Abstract
Summary: Aspartokinase (Ask) exists within a variable network that supports the synthesis of 9 amino acids and a number of other important metabolites. Lysine, isoleucine, aromatic amino acids, and dipicolinate may arise from the ASK network or from alternative pathways. Ask proteins were subjected to cohesion group analysis, a methodology that sorts a given protein assemblage into groups in which evolutionary continuity is assured. Two subhomology divisions, ASKα and ASKβ, have been recognized. The ASKα subhomology division is the most ancient, being widely distributed throughout the Archaea and Eukarya and in some Bacteria. Within an indel region of about 75 amino acids near the N terminus, ASKβ sequences differ from ASKα sequences by the possession of a proposed ancient deletion. ASKβ sequences are present in most Bacteria and usually exhibit an in-frame internal translational start site that can generate a small Ask subunit that is identical to the C-terminal portion of the larger subunit of a heterodimeric unit. Particularly novel are ask genes embedded in gene contexts that imply specialization for ectoine (osmotic agent) or aromatic amino acids. The cohesion group approach is well suited for the easy recognition of relatively recent lateral gene transfer (LGT) events, and many examples of these are described. Given the current density of genome representation for Proteobacteria, it is possible to reconstruct more ancient landmark LGT events. Thus, a plausible scenario in which the three well-studied and iconic Ask homologs of Escherichia coli are not within the vertical genealogy of Gammaproteobacteria, but rather originated via LGT from a Bacteroidetes donor, is supported.
INTRODUCTION
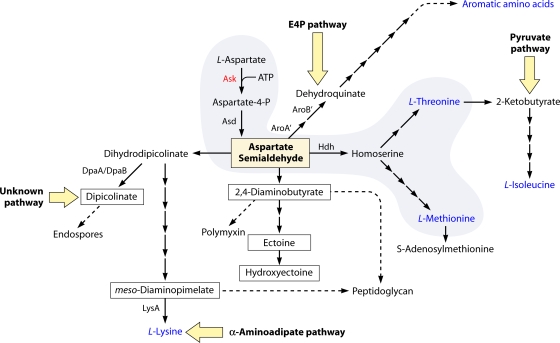
A multibranched web of biochemical pathways of central importance is initiated by the action of aspartokinase (Ask) in a reaction that combines aspartate and ATP. We will refer to the suite of pathways initiated by Ask as the ASK network. The aspartate-4-phosphate product of the Ask reaction is the immediate precursor of aspartate semialdehyde. Figure 1 illustrates the key position of aspartate semialdehyde at a metabolic hub, which serves as a point of divergence to a multitude of different end products. The figure is a composite illustration of known branches, not all of which coexist in any given organism. The biosynthetic origins of threonine, methionine, ectoine (or hydroxyectoine), meso-diaminopimelate (DAP), and diaminobutyrate all invariably track back to aspartate (so far). On the other hand, alternatives to the aspartate-derived pathways shown in Fig. 1 exist for isoleucine, lysine, dipicolinate, and the aromatic amino acids.
FIG. 1.
The ASK network of end products that may radiate from aspartate. The minimal ASK network is bounded with gray shading in a progression that begins with l-aspartate and ends with l-threonine and l-methionine. End product names shown in blue are essential metabolites for all organisms. Metabolites that are enclosed within rectangles are specialized compounds that have become essential, or at least important, in particular lineages. The dashed arrows indicate multiple enzymatic steps. Aromatic amino acids, isoleucine, dipicolinate, and lysine may or may not originate from the ASK network, and the alternative pathways that do not involve the ASK network are indicated by large yellow arrows. Enzyme abbreviations: Asd, aspartate-semialdehyde dehydrogenase; Hdh, homoserine dehydrogenase; DpaA/DpaB, alpha and beta subunits of dipicolinate synthase; LysA, DAP decarboxylase; AroA′, 2-amino-3,7-dideoxy-d-threo-hept-6-ulosonate synthase; and AroB′, dehydroquinate synthase II.
Crystal structures have been deposited in the Protein Data Bank (PDB) for Ask enzymes from a wide phylogenetic span of organisms, representing five phyla: Streptophyta, Euryarchaeota, Thermus, Actinobacteria, and Proteobacteria. Amino acid residues important for allostery have been documented for enzymes obtained from Arabidopsis thaliana (54), Methanocaldococcus jannaschii (51), Thermus thermophilus (92), Corynebacterium glutamicum (93), and Escherichia coli (47). Ask enzymes possess a C-terminal regulatory domain that is usually present in adjacent copies called ACT_1 and ACT_2. ACT domains are widespread regulatory elements that are often fused to catalytic domains, but they also exist as free-standing proteins. ACT domains were not easily detected by straightforward sequence homology, but more sophisticated techniques utilize secondary structure and similarity of residue types. Enzymes containing the amino acid binding ACT domains have been reviewed recently (16, 33, 50). All Ask enzymes appear to have in common the characteristic that ACT_1 is inserted into ACT_2 and that the effector-binding unit consists of one ACT domain on one chain interacting with another ACT domain on another chain (16).
Branches of the Ask Network
Canonical l-amino acids derived from l-aspartate.
Lysine, methionine, threonine, and isoleucine (together with asparagine and aspartate) comprise the classic “aspartate family” of amino acids. Although this is an apt descriptor for organisms such as E. coli, it has proved to be a partial misnomer in the cases of the many organisms subsequently discovered to use alternative nonaspartate pathways to lysine and/or isoleucine, as well as for organisms that derive aromatic amino acids from aspartate.
(i) l-Threonine.
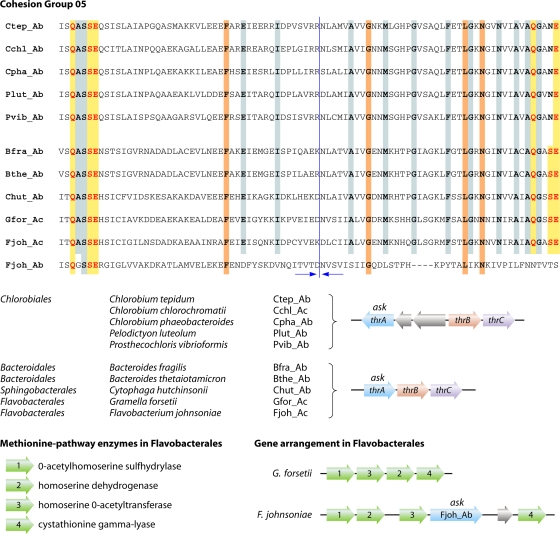
The threonine branch is a straightforward and universal two-step conversion beginning with the utilization of homoserine at the branch point. These steps are mediated by homoserine kinase (ThrB) and threonine synthase (ThrC). In some organisms, such as higher plants, ThrB also functions as an activation step of methionine biosynthesis, in which case the specific threonine branch is a one-step conversion. As a relatively rare alternative to ThrB, the phosphorylation of homoserine appears to be carried out in some organisms by a different enzyme analog that is homologous to cofactor-independent phosphoglycerate mutase. This variant phosphotransferase is found in Euryarchaeota, Bacteroides species, Chloroflexi, Deltaproteobacteria, and a few Clostridium spp. All three classes of the phylum Bacteroidetes contain genomes with a three-gene thr operon. However, it is interesting that whereas the classes Flavobacteria and Sphingobacteria possess an ask-hdh → thrB → thrC operon, organisms in the class Bacteroidetes have replaced thrB with the gene encoding the phosphoglycerate mutase analog in exactly the same spatial location.
(ii) l-Lysine.
The DAP pathway of lysine biosynthesis, which is aspartate derived, is widespread in Bacteria. However, considerable variation may nevertheless exist because the individual steps between dihydrodipicolinate and lysine can vary in different genome lineages. At least four variant pathway arrangements exist (39). Hence, different organisms that rely upon the same overall pathway may nevertheless utilize some different enzymes and intermediates in the process. The steps of the DAP pathway are synonymous with those of the DAP pathway of lysine biosynthesis, and only DAP decarboxylase (LysA) is unique to lysine biosynthesis (Fig. 1). The total flux throughout the lysine branch of the network can be significantly greater in some organisms that utilize lysine for purposes other than protein synthesis. For example, in stationary-phase metabolism, Streptomyces clavuligerus makes the antibiotic cephamycin C from lysine (57), and Streptomyces albulus makes the antibiotic ɛ-poly-l-lysine from lysine (36). In the latter case, the organism's single Ask is only weakly inhibited by the combination of Thr plus Lys, in contrast to the potent pattern of concerted feedback inhibition that is usually present in Streptomyces.
Lysine is not always derived from aspartate; sometimes it is synthesized from 2-ketoglutarate via the α-aminoadipate (AAA) pathway (63). Accordingly, in organisms such as fungi, euglenoids, a significant fraction of the Archaea, and some bacteria, lysine belongs to the glutamate family of amino acids. In such organisms, lysine biosynthesis has no specific relevance to the ASK network. The AAA pathway of lysine biosynthesis requires fewer ATP equivalents than the DAP alternative and was surely the ancestral pathway. Presumably, an evolved commitment of many organisms to make DAP as an important cross-linking amino acid agent of cell wall biosynthesis favored the relatively easy subsequent recruitment of the single additional gene (lysA) needed to make lysine from DAP, followed by abandonment of the subsequently unnecessary AAA pathway.
(iii) l-Methionine.
The particular pathway steps for methionine biosynthesis can also vary within similar overall transformations occurring between homoserine and methionine. Activation of homoserine may be accomplished via acetylation, succinylation, or phosphorylation in different lineages. Phosphorylation of homoserine to produce O-phosphohomoserine is always a step of threonine biosynthesis, but this activated intermediate is usually not accepted by the second enzyme of methionine biosynthesis. In those exceptional cases where O-phosphohomoserine does enter into both the threonine and methionine pathways (as in higher plants), the pathway branch point of divergence is therefore displaced to a more peripheral location at O-phosphohomoserine rather than at homoserine (not shown in Fig. 1). Incorporation of sulfur, the next enzymatic step, can also occur in three different ways. A bioinformatics analysis by Gophna et al. (29) provides a recent and detailed evolutionary perspective on the considerable enzymatic diversity deployed in nature for methionine biosynthesis, and they point out the amazing statistic that at least 18 variant methionine pathways can be drawn when combinations of substrate ambiguities and alternative steps are considered. In addition, it appears certain that completely unknown steps of methionine biosynthesis must still exist due to the absence of homologs of known genes in certain finished genomes, such as those of Desulfovibrio spp. Although methionine is one of the least abundant amino acids in proteins, the demand for methionine is heightened because it is the precursor of S-adenosyl-methionine (SAM). SAM is a generally important source of methylating power for many reactions, and it also contributes to the synthesis of polyamines. Indeed, higher plants use SAM rather than methionine as part of a synergistic combination to regulate one of the multiple Ask paralogs that are present (see below).
(iv) l-Isoleucine.
Isoleucine is best known to be derived from threonine in five steps, beginning with the formation of 2-ketobutyrate from threonine (catalyzed by threonine dehydratase). However, in yet another of nature's variations, isoleucine formation may be entirely disconnected from aspartate. An alternative pathway from pyruvate (the citramalate pathway) requires fewer ATP equivalents and likely was the ancestral pathway. This pathway, first described in Leptospira (10) as a seemingly anomalous variation, is increasingly becoming recognized as either the favored route or the sole route to isoleucine biosynthesis in nature (23). Many organisms have the potential to utilize either pathway, with the citramalate pathway likely being the preferential route. In such cases, threonine may be utilized only if exogenous threonine is available. The use of a threonine dehydratase with a low affinity (but a high capacity) for substrate and subject to feedback inhibition by isoleucine would fit the latter scenario of a biosynthetic function restricted to the presence of exogenous threonine. Whenever the citramalate pathway is the source of isoleucine, the function of Ask is directly relevant to threonine, but not to isoleucine.
Nonuniversal parts of the ASK network. (i) DAP.
The major amino acid branches shown in Fig. 1 can themselves contribute focal points of internal branching, as illustrated by the lysine branch. These internal branches are lineage specific. Thus, DAP, the immediate precursor of lysine, marks a branch point whereby DAP has an alternative fate of incorporation into the cross-linked matrix of cell wall peptidoglycan in most gram-negative and gram-positive bacteria. Archaea do not make peptidoglycan, but some of them make pseudomurein (32), and this metabolic variation is correlated with the presence of the DAP pathway to lysine. This correlation generates the expectation that DAP will be a component of pseudomurein. However, lysine, rather than DAP, is used as a cross-linking agent, so the selective pressure that accounts for the use of the DAP pathway for lysine biosynthesis in these Archaea is not readily apparent. Perhaps DAP originally had a cross-linking function in the ancestral state and was later replaced by lysine, or perhaps DAP (or one of the earlier precursors) has a yet-to-be-discovered function in some of the Archaea.
(ii) Dipicolinate.
Another lesser-known intermediate of the lysine pathway (2,3-dihydropicolinate) can be a branch point metabolite whenever it can be diverted to dipicolinate, which is a crucial and abundant metabolite for endospore differentiation by certain gram-positive bacteria. Dipicolinate synthase consists of two subunits known in the literature for many years by their stage V sporulation names (SpoVFA and SpoVFB). The genes have more recently been named dpaA and dpaB (17), and the corresponding enzyme subunits have been named DpaA and DpaB in Fig. 1. Although it is beyond the scope of this review, we note that dihydrodipicolinate synthase is encoded by multiple homolog genes in some organisms. This could represent a situation analogous to Hdh, where multiple enzyme species can be differentially regulated, a circumstance that effectively moves the branch point one step back.
(iii) Aromatic amino acids.
Some organisms lack the first two steps of the classic erythrose-4-phosphate (E4P) pathway for the synthesis of aromatic amino acids (75). Instead of utilizing E4P and phosphoenolpyruvate in the initial step, l-aspartate semialdehyde and 6-deoxy-5-ketofructose 1-phosphate (DKFP) are combined to form 2-amino-3,7-dideoxy-d-threo-hept-6-ulosonate via the action of an aldolase called AroA′. A subsequent step, catalyzed by AroB′, generates dehydroquinate, which then merges with the canonical pathway scheme. Thus, in these organisms, Ask is of fundamental importance for the biosynthesis of aromatic amino acids and other aromatic compounds.
Because the pathway of aromatic biosynthesis demands a large biochemical output, the already great significance of Ask as a provider of some or all of the aspartate family of amino acids would seem to be even more pronounced in some of the above-mentioned organisms. This burden may be somewhat counterbalanced in cases where isoleucine and lysine are derived from alternative pathways that are not dependent upon Ask (Fig. 1).
(iv) Ectoine.
A variety of organisms synthesize ectoine in three steps (EctABC) from aspartate semialdehyde. Ectoine is an osmotic agent (compatible solute) present in halophilic or halotolerant organisms. Ectoine can also serve as a protective agent for proteins in response to thermal stress (see Kuhlmann et al. [48] and references therein). Because cells can tolerate ectoine at high concentrations, it may also serve as a reserve source of carbon and energy. This is perhaps a more likely scenario in halotolerant organisms in transition from high-salt to moderate-salt conditions. It may also apply to conditions of transition from stationary-phase physiology to growing conditions in organisms like Halobacillus halophilus (81), which accumulate ectoine dramatically in stationary phase. Sometimes an additional ectoine hydroxylase (encoded by ectD) is present, and this converts ectoine to hydroxyectoine (see reference 8 for a complete biochemical diagram of the four-step pathway). The three or four ect genes are usually, but not always, present as a compact operon.
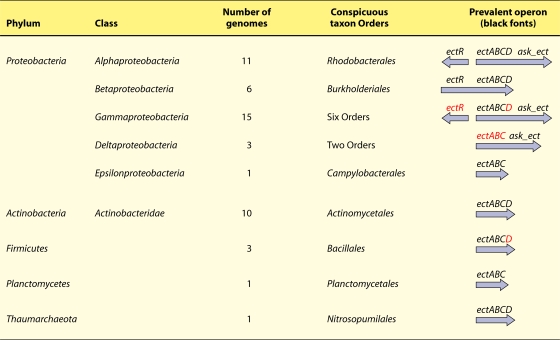
An analysis of 51 complete genomes that possess the ectoine biosynthetic pathway (ectABC) or the hydroxyectoine biosynthetic pathway (ectABCD) is summarized in Fig. 2. This pathway is abundant only in the phyla Proteobacteria and Actinobacteria. The genomic presence of ect genes is not distributed along cleanly delineated phylogenetic lines. This probably parallels the distribution of halophilic/halotolerant physiologies, which also exhibit erratic phylogenetic profiles (68). Thus, the ectoine operon is rather widely distributed, and yet its occurrence appears to be relatively sparse and quite erratic in any given taxon assemblage. Very recent losses of ectoine pathway genes and ectoine-associated Ask enzymes are apparent in some cases, e.g., in the Vibrionales. This may suggest its ubiquity in an ancient world that was largely marine, followed by loss of the ectoine operon in lineages that became adapted to a terrestrial environment. The widespread availability of exogenous ectoine for uptake may be an additional explanation for the loss of capability for ectoine biosynthesis. The gene encoding ectoine hydroxylase appears to have been a component of ancestral operons, although it has frequently been lost in individual lineages.
FIG. 2.
Phylogenetic distribution of ect operons. Operons are indicated on the right by continuous arrows in the direction of transcription. ask_ect is the designation for ask genes that are associated with ect operons. The most common contemporary gene arrangements are shown in black. The presumed ancestral gene arrangement is also shown, based upon the occasional presence in some genomes of the additional genes shown in red. Thus, in the Gammaproteobacteria, for example, the most widespread contemporary gene arrangement is ectABC → ask_ect, but ectD and the divergently oriented ectR are sometimes present. ectR is a putative regulatory gene for the ect operon, which belongs to the marR family of repressors. The order of enzyme reactions for ectoine biosynthesis is EctB (l-2,4-diaminobutyrate aminotransferase), EctA (l-2,4-diaminobutyrate acetyltransferase), and EctC (ectoine synthase). If present, hydroxyectoine is formed by EctD (ectoine hydroxylase).
Only a single archaeon (Nitrosopumilus maritimus) was found to possess the ectoine pathway, one encoded by an ectABCD operon. The single available member of the phylum Planctomycetes (Blastopirellula marina) possesses an ectABC operon. The three bacilli listed in Fig. 2 have an ectABC operon, but one of them possesses in addition an unlinked ectD gene. Those Actinobacteria capable of making ectoine all possess an ectABCD operon, except for Thermobifida fusca, which lacks ectD. Streptomyces coelicolor exemplifies a well-studied ectABCD system (7) that comprises one branch of an ASK network in which a single unlinked ask species supports the entire network.
The phylum Proteobacteria contains organisms that have an ect operon in all five of its classes (Fig. 2). The phylum is thus far unique in two ways. (i) A putative regulatory gene, here termed ectR, which encodes a gene product belonging to the MarR family of transcriptional regulators, is often present. (ii) A distinctive Ask gene is frequently associated with ect operons and is undoubtedly dedicated to coordination of aspartate semialdehyde availability with cellular demands for output of ectoine and/or hydroxyectoine. The details are covered below in a discussion of the various branch-specialized Ask enzymes.
(v) Diaminobutyrate.
The gene product of EctB, l-2,4-diaminobutyrate, is the first specific intermediate of the branch leading to ectoine/hydroxyectoine. In some organisms, e.g., Clavibacter michiganensis subsp. michiganensis NCPPB 382, it can also be used as a diamino component of peptidoglycan, and it is also known to be incorporated into certain peptide antibiotics (reference 66 and references therein). Polymyxin produced by Paenibacillus polymyxa is an antibiotic in which 6 of the 10 component amino acids are 2,4-diaminobutyrate. If at least two of the three possible functional roles leading from diaminobutyrate (as shown in Fig. 1) coexist in a given organism, then diaminobutyrate is a branch point metabolite in that ASK network.
Overview.
The foregoing indicates that Ask may potentially be irrelevant to any given end product shown in Fig. 1 for any of several reasons. First, an alternative pathway not involving Ask may be utilized, e.g., in the many cases where aromatic amino acid biosynthesis employs the E4P pathway. Second, an end product may be irrelevant to Ask simply because the end product is not synthesized at all. If so, the end product may be unimportant to a given lineage, and therefore, it is absent; e.g., dipicolinate is not important to gram-negative bacteria, which do not make endospores. On the other hand, even though an end product, such as one of the amino acids, may be absolutely essential throughout a given lineage, fastidious members of the lineage (including a variety of pathogens and endosymbionts) may nevertheless lack some or all of the ASK pathway network due to reliance upon environmental or host resources for the preformed end product.
A general consideration of interest (beyond the scope of this review) when comparing the ASK networks in different organisms is the variability in the demand of general protein synthesis for acidic residues, like aspartate, and basic residues, like lysine, due to differing codon preferences. Although it is not shown in Fig. 1, general protein synthesis represents a major avenue of competition for aspartate with respect to the ASK network. Thus, Salinibacter ruber, an extreme halophile, has proteins with high aspartate contents and low lysine contents (59).
Considerations Relevant to the Overall Regulation of the ASK Network
The aspartate pathway drawn in Fig. 1 begins with a short two-step common pathway, which then branches divergently at aspartate semialdehyde. There are multiple focal points of the usual feedback regulation—actually, a hierarchical array of them. In addition to Ask, sites of feedback control are potentially positioned at each step leading from aspartate semialdehyde, as well as at more peripheral branch points, e.g., the two enzymes competing for homoserine. An aspect of complexity is that aspartate semialdehyde is an unstable compound that polymerizes in solution (79). It has also been reported to be quite toxic (3). Thus, on one hand, the multiple enzymes competing for aspartate semialdehyde must access a substrate available at low concentrations and vulnerable to short half-life conditions, but on the other hand, regulatory restraints on the enzymes using the semialdehyde must not result in undue accumulation. Hence, the generation of aspartate semialdehyde (which is heavily impacted by Ask regulation) must be well matched to the utilization of aspartate semialdehyde (which is impacted by the regulation of multiple peripheral enzymes). Another aspect to consider is the bioenergetics of the ASK reaction. Aspartate-4-P probably has a more exergonic free energy of hydrolysis than ATP, making the forward reaction unfavorable under standard steady-state conditions. Thus, ATP/ADP and aspartate levels must contribute significantly to driving the reaction (5). These bioenergetic properties make Ask an excellent target for metabolic regulation.
Direct regulation of Ask activity.
Feedback regulation of Ask is generally of paramount importance, since Ask funnels a great amount of energy-expensive precursor to the various end products. The feedback principle of end product control of a key early enzyme in a linear pathway, whether exerted at a level of controlling catalytic activity or a level of controlling enzyme synthesis, is relatively straightforward compared to considerations evoked for a multibranched pathway, such as the ASK pathway. The regulatory dilemma for a key early enzyme such as Ask is the matter of how the activity of a single enzyme step can be adjusted by some kind of sensing mechanism tuned to the pace of the demand for multiple end products.
