bergmann, G. Fritz, E. Grohmann, and W. Keller. 2005. TraA and its N-terminal relaxase domain of the gram-positive plasmid pIP501 show specific oriT binding and behave as dimers in solution. Biochem. J. 387:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Krause, S., M. Barcena, W. Pansegrau, R. Lurz, J. M. Carazo, and E. Lanka. 2000. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. USA 97:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182:2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kumar, R. B., and A. Das. 2001. Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8. J. Bacteriol. 183:3636-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kumar, R. B., Y. H. Xie, and A. Das. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36:608-617. [DOI] [PubMed] [Google Scholar]
- 163.Kurenbach, B., J. Kopec, M. Magdefrau, K. Andreas, W. Keller, C. Bohn, M. Y. Abajy, and E. Grohmann. 2006. The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology 152:637-645. [DOI] [PubMed] [Google Scholar]
- 164.Kwok, T., D. Zabler, S. Urman, M. Rohde, R. Hartig, S. Wessler, R. Misselwitz, J. Berger, N. Sewald, W. Konig, and S. Backert. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862-866. [DOI] [PubMed] [Google Scholar]
- 165.Lai, E. M., O. Chesnokova, L. M. Banta, and C. I. Kado. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Larkin, C., S. Datta, A. Nezami, J. A. Dohm, and J. F. Schildbach. 2003. Crystallization and preliminary X-ray characterization of the relaxase domain of F factor TraI. Acta Crystallogr. D Biol. Crystallogr. 59:1514-1516. [DOI] [PubMed] [Google Scholar]
- 167.Larkin, C., R. J. Haft, M. J. Harley, B. Traxler, and J. F. Schildbach. 2007. Roles of active site residues and the HUH motif of the F plasmid TraI relaxase. J. Biol. Chem. 282:33707-33713. [DOI] [PubMed] [Google Scholar]
- 168.Lau-Wong, I. C., T. Locke, M. J. Ellison, T. L. Raivio, and L. S. Frost. 2008. Activation of the Cpx regulon destabilizes the F plasmid transfer activator, TraJ, via the HslVU protease in Escherichia coli. Mol. Microbiol. 67:516-527. [DOI] [PubMed] [Google Scholar]
- 169.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 170.Lee, C. A., J. M. Auchtung, R. E. Monson, and A. D. Grossman. 2007. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 66:1356-1369. [DOI] [PubMed] [Google Scholar]
- 171.Lee, C. A., and A. D. Grossman. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 189:7254-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lillestol, R. K., S. A. Shah, K. Brugger, P. Redder, H. Phan, J. Christiansen, and R. A. Garrett. 2009. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72:259-272. [DOI] [PubMed] [Google Scholar]
- 173.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 9:2644-2657. [DOI] [PubMed] [Google Scholar]
- 174.Lipps, G. 2006. Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus. Extremophiles 10:17-28. [DOI] [PubMed] [Google Scholar]
- 175.Liu, Z., and A. N. Binns. 2003. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 185:3259-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Llosa, M., J. Zupan, C. Baron, and P. Zambryski. 2000. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 182:3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lopez, J. E., G. H. Palmer, K. A. Brayton, M. J. Dark, S. E. Leach, and W. C. Brown. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect. Immun. 75:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lu, J., A. den Dulk-Ras, P. J. J. Hooykaas, and J. N. M. Glover. 2009. Agrobacterium tumefaciens VirC2 enhances T-DNA transfer and virulence through its C-terminal ribbon-helix-helix DNA-binding fold. Proc. Natl. Acad. Sci. USA. 106:9643-9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lu, J., and L. S. Frost. 2005. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J. Bacteriol. 187:4767-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lu, J., J. J. Wong, R. A. Edwards, J. Manchak, L. S. Frost, and J. N. Glover. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 70:89-99. [DOI] [PubMed] [Google Scholar]
- 182.Mahairas, G. G., J. A. Cao, and F. C. Minion. 1990. Genetic exchange of transposon and integrative plasmid markers in Mycoplasma pulmonis. J. Bacteriol. 172:2267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Marenda, M., V. Barbe, G. Gourgues, S. Mangenot, E. Sagne, and C. Citti. 2006. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J. Bacteriol. 188:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Marrero, J., and M. K. Waldor. 2007. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J. Bacteriol. 189:6469-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Marrero, J., and M. K. Waldor. 2005. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev. Cell 8:963-970. [DOI] [PubMed] [Google Scholar]