Alternative solutions are observed in nature with respect to this general dilemma that confronts the divergent multibranched pathway, as has been extensively documented for the initial enzyme step of the branched pathway of aromatic amino acid biosynthesis (30, 43). (i) The total activity of the enzyme reaction may be partitioned between specialized paralogs that are differentially regulated by particular end products. (ii) A single enzyme may be inhibited by a combination of multiple end products but not by individual end products (concerted feedback inhibition). (iii) Individual end products may exert partial inhibitory capabilities upon the early enzyme, whereas combinations of end products exert additive effects (cumulative feedback inhibition). (iv) The level of a pathway intermediate located at a branch point may rise and fall as the result of feedback control of individual terminal branches, and that branch point intermediate may in turn function in the feedback control of the early enzyme (sequential feedback inhibition).
In various organisms, the Ask step is usually driven by differentially regulated homolog isoenzymes (e.g., E. coli), by concerted feedback inhibition of a single enzyme (e.g., C. glutamicum), or by a combination of these (Bacillus subtilis). There is a single report in the literature describing the inhibition of a single Ask species in Rhodopseudomonas spheroides by aspartate semialdehyde (19), a pattern that would be one of sequential feedback inhibition. It is not clear whether screening for this allosteric pattern has been carried out very often.
Transcriptional and translational regulation of Ask levels.
Alternative regulatory mechanisms may parallel the variety of feedback inhibition patterns described above, namely, via (i) differential repression of specialized paralogs, (ii) multivalent (or concerted) repression of a single Ask enzyme by synergistic end product combinations, (iii) cumulative repression of a single Ask enzyme through additive end product effects, and (iv) sequential repression of a single Ask enzyme. These mechanisms may be exerted at both the RNA and DNA levels.
Pathway Partitioning via Specialized Ask Paralogs
The discussion above (as encapsulated in Fig. 1) delineates the major metabolite products whose biosynthesis may rely on Ask. At one extreme, a single Ask enzyme supports the entire existing ASK network. In other cases, specialized paralogs are coordinated with the production of a single end product or a few of the end products. Specialization is achieved by differential regulation, accomplished via allosteric control of enzyme activity and/or by transcriptional/translational control of enzyme synthesis.
Differential allosteric regulation.
In the case of Ask, allosteric regulation is accomplished by multiple ACT domains that exist in the C-terminal portions of Ask enzymes. Conserved domain (cd) signatures in the cd database (CDD) maintained by NCBI (53) allow some degree of prediction of allosteric specificities. If an ask gene is clustered with some or all of the genes encoding enzymes of a particular divergent branchlet, one can reasonably infer that the gene is specialized by regulation to support the production of that branchlet end product. Thus, the Ask activities of the three paralogs of E. coli are tuned to the output of single end products: ThrA is inhibited by threonine, LysC is inhibited by lysine, and the synthesis of MetL is repressed by methionine (72).
Use of gene context to infer specialized partitioning of function.
In Bacteria and Archaea, observations of the tendency of a given gene to cluster with one or more other genes (gene context) has been an extremely successful approach for inferring general functional roles and for deducing specialized partitioning of that function for paralog variants (70). A recent illustration of gene discovery that exploited the approach of gene context in combination with a battery of bioinformatics techniques, in addition to key experimental work, is provided by Yang et al. (91) (and references therein).
Clues to the differential functional roles of paralog ask genes can frequently be deduced by gene context because adjacent genes are subject to various mechanisms of cis coregulation. An ask gene that persists as a single ortholog will not generally be organized with genes relevant to a particular branch of the ASK network. In the case of the aforementioned E. coli paralogs, thrA and metL exist within operons containing genes encoding enzymes that function within the threonine and methionine branches, respectively. Since DAP pathway genes are synonymous with lysine pathway genes, it may not necessarily be clear whether ask-associated genes recognize DAP or lysine as an end product cue. However, the association of an ask paralog in an operon with lysA specifically implies regulatory tuning to lysine biosynthesis rather than to DAP synthesis, since lysA, encoding LysA (Fig. 1), is unique to lysine biosynthesis. There are many examples of Ask paralogs whose functional specializations have been tied to the threonine branch, to the methionine branch, and to the lysine branch. In this paper, we especially consider the extent to which other, lesser-known Ask homologs can be inferred to have functional specializations tuned to the additional branches of the ASK network shown in Fig. 1.
What dictates whether differentially regulated paralogs or lone Ask enzymes are used? (i) Differential impacts of environmental and physiological cues.
In some organisms, the ultimate end products that depend upon Ask may be synthesized more or less in proportion to growth without the need for dramatic upregulation and downregulation shifts, which in other organisms are occasioned by the presence or absence of some of the end products in the environment or where synthesis of a given end product may be triggered only by environmental cues or particular physiological states. Thus, the periodic feast-or-famine lifestyle of E. coli demands rapid and frequent pulses of upregulation and downregulation for the ASK pathway amino acids. Similarly, the demand for dipicolinate in B. subtilis is relatively abrupt and quantitatively significant during the unique physiological state of endospore formation. Therefore, it may be no accident that E. coli and B. subtilis exemplify organisms having multiple Ask isoenzymes subject to differential transcriptional and allosteric regulation. Likewise, halotolerant bacteria may express a specialized Ask paralog along with ectoine genes in transitional responses to exposure to high-salt environments. On the other hand, organisms such as cyanobacteria, which utilize very little in the way of exogenous organic nutrients and which have no specialized Ask-dependent pathways that are particularly responsive to environmental or physiological cues, can presumably support their ASK networks with a single Ask enzyme at a basal level. Sophisticated control patterns, such as concerted feedback inhibition, would still be anticipated to be critical for the achievement of a balanced network output.
(ii) Caveats.
Even if the outputs of different ASK network branches respond differently to environmental or physiological cues, the regulation in place could allow appropriate channeling of aspartate semialdehyde without the need for differential deployment of multiple Ask enzymes. For example, if ectoine were made primarily during stationary-phase physiology, then the amino acid branches would be essentially closed in this physiological state due to end product regulation by amino acids no longer being drawn into primary protein synthesis, thus allowing near-exclusive entry of aspartate semialdehyde into the ectoine channel. On the other hand, challenge by a new physiological or environmental state, even if the foregoing applies, may still require expression of an Ask homolog that has different physical properties, e.g., an Ask_ect species generated in response to heat stress might be heat resistant itself, thus allowing initial activity to generate the precursor needed for the synthesis of molecules (ectoine) that are protective for other proteins.
(iii) When specialized paralogs may not be absolutely specialized.
Consideration of what is known about the multibranched pathway of aromatic amino acid biosynthesis in E. coli with respect to the three differentially regulated paralogs of the initial pathway enzyme (3-deoxy-d-arabino-heptulosonate [DAHP] synthase) may provide a helpful analogy here. Tryptophan, tyrosine, and phenylalanine are each specific regulators of one of the paralogs via both feedback inhibition and repression control. In unsupplemented minimal medium, where all three amino acids must be synthesized during exponential growth, almost all of the DAHP synthase activity is present as the phenylalanine-sensitive paralog (42). This is because internal regulatory circuits within the terminal branchlets favor metabolite flow to tryptophan and tyrosine in preference to phenylalanine and the internal pool of phenylalanine is insufficient to inhibit and repress the phenylalanine-sensitive paralog. Thus, under these conditions, a single phenylalanine-sensitive DAHP synthase provides a basal level of activity that supports the network feeding all three amino acid branches. If the latter growth conditions were the usual nutritional growth regimen of E. coli, a single DAHP synthase would be generally adequate. (This condition of growth in minimal medium can be viewed as roughly comparable to the everyday nutrition of organisms such as cyanobacteria, which do not have a lifestyle that exploits exogenous amino acids and which usually possess a single DAHP synthase.) On the other hand, E. coli depends heavily upon the periodic provision of exogenous amino acids. In the presence of exogenous phenylalanine, the tyrosine-sensitive and tryptophan-sensitive DAHP synthases are capable of marked derepression in response to any limitation of tyrosine and/or tryptophan that might ensue.
The Cohesion Group Approach
The cohesion group approach for evolutionary analysis of biochemical pathways is illustrated by very detailed work done with the tryptophan (89, 90) and the tyrosine (6) branches of aromatic amino acid biosynthesis. Individual cohesion groups provide a manageable assemblage of subsets of a given protein (or protein concatenates) that have evolutionary continuity. This allows recognition both of lateral gene transfer (LGT) events and of character state changes in the vertical genealogy. Cohesion groups usually consist of functionally equivalent orthologous genes, and detailed knowledge about one member will usually apply to the other members. This is quite helpful. However, if on occasion they contain paralogs or xenologs having different functional roles and/or regulation, that is likely to facilitate even more valuable inferences. For example, S. coelicolor possess two paralogs in TyrCG-17 (6). One is an arogenate dehydrogenase; the other is PapC, an enzyme having a single substitution at position 154 that alters its substrate specificity and that is encoded by a gene regulated within the calcium-dependent antibiotic cluster. Since the paralogs are sufficiently similar to be within the same cohesion group, one can deduce that papC arose recently by gene duplication, followed for one paralog by a novel specialization of substrate specificity and by translocation to an antibiotic synthesis module. Once convincing conclusions are formulated for a set of cohesion groups, parsimonious deductions can be made about evolutionary changes that must have occurred at deeper phylogenetic levels. This approach employs a process in which groups of very similar sequences on an initial tree are recognized as instances of overrepresentation, and each group is then reduced to a single representative sequence. Cohesion groups are obtained by a conservative process of tree building, collapsing the tree at any nodes having high bootstrap values and rebuilding the tree using an arbitrarily chosen representative sequence for those positioned at collapsed nodes. This process is repeated until a tree that has no branches with high bootstrap values is obtained.
A final collapsed tree (in which each branch represents either an orphan sequence or a single arbitrarily chosen member of a cohesion group) is one in which the order of cohesion group branching is uncertain because, by definition, the bootstrap support at the set of collapsed nodes is low. However, within cohesion groups of sufficient size, evolutionary events can be deduced with confidence. In a general context of vertical genealogy, scenarios of LGT can be recognized whereby “intruder” sequences clearly originated from a donor within the lineage represented by the cohesion group but are currently present in an organism that is phylogenetically incongruent with the rest of the organisms hosting members of the cohesion group. In short, the presence of an intruder sequence in a cohesion group identifies both the recipient and the general donor lineage of an LGT event. Scenarios of LGT can still be incomplete due to a lack of experimental information about whether alterations of functional roles or regulation differ in comparisons of the recipient and donor organisms. Two complete examples of LGT of Trp pathway operons were recently described (58) in which the functional roles and regulation of these operons in the donor organisms (previously known in great detail [89]) were matched with thorough experimental information that subsequently became available about altered regulation and functional contexts in the recipient organisms. In this paper, we apply the approach of cohesion group analysis to address the evolutionary relationships of Ask in the highly variable and fascinating organismal contexts where different patterns of the aspartate-derived metabolic network exist.
Superorder Divisions of the Gammaproteobacteria
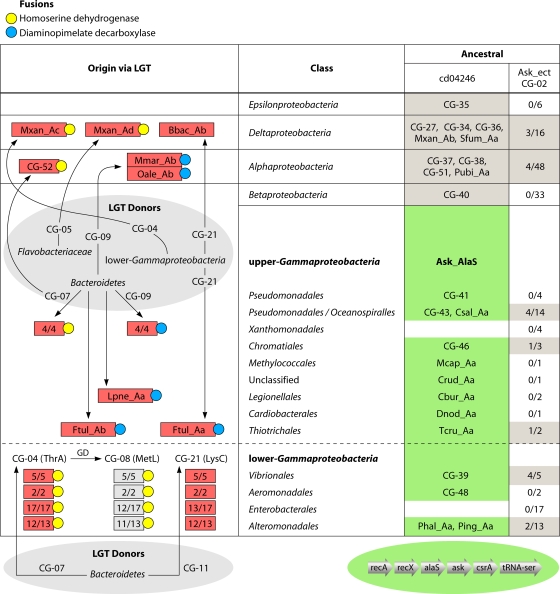
The Gammaproteobacteria currently enjoy a relatively high density of representation among finished genomes. This provides enhanced opportunities for much more extensive evolutionary interpretation than is possible in other lineages, and this focus on the Gammaproteobacteria (within the larger scope of the phylum Proteobacteria) is a highlight of this article. The formal taxons of Gammaproteobacteria at the hierarchical level of the order separate into two distinct groups based on the criteria of many character states of aromatic amino acid biosynthesis, as exemplified by several recent publications that employed the cohesion group approach (6, 89). Kleeb et al. (45) found two comparable subdivisions in the phylogenetic tree of the cyclohexadienyl dehydratase family, which they called Gammaproteobacteria I and Gammaproteobacteria II, and the same separation is evident in the bacterial-genome tree of Wu and Eisen (88). We found exactly the same division to be apparent in the current cohesion group analysis of Ask. These two informal “superorders” have been called the lower Gammaproteobacteria and the upper Gammaproteobacteria (6, 84, 89), and these designations are applied in the present analysis.
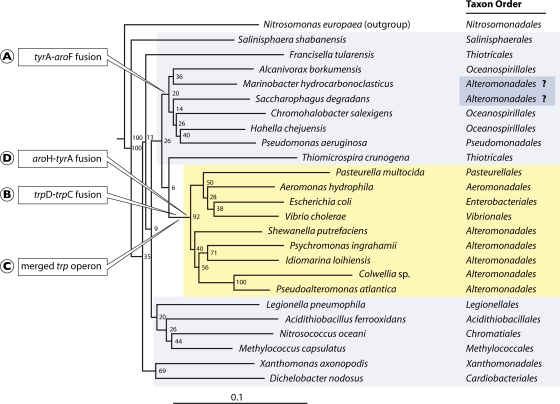
Figure 3 shows a 16S rRNA tree of organisms selected from various orders of Gammaproteobacteria. Five orders in the right-hand column belong to the lower Gammaproteobacteria, and the remaining orders are the upper Gammaproteobacteria. Gene fusions have been shown to have utility for identification of hierarchical slices of a given phylogeny that had a common ancestor (41). Figure 3 illustrates this approach, in which all lower Gammaproteobacteria, but none of the upper Gammaproteobacteria, have a set of two nested gene fusions in common. On the other hand, a small clade of the upper Gammaproteobacteria has another gene fusion that is absent in the lower Gammaproteobacteria.
FIG. 3.
16S rRNA tree of Gammaproteobacteria. The TreeBuilder tool at the ribosomal database (http://rdp.cme.msu.edu) was used. The sequence of N. europaea (Betaproteobacteria) was used as an outgroup. The lower-Gammaproteobacteria orders are highlighted in yellow, and the upper-Gammaproteobacteria orders are shown in gray. The evolutionary times for acquisition of four character states (circled letters A, B, C, and D) are shown at appropriate positions on the tree. Fusion A is for genes encoding cyclohexadienyl dehydrogenase (tyrA) and enolpyruvyl-shikimate-3-phosphate synthase (aroF), fusion B is for genes encoding indoleglycerol phosphate synthase (trpD) and phosphoribosyl-anthranilate isomerase (trpC), fusion D is for genes encoding chorismate mutase (aroH) and cyclohexadienyl dehydrogenase (tyrA), and evolutionary event C is the merging of the split-operon components of the trp operon (90). The current designations of two organisms at the upper right as Alteromonadales is considered highly doubtful, as indicated by question marks, because unlike the five Alteromonadales organisms shown in yellow, they possess character state A and they lack character states B, C, and D.
These character state fusions are pertinent to the consideration of a taxonomic discrepancy in which Marinobacter hydrocarbonoclasticus and Saccharophagus degradans (both belonging to the family Alteromonadaceae) are currently designated Alteromonadales at NCBI. However, they do not group in the tree with the other designated Alteromonadales in the lower Gammaproteobacteria. Furthermore, Marinobacter and Saccharophagus share the character state of having a tyrA-aroF fusion that exists throughout organisms belonging to the Oceanospirillales and Pseudomonadales (Fig. 3). In addition, the true Alteromonadales in the lower Gammaproteobacteria (but not Marinobacter and Saccharophagus) all possess the three character states depicted, which are present throughout all of the lower Gammaproteobacteria (except for some cases of recent wholesale reductive evolution in pathogens and endosymbionts). These character states are two fusions (aroHI-tyrA [6] and trpD-trpC [90]), as well as a fully merged trp operon from former split-operon components (90). It is noteworthy that our assertion that the taxonomic placement of Marinobacter and Saccharophagus is erroneous received strong confirmation from the results of Wu and Eison (88), in whose genome tree the orders Alteromonadales, Oceanospiralles, and Pseudomonadales were interspersed and paraphyletic. They concluded that “the taxonomy needs to be revisited and possibly revised in such cases.”
ASSEMBLY OF Ask COHESION GROUPS
Delineation of Cohesion Groups
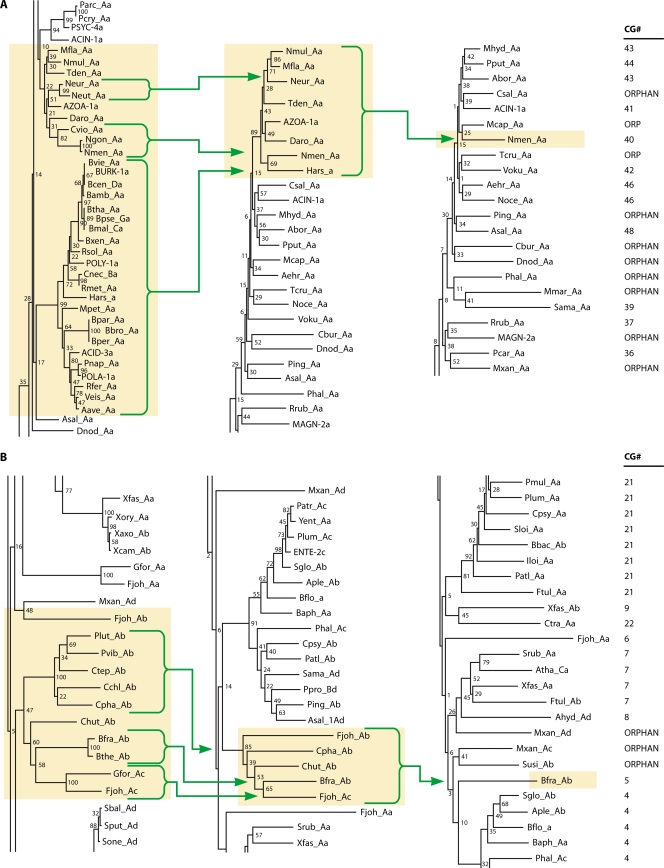
An initial inventory of Ask sequences was aligned and monitored to ensure that the K(F/Y/I)GG motif (see below) was present in the immediate region of the N terminus. In some cases, annotations having incorrect translational start sites were corrected. A variety of manual corrections were made in the alignment, and various fusions at either the N terminus or at the C terminus were trimmed. The subsequent process of collapsing nodes on progressively refined trees is illustrated in Fig. 4A. The 33 sequences that were eventually placed in cohesion group 40 are highlighted in Fig. 4A in the appropriate section of the tree, a snapshot of which is shown on the left. The sequences are all from the Betaproteobacteria, and no sequences from the Betaproteobacteria were found to belong to any other cohesion group after the final round of node collapsing was performed. The most overrepresented sequences (dense collections of very close homolog relatives) were a group of 23 that join at a node that was collapsed, and Hars_Aa was arbitrarily chosen to represent the collapsed group in the next round of tree building. Small groups of two and three sequences were similarly collapsed. A total of eight “representative” sequences were used in the second round of tree building, and this yielded a common node for all of them that was supported by a bootstrap value of 89%. The strong bias created by the 23 overrepresented sequences compared to the use of a single arbitrarily chosen “representative” sequence of that group is evident. Accordingly, Nmen_Aa was chosen to represent CG-40 for the alignment serving as input for the next tree (shown on the right). The cohesion groups or orphans to which the sequences in the tree snapshot on the right were eventually assigned are indicated on the far right. It can be seen that at this stage the CG-43 and CG-46 cohesion groups had yet to be completely consolidated.
FIG. 4.
Progression of tree-making steps to generate cohesion groups. (A) A “snapshot” of the large master tree of Ask sequences is shown in the vicinity of the ultimate CG-40 (highlighted). The three arrows leading from bracketed sequences on the left point to “representative” sequences that were arbitrarily chosen to represent collapsed nodes supported by very high bootstrap values. The three representative sequences plus the five remaining orphan sequences were included in a new multiple alignment that was used to build another tree. The appropriate portion of the rebuilt tree shown in the middle exhibits a common node with bootstrap support of 89, which then was collapsed at that node to yield a subsequent rebuilt tree having the Neisseria meningitidis sequence (Nmen_Aa) chosen as a single representative sequence for CG-40. The ultimate cohesion group assignments are shown on the far right. (B) A snapshot of the master tree of Ask sequences is shown in the vicinity of the ultimate CG-05 (highlighted). The three arrows leading from bracketed sequences point to “representative” sequences chosen to represent a collapsed node supported by very high bootstrap values. The rebuilt tree shown in the middle exhibits a common node with bootstrap support of 85, which then was collapsed at that node to yield a subsequent rebuilt tree having the B. fragilis sequence (Bfra_Ab) chosen as a single representative sequence for the 11 members (indicated on the far left) of CG-05.
A second example is illustrated in Fig. 4B. Here, the ultimate 11-member CG-05 is shown on the left in the initial tree. Collapsing the three nodes with 100% bootstrap support resulted in the five-member grouping shown in the middle tree. Since these were now all supported with a bootstrap value of 85%, Bfra_Ab became the representative sequence for cohesion group CG-05, as seen in the third tree. At this stage, CG-04, CG-07, and CG-21 had not yet been fully consolidated, as shown on the right.
Final totals of 52 cohesion groups and 30 orphans were obtained. The orphan Ask enzymes and a representative enzyme from each cohesion group are listed in Table 1. There, each enzyme's acronym is given, along with the complete genome name, the phylogenetic lineage, the cohesion group affiliation, and a variety of other features that have been analyzed in this paper. A sortable, multiply hyperlinked, and completely expanded version of this table (called the dynamic table) is available online (http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html). It is recommended that the reader access this online table while reading this article, as one can navigate with one click to such interesting tools as the cohesion group gene neighborhoods. The dynamic table is described more fully in the Appendix. The dynamic table also has a link (http://www.theseed.org/Papers/MMBR-Aspartokinase/representatives.html) for accessing a dynamic version (the representatives table) of Table 1, which has all of the columns and features of the dynamic table, except that only a single member of any given cohesion group is included.
TABLE 1.
Representative sequence members of aspartokinase cohesion groups and orphan sequences
| CG no. | Domain | Phylum | Class | Genome name | AroPath code | gi_no. | Fused domains | Indel group | Internal start site |
|---|---|---|---|---|---|---|---|---|---|
| 01 | Eukarya | Fungi | Ascomycota | Saccharomyces cerevisiae S288c | Scer_Aa | 6320893 | ASKα | Absent | |
| 02 | Bacteria | Proteobacteria | Alphaproteobacteria | Hyphomonas neptunium ATCC 15444 | Hnep_Aa | 114800224 | ASKα | Absent | |
| 03 | Bacteria | Firmicutes | Bacilli | Lactobacillus casei ATCC 334 | Lcas_Aa | 116495609 | ASKα | Absent | |
| 04 | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Sodalis glossinidius morsitans | Sglo_Ab | 85058382 | Ask-Hdh | ASKα | Absent |
| 05 | Bacteria | Bacteroidetes | Bacteroidetes | Bacteroides fragilis NCTC 9343 | Bfra_Ab | 60680126 | Ask-Hdh | ASKα | Absent |
| 06 | Bacteria | Bacteroidetes | Flavobacteria | Flavobacterium johnsoniae UW101 | Fjoh_Aa | 146298298 | ASKα | Absent | |
| 07 | Bacteria | Bacteroidetes | Sphingobacteriales | Salinibacter ruber DSM 13855 | Srub_Ac | 28199159 | Ask-Hdh | ASKα | Absent |
| 08 | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Aeromonas hydrophila subsp. hydrophila ATCC 7966 | Ahyd_Ad | 117621282 | Ask-Hdh | ASKα | Absent |
| 09 | Bacteria | Bacteroidetes | Sphingobacteriales | Salinibacter ruber DSM 13855 | Srub_Ad | 28198325 | Ask-LysA | ASKα | Absent |
| 10 | Bacteria | Chlorobi | Chlorobia | Chlorobium tepidum TLS | Ctep_Aa | 21672936 | ASKα | Absent | |
| 11 | Bacteria | Bacteroidetes | Flavobacteria | Gramella forsetii KT0803 | Gfor_Ab | 120435904 | ASKα | Absent | |
| 12 | Bacteria | Acidobacteria | Solibacteres | Solibacter usitatus Ellin6076 | Susi_Aa | 116626024 | ASKα | Absent | |
| 13 | Eukarya | Streptophyta | Rosids | Arabidopsis thaliana Columbia | Atha_b | 15240593 | ASKα | Absent | |
| 14 | Archaea | Euryarchaeota | Environmental sample | Uncultured methanogenic archaeon RC-I | Umet_Aa | 147919906 | ASKα | Absent | |
| 15 | Archaea | Euryarchaeota | Methanomicrobia | Methanoculleus marisnigri JR1 | Mmar-6_Aa | 126178173 | ASKα | Absent | |
| 16 | Archaea | Euryarchaeota | Methanococci | Methanocaldococcus jannaschii DSM 2661 | Mjan_Aa | 15668751 | ASKα | Absent | |
| 17 | Bacteria | Deinococcus | Deinococci | Deinococcus geothermalis DSM 11300 | Dgeo_Aa | 94985229 | ASKα | Absent | |
| 18 | Archaea | Euryarchaeota | Thermococci | Thermococcus kodakaraensis KOD1 | Tkod_Aa | 18893117 | ASKα | Absent | |
| 19 | Archaea | Crenarchaeota | Thermoprotei | Aeropyrum pernix K1 | Aper_Aa | 5104810 | ASKα | Absent | |
| 20 | Archaea | Crenarchaeota | Thermoprotei | Metallosphaera sedula DSM 5348 | Msed_Aa | 146304462 | ASKα | Absent | |
| 21 | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Escherichia coli K-12 | Ecol_Aa | 16131850 | ASKα | Absent | |
| 22 | Bacteria | Chlamydiae | Chlamydiae | Chlamydia trachomatis D/UW-3/CX | Ctra_Aa | 15605086 | ASKα | Absent | |
| 23 | Archaea | Euryarchaeota | Methanobacteria | Methanosphaera stadtmanae DSM 3091 | Msta_Aa | 84488937 | ASKα | Absent | |
| 24 | Bacteria | Firmicutes | Clostridia | Moorella thermoacetica ATCC 39073 | Mthe-3_Ab | 83589915 | ASKβ | Absent | |
| 25 | Bacteria | Actinobacteria | Actinobacteridae | Corynebacterium glutamicum ATCC 13032 (Kitasato) | Cglu_Aa | 19551502 | ASKβ | VEE | |
| 26 | Bacteria | Thermotogae | Thermotogae (class) | Thermotoga eningit MSB8 | Tmar_Ab | 15643313 | Hdh-Ask | ASKβ | VEN |
| 27 | Bacteria | Proteobacteria | Deltaproteobacteria | Anaeromyxobacter dehalogenans 2CP-C | Adeh_Aa | 86160007 | ASKβ | VVK | |
| 28 | Bacteria | Firmicutes | Clostridia | Clostridium botulinum A ATCC 3502 | Cbot_Ba | 148381279 | ASKβ | MNM | |
| 29 | Bacteria | Thermotogae | Thermotogae (class) | Thermotoga petrophila RKU-1 | Tpet_Aa | 148270404 | ASKβ | MVV | |
| 30 | Bacteria | Firmicutes | Bacilli | Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 | Ssap_Aa | 73662666 | ASKβ | MPQ | |
| 31 | Bacteria | Spirochaetes | Spirochaetes (class) | Leptospira interrogans serovar Copenhageni Fiocruz L1-130 | Lint_Aa | 45658739 | ASKβ | MEK | |
| 32 | Bacteria | Cyanobacteria | Cyanobacteria | Prochlorococcus marinus subsp. pastoris CCMP1986 | Pmar_Ca | 33862204 | ASKβ | Absent | |
| 33 | Bacteria | Firmicutes | Bacilli | Bacillus halodurans C-125 | Bhal-1_Aa | 15615658 | ASKβ | MEQ | |
| 34 | Bacteria | Proteobacteria | Deltaproteobacteria | Desulfovibrio desulfuricans G20 | Ddes_Aa | 78357091 | ASKβ | MEA | |
| 34 | Bacteria | Proteobacteria | Deltaproteobacteria | Lawsonia intracellularis PHE/MN1-00 | Lint_Aa | 94987445 | ASKβ | MEE | |
| 35 | Bacteria | Proteobacteria | Epsilonproteobacter | Campylobacter jejuni subsp. jejuni NCTC 11168 | Cjej_Aa | 15791942 | ASKβ | MEQ | |
| 36 | Bacteria | Proteobacteria | Deltaproteobacteria | Pelobacter carbinolicus DSM 2380 | Pcar_Aa | 77918612 | ASKβ | MET | |
| 37 | Bacteria | Proteobacteria | Alphaproteobacteria | Rhodospirillum rubrum ATCC 11170 | Rrub_Aa | 83592082 | ASKβ | VEK | |
| 38 | Bacteria | Proteobacteria | Alphaproteobacteria | Wolbachia endosymbiont of Drosophila melanogaster | Wend_a | 42520775 | ASKβ | VLH code | |
| 39 | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Shewanella amazonensis SB2B | Sama_Aa | 119774186 | ASKβ | Absent | |
| 40 | Bacteria | Proteobacteria | Betaproteobacteria | Neisseria meningitides FAM18 | Nmen_Aa | 121635172 | ASKβ | MER | |
| 41 | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Acinetobacter sp. strain ADP1 | ACIN-1a | 50084438 | ASKβ | MEQ | |
| 42 | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | “Candidatus Vesicomyosocius okutanii” HA | Voku_Aa | 148244688 | ASKβ | VEQ | |
| 43 | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Pseudomonas putida F1 | Pput_Aa | 148546682 | ASKβ | MEQ | |
| 44 | Bacteria | Chloroflexi | Dehalococcoidetes | Dehalococcoides ethenogenes 195 | Deth_Aa | 57233704 | ASKβ | MEI | |
| 45 | Bacteria | Firmicutes | Clostridia | Desulfotomaculum reducens MI-1 | Dred_Ab | 134299030 | ASKβ | MEN | |
| 46 | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Alkalilimnicola ehrlichei MLHE-1 | Aehr_Aa | 114320636 | ASKβ | MEE | |
| 47 | Bacteria | Bacteroidetes | Sphingobacteria | Salinibacter ruber DSM 13855 | Srub_Ab | 83815171 | ASKα | Absent | |
| 48 | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Aeromonas salmonicida subsp. salmonicida A449 | Asal_Aa | 145300679 | ASKβ | MEA | |
| 49 | Archaea | Crenarchaeota | Thermoprotei | Nitrosopumilus maritimus SCM1 | Nmar_Aa | 161527701 | ASKα | Absent | |
| 50 | Archaea | Crenarchaeota | Thermoprotei | Nitrosopumilus maritimus SCM1 | Nmar_Ab | 161529262 | ASKα | Absent | |
| 51 | Bacteria | Proteobacteria | Alphaproteobacteria | Maricaulis maris MCS10 | Mmar_Aa | 114570809 | ASKβ | VVS | |
| 52 | Bacteria | Proteobacteria | Alphaproteobacteria | Maricaulis maris MCS10 | Mmar_Ac | 114569649 | Ask-Hdh | ASKα | Absent |
| ORPa | Archaea | Euryarchaeota | Methanopyri | Methanopyrus kandleri AV19 | Mkan_Ab | 19886393 | ASKα | Absent | |
| ORP | Bacteria | Acidobacteria | Solibacteres | Solibacter usitatus Ellin6076 | Susi_Ab | 116619197 | ASKα | Absent | |
| ORP | Bacteria | Actinobacteria | Actinobacteridae | Frankia sp. strain CcI3 | FRAN-2a | 86740773 | ASKβ | absent | |
| ORP | Bacteria | Actinobacteria | Actinobacteridae | Frankia alni ACN14a | Faln_Aa | 111221839 | ASKβ | absent | |
| ORP | Bacteria | Actinobacteria | Actinobacteridae | Tropheryma whipplei TW08/27 | Twhi_Aa | 28572866 | ASKβ | MEE | |
| ORP | Bacteria | Actinobacteria | Actinobacteridae | Streptomyces griseus subsp. griseus NBRC 13350 | Sgri_Ab | 182438043 | ASKβ | MLE | |
| ORP | Bacteria | Actinobacteria | Rubrobacteridae | Rubrobacter xylanophilus DSM 9941 | Rxyl_Aa | 108803931 | ASKα | Absent | |
| ORP | Bacteria | Actinobacteria | Rubrobacteridae | Rubrobacter xylanophilus DSM 9941 | Rxyl_Ab | 108802953 | ASKβ | VEH | |
| ORP | Bacteria | Aquificae | Aquificae (class) | Aquifex aeolicus VF5 | Aaeo_Aa | 15606405 | ASKβ | MEK | |
| ORP | Bacteria | Chloroflexi | Chloroflexi (class) | Roseiflexus sp. strain RS-1 | ROSE-4a | 148657187 | ASKα | Absent | |
| ORP | Bacteria | Deinococcus-Thermus | Deinococci | Thermus thermophilus HB27 | Tthe_Aa | 46198474 | ASKβ | MEM | |
| ORP | Bacteria | Firmicutes | Bacilli | Symbiobacterium thermophilum IAM 14863 | Sthe-17_Ab | 51892824 | ASKβ | VVA | |
| ORP | Bacteria | Firmicutes | Clostridia | Carboxydothermus hydrogenoformans Z-2901 | Chyd_Aa | 78044447 | ASKβ | LEK | |
| ORP | Bacteria | Firmicutes | Clostridia | Syntrophomonas wolfei subsp. wolfei Goettingen | Swol_Aa | 114566842 | ASKβ | MEN | |
| ORP | Bacteria | Firmicutes | Clostridia | Moorella thermoacetica ATCC 39073 | Mthe-3_Aa | 83590151 | ASKβ | MER | |
| ORP | Bacteria | Planctomycetes | Planctomycetacia | Rhodopirellula baltica SH 1 | Rbal_Aa | 32475630 | ASKβ | MIV | |
| ORP | Bacteria | Proteobacteria | Alphaproteobacteria | “Candidatus Pelagibacter ubique” HTCC1062 | Pubi_Aa | 71083222 | ASKβ | Absent | |
| ORP | Bacteria | Proteobacteria | Deltaproteobacteria | Myxococcus xanthus DK 1622 | Mxan_Ab | 108760667 | ASKβ | MED | |
| ORP | Bacteria | Proteobacteria | Deltaproteobacteria | Myxococcus xanthus DK 1622 | Mxan_Ac | 108759004 | Ask-Hdh | ASKβ | Absent |
| ORP | Bacteria | Proteobacteria | Deltaproteobacteria | Myxococcus xanthus DK 1622 | Mxan_Ad | 108762994 | Ask-Hdh | ASKβ | Absent |
| ORP | Bacteria | Proteobacteria | Deltaproteobacteria | Syntrophobacter fumaroxidans MPOB | Sfum_Aa | 116749603 | ASKβ | MEK | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Pseudoalteromonas haloplanktis TAC125 | Phal_Aa | 77359490 | ASKβ | MPG | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (lower) | Psychromonas ingrahamii 37 | Ping_Aa | 119946984 | ASKβ | MES | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Dichelobacter nodosus VCS1703A | Dnod_Aa | 146329519 | ASKβ | VEG | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Coxiella burnetii RSA 493 | Cbur_Aa | 29654360 | ASKβ | Absent | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Methylococcus capsulatus Bath | Mcap_Aa | 53802422 | ASKβ | VEK | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Chromohalobacter salexigens DSM 3043 | Csal_Aa | 92112759 | ASKβ | MEE | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria (upper) | Thiomicrospira crunogena XCL-2 | Tcru_Aa | 78485930 | ASKβ | MEK | |
| ORP | Bacteria | Proteobacteria | Gammaproteobacteria | “Candidatus Carsonella ruddii” PV | Crud_Aa | 116335010 | ASKβ | MER | |
| ORP | Bacteria | Proteobacteria | Unclassified | Magnetococcus sp. strain MC-1 | MAGN-2a | 117924273 | ASKβ | MES | |
| ORP | Bacteria | Chloroflexi | Chloroflexi (class) | Roseiflexus sp. strain RS-1 | ROSE-4b | 148656161 | ASKα | Absent |
ORP, orphan sequence.
An Ancient Subhomology Divide Separates ASKα and ASKβ
Indel structuring.
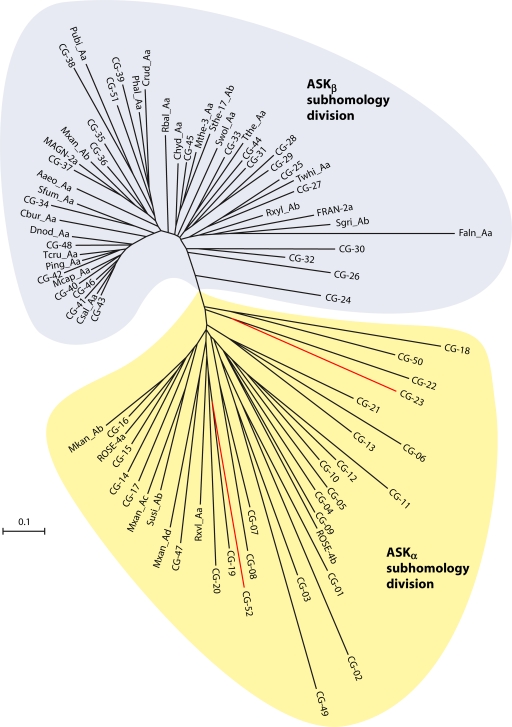
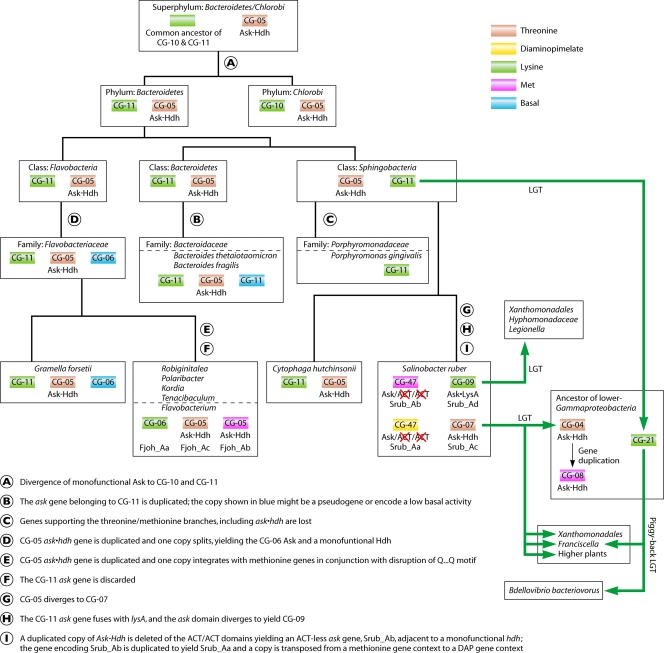
Careful examination of the multiple alignment (available in supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG_representatives_aln.html) of the 85 sequences from the genomes listed in Table 1 revealed a clear separation of two subhomology divisions. They are denoted ASKα and ASKβ in Fig. 5. The greater number of conserved regions in the ASKβ portion of the alignment and the correspondingly more compact portion of the tree in the ASKβ region suggest that ASKα must be the most ancient subhomology division. All of the Archaea and Eukarya sequences belong to the ASKα subhomology division, as well as many Bacteria. In contrast, the ASKβ subhomology group contains only Bacteria. The Faln_Aa orphan (from Frankia alni) within the ASKβ subhomology group exemplifies a likely pseudogene, judging from the many deviations from residues that are otherwise absolutely conserved (especially in the region for nucleotide binding). This is also consistent with its long branch length. Well-characterized Ask enzymes have been described as either homo-oligomeric molecules or as heterotetramers (16), and it appears that these two structural types correspond to the ASKα and ASKβ subhomology divisions.
FIG. 5.
Distribution of cohesion groups on a phylogenetic tree displayed in radial form. Orphan sequences and representative Ask sequences from each cohesion group (listed in Table 1) were aligned, with some manual adjustments, and submitted to a tree-building program as described in the text. The two divergent subhomology divisions (ASKα and ASKβ) are indicated in yellow and blue, respectively. CG-23 and CG-52, marked in red, lack the indel insertions that are otherwise uniquely present in the ASKα subhomology grouping.
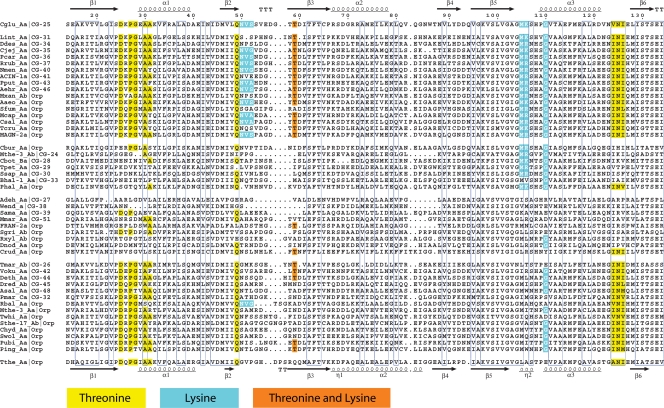
The two subhomology divisions differ most conspicuously in indel structuring. The ASKα subhomology group possesses a region of approximately 50 amino acid residues that has no counterpart in ASKβ sequences and which therefore defines the indel region. This occurs in the vicinity of 70 residues past the N terminus. The indel is between two catalytic residues that in ASKβ sequences are separated by about 25 residues (e.g., T47 and E74 in C. glutamicum, as illustrated in Fig. 6). The sequences belonging to CG-23 and CG-52 (Fig. 5) are unusual in that they clearly belong to ASKα but nevertheless lack the 50-amino-acid regions, thus being similar to ASKβ in this respect.
FIG. 6.
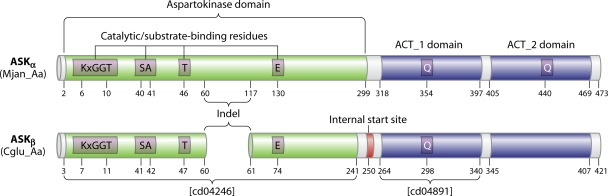
Schematic depicting the essential differences between an ASKα enzyme and an ASKβ enzyme. The enzyme selected to represent each subhomology division has been studied by X-ray crystallography and is from M. jannaschii (Mjan_Aa) or C. glutamicum (Cglu_Aa), respectively. Catalytic and substrate-binding residues are shown only as far as the first catalytic residue past the indel region. An unknown query can readily be recognized unambiguously as belonging to ASKα or to ASKβ by obtaining the cd identifier (at the top of the cd hierarchy). If the cd for the Ask domain is cd04246, then the enzyme is in the ASKβ division, because all ASKβ enzymes have that cd (Fig. 7). If the cd identifier is anything else, the enzyme is in the ASKα division, which has five cd numbers (Fig. 7). Likewise, if the cd for the ACT_1 domain is cd04891, it is in the ASKβ division, because all members of that division have this cd. If the cd is anything else, the enzyme belongs to the ASKα division. Internal start sites are shown in Fig. 12 for all ASKβ cohesion group representatives or orphans. The Q…Q motif is shown with one glutamine residue in ACT_1 and the other in ACT_2 for ASKα enzymes that exhibit threonine allostery. A glutamine residue is present in the homologous position of the ACT_1 domain in ASKβ enzymes that are inhibited by threonine or by Thr plus Lys. However, ACT_2 domains of ASKβ enzymes never possess a Q residue in the homologous position.
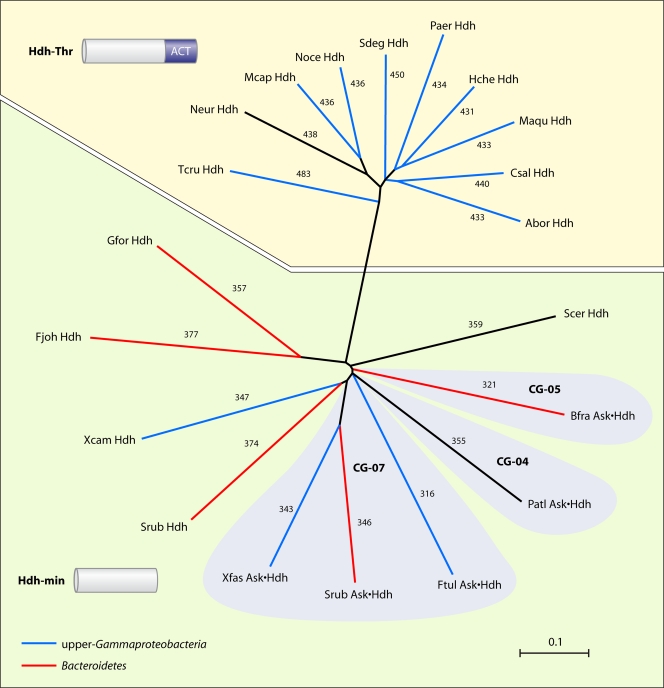
Distribution of multiple homoserine dehydrogenases between ASKα and ASKβ.
It is interesting that organisms harboring ASKα Ask enzymes typically possess a short Hdh of minimal length (Hdh-min), whereas ASKβ organisms characteristically have an Hdh that possesses an approximately 70-residue extension at the C terminus. This extension is in fact an ACT domain, which is represented by a single cd, cd04881, in the CDD of NCBI. In all cases where it has been experimentally determined, ACT-containing homoserine dehydrogenases are feedback inhibited by threonine (reference 61 and references therein). We refer to this class as Hdh-thr. We have noticed that there is a third group of Hdh enzymes that has a C-terminal extension that is not recognized at NCBI as an ACT domain. These are most often seen in the phyla Firmicutes and Actinobacteria. It seems likely that this group may be methionine-inhibited Hdh enzymes (which we denote Hdh-met). Consistent with this assertion, Lactobacillus plantarum has an hdh-met gene organized with other enzymes of methionine biosynthesis, Bacillus clausii and Bacillus halodurans exhibit an SAM riboswitch upstream of hdh-met, and Thermoanaerobacter tengcongensis possesses an SAM riboswitch ahead of hdh-met, followed by a gene for methionine biosynthesis. The three classes of Hdh enzymes are sorted into different columns of the dynamic table online (http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html) (Hdh-thr, Hdh-met, and Hdh-min) and are hyperlinked to a gene detail page. Some genomes support multiple hdh genes. For any genome included in this study, one can conveniently navigate to one or more hdh gene positions and their surrounding gene neighborhoods directly from “Ask Dynamic Table.”