- 187.Massey, T. H., C. P. Mercogliano, J. Yates, D. J. Sherratt, and J. Lowe. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23:457-469. [DOI] [PubMed] [Google Scholar]
- 188.Masui, S., T. Sasaki, and H. Ishikawa. 2000. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J. Bacteriol. 182:6529-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Meyer, R. 2009. The R1162 Mob proteins can promote conjugative transfer from cryptic origins in the bacterial chromosome. J. Bacteriol. 191:1574-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Middleton, R., K. Sjolander, N. Krishamurthy, J. Foley, and P. Zambryski. 2005. Predicted hexameric structure of the Agrobacterium VirB4 C-terminus suggests VirB4 acts as a docking site during type IV secretion. Proc. Natl. Acad. Sci. USA 102:1685-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mojica, F. J., C. Diez-Villasenor, J. Garcia-Martinez, and C. Almendros. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defense system. Microbiology 155:733-740. [DOI] [PubMed] [Google Scholar]
- 192.Moncalian, G., and F. de la Cruz. 2004. DNA binding properties of protein TrwA, a possible structural variant of the Arc repressor superfamily. Biochim. Biophys. Acta 1701:15-23. [DOI] [PubMed] [Google Scholar]
- 193.Monzingo, A. F., A. Ozburn, S. Xia, R. J. Meyer, and J. D. Robertus. 2007. The structure of the minimal relaxase domain of MobA at 2.1 Å resolution. J. Mol. Biol. 366:165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nagai, H., E. D. Cambronne, J. C. Kagan, J. C. Amor, R. A. Kahn, and C. R. Roy. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. USA 102:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nagai, H., and C. R. Roy. 2003. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell. Microbiol. 5:373-383. [DOI] [PubMed] [Google Scholar]
- 196.Navarre, W. W., M. McClelland, S. J. Libby, and F. C. Fang. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456-1471. [DOI] [PubMed] [Google Scholar]
- 197.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 198.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nelson, C. M., M. J. Herron, R. F. Felsheim, B. R. Schloeder, S. M. Grindle, A. O. Chavez, T. J. Kurtti, and U. G. Munderloh. 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nguyen, K. T., K. Piastro, and K. M. Derbyshire. 2009. LpqM, a mycobacterial lipoprotein-metalloproteinase, is required for conjugal DNA transfer in Mycobacterium smegmatis. J. Bacteriol. 191:2721-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ninio, S., J. Celli, and C. R. Roy. 2009. A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 5:e1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15:372-380. [DOI] [PubMed] [Google Scholar]
- 203.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55:912-926. [DOI] [PubMed] [Google Scholar]
- 204.Novick, R. P. 1987. Plasmid incompatibility. Microbiol. Rev. 51:3813-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ogata, H., B. La Scola, S. Audic, P. Renesto, G. Blanc, C. Robert, P. E. Fournier, J. M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ogata, H., P. Renesto, S. Audic, C. Robert, G. Blanc, P. E. Fournier, H. Parinello, J. M. Claverie, and D. Raoult. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ohashi, N., N. Zhi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70:2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Omsland, A., D. C. Cockrell, D. Howe, E. R. Fischer, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, and R. A. Heinzen. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. USA 106:4430-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Osborn, A. M., and D. Boltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202-212. [DOI] [PubMed] [Google Scholar]
- 210.Ozbek, E., S. A. Miller, T. Meulia, and S. A. Hogenhout. 2003. Infection and replication sites of Spiroplasma kunkelii (class: Mollicutes) in midgut and Malpighian tubules of the leafhopper Dalbulus maidis. J. Invertebr. Pathol. 82:167-175. [DOI] [PubMed] [Google Scholar]
- 211.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 212.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 214.Parker, C., and R. J. Meyer. 2007. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol. Microbiol. 66:252-261. [DOI] [PubMed] [Google Scholar]
- 215.Pattis, I., E. Weiss, R. Laugks, R. Haas, and W. Fischer. 2007. The Helicobacter pylori CagF protein is a type IV secretion chaperone-like molecule that binds close to the C-terminal secretion signal of the CagA effector protein. Microbiology 153:2896-2909. [DOI] [PubMed] [Google Scholar]
- 216.Pettis, G. S., and S. N. Cohen. 2001. Unraveling the essential role in conjugation of the Tra protein of Streptomyces lividans plasmid pIJ101. Antonie van Leeuwenhoek 79:247-250. [DOI] [PubMed] [Google Scholar]