The K(F/Y/I)GG Motif
Very close to the near end of the N terminus, all Ask proteins possess a highly conserved motif that is distributed in three variations. Bareich and Wright (4) have referred to VXKFGG(T/S)SV as “the AK signature motif.” It is best known in the literature as the KFGG motif because that happened to be the variation present in the earliest well-studied enzymes. It is in fact the most widespread motif, but a KYGG motif is also quite abundant. A KIGG motif has limited distribution and has not been reported before. The general motif could be more fully described as the K(F/Y/I)GGTS motif, where the KXGGT amino acids comprise one catalytic residue (K) and three substrate-binding residues for ATP (GGT). In the Ask enzymes having either a KIGG or KYGG motif, there are no amino acid residue substitutions. In the list of Ask enzymes having the KFGG motif, there are five variants: KFGK in Bfra_Ac (CG-11), KFEK in Bthe_Ac (CG-11), KFGA in ROSE-4b (orphan), and KFGS in Csym_Aa (CG-49) and Mmar_Ac (CG-52). The five exceptions are all from ASKα division members. The two CG-11 enzymes are encoded by genes that arose as extraneous paralogs with alterations in important conserved codons throughout the genes and are likely pseudogenes. The last three variants have amino acid substitutions in the motif that are probably tolerated, and important catalytic and substrate-binding residues are otherwise conserved.
The KIGG motif is uniformly present in all members of CG-02 and CG-20, and both of these cohesion groups belong to the ASKα division. Otherwise, ASKα Ask enzymes almost always possess a KFGGTS motif, the only exceptions so far being the KYGG motifs present in Fneo_Aa of CG-01 and in Nmar_Ab of CG-50. ASKβ Ask enzymes usually possess a QK(F/Y/)GGTS motif. It is quite striking that the Q residue is almost always present in ASKβ Ask enzymes and is almost never present in ASKα Ask enzymes. (See the multiple alignment of representative cohesion group members and orphans in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG_representatives_aln.html). All of the members of a given ASKβ cohesion group usually have identical motifs: either KFGG or KYGG, but occasional cohesion groups exhibit a mixture of motifs. For example, CG-32 has six members with a KFGG motif and five members with a KYGG motif.
Ask Fusions
The ask gene is sometimes fused to one of two enzymes in the ASK network, namely, the gene encoding homoserine dehydrogenase (hdh) or the gene encoding DAP decarboxylase (lysA). The fusion of ask with hdh implies a functional specialization of ask for threonine and/or methionine biosynthesis (Fig. 1). The fusion of ask with lysA implicates a functional specialization for lysine biosynthesis (rather than for DAP biosynthesis), since LysA is uniquely used for lysine production. Nine cohesion groups are uniformly populated with ask fusions. Ask-LysA sequences are present in a single cohesion group (CG-09), which implies a recent common origin. However, these sequences are from phylogenetically incongruent organisms, which suggests multiple and recent LGT events, as discussed below. The remaining eight cohesion groups contain Ask enzymes that are fused to Hdh. CG-26 possesses Hdh-Ask fusions from two species of Thermotoga. This fusion (Hdh-Ask) clearly evolved independently of the others, since it is the only instance in which Hdh is fused to the N terminus of Ask. It is also the only fusion joining the ask and hdh genes in the ASKβ division. The remaining five cohesion groups and two orphans (CG-04, CG-05, CG-07, CG-08, and CG-52 and Mxan_Ac and Mxan_Ad) having Ask-Hdh fusions are ones in which Hdh is fused to the C terminus of Ask. The Ask components all belong to the ASKα subdivision. All of them are proposed to have radiated from a single ancient fusion that arose in the superphylum Bacteroidetes/Chlorobi, as discussed below. Han et al. (37) have noted that the sequential order of domain combinations in fusions is usually in one of the possible orders, i.e., if the order A-B occurs, then the order B-A is rarely found. The Thermotoga fusion appears to be one of these rare exceptions.
APPLICATION OF THE CDD FOR EVOLUTIONARY ANALYSIS
The CDD at NCBI is a powerful tool to analyze protein sequences in the context of domain family hierarchies, which are related by common descent and hence reflect evolutionary relationships (53). We have found the application of this tool to the current cohesion group analysis to be fruitful. Ask sequences possess an N-terminal Ask domain and usually have two adjacent ACT domains at the C terminus. It is a reassuring affirmation of the power of the cohesion group approach that the cd signatures for the aforementioned ASKα and ASKβ subhomology divisions are mutually exclusive for both the Ask and the ACT domains. Thus, if any new query sequence is scanned at the CDD for the cd signatures, assignment to ASKα or to ASKβ will be unambiguous.
Phylogenetic Distribution of CDD Domains for Ask
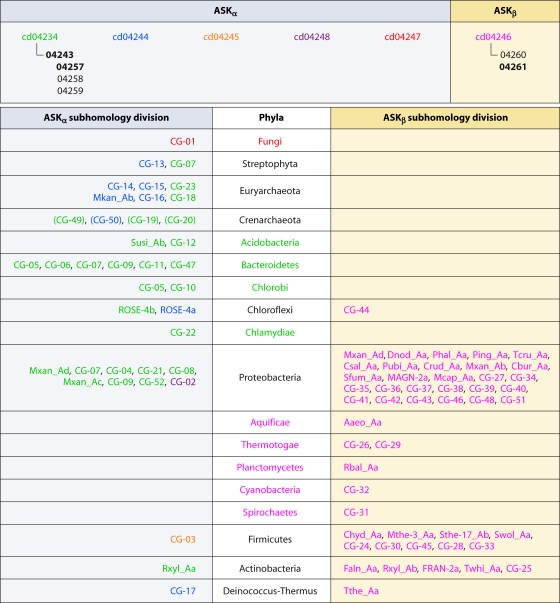
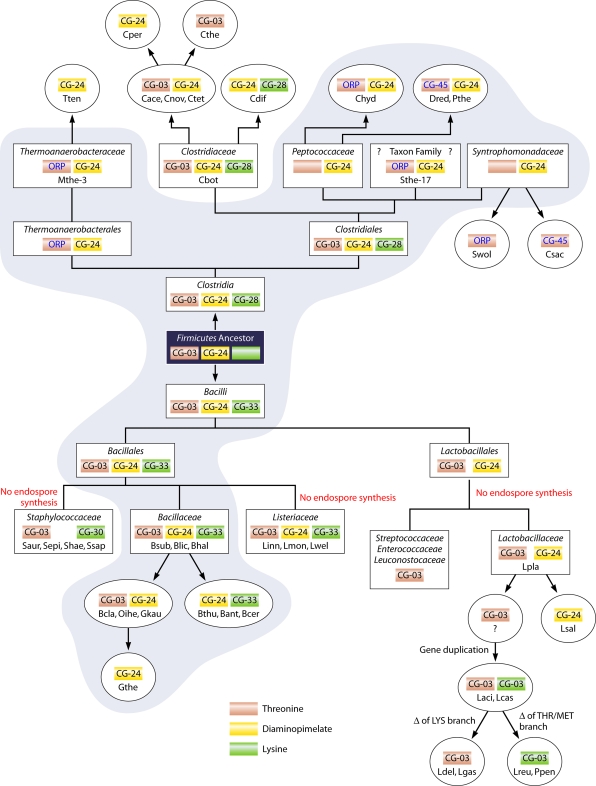
The Ask cds are hierarchical, and they are shown at the highest hierarchical level across the top of Fig. 7. Cohesion groups and orphan sequences are shown on the right and left sides across from the centrally listed phyla containing them. The ASKβ sequences fall under a single cd signature (cd04246), with cd0426l being by far the most common subsignature in this hierarchy. The ASKα sequences, on the other hand, are partitioned between the remaining five cd signatures in Fig. 7. Sequences belonging to CG-01, CG-02, and CG-03 are each restricted to a single phylum: Fungi, Proteobacteria, and Firmicutes, respectively.
FIG. 7.
Relationship of the six major NCBI Ask cds in the CDD to the ASKα and ASKβ subhomology divisions. The NCBI assemblage of cds is hierarchical, and some of the six have further subdivisions, as shown across the top. For example, cd04246 includes the “children” cd04260 and cd04261. Among the “children,” cd04261 is shown in boldface to indicate that it occurs as a specific hit or at least as the best hit when queried in the CDD. Similarly, cd04243 and cd04257 are the most frequent specific hits within the cd04234 family. CG groups and orphans belonging to the ASKα subhomology division are listed on the left, and those belonging to the ASKβ subhomology division are listed on the right. The CG group and orphan designations are color coded to match the cd assignments. Phyla shown in the middle column are color coded in all cases where every cohesion group and orphan uniformly carries the same cd assignment. In cases where there are no specific hits, the best nonspecific hit is color coded and the CG designation is enclosed in parentheses.
CDD Domains for ACT: Functional and Evolutionary Implications
Varied multiplicities of ACT domains in Ask enzymes.
Ask enzymes may (rarely) lack ACT domains altogether, may (almost always) possess two adjacent ACT domains at the C terminus in the configuration ACT_1/ACT_2, or may (rarely) possess four adjacent ACT domains in the configuration ACT_1/ACT_2/ACT_1/ACT_2. In the two paralog members of CG-47 and in the two members of CG-52, there are no ACT domains at all. In many cases one (or both) ACT domain has been sufficiently disrupted that it is not recognized at all on the BLAST graphic, or it is identified as a “nonspecific hit.” Quite often, this is typical of an evolutionary shift from an Ask specialization for threonine responsivity to a specialization for methionine responsivity. In almost all of these cases, the sequence region is largely retained, probably because of structurally important properties of the twin ACT domain architecture. Four ACT domains are present in the Ask proteins of all members of the phylum Cyanobacteria, which populate a single highly conserved cohesion group (CG-32). The single orphan member (Rbal_Aa) of the phylum Planctomycetes also has four ACT domains. The members of CG-32 and Rbal_Aa may have had a common origin, but if so, the divergence is too great to make any conclusions without the availability of additional genome representation. However, the possibility that the acquisitions of four ACT domains in these two phyla were independent events is consistent with the presence of an in-frame internal start site upstream of the ACT domains in Rbal_Aa but not in any members of CG-32 (see below). Detailed experimental study of Ask enzymes having these two structure variations would clearly be of great interest.
Phylogenetic distribution.
There are many different cds for ACT in the CDD, and they reflect evolutionary descent from a common ancestor (53). These domains are arranged hierarchically in the CDD, and at the most elevated hierarchical level, there are three ACT domains (cd04890, cd04891, and cd04892). ACT_1 may be any of the three cds, whereas ACT_2 is always cd04892. In the ASKβ subhomology group, ACT_1 is cd04891. In the ASKα subhomology grouping, ACT_1 is either cd04890 or cd04892. Thus, not only does the cd signature for the Ask domain clearly distinguish ASKα from ASKβ, but the cd signature for ACT_1 also distinguishes ASKα from ASKβ. The archaeal sequences within the ASKα subhomology group always possess an ACT_1 domain, which has the cd04892 signature (or a degenerate version of it).
Tandem ACT domains undoubtedly originated by gene duplication. Since ACT_2 always has the cd04892 signature, the original ACT domain pair must have been ACT_1 (cd04892)-ACT_2 (cd04892). This domain combination is present in all contemporary Archaea and some bacteria, and they are listed in the top and middle sections of Fig. 8A and B. Since the above cd signature combination is required for an intact Q…Q motif, which also has relevance for Hdh allostery (see below), it is interesting to consider the possibility that the selective pressure favoring the gene duplication was to achieve an allosteric region that could function not only for Ask regulation by threonine, but also for regulation of an Hdh domain that participates in a protein-protein interaction with Ask. The protein-protein interaction could potentially occur between fused domains or between unfused but complexed domains.
FIG. 8.
Overall features of regulation within the ASKα subhomology division. (A) Alignment of the Q…Q region of the ACT_1-ACT_2 regulatory domains. This region corresponds to the nonequivalent threonine-binding sites described by Paris et al. (71). A cd04892-cd04892 organization is associated with regulation by threonine. At the top, the 10 cohesion groups and orphans exhibiting perfect QXXSE…QXXSE threonine configuration presumably possess a threonine-inhibited Ask and a threonine-inhibited Hdh, as suggested at the right of panel B in the corresponding section. The next block of four possess a recognizable threonine configuration, but with disruption of the motif. The CG-52 Ask enzymes lack the ACT domains but are included because the fusion with Hdh and the gene context (panel B) clearly indicate functional specialization for threonine. The next block of 10 have diverged away from threonine specialization, and in most cases, the new specialization can be inferred (see the text). At the bottom, a different cd organization (cd04890-cd04892) is associated with a pattern of lysine regulation. The green-shaded sequences indicate a perfect “lysine configuration” of allostery, based upon X-ray crystal studies (47), and important residues are boxed and shown in blue. Although the four cohesion groups above the lysine configuration are derivatives of it, they have unknown allosteries (CG-02) or different allosteries (CG-01 members are threonine inhibited, CG-03 members are inhibited synergistically by lysine-plus-threonine combinations, and CG-13 members are inhibited synergistically by lysine-plus-SAM combinations). Amino acid residues shown to be important for threonine inhibition of the yeast Scer_Aa Ask (2) are shown with white letters against a blue background on the dimmed line, and amino acid residues shown to be important for SAM binding in Arabidopsis (54) are shown in magenta at the bottom. (B) The same Ask sequences as in panel A are displayed in a context of operation within the total ASK network and the regulatory implications. The presence of one or another of alternative biosynthetic pathways for lysine, aromatic amino acids, and isoleucine are indicated by shaded blocks. Light-blue shading is used for the Ask-relevant alternatives, and dark-blue shading is used for Ask-irrelevant alternatives. The descending arrows indicate the origins of functionally divergent paralogs in the middle section from the putative ancestral threonine-regulated Ask enzymes in the upper section. Srub_Ab in CG-47 was derived by gene duplication and then underwent another gene duplication to produce another paralog (Srub_Aa) in CG-47.
The Q…Q motif specifies nonequivalent threonine-binding sites.
Each of the twin ACT domains in the cd04892-cd04892 configuration often possesses a QXXSE motif to yield a tandem combination (QXXSE…QXXSE), which we refer to as the Q…Q motif. In A. thaliana, it has been shown (71) that an Ask-Hdh fusion (belonging to CG-07) possesses a Q443…Q524 region in which these glutamine residues are targeted by threonine for allosteric binding. Interestingly, Q443 and Q524 are key elements of nonequivalent threonine-binding sites. Only Q443 is essential for threonine inhibition of Ask activity, whereas both Q443 and Q524 are essential for threonine inhibition of homoserine dehydrogenase activity. Q443 contributes to a high-affinity binding site for threonine, and binding of threonine there activates the binding of threonine to the second Q524 site.
Figure 9 illustrates a conserved Q…Q motif in members of CG-05. Members of CG-05 have Ask-Hdh fusion proteins whose Ask and Hdh activities are both likely to be inhibited by threonine, and the corresponding genes are positioned in context with the other threonine pathway genes. Flavobacterium johnsoniae possesses two paralog members of CG-05, shown at the bottom of Fig. 9. Fjoh_Ac exhibits an intact Q…Q motif (aligning well with the sequences above it) and is adjacent to other threonine pathway genes. In contrast, the Fjoh_Ab paralog has migrated to a gene context of methionine biosynthesis and exhibits a disrupted ACT_2 region. This indicates that the Ask of the new Ask-Hdh fusion protein, derived by gene duplication, is probably still inhibited by threonine, but the homoserine dehydrogenase activity is not. Starvation for methionine may derepress the new Ask-Hdh fusion protein or at least provide sufficient residual activity, even in the presence of threonine, to support methionine biosynthesis. A number of other genera in the same family as F. johnsoniae (not shown in Fig. 9) possess the same ask gene duplicate, having characteristics suggesting specialization for methionine biosynthesis. Perhaps this clade has an alteration of methionine metabolism that has selected for a greater output of methionine from the ASK network.
FIG. 9.
Example of paralog deregulation and recruitment to methionine pathway specialization in one functionally divergent member of CG-05. CG-05 contains 11 Ask-Hdh sequence members from the phyla Chlorobi and Bacteroidetes. All of these must be tied functionally to the threonine branch because of the gene context shown, where ask (thrA) is near thrB and thrC, as shown directly under the alignment. F. johnsoniae contributes two paralogs to CG-05, one of which (Fjoh_Ac) shows the same conservation of the Q…Q motif in the ACT_1/ACT_2 regions (shown in yellow) as do the other sequences. The other paralog (Fjoh_Ab) exhibits disruption of the motif that would be consistent with loss of regulation by threonine. At the bottom is shown the methionine operon of G. forsetii for comparison with the F. johnsoniae methionine operon, which contains the newly inserted Fjoh_Ab paralog. This methionine-specialized paralog originated in the common ancestor of F. johnsoniae, Polaribacter irgensii, Tenacibaculum sp., Kordia algicida, and Robiginitalea biformata (Fig. 13).
The rationale for interpretation of evolutionary events with respect to the ASKα subhomology group.
In Fig. 8A and B are companion diagrams that attempt to equate sequence motif and cd signature information (Fig. 8A) of the twin ACT domain region with ASK network, gene fusion, and gene context (Fig. 8B) information. At the top of the alignment is a “threonine configuration” (cd04892-cd04892). In this analysis, we assume that the presence of a signature Q…Q motif across the ACT_1-ACT_2 region indicates that Ask is feedback inhibited by threonine and Hdh is feedback inhibited by threonine. In cases where Ask and Hdh are not fused, we suggest that Ask and Hdh form a protein-protein complex, a reasonable assumption, since many enzyme complexes are known to have fusion counterparts, e.g., anthranilate synthase and tryptophan synthase (90). The middle block of cohesion groups consist of disrupted or altered twin ACT domains, which are derived from the cd04892-cd04892 configuration. They are generally associated with specialization with a branch other than the threonine branch. At the bottom is a “lysine configuration” (cd04890-cd04892), from which are derived (i) a threonine-sensitive Ask (CG-01); (ii) an Ask species having unknown, if any, allostery (CG-02); (iii) an Ask species subject to allostery by a synergistic combination of lysine and threonine (CG-03); and (iv) an Ask species subject to allostery by a synergistic combination of lysine and SAM (CG-13).
SUGGESTED EVOLUTIONARY PROGRESSION OF THE ASK NETWORK
The Likely Ancestral Network
An ancient minimal network.
ASKα was probably the most ancient species of Ask, since the divergence of multiple cd signatures for the Ask domain in ASKα sequences, but not in ASKβ sequences (Fig. 7), and the greater sequence divergence of ASKα sequences than ASKβ sequences (see the alignment in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG_representatives_aln.html) imply a greater elapsed evolutionary time. The earliest organisms probably used the AAA pathway for lysine biosynthesis and the pyruvate pathway for isoleucine biosynthesis because these Ask-independent pathways require fewer ATP equivalents. If so, a relatively simple ASK network was probably dedicated to threonine as a quantitatively major end product and methionine as a quantitatively minor end product. This ancestral core network (the minimal network) is shown in Fig. 1. Note that in this restricted pathway scheme, aspartate semialdehyde is not located at a metabolic branch point and is merely an intermediate in a straight path to homoserine. As such, there is no expectation that Hdh would be subject to feedback inhibition. In fact, any accumulation of intermediates would preferably be in the form of homoserine rather than aspartate semialdehyde, which is unstable and toxic (3, 79). Regulation of Ask and ThrB by threonine may have been sufficient, with residual Ask activity being adequate to feed the minor methionine branch. The presence or absence of the DKFP branch to supply aromatic biosynthesis may have been a very ancient dichotomy dependent upon variations of carbohydrate metabolism that dictated the availability of E4P or DKFP. In any event, the presence of a DKFP branch represents increased network complexity, which would be expected to create selective pressure for regulation of homoserine dehydrogenase, which competes with AroA′ at the central branch point. Likewise, selective pressure for regulation of Hdh would have coincided with the addition of any other branches that compete with Hdh for aspartate semialdehyde to the network, e.g., the DAP pathway of lysine biosynthesis or the ectoine pathway.
New branches can be appended to the ASK network without making a new branching connection at the level of aspartate semialdehyde. Thus, addition of a new branch extending from threonine to isoleucine transforms threonine into a new branch point metabolite, now having the alternative fates of entering protein synthesis or continuing on to isoleucine. This still indirectly has an impact at the level of aspartate semialdehyde, since the quantitative demand for substrate entry into the threonine branch is greatly increased. In the latter case, threonine and isoleucine are needed during a common regimen of vegetative growth. In another case, the biosynthetic pathway to cephamycin C extends from lysine in S. clavuligerus (57), but here, the distribution of lysine to general protein synthesis on one hand and to antibiotic biosynthesis on the other hand occurs in different temporal modes of physiology.
Response of Hdh to increased complexity.
New selective pressures upon Hdh for refinements in regulation have generated a number of recognizably different Hdh enzymes. Four structural types of Hdh can be recognized: (i) the minimal catalytic domain, which lacks an attached allosteric domain (Hdh-min) and typically is present in organisms having ASKα Ask enzymes, (ii) an Hdh-min domain that is fused to an ASKα Ask (Ask-Hdh) (note that as an isolated novelty, Hdh-min is fused to an ASKβ Ask [Hdh-Ask] in the opposite fusion orientation in Thermotoga spp.), (iii) a sequence having a C-terminal ACT domain (cd04881) responsible for feedback inhibition by threonine (Hdh-thr) and that is typically present in organisms utilizing ASKβ Ask enzymes, and (iv) a sequence having a C-terminal extension that is not recognized at NCBI as an ACT domain and that is here suggested to be an allosteric domain for regulation by methionine (Hdh-met).
Hdh-min has no allosteric region and as such may simply exist as a feedback-insensitive step, as is indeed expected for a minimal network. It was impressive insight on the part of Parsot and Cohen when they suggested over 20 years ago (72) that “one can speculate that the ancestral homoserine dehydrogenase was not regulated and that two different strategies have arisen to ensure regulation of this enzyme activity, i.e., fusion to the C-terminal end of an already regulated aspartokinase or addition of a regulatory domain to the C terminus of homoserine dehydrogenase.” However, in addition to the eventual verification of the last two strategies, there are two contexts where Hdh-min may achieve allostery via protein-protein interaction: one that is relevant to threonine and one that is (surprisingly) relevant to lysine. In Ask enzymes with the ACT_1(cd04892)-ACT_2(cd04892) signature and endowed with the Q…Q motif, complex formation between Ask and Hdh may occasion a protein-protein configuration allowing Hdh to “share” the allosteric region of Ask. This is consistent with the experimentally demonstrated involvement of the Q…Q motif spanning ACT_1 and ACT_2 with threonine allostery of both Ask and Hdh activities in the Ask-Hdh species of Arabidopsis (71). It is also consistent with the correlated coexistence of ASKα monofunctional Ask enzymes and “short” Hdh enzymes (Hdh-min) lacking an allosteric domain. If correct, the very earliest Ask species may have possessed a single ACT domain that was dedicated to threonine allostery for Ask. Gene duplication to create adjacent ACT domains may have coincided with competence for complex formation between Ask and Hdh.
A second hypothetical context where Hdh-min may achieve allostery via protein-protein interaction is one in which Hdh-min is adjacent to a free-standing ACT domain (Fig. 8B), which is proposed to interact with Hdh-min to yield a complex that is allosterically responsive to lysine and perhaps to one or more aromatic amino acids. The act gene is translationally coupled to hdh-min in those Archaea that have a DAP pathway of lysine biosynthesis. How a free-standing ACT domain might function as a regulatory subunit of an Hdh-ACT complex that responds to lysine in a way that appropriately impacts threonine/methionine biosynthesis is considered below. In the class Clostridia, an act-hdh gene cluster proposed to respond to lysine is usually embedded in a larger threonine/methionine pathway gene cluster (see “Firmicutes” below).
Evolution of the ASKα Assemblage
Contemporary minimal networks leading only to threonine and methionine.
The simplest network restricted to threonine and methionine biosynthesis (Fig. 1) and utilizing a single species each of Ask and Hdh-min is exemplified by the Crenarchaeota organism Aeropyrum pernix (CG-19) and by the four Crenarchaeota species whose single Ask sequences populate CG-20 (Fig. 8B). The QXXSE motif is intact for ACT_1 but not for ACT_2 (Fig. 8A), suggesting that Ask is feedback inhibited by threonine but that Hdh is not. In the presence of exogenous threonine, residual Ask activity may be sufficient to feed the quantitatively less demanding methionine branch.
The Euryarchaeota members of CG-18 exhibit a similar simplicity associated with a minimal network, except that both the ACT_1 and ACT_2 domains of Ask are disrupted and highly truncated; this suggests that neither Ask nor Hdh is feedback inhibited by threonine. In this case, as exemplified by Thermococcus kodakaraensis, ask, hdh, and other genes of both threonine and methionine biosynthesis are grouped together. This suggests that the two-branch network is coregulated as one unit. The three Pyrococcus spp. in CG-18 differ from T. kodakaraensis in that adjacent intraoperon paralogs of Ask exist in concert with indications of recent gene scrambling. It is not clear how the adjacent paralogs might have any differential regulation.
Species of Deinococcus in CG-17 contribute members of the single bacterial cohesion group that is associated with a minimal ASK network of the ancestral type envisioned. In Deinococcus spp., the cd signature (Fig. 8A) is somewhat imperfect, but it seems quite possible that both Ask and Hdh might be subject to feedback inhibition. If so, then Ask and Hdh-min must form a complex to allow the ACT_2 domain of Ask to function for allostery of Hdh-min. Since Deinococcus apparently requires obligate isoleucine biosynthesis from threonine, the quantitatively greater flux to threonine, relative to methionine, may accentuate the importance of threonine as a feedback signal in this case.
Reductively evolved minimal networks lacking the threonine and methionine branches.
The modern multibranched and more complex ASK networks are presumed to have arisen from a simpler minimal network that originally led only to threonine and methionine, as described above. Following replacement, in some organisms, of the AAA pathway to lysine with the Ask-relevant DAP pathway to lysine, it is interesting that simple contemporary ASK networks exist that have lost the ancestral threonine/methionine arms of the network with retention of only the more recently acquired DAP/lysine branch. These are cases of reductive evolution in pathogenic organisms that now rely upon host resources for threonine and methionine. Three examples of such reductive evolution in ASK networks that involve ASKα enzymes can be cited. Porphyromonas gingivalis is a Bacteroidetes organism having a single Ask species that is a member of CG-11, all members of which exhibit motifs consistent with feedback inhibition by lysine (Fig. 8A). Legionella pneumophila has a simple ASK network consisting of only the lysine biosynthesis branch. The organism is in a lineage (upper Gammaproteobacteria) that would not usually contain an ASKα species, but in this, case Lpne_Aa is a novel Ask fused with LysA that originated via LGT from a Bacteroidetes donor (see below). Third, in the entire phylum Chlamydiae, only a linear pathway extends from aspartate to DAP, since the enzyme that converts DAP to lysine is missing. Thus, in this case, the host is also the provider of lysine to the pathogen. In all of these examples, dihydrodipicolinate synthase has no enzymatic competitors for the aspartate semialdehyde substrate, and one would not expect this enzyme to be a focal point of regulation as it probably is in more complex networks. Various examples of reductive evolution in ASK networks involving ASKβ enzymes are enumerated below.
Adjustments of Hdh in response to the addition of competing branches to the minimal network. (i) Generation of differentially regulated paralogs in Archaea.
N. maritimus and Cenarchaeum symbiosum both possess the DKFP branch of aromatic amino acid biosynthesis, which thus puts Hdh in competition with AroA′ (Fig. 1). These organisms each possess two Ask paralogs belonging to different cohesion groups, one of the few examples of Ask paralogs in Archaea. The pair of sequences in CG-50 each has a perfect Q…Q motif signature, indicating that both Ask and Hdh in each organism are inhibited by threonine, presumably via complex formation between Ask and Hdh-min. Each organism has a second Ask paralog (CG-49), and these have a disrupted ACT_1-ACT_2 region, which suggests either lack of feedback regulation or sensitivity to something other than threonine. It seems quite feasible that the CG-49 Ask species might prove to be regulated by one or more aromatic amino acids. N. maritimus also has an ectoine pathway, and the CG-49 enzyme might instead be responsive to ectoine specialization. However, since C. symbiosum lacks an ectoine pathway, the specialized tuning of the two CG-49 Ask species to aromatic biosynthesis seems most likely.
(ii) Recruitment of a free-standing ACT domain in Archaea.
All of the Euryarchaeota, except the orders Thermococcales and Thermoplasmatales (the latter is not shown in Fig. 8A and B), have acquired the DAP pathway of lysine biosynthesis. A quantitatively great demand must be imposed upon the ASK network in these Archaea, since the DKFP branch of aromatic amino acid biosynthesis is also present (Fig. 8B). Each organism possesses a single Ask and a single Hdh-min species, with the Ask sequences belonging to CG-14, CG-15, CG-16, and CG-23. Methanopyrum kandleri also has the latter ASK network configuration, its ancestral orphan sequence being Mkan_Ab. (It has an additional Ask xenolog present in CG-19; see “LGT” below for more details.) Except for members of CG-23, the Ask species all exhibit a cd signature, suggesting that both Ask and Hdh are feedback inhibited by threonine (Fig. 8A). How, then, would starvation for lysine and aromatic amino acids be prevented in the presence of excess threonine? A clue to the answer might lie in the observation that most orders of the Euryarchaeota (but not Thermoplasmatales or Thermococcales) possess a gene adjacent to hdh that is translationally coupled and possesses an ACT domain (COG2061). In the COG database (http://www.ncbi.nlm.nih.gov/COG), COG2061 is described as a “predicted regulator of homoserine dehydrogenase.” The CDD assigns this ACT domain to cd04886. It corresponds to the C-terminal domain of catabolic threonine deaminase, but the allosteric specificities of the latter seem distinctly inappropriate to Hdh. There is a perfect correlation between the presence of the DAP pathway of lysine biosynthesis, the presence of the DKFP pathway of aromatic amino acid biosynthesis, and the presence of the act-hdh gene couple in Archaea (Fig. 8B). There is thus a correlation between the absence of specialized Ask enzymes to coordinate both lysine biosynthesis and aromatic biosynthesis and the unexpected presence of a highly conserved amino acid binding protein (ACT) associated with Hdh. A novel and indirect mechanism of control is implicated whereby insufficiency of lysine and aromatic amino acids triggers decreased function of Hdh.
Various mechanisms could accomplish this. For illustrative purposes, one possibility is advanced. Perhaps, under conditions of starvation for lysine and/or aromatic amino acids, Hdh is essentially sequestered by association with the ACT protein, and therefore, Hdh is largely prevented from complexing with Ask. If uncomplexed Hdh has poor activity, aspartate semialdehyde can be preferentially channeled to lysine and/or aromatic amino acids. On the other hand, in the presence of adequate lysine and aromatic amino acids, lysine and at least one aromatic amino acid bound to ACT may promote dissociation of Hdh and the ACT domain protein, in which case the transiently free Hdh is able to complex with Ask. Nutritional sufficiency for all end products of the ASK network would then produce a state in which both Ask and Hdh-min are sensitive to threonine inhibition. Note that in a remote lineage (the class Clostridia) the act-hdh gene pair is also highly conserved in correlation with the absence of a lysine-specialized Ask (see “Firmicutes” below). This mechanism seems to have evolved independently in the two lineages.
The foregoing addresses the dilemma of how threonine regulation of a single Ask species is prevented from blocking lysine and aromatic biosynthesis. It is suggested that ACT and Ask compete with one another for complex formation with Hdh. The Hdh-ACT pair is the preferred complex, and Hdh has low activity when complexed with ACT. Sufficiency of lysine and aromatic amino acids results in saturation of ACT with these amino acids, perhaps in synergistic combination, and promotes dissociation of ACT from Hdh and association of Hdh with Ask. The Ask-Hdh complex equates with a maximally active Hdh, which is sensitive to feedback inhibition by threonine. This proposed mechanism would work best if uncomplexed Ask (equated with insufficient lysine and/or aromatic amino acids) were catalytically active but insensitive to inhibition by exogenous threonine. Such a capability of ACT seems feasible because ACT proteins are known to be quite versatile in binding multiple amino acids. Indeed, the ACT domain of Hdh in Methylobacillus glycogenes has been reported to be as sensitive to l-phenylalanine as to l-threonine (61). The ACT domain of prephenate dehydratase in B. subtilis is sensitive to inhibitory or activating effects of phenylalanine, tyrosine, tryptophan, methionine, and leucine (77).
(iii) Generation of threonine-insensitive paralogs in ASKα Bacteria.
Roseiflexus sp. possesses a threonine-specialized paralog of Ask (ROSE-4a) whose encoding gene is located within a threonine context and which presumably complexes with Hdh-min to accomplish feedback inhibition by threonine of both Ask and Hdh. A second paralog of Hdh-min may help provide uncomplexed molecules of Hdh to facilitate methionine biosynthesis. The organism possesses a DAP pathway of lysine biosynthesis, and therefore, Ask activity is also required for lysine biosynthesis. A second copy of Ask (ROSE-4b) possesses a disrupted Q…Q motif and probably is not regulated by threonine. Although no aspect of regulation by lysine is apparent, this Ask paralog should provide support for lysine biosynthesis in the presence of threonine.
(iv) Generation of lysine-responsive paralogs in ASKα Bacteria and higher plants.
Some Bacteria (notably Chlorobia, Bacteroidetes, and Acidobacteria) that possess the ancestral ASKα type of Ask responded to the DAP pathway replacement of the AAA pathway of lysine biosynthesis by gene duplication and replacement of ACT_1(cd04892) with ACT_1(cd04890) in one of the paralogs. The bottom section of Fig. 8A shows conserved residues that are important for lysine allostery in the E. coli member of CG-21 (47). The Arabidopsis Ask in CG-13 is inhibited by the combination of lysine and SAM (54), and the bottom of Fig. 8A shows that the key residues for lysine allostery are perfectly maintained, in addition to the residues that confer SAM allostery. Solibacter usitatus (phylum Acidobacteria) possesses a degraded threonine-sensitive Ask paralog (Fig. 8A, Susi_Ab orphan), which probably is not inhibited by threonine. It has an Hdh-thr, however, that should be inhibited by threonine, as well as an Hdh-min species that may facilitate methionine biosynthesis. A lysine-responsive Ask paralog is evident, both from the lysine pathway gene context and from the conserved residues, indicating lysine feedback inhibition (Fig. 8A, Susi_Aa in CG-12). It is interesting that another Acidobacteria organism (Korebacter versatilis) has lost the threonine-sensitive paralog (inferred from lack of a cd04892 signature for ACT_1) but has two lysine-sensitive paralogs (defined by the presence of an ACT_1 domain having a cd04890 signature) that belong to CG-12. One of these (ACID-2a) is encoded by a gene with a lysine pathway context and has the motif signature for lysine feedback inhibition. The other paralog (ACID-2b) is encoded by a gene placed in a threonine gene context, including a gene encoding Hdh-thr. This paralog exhibits a poor match for the signature for lysine allostery. A second Hdh-min is also present, which is encoded by a gene having a methionine context. Thus, the original threonine-regulated paralog has been lost and functionally replaced by a duplicate of the lysine-regulated paralog.
Novel Ask members derived from the ACT_1 (cd04890) signature.
The ACT_1 (cd04890) signature is one that specifies lysine allostery in Bacteria and higher plants that have the DAP pathway of lysine biosynthesis, as indicated in the bottom sections of Fig. 8A and B. However, Ask members of CG-01, CG-02, and CG-03 are not inhibited by lysine, but they appear to have originated from lysine-sensitive Ask enzymes. CG-01 is populated by Ask enzymes of fungi that do not even have the DAP pathway of lysine biosynthesis. Fungi possess a single Ask species inhibited by threonine, and it can be seen that the important residues for lysine allostery are imperfectly conserved (Fig. 8A). The critical residues for threonine inhibition of Saccharomyces cerevisiae Ask (2) are shown for ACT_1 and ACT_2 in CG-01 in Fig. 8A. It appears that in this case additional residues important for threonine allostery are located remote from the ACT domains in the catalytic domain (87). Ectoine-associated Ask species are all present in CG-02, and they also exhibit remnants of residues deemed important for lysine allostery. It is unknown whether members of CG-02 are inhibited by ectoine. Members of CG-03 belong to Firmicutes bacteria and exhibit only a faint match for lysine allostery residues. This group is known to be subject to synergistic inhibition by lysine and threonine (46), so derivation from a lysine-sensitive Ask seems reasonable. Finally, the A. thaliana paralogs present in CG-13 exhibit a perfect match for lysine allostery. Two of them, Ath_a and Ath_c, are indeed feedback inhibited by lysine, whereas one of them, Ath_b, is known to be inhibited synergistically by a combination of lysine and SAM (54). Residues important for recognition of SAM are shown at the bottom of Fig. 8A.
Divergence of an ASKβ Subhomology Division
The ASKβ subhomology division is distinguished from the ancestral ASKα division by a number of character states (Fig. 6): (i) a region of up to 70 amino acids has been deleted from the common ancestor of ASKβ enzymes, thus generating an indel in ASKα/ASKβ alignments; (ii) a unique Ask domain cd (cd04246) has diverged for ASKβ; (iii) a unique ACT_1 regulatory domain cd (cd04891) has diverged for ASKβ, which seems to be generally associated with synergistic allostery by threonine-lysine combinations; and (iv) an internal in-frame translational start site has emerged just upstream of the ACT_1-ACT_2 region, which results in overlapping ask genes. It would be intriguing to pursue the extent to which some or all of these character states might be related to one another.
The indel region of ASKβ must be a deletion.
An indel is a region of a sequence alignment in which a section of one sequence has no match in the other sequence. This can be explained by the acquisition of an insertion in the longer sequence or by the occurrence of a deletion in the shorter sequence. If our assertion that ASKα represents the ancestral Ask is correct, it is likely that a common ancestor of ASKβ organisms experienced a deletion. Note that two widely separated ASKα cohesion groups (CG-23 and CG-52) (Fig. 5) are exceptional in having similar deletions of the indel region. These appear to have been two independent deletions that occurred relatively recently compared to the more ancient deletion that occurred in the common ancestor of all ASKβ organisms.
The ASKβ Ask domain.
The Ask domain of all ASKβ proteins exhibits the single cd signature cd04246 at the top of the cd hierarchy (Fig. 7). There are two subsignatures: cd04260 and cd04261. This cd hierarchy alone is sufficient to identify a given Ask sequence as a member of the ASKβ subhomology division. The Ask members of CG-24, which contains the DAP-regulated Ask enzymes of the Firmicutes, always carry the cd04260 subsignature. No other cohesion group members or orphans possess the cd04260 subsignature. For the remaining ASKβ proteins, cd04261 is by far the most common specific hit in response to a CDD query.
The ASKβ pattern of allosteric regulation. (i) The twin ACT domain and allostery.
The ACT_1-ACT_2 region of ASKβ sequences exhibits much more overall conservation of amino acid residues than the corresponding region of the ASKα sequences, partly because ACT_1 in ASKβ sequences has only one cd signature (cd04891). We note that the Q residue that resides in ACT_1 as part of the Q…Q motif in the “threonine configuration” assemblage of ASKα (Fig. 8A) is a highly conserved residue throughout ASKβ sequences and has been implicated as part of the threonine-binding region (93). On the other hand, the Q residue that resides in ACT_2 is highly conserved in ASKα, whether in sequences having the threonine configuration or the lysine configuration (Fig. 8A), but the Q residue is absolutely abolished in all ASKβ sequences (Fig. 6). In fact, a small deletion appears to have removed the Q residue in a region of ASKβ marked by an almost completely conserved STSE motif, which is absent in ASKα sequences (see the alignment in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG_representatives_aln.html).
(ii) Concerted feedback inhibition.
Most organisms that possess an ASKβ Ask that has been experimentally characterized exhibit a pattern of concerted feedback inhibition by the combination of threonine and lysine (Thr plus Lys). Rhodospirillum rubrum, Rhodopseudomonas palustris, and Gluconobacter oxydans all have single Ask proteins in CG-37 that are subject to concerted feedback inhibition by Thr plus Lys (18, 78). Characterization of various Ask enzymes as ones that fit the pattern of concerted feedback inhibition by Thr plus Lys is widely used as a shortcut description because it concisely communicates the essence of what is thought to be a logical mode of allostery, but additional effector results are common. For example, C. glutamicum Ask (CG-25) is also inhibited (but less effectively) by threonine or by lysine alone, and isoleucine is an activator (82). Azotobacter vinelandii Ask (CG-43) is subject to concerted feedback inhibition by Thr plus Lys, with lysine and threonine individually also producing some inhibition (25). Pseudomonas fluorescens Ask (CG-43) is not only subject to concerted inhibition by the Thr-plus-Lys combination, but also by the Thr-plus-Met combination; threonine alone causes weak inhibition, whereas both methionine and lysine are weak activators (24). Rhodocyclus tenue Ask is also subject to concerted feedback inhibiton by both the Thr-plus-Lys and the Thr-plus-Met combinations. Lysine and threonine, individually, are inhibitors. Glycine, isoleucine, methionine, or phenylalanine reverses inhibition by lysine, whereas glycine, isoleucine, or phenylalanine reverses Thr-plus-Lys concerted feedback inhibition. At least Azoarcus sp. and Dechloromonas aromatica in CG-40 can be inferred to have the same complex pattern of allostery as the closely related R. tenue. Leptolyngbya boryanum (a cyanobacterium) is subject to concerted feedback inhibition by both the Thr-plus-Lys and the Thr-plus-Met combinations. In addition, threonine, isoleucine, and homoserine are individual feedback inhibitors (52). Since the cyanobacteria in our collection comprise a large and uniform cohesion group (CG-32), it is possible that all of the cyanobacterial Ask enzymes will prove to have the same complex feedback pattern as does L. boryanum (whose genome is not currently sequenced). It is not always clear to what extent the various effectors or effector combinations have been tested.
(iii) Other allosteric specificities.
T. thermophilus possesses a single Ask protein that is known to be inhibited only by threonine (64). The incompetence of lysine as an effector makes sense, since the alternative AAA pathway is used for lysine biosynthesis in the bacterium. On the other hand, the Firmicutes possess paralogs in CG-24 and CG-33 that are inhibited only by DAP or Lys, respectively, as documented most fully by the pioneering work of Chen and Paulus with B. subtilis (13). Clearly, against a general background of concerted feedback inhibition by a Thr-plus-Lys combination in ASKβ enzymes, the twin ACT region is able to accommodate a number of allosteric variations.
(iv) X-ray crystal structures as a guide to allosteric specificities.
At this time, there are only two X-ray crystal studies that elucidate amino acid residues that are important for allostery in the ASKβ subhomology division. One is the threonine-sensitive enzyme of T. thermophilus (92), and the other is the Thr-plus-Lys-sensitive enzyme of C. glutamicum (93). The X-ray crystal information from the enzymes of these two organisms accommodates sorting of ASKβ enzymes into four groups with different allosteric patterns: sensitive to Thr plus Lys, sensitive to threonine, sensitive to lysine, and no sensitivity to threonine or lysine. We suggest (as discussed below) that lysine-sensitive enzymes and threonine-sensitive enzymes possess homologous residues that parallel those in Thr-plus-Lys-sensitive enzymes (concerted feedback inhibition). However, in addition, the Thr-plus-Lys-sensitive enzymes possess a central region containing both threonine- and lysine-binding residues, and this region may dictate the synergy mechanism.
Figure 10 shows an alignment of ASKβ orphan and cohesion group representatives in the twin ACT region in which the four groups have been sorted out. At the top is the C. glutamicum sequence in which the threonine-binding residues and lysine-binding residues that are conserved are marked by color coding. The first group shown below the Cglu_Aa sequence is very likely to be subject to the same pattern of concerted feedback inhibition, and in fact, all enzymes that have been reported experimentally to have this pattern of allosteric control are located in the top grouping. At the bottom is the T. thermophilum sequence in which the threonine-binding residues match up well with the threonine-binding residues of C. glutamicum. The group immediately above it (group 4) is comprised of putative threonine-sensitive Ask enzymes. Group 2 members consist of putative lysine-sensitive enzymes, and group 3 members appear to be insensitive to the allosteric effects of threonine and lysine, either singly or in combination. This does not mean that there is not some other allosteric specificity that remains to be established. For example, the Sgri_Ab and FRAN-2a orphans, which are in group 3, have an ask_aro specialization gene context, and it would be interesting to know whether one or more aromatic amino acids might be feedback inhibitors.
FIG. 10.
Different allosteric-specificity groupings in ASKβ enzymes. An alignment of representative orphan and cohesion group Ask sequences was trimmed to the twin ACT region deemed to be relevant to allostery. The ESPript/ENDscript software program (31) was used to coordinate with PDB identifiers in order to create a multiple-sequence alignment that carries (at the top and bottom) secondary structure elements of the two sequences of known three-dimensional structures. In addition to the trimmed sequence alignment file, two PDB files (2DTJ for Cglu_Aa and 2DT9 for The_Aa) were used as data input. Using an advanced mode of the web tool, color features were removed, except for the blue frames. Residues were marked with yellow, blue, and orange to indicate relevance to threonine binding, lysine binding, or the binding of both, respectively.
The key residues for threonine binding appear to be the 25DXPGXA30 motif, Q49, and N125. N125 is flanked by I/V/L/M residues. The key motif for lysine binding appears to be 105M(K/R)XXXG110. Although the members of CG-33 deviate from this motif slightly, experimental work has shown that these enzymes are feedback inhibited by lysine. The foregoing threonine-binding residues of threonine-sensitive enzymes and the lysine-binding residues of lysine-sensitive enzymes seem to be very similar in alignment placement to those present in organisms that are subject to concerted feedback inhibition by the Thr-plus-Lys combination (the top group in Fig. 10). If the allosteric regions for lysine and threonine were completely independent, one might expect the effector combination to exhibit cumulative inhibitory effects. However, there is a region between Q49 and D60 of the members of the top group that is absent in the other groups, which we suggest involves an interplay of threonine- and lysine-binding residues that might be key to the synergy phenomenon. T59 has been implicated in both threonine and lysine binding, and the three residues following Q49 have been implicated in lysine binding (93). As is typical of ACT domains (33), the various allosteric regions that are highlighted in Fig. 10 tend to be located in the loops between alpha helices and beta sheets.