- 217.Prangishvili, D., S. V. Albers, I. Holz, H. P. Arnold, K. Stedman, T. Klein, H. Singh, J. Hiort, A. Schweier, J. K. Kristjansson, and W. Zillig. 1998. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid 40:190-202. [DOI] [PubMed] [Google Scholar]
- 218.Pulliainen, A. T., and C. Dehio. 2009. Bartonella henselae: subversion of vascular endothelial cell functions by translocated bacterial effector proteins. Int. J. Biochem. Cell Biol. 41:507-510. [DOI] [PubMed] [Google Scholar]
- 219.Rabel, C., A. M. Grahn, R. Lurz, and E. Lanka. 2003. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 185:1045-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Raivio, T. L. 2005. Envelope stress responses and gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 221.Rakita, R. M., N. N. Vanek, K. Jacques-Palaz, M. Mee, M. M. Mariscalco, G. M. Dunny, M. Snuggs, W. B. Van Winkle, and S. I. Simon. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 67:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Rambow-Larsen, A. A., and A. A. Weiss. 2002. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 184:2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Rances, E., D. Voronin, V. Tran-Van, and P. Mavingui. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J. Bacteriol. 190:5020-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Roberts, M. C., and G. E. Kenny. 1987. Conjugal transfer of transposon Tn916 from Streptococcus faecalis to Mycoplasma hominis. J. Bacteriol. 169:3836-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rocco, J. M., and G. Churchward. 2006. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J. Bacteriol. 188:2207-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rohde, M., J. Puls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 228.Rosenshine, I., R. Tchelet, and M. Mevarech. 1989. The mechanism of DNA transfer in the mating system of an archaebacterium. Science 245:1387-1389. [DOI] [PubMed] [Google Scholar]
- 229.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sagulenko, Y., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Saillard, C., P. Carle, S. Duret-Nurbel, R. Henri, N. Killiny, S. Carrere, J. Gouzy, J. M. Bove, J. Renaudin, and X. Foissac. 2008. The abundant extrachromosomal DNA content of the Spiroplasma citri GII3-3X genome. BMC Genomics 9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Salgado-Pabon, W., S. Jain, N. Turner, C. van der Does, and J. P. Dillard. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol. Microbiol. 66:930-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Samuels, A. L., E. Lanka, and J. E. Davies. 2000. Conjugative junctions in RP4-mediated mating of Escherichia coli. J. Bacteriol. 182:2709-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schandel, K. A., M. M. Muller, and R. E. Webster. 1992. Localization of TraC, a protein involved in assembly of the F conjugative pilus. J. Bacteriol. 174:3800-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schleper, C., I. Holz, D. Janekovic, J. Murphy, and W. Zillig. 1995. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177:4417-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schroder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 238.Schroder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185:4371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schulein, R., P. Guye, T. A. Rhomberg, M. C. Schmid, G. Schroder, A. C. Vergunst, I. Carena, and C. Dehio. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. USA 102:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Seth-Smith, H., and N. J. Croucher. 2009. Genome watch: breaking the ICE. Nat. Rev. Microbiol. 7:328-329. [DOI] [PubMed] [Google Scholar]
- 241.Seubert, A., R. Hiestand, F. de la Cruz, and C. Dehio. 2003. A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49:1253-1266. [DOI] [PubMed] [Google Scholar]
- 242.Shah, S. A., N. R. Hansen, and R. A. Garrett. 2009. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem. Soc. Trans. 37:23-28. [DOI] [PubMed] [Google Scholar]
- 243.She, Q., H. Phan, R. A. Garrett, S. V. Albers, K. M. Stedman, and W. Zillig. 1998. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2:417-425. [DOI] [PubMed] [Google Scholar]
- 244.Siefert, J. L. 2009. Defining the mobilome. Methods Mol. Biol. 532:13-27. [DOI] [PubMed] [Google Scholar]
- 245.Silverman, P. M. 1997. Towards a structural biology of bacterial conjugation. Mol. Microbiol. 23:423-429. [DOI] [PubMed] [Google Scholar]
- 246.Simone, M., C. A. McCullen, L. E. Stahl, and A. N. Binns. 2001. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 41:1283-1293. [DOI] [PubMed] [Google Scholar]
- 247.Sirand-Pugnet, P., C. Lartigue, M. Marenda, D. Jacob, A. Barre, V. Barbe, C. Schenowitz, S. Mangenot, A. Couloux, B. Segurens, A. de Daruvar, A. Blanchard, and C. Citti. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 3:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Soppa, J., A. Baumann, M. Brenneis, M. Dambeck, O. Hering, and C. Lange. 2008. Genomics and functional genomics with haloarchaea. Arch. Microbiol. 190:197-215. [DOI] [PubMed] [Google Scholar]
- 249.Spudich, G. M., D. Fernandez, X. R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. USA 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Stedman, K. M., Q. She, H. Phan, I. Holz, H. Singh, D. Prangishvili, R. Garrett, and W. Zillig. 2000. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in crenarchaeota. J. Bacteriol. 182:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Steen, J. A., T. L. Bannam, W. L. Teng, R. J. Devenish, and J. I. Rood. 2009. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J. Bacteriol. 191:2926-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Stoebel, D. M., A. Free, and C. J. Dorman. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in gram-negative enteric bacteria. Microbiology 154:2533-2545. [DOI] [PubMed] [Google Scholar]
- 253.Street, L. M., M. J. Harley, J. C. Stern, C. Larkin, S. L. Williams, D. L. Miller, J. A. Dohm, M. E. Rodgers, and J. F. Schildbach. 2003. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim. Biophys. Acta 1646:86-99. [DOI] [PubMed] [Google Scholar]
- 254.Szpirer, C. Y., M. Faelen, and M. Couturier. 2000. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol. Microbiol. 37:1283-1292. [DOI] [PubMed] [Google Scholar]
- 255.Szpirer, C. Y., M. Faelen, and M. Couturier. 2001. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 183:2101-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tanaka, J., T. Suzuki, H. Mimuro, and C. Sasakawa. 2003. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell. Microbiol. 5:395-404. [DOI] [PubMed] [Google Scholar]
- 257.Tato, I., I. Matilla, I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 282:25569-25576. [DOI] [PubMed] [Google Scholar]
- 258.Teachman, A. M., C. T. French, H. Yu, W. L. Simmons, and K. Dybvig. 2002. Gene transfer in Mycoplasma pulmonis. J. Bacteriol. 184:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Teng, W. L., T. L. Bannam, J. A. Parsons, and J. I. Rood. 2008. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens. J. Bacteriol. 190:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.te Poele, E. M., H. Bolhuis, and L. Dijkhuizen. 2008. Actinomycete integrative and conjugative elements. Antonie van Leeuwenhoek 94:127-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Terradot, L., R. Bayliss, C. Oomen, G. A. Leonard, C. Baron, and G. Waksman. 2005. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc. Natl. Acad. Sci. USA 102:4596-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Thomas, J., and D. W. Hecht. 2007. Interaction of Bacteroides fragilis pLV22a relaxase and transfer DNA with Escherichia coli RP4-TraG coupling protein. Mol. Microbiol. 66:948-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tomich, M., P. J. Planet, and D. H. Figurski. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363-375. [DOI] [PubMed] [Google Scholar]
- 264.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of gram-positive bacteria. Biochim. Biophys. Acta 1694:269-278. [DOI] [PubMed] [Google Scholar]
- 265.Toro, N., A. Datta, O. A. Carmi, C. Young, R. K. Prusti, and E. W. Nester. 1989. The Agrobacterium tumefaciens virC1 gene product binds to overdrive, a T-DNA transfer enhancer. J. Bacteriol. 171:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Vanek, N. N., S. I. Simon, K. Jacques-Palaz, M. M. Mariscalco, G. M. Dunny, and R. M. Rakita. 1999. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol. Med. Microbiol. 26:49-60. [DOI] [PubMed] [Google Scholar]
- 267.VanRheenen, S. M., Z. Q. Luo, T. O'Connor, and R. R. Isberg. 2006. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect. Immun. 74:3597-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Varsaki, A., G. Moncalián, M. del Pilar Garcillán-Barcia, C. Drainas, and F. de la Cruz. 2009. Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J. Bacteriol. 191:1446-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Vergunst, A. C., M. C. van Lier, A. den Dulk-Ras, T. A. G. Stuve, A. Ouwehand, and P. J. Hooykaas. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA 102:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Vergunst, A. C., M. C. Van Lier, A. Den Dulk-Ras, and P. J. Hooykaas. 2003. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 133:978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Vincent, C. D., and J. P. Vogel. 2006. The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol. 61:596-613. [DOI] [PubMed] [Google Scholar]