The assignments of likely allostery in Fig. 10 are pleasingly in concert with expectations that organisms having a single Ask exercise allosteric control by concerted feedback inhibition and that the ask gene would be isolated in the genome, remote from genes encoding function in a particular branch. Most genomes represented in Fig. 10 that possess only a single Ask enzyme fall into the group of enzymes subject to concerted feedback inhibition, e.g., Aquifex, Leptospira, and most upper Gammaproteobacteria. An exception, such as the solitary T. thermophilus Ask, which is feedback inhibited by threonine alone (64, 92), makes sense because it uses the AAA pathway for lysine biosynthesis and therefore does not need to deploy lysine as an Ask effector. On the other hand, organisms that possess multiple Ask enzymes are expected to deploy them for differential specialization with respect to some portion of the overall network. These specializations are reflected by different shared gene contexts and by different allosteric specificities. A perfect example is the genus Thermotoga, which has generated two differentially controlled Ask enzymes. The two Ask members of CG-26 are encoded by genes that are within a threonine operon, and the ask gene is fused at the N terminus with the gene encoding homoserine dehydrogenase. Figure 10 shows a profile for CG-26 that strongly indicates threonine allostery, but not lysine allostery. The second ask paralog of Thermotoga spp. (in CG-29) is part of an extensive lysine pathway operon. In addition, the latter gene context in Fig. 10 indicates lysine allostery, but not threonine allostery.
Experimental studies of some members of CG-33 allow the bioinformatic projection that the enzymes of this group are feedback inhibited by lysine, and this is reinforced by examination of Fig. 10. Members of CG-28 and CG-30, which were initially deduced to have lysine pathway specialization based on the criterion of gene context, fall into the lysine allostery group in Fig. 10. Members of CG-24 are known to be inhibited specifically by DAP, and it is not surprising, in view of the structural similarity between DAP and lysine, that CG-24 is in the lysine group in Fig. 10. On a case-by-case basis, most of the proposed specificities of Fig. 10 can be explained in a rational way. For example, Maricaulis maris (and the closely related Oceanicaulis alexandrii) is unusual among the Alphaproteobacteria in having two ASKα genes that originated in the genome via LGT (Fig. 11). One, in CG-52, is fused with hdh and is clearly threonine pathway specialized; the other, in CG-09, is fused with lysA and is clearly lysine pathway specialized. The third ask gene in the genome encodes an ancestral ASKβ enzyme, which in the vast majority of Alphaproteobacteria (CG-37) is the sole Ask and one that is subject to concerted feedback inhibition. However, the M. maris enzyme (consistent with divergence to membership in CG-51) exhibits no indication of an ability to bind either threonine or lysine (Fig. 10). Very likely it has a new functional role to support methionine biosynthesis, in coordination with the emergence of a novel Hdh-min species that is not otherwise present in the Alphaproteobacteria. (Alphaproteobacteria typically possess a single threonine-inhibited Hdh-thr.) It is clear from pairwise identity comparisons that the monofunctional hdh-min originated as a gene duplicate of the hdh domain of the CG-52 xenolog fusion (ask-hdh). Thus, acquisition by LGT of the two different ask fusions specialized for threonine (ask-hdh-min) and lysine (ask-lysA) biosynthesis was followed by selection for key methionine specialist enzymes consisting of an Ask enzyme (obtained by allosteric desensitization of the ancestral Ask) and an allosterically insensitive Hdh-min (obtained via duplication of the C-terminal domain of the ask-hdh-min zenolog). This new hdh was inserted into a methionine operon.
FIG. 11.
Events of LGT within the vertical genealogy of the phylum Proteobacteria. Taxa at the level of class are shown down the middle. Gammaproteobacteria are divided with a dashed line into upper Gammaproteobacteria and lower Gammaproteobacteria (Fig. 3), and most of the orders within the Gammaproteobacteria are listed. In the vertical genealogy on the right, two ancestral ask genes are proposed. ask_ect genes encode Ask enzymes belonging to CG-02 (ASKα), and these have a broad but erratic distribution, as indicated in the far right column. Thus, 4 of 5 Vibrionales genomes have Ask_ect, whereas 3 of 16 Deltaproteobacteria genomes and 0 of 6 Epsilonproteobacteria genomes have Ask_ect. The remaining (and most abundant) Ask enzymes in the vertical genealogy are recognized as members of cd04246 and belong to ASKβ (Fig. 7). The Ask enzymes in this column from the Gammaproteobacteria (shown with green shading) are distinctive in being encoded by an ask gene that has a highly conserved context of surrounding genes (shown at the bottom). LGT acquisitions are shown on the left (see the text). The proposed donor lineages are indicated within two shaded ovals. Many of the LGT events involve Ask enzymes having C-terminal fusions that are indicated by yellow or blue circles as defined at the top middle. At the bottom left, CG-04 members originated via LGT from a donor in the Bacteroidetes lineage. Members of CG-04 and CG-08 comprise two sets of paralogs within the lower Gammaproteobacteria as the result of a gene duplication (GD) of an early CG-04 gene ancestor, as diagrammed. Thus, CG-08 members abandoned their threonine pathway specialization evolved in the vertical genealogy via steps in which threonine pathway specialization was abandoned and functional specialization for methionine biosynthesis was acquired. Note that the Enterobacterales and the Xanthomonadales have experienced complete displacement of phylum-ancestral ask genes by LGT.
Emergence of an internal start site within the Ask domain.
Just upstream of the ACT domain regulatory region in ASKβ Ask enzymes is positioned an in-frame internal translational start site (diagrammed in Fig. 6), which results in overlapping genes. This produces a small subunit that is identical to the C-terminal portion of the large subunit and thus magnifies the regulatory potential by doubling the number of ACT units. This was first demonstrated by the seminal work of Chen and colleagues for a lysine-inhibited Ask paralog of B. subtilis (11), which belongs to CG-33. Similar overlapping genes have been asserted for C. glutamicum (CG-25) (27). Figure 12, left, shows an amino acid alignment of the internal start site region for all of the orphans and one representative from each cohesion group, which comprise the set of ASKβ enzymes. Start codons (ATG, GTG, or TTG) are shown in what appears to be slightly different positions in the overall alignment. Four orphans and three cohesion groups appear to lack an internal start site (shown on the left). On the right is an enlarged portrayal of the alignment of all members of CG-38 present in a group of pathogenic Alphaproteobacteria. This group of organisms lacks the threonine and methionine branches of the ASK network. Amar_Aa and Ecan_Aa (near the top) appear to have two closely spaced alternative start sites. Interestingly, these match up with the two positions of internal start sites in the overall alignment on the left.
FIG. 12.
In-frame internal start sites within the ASKβ subhomology division. An appropriate section of the multiple alignment of representative sequences of cohesion groups and orphans within ASKβ members is shown on the left, with putative internal-start-site residues indicated in blue or green. Cohesion groups or orphans lacking an internal start site are indicated in red at the far left. On the right is an expansion of the entire cohesion group CG-38. The two sequences at the bottom contain insertions (highlighted in yellow) that disrupt the spacing of otherwise recognizable ribosome binding sites that are indicated in boldface. The second and third sequences from the top have two alternative in-frame internal start sites, which are shown in green or blue. Uppercase letters are amino acid residues, and lowercase letters are DNA nucleotides (e.g., atg encoding M at the bottom right in blue).
Evolved abolition of overlapping genes.
Since 42 of 49 Ask cohesion groups and orphans appear to have overlapping genes (Fig. 12), it seems likely that the evolved acquisition of an internal start site occurred coincident with or soon after the divergence of ASKβ Ask enzymes. If so, the four orphans and three cohesion groups shown in Fig. 12 must have subsequently lost the internal start site. At least in the case of the three cohesion groups, this must have been a fairly ancient event. Thus, all members of the phylum Cyanobacteria contribute Ask species to CG-32 that uniformly lack internal start sites. Likewise, all DAP-sensitive Ask species belonging to CG-24, all from Firmicutes, uniformly lack internal start sites.
The membership of all cohesion groups is uniform with respect to the presence or absence of an internal start site. The only exception is a very recent loss of the internal start site in the common ancestor of Rickettsia prowazekii and Rickettsia typhi, as illustrated in the right portion of Fig. 12. CG-38 contains 11 sequences from pathogenic Alphaproteobacteria. As shown by the enlargement on the right, nine of these, including the representative sequence Wend_a, possess overlapping genes. The two bottom sequences illustrate a very recent loss of the internal start site as a result of an insertion between the ancestral start codon and the still-recognizable ribosome binding site.
Overview of Key Differences between ASKα and ASKβ Ask Enzymes
Figure 6 provides a schematic graphic to summarize the key differences between enzymes that belong to the ASKα or to the ASKβ subhomology division. Both groups have an Ask domain followed by two ACT domains: ACT_1 and ACT_2. Occasionally the twin ACT region has been lost or highly degraded. In two unusual ASKβ cases, four ACT domains are present. With two exceptions, ASKα members always have a region with no counterpart in ASKβ (an indel region). It was asserted earlier that the indel region represents a deletion in the more recent ASKβ division. One wonders if the indel region in ASKα enzymes might be important for protein-protein contacts between Ask and Hdh, regardless of whether these enzymes are fused or complexed with one another. The indel region might also interact with the twin ACT domain. The last two possibilities are not necessarily mutually exclusive. It is suggestive that seemingly independent deletions of the indel region, which occurred in the common ancestors of CG-23 and CG-52 in the ASKα subhomology division (Fig. 5), correlate with either the complete loss of the twin ACT region in CG-52 or the extensive disruption of the Q…Q motif of the twin ACT region in CG-23 (Fig. 8A).
Catalytic and/or substrate-binding residues that are nearly invariant throughout both subhomology divisions are indicated in Fig. 6. Additional catalytic or substrate-binding residues that are not as highly conserved exist in the C-terminal portion of the Ask domain and are not shown in Fig. 6. Six cds exist at the highest hierarchical level in the CDD for the total population of Ask domains (Fig. 7), but one of them, cd04246, uniquely describes ASKβ enzymes. Therefore, any cd that is not cd04246 belongs to the ASKα subhomology division. Similarly, cd04891 uniquely identifies ACT_1 domains of ASKβ enzymes. Most ASKβ enzymes support an internal translation start site that generates a small subunit, essentially doubling the repertoire of regulatory ACT domains. The internal translation start site has been lost in three cohesion groups and in four orphans (Fig. 12). The Mjan_Aa enzyme is shown in Fig. 6 to possess a Q…Q motif. These glutamine residues are located near the midpoints of ACT_1 and ACT_2, and they are nonequivalent threonine-binding residues. The ACT_1 Q residue is positioned in a high-affinity binding region and is essential for threonine inhibition of Ask. The ACT_2 Q residue is positioned in a low-affinity binding region in which the binding is greatly enhanced by occupation of the first binding region. Occupation of both binding regions is essential for inhibition of Hdh in cases where Ask and Hdh are fused. The likelihood that monofunctional Ask and Hdh enzymes are complexed in organisms such as M. jannaschii and that the Q…Q motif has the same significance for threonine regulation of both enzymes was discussed extensively earlier. The Q residue in ACT_1 is essential for threonine allostery in ASKα enzymes (Fig. 8A) and is also important for threonine allostery in ASKβ enzymes (Fig. 10). The Q residue is always absent in the ACT_2 domain of ASKβ enzymes, where a very small deletion appears to have occurred.
INFERENCE OF FUNCTIONAL SPECIALIZATION FROM GENE CONTEXT
When No Suggestive Gene Context Exists
Solitary ask genes.
In most cases where a single ask gene governs precursor availability to whatever ASK network exists in a given organism, ask is not adjacent to genes that are relevant to any of the divergent ASK branches. This does not necessarily mean that the lone ask is constitutive and insensitive to end product regulation. Indeed, elaborate regulatory specificities are possible, but whatever regulation exists senses the overall gestalt of end products, e.g., mechanisms such as concerted feedback inhibition and multivalent (cumulative) repression. Single Ask enzymes seemingly are more feasible in organisms in which the balance of end products produced by the network is relatively constant, regardless of physiological or environmental conditions. Exceptions do exist where solitary ask genes are adjacent to genes of a given branch, but these are cases where the ASK network is relatively uncomplicated. For example, all species of the phylum Chlamydiae (CG-22) possess a single ask gene that is adjacent to some enzymes of DAP biosynthesis, but these pathogens use ask exclusively for DAP biosynthesis, having lost the capability to transform DAP into lysine and having lost all other branches via reductive evolution. In another case where a relatively simple network exists, T. kodakaraensis in CG-18 does not use the DAP pathway to lysine or the DKFP pathway to aromatic amino acids, and Ask supports only the methionine and threonine branches of biosynthesis. Here, the solitary ask gene is clustered with and presumably coregulated with most of the individual genes for methionine and threonine biosynthesis.
Distribution of solitary ask genes in the ASKα and ASKβ subhomology divisions.
Ask proteins of Archaea always belong to the ASKα subhomology division, and they are nearly always encoded by solitary genes in any given genome. Bacterial genomes that possess ASKα enzymes generally possess multiple homologs with different specializations. In contrast, those Bacteria that possess ASKβ enzymes are most frequently represented by genomes that have solitary ask genes.
Contexts of ask Genes That Imply Functional Specialization
Whenever the total Ask activity is partitioned among two or more homologs in a given organism, one or more of the corresponding genes are often associated with genes encoding enzymes that function in one of the network branches. Observations of gene context are well known to assist the assignment of functional roles (70), and we have found that the gene context of ask genes sometimes provides important clues that imply a specialized role. Examples exist in which a given Ask is largely specialized for one of the following: aromatic biosynthesis (Ask_aro), ectoine biosynthesis (Ask_ect), DAP biosynthesis (Ask_dap), both DAP and dipicolinate biosynthesis (Ask_dap+dpl), lysine biosynthesis (Ask_lys), threonine biosynthesis (Ask_thr), methionine biosynthesis (Ask_met), and both threonine and methionine biosynthesis (Ask_thr+met). Even if an ask gene is adjacent to only one of the various genes encoding the enzymes of a network branch, we consider it to have a gene context implying specialization for that branch. If an ask gene is adjacent to an hdh gene (and it is the sole hdh gene in the genome), the context is considered to imply Ask_thr+met specialization.
The Firmicutes typically possess multiple ask homologs with a number of specializations. The B. subtilis member of CG-03 is known to be subject to concerted feedback inhibition by Thr plus Lys, but the individual alignment of the twin ACT region seems sufficiently variable (see the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG03_aln.html) that other allosteric patterns may exist for enzymes in CG-03. This is consistent with the observation that CG-03 is a very large cohesion group that contains ask genes with a great variety of gene context arrangements, i.e., with lysine gene contexts, with threonine gene contexts, or with no ASK network gene contexts. Curiously, it is quite common to find partial or complete lysine pathway operons or partial or complete threonine pathway operons adjacent to a divergently oriented ask gene. Indeed, even adjacent genes for threonine biosynthesis or lysine biosynthesis may have opposite orientations. Such gene scrambling is often an indication of ongoing reductive evolution. These gene neighborhoods for CG-03 ask genes can be viewed in the cohesion group gene neighborhood column (the second column) of the dynamic table (http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html). Other ask genes in Firmicutes belong to the ASKβ division, and many of these have specialized gene contexts as summarized in Table 2. Since the ASK network of the Firmicutes is discussed comprehensively below, the various ask gene contexts of the Firmicutes either are not enumerated directly here or are referred to only briefly.
TABLE 2.
Distribution of Ask cohesion groups in the phylum Firmicutes
| Cohesion group | Functional specialization | No. of members | Subhomology division | Internal start site present | Allosterya | Gene contextb | Phylogenetic breadth of distribution |
|---|---|---|---|---|---|---|---|
| CG-03 | Threonine | 37 | ASKα | No | Lysine-threonine synergism | thr+met; thr; lys or null | Phylum: Firmicutes |
| CG-24 | DAP | 27 | ASKβ | No | Diaminopimelate | dap | Phylum: Firmicutes |
| CG-28 | Lysine | 2 | ASKβ | Yes | Lysine (Fig. 10) | lys | Genus: Clostridium |
| CG-30 | Lysine | 4 | ASKβ | Yes | Lysine (Fig. 10) | lys | Genus: Staphylococcus |
| CG-33 | Lysine | 9 | ASKβ | Yes | Lysine | Family: Bacillaceae; Listeriaceae | |
| CG-45 | Threonine | 3 | ASKβ | Yes | Threonine (Fig. 10) | thr+met | Order: Clostridiales |
| Orphan | Threonine | 1 | ASKβ | Yes | Threonine (Fig. 10) | thr+met | Syntrophomonas wolfei |
| Orphan | Threonine | 1 | ASKβ | Yes | Threonine (Fig. 10) | met; SAM riboswitch | Symbiobacterium thermophilum |
| Orphan | Threonine | 1 | ASKβ | Yes | Threonine (Fig. 10) | thr+met | Carboxydothermus hydrogenoformans |
| Orphan | Threonine | 1 | ASKβ | Yes | Threonine (Fig. 10) | thr+met | Moorella thermoacetica |
The indicated pattern of allostery is known to typify some members of CG-03, but is not necessarily present in all members.
The indicated context is not always present with respect to every cohesion group member.
ask_aro gene contexts.
Organisms that utilize the DKFP pathway rather than the E4P pathway of aromatic biosynthesis are commonly found in Archaea but are rarely found in Bacteria. Examples of DKFP occurrence among the Bacteria are Aquifex aeolicus and some Deltaproteobacteria (Desulfovibrio vulgaris, Desulfovibrio desulfuricans, and Desulfococcus oleovorans). The latter are quite remarkable in that about a dozen genes representing most of the common trunk of aromatic biosynthesis, as well as all three terminal amino acid branches, cocluster with aroA′ and aroB′, the initial two genes that are unique to the DKFP pathway. Among Archaea, the DKFP pathway is mostly known to be present in the Euryarchaeota, although some members of the Crenarchaeota, such as N. maritimus and C. symbiosum, also have the DKFP pathway (Fig. 8B). The single genome of Korarchaota that is currently available possesses the DKFP pathway, as well. The E4P pathway, rather than the DKFP pathway, is so far known to be present in some of the Crenarchaeota (e.g., A. pernix, Metallosphaera sedula, Solfolobus spp., Caldiverga maquilingensis, and Pyrobaculum aerophilum) and some of the Euryarchaeota (e.g., Pyrococcus spp., Thermococcus spp., Picrophilus spp., and Thermoplasma spp.). Those Euryarchaeota that rely exclusively upon the DKFP variation of aromatic biosynthesis generally possess a single Ask species. Only Methanopyrus kandleri possesses two Ask enzymes (one in CG-19 and the other an orphan). The M. kandleri Ask in CG-19 appears to be a xenolog intruder obtained from a distant archaeon relative (see “LGT” below). On the other hand, those Crenarchaeota (N. maritimus and C. symbiosum) that rely exclusively upon the DKFP pathway so far possess two Ask enzymes. The single Korarchaota genome available also possesses two Ask enzymes. Organisms that rely upon the DKFP pathway variation possess Ask enzymes that either are orphans or belong to cohesion groups CG-14, CG-15, CG-16, CG-23, and CG-50. Of the few Bacteria that rely exclusively upon the DKFP pathway, the A. aeolicus genome has a single ask orphan gene, and five members of the Deltaproteobacteria (representing three orders) each have Ask enzymes that belong to CG-34. Of these, D. oleovorans is exceptional in having a second ask gene (CG-02) that is associated with an ectoine operon.
So far, no organism that relies exclusively upon the DKFP pathway for aromatic biosynthesis possesses an ask gene that is associated with aromatic-pathway genes. However, some bacteria in the order Actinomycetales possess both the E4P and DKFP pathways, namely, Frankia sp., Streptomyces griseus, Streptomyces pristinaespiralis, and Streptomyces scabiei. In two of these organisms, aroA′ and aroB′ are associated with an ask paralog (which is thus denoted ask_aro) in Frankia sp. and in S. griseus. The two Ask_aro enzymes (FRAN-2a and Sgri_Ab) do not belong to the same cohesion group and are orphan sequences. However, these two orphan sequences are most closely related to one another on our master phylogenetic tree at a node having bootstrap support of 51%. Curiously, Frankia sp. has two copies of adjacent aroA′ and aroB′ genes, only one pair of which is associated with ask_aro. F. alni lacks aroA′ and aroB′ altogether.
In summary, this is a previously unappreciated Ask specialization, and there are so far only two examples of an Ask that has an implied specialization for aromatic amino acid biosynthesis. In the cases of ask_aro in Frankia sp. and S. griseus, we suspect that the DKFP pathway may be specifically associated with some form of secondary metabolism, such as antibiotic synthesis, in which aromatic amino acids are often incorporated as components. If so, that would broaden the range of the ASK network output beyond what is portrayed in Fig. 1.
ask_ect gene contexts.
Only the Proteobacteria so far exhibit an ectoine operon that is sometimes linked to a putative regulatory gene (ectR) and/or linked to a paralog of Ask (ask_ect), as summarized in Fig. 2. Each class of the Proteobacteria has at least a limited occurrence of ect genes. Neither ask_ect nor ectR has been seen in the class Epsilonproteobacteria, in which only Wolinella succinogenes has an ect operon. In the Betaproteobacteria, genes of an ect operon seem to be confined to the order Burkholderiales (and there to the families Oxalobacteraceae and Alcaligenaceae). Here, a single operon is present in the order ectR → ectABCD. Since the ectR regulatory gene is within the operon, it presumably is autoregulated. This gene arrangement is unique to the Betaproteobacteria. All Betaproteobacteria possess a single Ask gene, regardless of whether the ectoine pathway is present or not. Hence, it is not surprising that this Ask gene is not associated with genes of the ectoine branch (or any other branch) of the network. Examples of specialized ectoine-associated Ask enzymes can be found in the remaining three classes of Proteobacteria as outlined below.
(i) Deltaproteobacteria.
Three members of the Deltaproteobacteria possess ask_ect. In one case (D. oleovorans), ask_ect exhibits the expected linkage to an ectABC operon. Curiously, the other two organisms (Anaeromyxobacter dehalogenans and Desulfotalea psychrophila) each have a stand-alone ask_ect that is not linked to ect genes, and in fact, ect genes are missing from these genomes entirely. The Ask_ect Ask of D. psychrophila is extraordinary in that it is the sole Ask supporting the divergent pathways leading from aspartate semialdehyde, and yet, the organism lacks the ectoine pathway for which this paralog is generally specialized elsewhere. This exemplifies a case in which a specialized paralog has abandoned its specialized role, instead replacing the generalized function of the ancestral ask species that is typical of Proteobacteria.
(ii) Alphaproteobacteria.