- 272.Voelker, L. L., and K. Dybvig. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 178:6078-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Voth, D. E., and R. A. Heinzen. 2009. Coxiella type IV secretion and cellular microbiology. Curr. Opin. Microbiol. 12:74-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Voth, D. E., D. Howe, P. A. Beare, J. P. Vogel, N. Unsworth, J. E. Samuel, and R. A. Heinzen. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J. Bacteriol. 191:4232-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Walker, D. H., and N. Ismail. 2008. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat. Rev. Microbiol. 6:375-386. [DOI] [PubMed] [Google Scholar]
- 276.Wang, A., and F. L. Macrina. 1995. Characterization of six linked open reading frames necessary for pIP501-mediated conjugation. Plasmid 34:206-210. [DOI] [PubMed] [Google Scholar]
- 277.Wang, J., and K. M. Derbyshire. 2004. Plasmid DNA transfer in Mycobacterium smegmatis involves novel DNA rearrangements in the recipient, which can be exploited for molecular genetic studies. Mol. Microbiol. 53:1233-1241. [DOI] [PubMed] [Google Scholar]
- 278.Wang, J., P. K. Karnati, C. M. Takacs, J. C. Kowalski, and K. M. Derbyshire. 2005. Chromosomal DNA transfer in Mycobacterium smegmatis is mechanistically different from classical Hfr chromosomal DNA transfer. Mol. Microbiol. 58:280-288. [DOI] [PubMed] [Google Scholar]
- 279.Wang, J., L. M. Parsons, and K. M. Derbyshire. 2003. Unconventional conjugal DNA transfer in mycobacteria. Nat. Genet. 34:80-84. [DOI] [PubMed] [Google Scholar]
- 280.Wang, Y. A., X. Yu, P. M. Silverman, R. L. Harris, and E. H. Egelman. 2009. The structure of F-pili. J. Mol. Biol. 385:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ward, J. E., Jr., E. M. Dale, and A. N. Binns. 1991. Activity of the Agrobacterium T-DNA transfer machinery is affected by virB gene products. Proc. Natl. Acad. Sci. USA 88:9350-9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Watarai, M., H. L. Andrews, and R. Isberg. 2000. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 39:313-329. [DOI] [PubMed] [Google Scholar]
- 284.Waters, C. M., and G. M. Dunny. 2001. Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance. J. Bacteriol. 183:5659-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Waters, C. M., H. Hirt, J. K. McCormick, P. M. Schlievert, C. L. Wells, and G. M. Dunny. 2004. An amino-terminal domain of Enterococcus faecalis aggregation substance is required for aggregation, bacterial internalization by epithelial cells and binding to lipoteichoic acid. Mol. Microbiol. 52:1159-1171. [DOI] [PubMed] [Google Scholar]
- 286.Waters, C. M., C. L. Wells, and G. M. Dunny. 2003. The aggregation domain of aggregation substance, not the RGD motifs, is critical for efficient internalization by HT-29 enterocytes. Infect. Immun. 71:5682-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Winans, S. C., and G. C. Walker. 1985. Conjugal transfer system of the IncN plasmid pKM101. J. Bacteriol. 161:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]
- 290.Yeo, H. J., Q. Yuan, M. R. Beck, C. Baron, and G. Waksman. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. USA 100:15947-15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yoshida, H., N. Furuya, Y. J. Lin, P. Guntert, T. Komano, and M. Kainosho. 2008. Structural basis of the role of the NikA ribbon-helix-helix domain in initiating bacterial conjugation. J. Mol. Biol. 384:690-701. [DOI] [PubMed] [Google Scholar]
- 292.Yuan, Q., A. Carle, C. Gao, D. Sivanesan, K. A. Aly, C. Hoppner, L. Krall, N. Domke, and C. Baron. 2005. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 280:26349-26359. [DOI] [PubMed] [Google Scholar]
- 293.Zahrl, D., A. Wagner, M. Tscherner, and G. Koraimann. 2007. GroEL plays a central role in stress-induced negative regulation of bacterial conjugation by promoting proteolytic degradation of the activator protein TraJ. J. Bacteriol. 189:5885-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. E. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455-3467. [DOI] [PubMed] [Google Scholar]
- 295.Zahrl, D., M. Wagner, K. Bischof, and G. Koraimann. 2006. Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. J. Bacteriol. 188:6611-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965-976. [DOI] [PubMed] [Google Scholar]
- 297.Zhang, S., and R. Meyer. 1997. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol. Microbiol. 25:509-516. [DOI] [PubMed] [Google Scholar]
- 298.Zhao, Z., E. Sagulenko, Z. Ding, and P. J. Christie. 2001. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J. Bacteriol. 183:3855-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ziegelin, G., J. P. Furste, and E. Lanka. 1989. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J. Biol. Chem. 264:11989-11994. [PubMed] [Google Scholar]
- 300.Zupan, J., C. A. Hackworth, J. Aguilar, D. Ward, and P. Zambryski. 2007. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol. 189:6551-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]