A particularly large number of complete genomes are available for the Alphaproteobacteria, and the instability of the ectoine operon is quite apparent. No Alphaproteobacteria have ect genes, except the relatively few that possess an ect_ask operon. Those Alphaproteobacteria that have ect genes generally possess an ectABCD → ask_ect operon positioned next to a divergently oriented ectR regulatory gene (COG1846), and this most likely is the ancestral gene organization for this class. In comparison with the presumed ancestral operon, a number of genomes exhibit operon degradation. Three genomes have lost ectD, two genomes have transposed ectD out of the operon, one genome has lost ask_ect, and one genome has lost both ectD and ask_ect. The phylogenetic distribution of the Ask-associated ectoine operons within the class Alphaproteobacteria is curious. Only one (Acidiphilium cryptum) of five representatives of the order Rhodospiralles possesses an ect_ask operon, only two (Hyphomonas neptunium and Silicibacter sp.) of nine representatives of the order Rhodobacterales possess an ect_ask operon, and only one (Sphingopyxis alaskensis) of five representatives of the Sphingomonadales possesses an ect_ask operon. It seems that either an ect_ask operon was present in the common ancestor of Alphaproteobacteria and has been retained only in scattered clades (most likely) or that the ect_ask operon has been acquired on at least three independent occasions (less likely).
(iii) Gammaproteobacteria.
Both upper Gammaproteobacteria (five orders) and lower Gammaproteobacteria (only the order Vibrionales) are known to possess ectoine pathway genes. Those Gammaproteobacteria that possess an ect operon usually exhibit an ectABC → ask_ect operon, as indicated in Fig. 2. However, it seems likely that the lineage initially possessed ectD and the divergently oriented ectR, since these genes remain in various organisms. For example, Pseudomonas stutzeri and Marinomonas sp. have the ectABCD → ask_ect gene configuration, whereas S. degradans has the ← ectR → ectABC → ask_ect gene configuration. Thus, assuming ← ectR → ectABCD → ask_ect to be the ancestral gene organization of Gammaproteobacteria, P. stutzeri has lost (or translocated) ectR and S. degradans has lost ectD. It is interesting that the genes that flank the five-gene Ask-associated ect operon in P. stutzeri are adjacent to one another in the closely related P. aeruginosa. These genes encode a protease (COG0826 collagenase and related proteases) and glucose-6-P 1-dehydrogenase. This likely means that the Ask-associated ect operon was present in the common ancestor of P. aeruginosa strains prior to a deletion event. This is consistent with the overall instability and erratic distribution of ectoine operons in different lineages. The alternative, that the operon was obtained recently via LGT at a time after the divergence of P. aeruginosa and P. stutzeri and inserted between the two genes in P. stutzeri, is possible. However, parametric data in support of this were not obtained, and the operonic gene products of P. stutzeri were not strikingly similar to any known gene products in the current databases that might have implicated an LGT donor.
As with the Alphaproteobacteria, the ect-associated genes of Gammaproteobacteria seem quite unstable. Thus, one genome has ectR divergently adjacent to ectABC, but ectD has been transposed elsewhere. One genome has ectR divergently adjacent to ectC → ask_ect, and ectAB has been transposed elsewhere. One genome has ectAB convergently oriented to ectC, and ectD, asp_ect, and ectR are absent. One genome has only ectABCD, and another has ectABC with ectD transposed elsewhere. The Vibrionales exemplify the genetic instability of the ectoine system. Members of this order usually possess five Ask genes, three of them being the classic genes studied in E. coli (and originating via LGT), one being a gene persisting in the vertical genealogy, and the fifth being the ectoine-specialized homolog. However, Vibrio vulnificus (in contrast to Vibrio fischeri, Vibrio cholerae, and Vibrio parahaemolyticus) has lost both the ect operon and ask_ect. Photobacterium profundum exemplifies an even more recent destabilization, as illustrated in Fig. 1S in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/figure-1S.html. This figure is a snapshot of an annotation overview in the SEED database. Strain 3TCK (shown on the second line of the SEED viewer) possesses the ectABC → ask_ect operon (genes 4 → 3 → 2 → 1) that is typical of most other Vibrionales. However, strain SS9 (top line) has experienced a transposase insertion that has fragmented ask_ect and that is associated with the complete loss of ectABC. On the N-terminal-flanking side, a four-gene region encoding glutathione synthase (gene 16) and two conserved hypothetical proteins (genes 14 and 15), along with a divergently oriented lysR transcriptional regulator (gene 13), have additionally been inverted as a unit in strain SS9.
ask_dap gene contexts. (i) Within the ASKα subhomology division.
Members of the phylum Chlamydiae possess an extremely simple ASK network that contains a single branch and therefore is, in fact, a linear biochemical pathway beginning with aspartate and ending with DAP. DAP is not converted to lysine, and therefore, DAP is significant per se as an end product. This was a surprising observation for many years because these intracellular parasites were not thought to form peptidoglycan. However, a growing body of evidence indicates that peptidoglycan and some version of a cell wall are critical for at least some stage of the cell cycle (56, 73). The single Ask enzyme of each member of the entire phylum belongs to the ASKα division and populates CG-22. In each case, ask resides within a four-gene operon that encodes the first four steps of the pathway. Genes encoding the l,l-DAP aminotransferase (a pathway variation discovered recently [38, 39]) and DAP epimerase (the final enzymatic step) are dispersed elsewhere in the genome.
The S. ruber paralog Srub_Aa in CG-47 is also suggested to be specialized for DAP biosynthesis (Fig. 8B), although the rationale is weakly based only on a process of elimination. The encoding gene is adjacent to one gene of DAP/lysine biosynthesis. Specialization for DAP biosynthesis (rather than for lysine) is inferred indirectly, since the other possible functional roles of specialization are already covered by other ask paralogs. These three additional Ask enzymes have features of allostery and gene context that strongly imply specialization for lysine, threonine, and methionine (Fig. 13).
FIG. 13.
Vertical genealogy of ask paralogs in Bacteroidetes and proposed role as an LGT donor to Proteobacteria. Gene contexts that imply functional specializations are color coded as shown at the upper right. Gene fusions resulting in fusions of Ask with homoserine dehydrogenase (Hdh) or with DAP decarboxylase (LysA) are indicated. Evolutionary events that are deduced are indicated by circled letters that correspond to the descriptions at the bottom. LGT relationships are depicted by green arrows on the right.
(ii) Within the ASKβ subhomology division.
CG-24 contains a large group of ASKβ Ask enzymes from the phylum Firmicutes. These generally are encoded by ask genes positioned near DAP/lysine/peptidoglycan pathway genes. The CG-24 Ask enzymes are quite distinctive and highly conserved. It is likely that most members are sensitive to feedback inhibition by DAP.
Contexts with nested dpaA and dpaB genes.
A subset of the foregoing CG-24 Ask enzymes belongs to those Firmicutes that are able to make endospores. These are Bacillus and Clostridia species that additionally possess two genes encoding the subunits of dipicolinate synthase (Fig. 1). They are the first two genes of a complex operon from which multiple transcripts are made, depending upon the physiological state (12). (Click the gene neighborhood icon for CG-24 in the dynamic table [http://www.theseed.org/Papers/MMBR-Aspartokinase/figure-1S.html, second column] to view the gene subset nested within the larger gene set.)
ask_lys gene contexts. (i) Within the ASKα subhomology division.
It is straightforward that many Ask enzymes in the ASKα division whose twin ACT regions exhibit the “lysine configuration” that predicts lysine allostery (Fig. 8A and B) would also be organized with lysine pathway genes, e.g., members of CG-09, CG-11, and CG-12. The fusion of LysA with Ask in CG-09 members is a special case of lysine pathway context. These lysine-specialized Ask enzymes are all present in organisms in which at least one other Ask homolog is present to accommodate the biosynthesis of threonine and methionine. The Ask enzymes of members of CG-21, such as E. coli, are clearly lysine specialized because of their lysine allostery, but a lysine pathway gene context is absent. Interestingly, the one exception is Ftul_Aa from Francisella tularensis, which is encoded by a xenolog intruder member of CG-21 obtained by F. tularensis via LGT and inserted into a lysine pathway operon. Lactobacillus spp. have generated unusual paralog copies in CG-03 that have been transposed to a lysine pathway context.
The inference of end product specialization from the informative gene contexts present in some members of a cohesion group can reasonably be extrapolated to other members in which that context is not present. For example, CG-11 contains sequences from five genomes. The ask genes from two different species of Bacteroides and from P. gingivalis are associated with lysA. It is further evident the P. gingivalis function must be oriented to lysine biosynthesis because the organism lacks homoserine dehydrogenase and therefore is incompetent for threonine and lysine biosynthesis. Therefore, CG-11 is labeled with a gene context of ask_lys in Fig. 8B, even though some members of the cohesion group exhibit no lysine pathway context. Note that an inference of specialization derived from a particular context does not necessarily exclude additional functional complexities. For example, an ask gene surrounded by a threonine gene context might still be subject to feedback inhibition by lysine, in which case its specialization is broadened to both end products.
The members of CG-23 are currently from either the Methanobacteria or Halobacteria taxons (Fig. 8B). ask genes present in the Methanobacteria, but not the Halobacteria, are encoded by genes positioned with a lysine pathway context. This is unusual, since such solitary ask genes usually exhibit no gene context with genes of individual branches. The Methanobacteria do not have an AAA pathway of lysine biosynthesis, whereas the Halobacteria appear to have both the AAA and DAP pathways. The CG-23 members are ASKα enzymes that are derived from the “threonine configuration” but that have a disrupted twin ACT domain. Thus, the allosteric specificity is uncertain. CG-23 is further distinctive in that it is one of only two cohesion groups in the ASKα homology division to lack the indel region (Fig. 5). (The possibility that the indel region might interact with the twin ACT region is discussed below.)
(ii) Within the ASKβ subhomology division.
The relatively few Bacteria having multiple ask homologs belonging to the ASKβ division can be expected to exhibit functional specialization, and one clue implying a particular specialization can be gene context. The Rxyl_Ab ask from Rubrobacter xylanophilus has a lysine pathway gene context (but lysine allostery is not indicated in Fig. 10). Although R. xylanophilus does have another ask homolog (encoding Rxyl_Aa), it belongs to the ASKα homology subdivision and exhibits a threonine gene context (with no threonine allostery indicated in Fig. 8A). Two species of Thermotoga have two ask genes, one of which resides within an extensive lysine pathway gene context. The Firmicutes possess two cohesion groups (CG-28 and CG-30) whose gene members exist within a lysine gene context, as summarized in Table 2.
ask_thr gene contexts. (i) Within the ASKα subhomology division.
The relatively few Bacteria that possess ASKα Ask enzymes usually possess more than one ask gene, and this multiplicity is generally associated with specialization. A context of threonine pathway genes is quite widespread for ask genes in these Bacteria (Fig. 8B, top). In one interesting case, Acidobacteria sp. (synonymous with K. versatilis Ellin345) has generated a recent gene duplicate of a lysine-specialized gene present in CG-12 and has transformed it to threonine specialization. One Ask (ACID-2a) is presumed to have retained the original ancestral features and has the lysine configuration associated with lysine allostery in the twin ACT region, as well as exhibiting a lysine pathway gene context (Fig. 8A and B). The altered paralog (ACID-2b) has a somewhat disrupted signature for lysine allostery in the twin ACT region and likely has a reduced sensitivity to inhibition by lysine. More dramatically, the gene has been translocated to a gene neighborhood that includes the threonine pathway genes. This new context includes hdh-thr. Although the context includes a gene for homoserine dehydrogenase, it is considered to be an ask_thr context because a second hdh gene (hdh-min) is present in context with a methionine metabolism gene.
Members of CG-20 are from genomes having solitary ask genes, and they are clustered with thrC. This is unusual because the solitary ask genes that are typically found in Archaea are rarely associated with genes encoding enzymes of particular network branches. In this group of Archaea, Ask supports only the threonine and methionine branches. Perhaps the coregulation of ask and thrC is balanced somehow by the association and presumed coregulation of thrB with metB in another chromosomal location.
The two members of CG-52 are present in two Alphaproteobacteria genomes that have genes encoding Ask-homoserine dehydrogenase fusions comprising part of a threonine operon that has been obtained intact from a Bacteroidetes member of CG-07 (see “LGT” below). Although the fused hdh gene is, of course, part of the operon, this is deemed to be an ask_thr gene context because a second hdh (hdh-min) is present in the genome and located in context with methionine branch genes. Regulation of gene expression, rather than allostery, is probably critical because CG-52 members lack the twin ACT region altogether (Fig. 8A).
(ii) Within the ASKβ subhomology division.
Most Bacteria have solitary ask genes belonging to the ASKβ division, and these usually do not exist in a physical context with genes encoding enzymes of particular network branches. The ask genes of the two Chloroflexi species in CG-44 and the ask orphan of Rhodopirellula baltica have in common the fact that they are adjacent to the novel “predicted functional analog of threonine kinase” described earlier that appears to have replaced thrB.
ask_met gene contexts. (i) Within the ASKα subhomology division.
In the ASKα subdivision, methionine-specialized Ask enzymes are all found in the middle of Fig. 8A and B. These Ask enzymes are derived from the threonine-specialized cd04892-cd04892 signature of the twin ACT domain region, but the Q…Q motif has become completely disrupted (Fig. 8A). In a number of cases, a threonine-specialized ask (Fig. 8A, top) has undergone gene duplication, with one copy becoming disrupted in the twin ACT region for threonine allostery in order to allow a new specialization. These gene duplications are indicated in Fig. 8B. Some of them are copies that were translocated to a methionine pathway gene context. Such duplication-origin pairs of threonine/methionine specializations for ask genes include CG-04-CG-08, CG-07 (Srub_Ac)-CG-47 (Srub_Ab), and Mxan_Ac orphan-Mxan_Ad orphan.
(ii) Within the ASKβ subhomology division.
It is rare for ASKβ Ask enzymes to be clustered with one or more genes encoding enzymes of the methionine branch unless genes encoding enzymes of the threonine branch are also included (see below). One exception is Symbiobacterium thermophilum Sthe-17_Ab, an orphan that is encoded by an ask gene clustered with a SAM riboswitch and one gene of methionine biosynthesis (Table 2). However, since the other ask gene of the genome belongs to CG-24, which is highly dedicated to DAP and lysine biosynthesis, Sthe-17_Ab must by default have a functional specialization for threonine biosynthesis, as well. Indeed, as indicated in Fig. 10, Sthe-17 is likely to be feedback inhibited by threonine. Furthermore, the Sthe-17 orphan can be inferred to have threonine pathway functionality because it belongs to a group of novel ASKβ enzymes (in CG-45 and three other orphans) that have replaced CG-03 threonine specialists (ACTα enzymes). This is discussed more fully below. All of the other ASKβ replacements are ask genes within a threonine pathway gene context.
ask_thr+met gene contexts. (i) Within the ASKα subhomology division.
T. kodakaraensis, an archaeon, possesses a single Ask enzyme belonging to CG-18. Although lone ask genes are rarely associated with genes encoding branch segments of the ASK network, the ask gene of T. kodakaraensis is associated with genes of both the threonine and methionine branches, including homoserine dehydrogenase. This can be understood given the extreme simplicity of the ASK network in the CG-18 organisms, where nonnetwork pathways are used for lysine, isoleucine, and aromatic amino acid biosynthesis. The synthesis of Ask and the enzymes of both the threonine and methionine pathways thus appear to be coregulated. The twin ACT region is disrupted and largely truncated, indicating that Ask may not be under allosteric control. Three species of Pyrococcus also have ask genes that belong to CG-18. Although each has two recent paralogs, they are adjacent and associated with genes of both methionine and threonine biosynthesis (albeit with some scrambling).
R. xylanophilus (phylum Actinobacteria) has two ask homologs, one (Rxyl_Aa) encoded by an ASKα gene with a methionine gene context. Since one of the clustered genes encodes homoserine dehydrogenase (Hdh-min) and this is the only hdh gene in the genome, this ask gene can be considered to have a gene context for both the threonine and methionine branches. In terms of specialization, this makes sense, because the other ask gene, encoding Rxyl_Ab, is embedded within a lysine gene context.
The Firmicutes typically have multiple Ask enzymes. Of the two Ask species present in Staphylococcus spp., the one belonging to CG-03 (ASKα) exhibits a threonine pathway gene context. This includes a threonine operon that contains the gene encoding Hdh-met, the sole Hdh present in Staphylococcus. Presumably, this Hdh species is transcriptionally regulated by threonine but allosterically regulated by methionine. Lactobacillus exhibits great variation among species, as discussed further in “Firmicutes” below. Among these, Lactobacillus casei and Lactobacillus acidophilus possess two paralogs within CG-03. One of them is clustered with all of the threonine branch genes and hdh-met. Interestingly, these genes are not in the same order in the two species, and gene scrambling has resulted in some of the genes being divergently oriented with respect to ask. (Click the gene neighborhood icon in the second column of the dynamic table [http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html] within the CG-03 section to view these gene organizations.)
(ii) Within the ASKβ subhomology division.
The Firmicutes usually have multiple Ask genes in the ASKβ division, and they are distributed among five cohesion groups and four orphans. Members of CG-45 and three orphans exhibit a thr+met gene context, as summarized in Table 2. For example, Moorella thermoacetica (Clostridia) possesses an Ask orphan (Mthe-3_Aa) that is encoded by the last member of a large operon that contains hdh-thr, thrC, thrB, and two genes of the methionine pathway. hdh-thr is the sole hdh gene in the genome and hence must function for both methionine and threonine biosynthesis. Members of CG-45 and all four orphans appear to be subject to feedback inhibition by threonine (Fig. 10).
The genus Thermotoga is represented by two available species that each have a pair of ask genes. One of them is not only fused with hdh-min (a novel variation in that the latter is fused to the N-terminal domain of ask), but is localized with the remaining threonine pathway genes. Since no other hdh genes exist in the genome, this is considered to be an ask_thr+met gene context.
Less Intuitively Obvious Gene Contexts
ask_alaS gene contexts.
The phylum Gammaproteobacteria generally possesses a single Ask (denoted ask_alaS) that is associated with a string of closely linked genes, recA → recX → alaS → ask → csrA → tRNA-ser. The gene linkage alaS → ask → csrA is especially conserved. The various ask genes having an alaS context have diverged to produce 27 different cohesion groups and orphans. Hence, in spite of substantial divergence of the ancestral Ask enzyme, the gene neighborhood has been conserved to a remarkable extent. In the Gammaproteobacteria, ask_alaS is never associated with genes encoding enzymes of an individual branch of the ASK network. The specific significance of the ask-alaS relationship is unknown, but perhaps it reflects broad metabolic interrelationships, as reflected by one of the KEGG metabolic maps (http://www.genome.ad.jp/kegg/pathway/map/map00250.html).
ask_tilS gene contexts.
The Betaproteobacteria usually have a single Ask gene that follows a gene encoding tRNAIle-lysidine synthetase, an enzyme involved in tRNA processing (83). Here, the ask_tilS gene is usually followed by a gene encoding tRNASer, reminiscent of the last gene in a typical ask_alaS cluster described above. It is remarkable that Methylobacillus flagellatus possesses a single Ask (Mfla_Aa) that belongs to CG-40, as is typical of Betaproteobacteria, and yet its surrounding gene context is typical of ask_alaS for Gammaproteobacteria. Its gene cluster is the same as that shown at the bottom of Fig. 11, except for the absence of csrA. An LGT explanation for this does not seem to be supported.
ask_ptsP gene contexts.
King and O'Brian (44) have reported an intriguing direct interaction between Ask and enzyme INtr (encoded by ptsP) in Bradyrhizobium japonicum. The ability of INtr to transport oligopeptides requires the presence of Ask, which apparently is involved in regulating the phosphorylation state of enzyme INtr. B. japonicum is a member of the Alphaproteobacteria, and it is striking that the ask and ptsP genes, which are adjacent in B. japonicum, are also adjacent (or nearby) in most of the Alphaproteobacteria that make up CG-37 (click the gene neighborhood icon for CG-37 in the second column of the dynamic table [http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html]).
ASK NETWORKS SUPPORTED BY A SINGLE ask GENE
The substantial number of phyla (or classes) in which there is usually one ask gene and most or all of the Ask gene products fall within a single cohesion group that is united by conserved amino acid residues specific to the cohesion group, by a common cd signature, etc., are enumerated below.
Archaea
It is rather striking that almost all of the Archaea possess a single ask gene to support the ASK network. Two exceptions are the N. maritimus and C. symbiosum paralog pairs that belong to CG-49 and CG-50 (shown in the gene duplication column of Fig. 8B). Another exception is M. kandleri (Euryarchaeota), which in addition to its ancestral enzyme (Mkan_Ab) possesses a xenolog obtained from the A. pernix (Crenarchaeota) lineage. Ask enzymes from Archaea populate eight cohesion groups, and so far, only Mkan_Ab is an orphan sequence. All of the sequences belong to the ASKα subhomology division. None of them have a biosynthetic threonine dehydratase, and isoleucine is primarily or completely derived from the pyruvate pathway. The DAP pathway of lysine biosynthesis is present in most of the Euryarchaeota (exceptions are the organisms having Ask members in CG-18, as exemplified by Pyrococcus furiosis). On the other hand, all of the Crenarchaeota rely upon the AAA pathway for lysine biosynthesis. The DKFP pathway of aromatic biosynthesis is present in most of the Euryarchaeota, again with the exception of organisms having CG-18 Ask members. The latter deploy the E4P pathway instead. Some of the Crenarchaeota possess the DKFP pathway (CG-49 and CG-50), whereas the remainder (CG-19 and CG-20) utilize the E4P pathway. The distribution of alternative pathways for lysine, aromatic amino acids, and isoleucine for the organisms possessing ASKα enzymes is summarized in Fig. 8B. With respect to the twin ACT region and allostery, all of the archaeal Ask enzymes are derived from the “threonine configuration” (Fig. 8A) or a degraded version of it in which both ACT_1 and ACT_2 carry cd04892 identifiers.
Fungi
Fungi have a single Ask enzyme inhibited by threonine, and these proteins populate CG-01 in the ASKα subhomology grouping. Fungi use the AAA pathway to lysine, the E4P pathway to aromatic amino acids, and the threonine pathway to isoleucine. Since lysine is irrelevant to the ASK network in fungi, it is quite reasonable that the S. cerevisiae Ask is known to be subject to feedback inhibiton by threonine (2). However, it is curious that the two ACT domains possess cd signatures that fit expectations for an Ask that is sensitive to inhibition by lysine quite well (Fig. 8A). Perhaps that is somehow related to the report that in S. cerevisiae a prolyl isomerase called FKBP12 enters into a protein-protein interaction with Ask in such a way as to influence sensitivity to feedback inhibition by threonine (3). The single member available from the class Basidiomycota exhibits a KYGG motif in contrast to the KFGG motif of the three members of the class Ascomycota in our collection.
Cyanobacteria
Eleven genomes of cyanobacteria are quite similar in having a single ASKβ species that is represented in CG-32. They are distinctive Ask species in having four tandem ACT domains (ACT_l-ACT_2-ACT_1-ACT_2), as well as in their apparent lack of an in-frame internal translation start site. Since the internal start site generates a small subunit that, in combination with the primary translation start site, essentially yields a total of four ACT domains per round of translation, one might think that the increased multiplicity of ACT domains and the lack of an internal start site are directly related. However, the Planctomycetes (Rbal_Aa orphan) appear to possess an internal start site (Fig. 12), even though they also possess four tandem ACT domains. Cyanobacterial genomes exhibit a mixture of KFGG and KYGG motifs. Cyanobacteria uniformly possess a variant of the DAP pathway of lysine biosynthesis that bypasses acyl intermediates through the utilization of an l,l-DAP aminotransferase (38). Figure 10 suggests threonine allostery for members of CG-32.
Spirochaetes
CG-31 contains the solitary Ask enzymes present in Leptospira interrogans and Leptospira borgspetersenii. These organisms lack a metabolic connection of threonine to isoleucine and do not support ectoine biosynthesis. Specific CDD hits for the catalytic domain (cd04261) and the twin ACT domains (cd04913 and cd04936) suggest a regulation mechanism of concerted feedback inhibition controlled by Thr plus Lys. Indeed, this is well supported in Fig. 10. A KYGG motif is typical of this cohesion group. Some species of Leptospira (e.g., Leptospira biflexa) that do possess threonine deaminase are known, and therefore, these species are potentially able to use threonine as a precursor of isoleucine under some conditions.
Aquificae
A. aeolicus was the single genome originally available in the phylum Aquificae. Its Askβ enzyme supports a large network in which the DKFP pathway to aromatic amino acids is used. This may be somewhat spared in that no threonine dehydratase is present, and therefore, the pyruvate pathway to isoleucine must be used. We note that recently available complete genomes (Hydrogenobaculum sp. and Sulfurihydrogenibium sp.) appear to be quite similar to that of A. aeolicus. A Thr-plus-Lys pattern of concerted feedback inhibition is indicated by the analysis shown in Fig. 10.
Chlamydiae
Chlamydial organisms are among the few bacteria that possess an ASKα Ask. They all belong to CG-22. These pathogens possess a minimal ASK network that in fact exhibits the simplicity of a linear pathway extending from aspartate to DAP. LysA is absent, and therefore, lysine is not made. The three steps involving acyl intermediates are also absent, but it has been found that all Chlamydia spp. bypass these steps with an l,l-DAP aminotransferase (38). For many years, peptidoglycan and the cross-linking component, DAP, were thought to be absent from these intracellular pathogens. However, a number of laboratories have recently developed a basis for the conclusion that peptidoglycan (and DAP) are in fact crucial for the lifestyle of chlamydial organisms (reference 73 and references therein.). Figure 8A indicates a twin ACT region derived from a threonine signature, but one that has been degraded so that it lacks a specific cd hit in the CDD. It could possess a DAP allostery that has yet to be established experimentally.
Planctomycetes
R. baltica is the only genome of the phylum Planctomycetes available. The Rbal_Aa orphan is a typical Askβ species, except that four tandem ACT domains are present, similar to cyanobacteria. Like cyanobacteria (CG-32), Fig. 10 suggests threonine-mediated allostery for Rbal_Aa. Unlike cyanobacteria, the R. baltica enzyme appears to have an internal start site (Fig. 12).
Actinobacteria
The phylum Actinobacteria has a large number of sequenced genomes, 29 of which populate CG-25. One member of this cohesion group is C. glutamicum, which has a single Ask species that has been well studied and for which X-ray crystal structures (93) have elucidated important amino acid residues for catalysis and allosteric regulation (concerted feedback inhibition by Thr plus Lys). Six additional orphan Ask species are contributed by our collection of Actinobacteria Ask enzymes. Four are proposed to be xenologs (see “LGT” below) that coexist with the homologs retained in the vertical genealogy. The remaining two have diverged to orphan status from CG-25 in correlation with apparent alteration of Thr-plus-Lys feedback inhibition. Thus, the Twhi_Aa enzyme from Tropheryma whipplei has been transformed from Thr-plus-Lys sensitivity to inhibition by threonine alone (Fig. 10). This is understandable, in that T. whipplei is a pathogen with extreme reductive evolution that has lost both the lysine and methionine pathways (76). The Rxyl_Ab enzyme from R. xylanophilus appears to have lost sensitivity to feedback inhibition by both threonine and lysine (Fig. 10).
Betaproteobacteria and Epsilonproteobacteria
At the level of the phylum Proteobacteria, there have been some very dynamic events of LGT and reductive evolution, as summarized in Fig. 11. However, every genome from the classes Betaproteobacteria and Epsilonproteobacteria possesses a single Ask species that resides in a common cohesion group (CG-40 and CG-35, respectively). Although these classes on occasion possess ectoine biosynthesis genes (Fig. 2), they lack the ectoine-specialized ask paralogs present in other classes of Proteobacteria. The Ask enzymes of both classes usually exhibit the KYGG motif variation. They undoubtedly are subject to concerted feedback inhibition by Thr plus Lys, judging from Fig. 10.
ASK NETWORKS SUPPORTED BY MULTIPLE ask GENES
Straightforward Cohesion Group Combinations
Thermotoga.
Thermotoga maritima and Thermotoga petrophila each possess a similar pair of Ask paralogs, both being members of the ASKβ assemblage that separate into CG-26 and CG-29. The members of CG-26 are distinctive in being the only ASKβ enzymes that are fused with another enzyme domain. They are additionally distinctive among all Ask enzymes in that Hdh is fused on the N-terminal side of Ask, in contrast to the various Ask-Hdh fusions in the ASKα grouping. The fused Hdh (Hdh-min) is the only Hdh present in the genus. The genes encoding the Ask enzymes in CG-26 are located adjacent to genes of the threonine branch. Because of the inclusion of hdh, the gene context can be considered to be ask_thr+met. Figure 10 indicates that the CG-26 enzymes must be feedback inhibited by threonine.
The Ask members of CG-29, on the other hand, are monofunctional enzymes that are encoded by genes that are located within a large lysine biosynthesis operon. Figure 10 indicates that the CG-29 enzymes must be feedback inhibited by lysine. The transcriptional and allosteric regulation exerted by lysine at the level of Ask must be sufficient to keep the network balanced because dihydrodipicolinate synthase, the first committed step of lysine biosynthesis, is not feedback inhibited by lysine (74). Of course, one caveat is that other mechanisms, such as the possibility of regulation of dihydrodipicolinate synthase by a leader RNA element, as occurs in B. subtilis (35), have not been evaluated.
Acidobacteria.
Acidobacteria is one of the relatively few bacterial phyla that possess ASKα division Ask enzymes within their vertical genealogies (Fig. 7). S. usitatus possesses two Ask paralogs. One (Susi_Aa), in CG-12, exhibits a good match for the lysine configuration of feedback inhibition (Fig. 8A), and the ask gene does indeed colocalize with lysine pathway genes. The other paralog is an orphan (Susi_Ab) that exhibits a disrupted, but recognizable, threonine configuration (Fig. 8A). This ask gene is not associated with the genes of any ASK network branch. Another genome, that of K. versatilis, also possesses two Ask paralogs, but they both belong to CG-12. One (ACID-2a) is encoded by a gene having a lysine pathway gene context and exhibits a very good match for the lysine configuration of feedback inhibition in the twin ACT domain region. The other (ACID-2b) displays a somewhat disrupted match for the lysine configuration. It appears that the latter paralog is encoded by a gene duplicate that has lost (or is losing) specialization for lysine biosynthesis and has gained specialization for threonine and methionine biosynthesis, as implied by the inclusion of genes encoding Hdh-thr and threonine branch enzymes in its gene neighborhood.
Each of the above-mentioned genomes possesses an ask paralog that can be clearly equated with the lysine branch, both in terms of likely feedback inhibition by lysine and likely coregulation with genes of lysine biosynthesis. Neither the Susi_Ab nor the ACID-2b paralog is likely to be feedback inhibited by threonine, although by default these paralogs would appear to be needed for threonine and methionine biosynthesis. In addition to the Hdh-min variant that is expected to be present in ACTα organisms, both organisms possess an additional Hdh species, Hdh-thr. Thus, allostery mediated by threonine may be largely targeted to Hdh-thr in this phylum.
Firmicutes
Specializations for individual network branches.
Multiple Ask homologs with different functional specializations are ubiquitous in the phylum Firmicutes, where they either are orphans (four) or are distributed among six cohesion groups (Table 2). Of the 50 Firmicutes genomes in our collection, the number of Ask enzymes per genome varies from one (21 genomes) to two (22 genomes) or three (7 genomes). The six cohesion groups can generally be equated with particular network branch specializations in consideration of gene context observations, published allosteric patterns that can be projected to closely related organisms, and an inspection of Fig. 10 for ASKβ enzymes. (i) DAP branch specialists are unique to the membership of CG-24. (ii) Lysine branch specialists are members of CG-28, CG-30, and CG-33. These cohesion groups probably radiated from a common ancestor. (iii) Threonine/methionine branch specialists include members of CG-03 and CG-45 and the four orphans. Unlike members of CG-03, members of CG-45 and the orphans belong to the ASKβ subdivision. These ASKβ sequences are restricted to the class Clostridia and probably diverged from a common ancestor, as discussed below. Three of the orphans have an ask gene associated with a thr+met gene context. Note that an ask-adjacent hdh gene, if present as the only hdh in the genome, is sufficient grounds to assert a thr+met gene context. The fourth orphan is an ask gene associated with a gene of methionine biosynthesis, in addition to a SAM riboswitch. Members of CG-45 and the four orphans are all sensitive to allosteric control by threonine based on the criterion of their position in the lower section of Fig. 10 (where the orphan acronyms are Mthe-3_Aa, Sthe-17-Ab, Chyd_Aa, and Swol_Aa).
The B. subtilis precedent.
The three well-studied B. subtilis homologs (12, 13, 34, 94) comprise a gold standard model underlying comparative analyses to understand the differential regulation of Ask enzymes in the complex physiological and developmental contexts existing in the Firmicutes. In B. subtilis, these three Ask species belong to CG-03 (feedback inhibited by a synergistic combination of lysine and threonine), CG-24 (feedback inhibited by DAP), and CG-33 (feedback inhibited by lysine). Figure 1 of reference 95 summarizes the multiple circuits of feedback inhibition that control the overall ASK network. The DAP-sensitive CG-24 Ask is almost totally dedicated to DAP synthesis and cannot support the rest of the network unless the Ask species is altered to be desensitized to feedback inhibition by DAP (95). Conversely, the other two Ask species cannot support DAP synthesis in stationary-phase physiology, when endospore formation occurs, because they are unstable under these physiological conditions (34, 95). Lysine is a feedback inhibitor for two enzyme targets, one being the CG-33 Ask enzyme. The other target is LysA, the last enzyme step of lysine biosynthesis. The latter, coupled with the fact that the first enzyme specifically committed to DAP/lysine biosynthesis (dihydrodipicolinate synthase) is not feedback inhibited by lysine, ensures that DAP is always available for peptidoglycan and/or dipicolinate synthesis, regardless of the presence of lysine. However, a degree of control of dihydrodipicolinate synthase is exercised at the transcriptional level by lysine, which interacts directly with a leader RNA element (L box). Such L box leader regions are also positioned ahead of the CG-33 ask gene and the lysA gene (35). In organisms such as higher plants, where DAP is merely relevant as an intermediate of lysine biosynthesis, dihydrodipicolinate synthase is very sensitive to feedback inhibition by lysine (21). The details of transcriptional control are an important aspect of specialization. In B. subtilis, the DAP-sensitive enzyme (CG-24) is constitutive (95), the lysine-sensitive enzyme (CG-33) is repressed by exogenous lysine (34, 35), and the Thr-plus-Lys-sensitive enzyme (CG-03) is repressed by exogenous threonine (34).
Reductive evolution.
Figure 14 presents the assertion that the three-Ask set of specialized Ask enzymes had emerged in the common ancestor of Bacilli and Clostridia. If so, what is the explanation for the high frequency of Firmicutes organisms in which only one or two Ask homologs support the network? Many Bacilli and Clostridia are pathogens that have evolved to exploit their hosts as a source of amino acids and other nutrients in a phenomenon known as reductive evolution. On the broad scale of evolutionary time, these host-pathogen relationships are relatively recent (since the mammalian hosts are relatively recent). Indeed, the abandonment of amino acid pathways can be seen as ongoing events, judging from varied losses in closely related species and the erratic retention of pathway gene remnants (e.g., as shown in Fig. 14, lower right). In addition, the loss of the need to make dipicolinate, as well as a lesser need for DAP in Bacillus families that have lost the ability to make endospores, greatly reduces the need for the DAP-regulated CG-24 Ask specialist. Elimination of the unique physiological stage associated with sporulation simplifies the otherwise mismatched temporal relationship of DAP and lysine because they then endure in a common temporal time frame of vegetative growth. A single Ask species could then accommodate the need for both DAP and lysine, as is frequently the case in nonsporulating organisms. A single CG-03 Ask enzyme subject to concerted feedback inhibition by the Thr-plus-Lys combination could then be an effective mechanism for overall pathway control. This, in fact, describes the current state of reductive evolution in the contemporary families Streptococcaceae, Enterococcaceae, and Leuconostocaceae (Fig. 14).
FIG. 14.
Suggested evolutionary scenario for functional specialization of Ask enzymes in Firmicutes. The Firmicutes common ancestor is shown in the middle, with the divergence of the class Clostridia and the class Bacilli depicted with dendrogram progressions upward or downward, respectively. Taxon ranks of class, order, and family are enclosed within rectangles, and the deduced Ask composition of that taxon rank, ranging from one to three Ask enzymes, is indicated with color-coded CG or orphan (ORP) designations. The color codes for functional specializations are given at the bottom. Thus, for example, the common ancestor of the class Bacilli had three Ask homologs, belonging to CG-03, CG-24, and CG-33. Individual genomes that have the same homolog assemblage as deduced for the family ancestor are also included in the box, using the AroPath genome acronyms. Individual genomes that have diverged from the proposed ancestral composition due to loss or gain of one or more ask homologs are shown within ovals at the top and bottom. Placement within the blue shading indicates the retention of the ancestral five-gene operon in the class Bacilli and retention of the closely related six-gene operon in the class Clostridia (see the text). Cper, Clostridium perfringens; Cthe, Clostridium thermocellum; Tten, Thermoanaerobacter tengcongensis; Cace, Clostridium acetobutylicum; Cnov, Clostridium novyi; Ctet, Clostridium tetani; Cdif, Clostridium difficile; Chyd, Carboxydothermus hydrogenoformans; Dred, Desulfotomaculum reducens; Pthe, Pelotomaculum thermopropionicum; Mthe-3, Moorella thermoacetica; Cbot, Clostridium botulinum; Sthe-17, Symbiobacterium thermophilum; Swol, Syntrophomonas wolfei subsp. wolfei; Csac, Caldicellulosiruptor saccharolyticus; Saur, Staphylococcus aureus; Sepi, Staphylococcus epidermidis; Ssap, Staphylococcus saprophyticus subsp. saprophyticus; Shae, Staphylococcus haemolyticus; Bsub, Bacillus subtilis; Blic, Bacillus licheniformis; Bhal, Bacillus halodurans; Linn, Listeria innocua; Lmon, Listeria monocytogenes; Lwel, Listeria welshimeri; Lpla, Lactobacillus plantarum; Lsal, Lactobacillus salivarius; Laci, Lactobacillus acidophilus; Lcas, Lactobacillus casei; Ldel, Lactobacillus delbrueckii subsp. bulgaricus; Lgas, Lactobacillus gasseri; Lreu, Lactobacillus reuteri; Ppen, Pediococcus pentosaceus; Bcla, Bacillus clausii; Oihe, Oceanobacillus iheyensis; Gkau, Geobacillus kaustophilus; Bthu, Bacillus thuringiensis serovar konkukian; Bant, Bacillus anthracis; Bcer, Bacillus cereus; Gthe, Geobacillus thermodenitrificans.
It is shown in Fig. 14 that the family Listeriaceae has lost the capability for endospore synthesis. In spite of the loss of a developmental stage for which the CG-24 ask gene is attuned, the Listeriaceae possess the same Ask trio as B. subtilis and many relatives. In this connection, it is interesting that Onyenwoke et al. (67) have found the Listeriaceae to be “asporogenic.” In contrast to “non-spore-forming” organisms, asporogenic organisms possess a nearly complete repertoire of sporulation-specific genes and presumably could regain competence for sporulation with the acquisition of relatively few genes. If so, the Listeriaceae are similarly poised, with a full complement of Ask genes.
The Firmicutes currently consist of two additional recently named classes other than the Bacilli and Clostridia, but knowledge about these is marginal at present. If the newly found classes or ones yet to be named prove to predate the emergence of endospore capability, one could imagine that a single CG-03 Ask enzyme subject to concerted feedback inhibition by the Thr-plus-Lys combination might equate with the ancestral state. Thus, loss of endospore capability in, e.g, the modern Streptococcaceae may essentially constitute a reversion to the ancestral state that preceded endospore capability. Viewed this way, competence to make endospores created a developmental stage in which DAP and dipicolinate were needed at a unique developmental time. This occasioned the emergence of a DAP-regulated Ask for function in a different temporal mode. More complexly, since DAP synthesis is additionally an obligatory part of lysine biosynthesis and therefore must be synchronized intimately with lysine biosynthesis during vegetative growth, another lysine-regulated Ask species that could be tuned to the physiological pace of growing cells was needed.
Threonine branch specialization.
As summarized in Table 2, two cohesion groups (CG-03 and CG-45) and all four orphans belonging to Firmicutes can be considered to be essentially specialized for threonine (and methionine) biosynthesis. Members of CG-03 (ASKα) are quite distinct from the remaining Ask sequences (all ASKβ).
(i) The ACTα CG-03 type.
CG-03 has the highest member tally among Firmicutes sequences. It is the only Firmicutes cohesion group belonging to the ASKα subhomology division (Fig. 7). The concerted pattern of feedback inhibition exerted by the Thr-plus-Lys combination is well documented for the B. subtilis CG-03 enzyme (34, 46). The particular mechanism surely must be completely different from the concerted feedback inhibition pattern that has been well studied in a number of ASKβ organisms, where this feedback mechanism is probably the most prevalent. The ACT_1-ACT_2 domain region of CG-03 members is derived from a cd organization (cd04890-cd04892) that characteristically has a “lysine configuration” of residues that denote lysine allostery (Fig. 8A). Although the Lcas_Aa sequence, which was arbitrarily chosen to represent CG-03 in Fig. 8A, gives little indication of lysine sensitivity, most of the other sequences in CG-03 exhibit a suggestive, albeit imperfect, conformation with a lysine configuration (see the individual CG alignment in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG03_aln.html). The L. casei enzyme (Lcas_Aa) happens to be the paralog associated with threonine pathway genes in the lower-right portion of Fig. 14, and one would thus expect loss of lysine binding for this paralog.
Since the B. subtilis CG-03 Ask cannot function during sporulation (34, 95), it is interesting to consider how an organism possessing only a CG-03 Ask species can sporulate. Clostridium thermocellum (Fig. 14) has lost both the DAP-regulated CG-24 Ask and the lysine-regulated CG-28 Ask, leaving the CG-03 Ask to support the entire ASK network. This means that it has to function during both the sporulation and vegetative-growth phases. Clearly, it has to differ from the B. subtilis enzyme in being stable and active during sporulation. It could still be subject to concerted feedback inhibition, which would be appropriate during vegetative growth. Such regulation would be inappropriate during sporulation. However, if certain key enzymes (LysA and homoserine dehydrogenase) were hyperlabile to the onset of sporulation, then in that physiological state lysine and threonine might not be available as feedback inhibitors that might interefere with DAP synthesis.
In the family Lactobacillaceae of Bacilli, the CG-03 Ask is frequently the only remaining Ask. The scheme at the lower right of Fig. 14 depicts a very active pattern of reductive evolution that is consistent with published reports about these organisms (86). This group has lost endospore-forming capability, and most species have lost the CG-24 DAP-inhibited Ask species. A gene duplication occurred in the common ancestor of L. acidophilus and L. casei to yield two recent paralogs. One CG-03 ask gene is associated with a complete threonine operon, and the other CG-03 ask gene is adjacent to multiple lysine pathway genes, most of which are divergently oriented. In the common ancestor of two species, Lactobacillus delbrueckii and Lactobacillus gasseri, the entire group of lysine pathway genes, together with the associated CG-03 ask gene, has been deleted from the genome. Incredibly, a complementary deletion in the common ancestor of Lactobacillus reuteri and Pediococcus pentosaceus has instead eliminated the threonine operon and the associated CG-03 ask gene. Thus, in one case, both the lysine pathway and its ask specialist have been eliminated from the genome, whereas in another case, the threonine/methionine pathway section, together with its ask specialist, has been discarded.
CG-03 has a large Ask membership (click the cohesion group gene neighborhood button on the dynamic table [http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html, second column] to view whether genes encoding any enzymes of the network branches are associated with a given CG-03 ask gene). The example discussed above describes a case where a CG-03 gene is associated with a lysine gene context, but this is highly unusual. Genes encoding CG-03 Ask members are not necessarily associated with any genes encoding network branches. Indeed, the classic B. subtilis member displays no relevant network context. Some genera (e.g., Streptococcus, Lactococcus, and Enterococcus) possess an Ask member of CG-03 that is encoded by the only remaining ask gene in the genome, which therefore supports the entire ASK network (Fig. 14). As would be generally expected, these ask genes are not associated with any ASK network genes. They are probably feedback inhibited by Thr plus Lys, and indeed, Streptococcus mutans exemplifies a case where the single CG-03 Ask has been shown experimentally to be subject to concerted feedback inhibition by the Thr-plus-Lys combination (55). Clostridium botulinum, shown in Fig. 14 to have maintained the ancestral set of three Ask enzymes, possesses a CG-03 ask species that is a component of a complete threonine branch operon. Although this operon includes Hdh-thr, it is functionally separate from methionine biosynthesis, since C. botulinum has a second Hdh enzyme, Hdh-met. The gene encoding Hdh-met is adjacent to several methionine pathway genes. In Staphylococcus spp., the CG-03 ask gene is adjacent to a similar threonine operon, with the following differences. ask is divergently oriented to the threonine operon, the hdh gene in the operon is hdh-met, and hdh-met is the only hdh gene in the genome. In this case, Hdh-met may be transcriptionally controlled by threonine and perhaps is allosterically controlled by methionine.
(ii) A novel ASKβ Ask replaces the CG-03 ASKα enzyme in Clostridia.
A new specialized Ask that belongs to the ASKβ subhomology division has emerged in correlation with the loss of CG-03 Ask enzymes and in correlation with the loss of CG-28 Ask enzymes. Three members of CG-45 and four orphans occur in the class Clostridia, with no counterparts so far known in the class Bacilli (Table 2 and Fig. 14). The new threonine/methionine ask specialists in Clostridia may, in fact, be derived from the lysine specialists belonging to CG-28, since there is a perfect correlation of loss of the CG-28 ask gene and appearance of both the CG-45 ask genes and the four orphan genes (Fig. 14). If so, the former CG-28 ask gene has migrated from a lysine gene context to a threonine/methionine gene context. Furthermore, the lysine allostery of a CG-28 Ask has not been retained, since threonine allostery is strongly implied in Fig. 10 for CG-45 members and the four orphans. For those Clostridia organisms that have lost both the Thr-plus-Lys-specialized enzyme (CG-03) and the lysine-specialized enzyme (CG-28), the threonine pathway specialization has been replaced by Ask members of CG-45 and the four related orphans. However, this raises the question of how lysine is sensed in the absence of any lysine-responsive Ask specialist.
CG-45 is currently populated by three members from the Clostridiales. Each one of these ask genes exhibits the following gene neighborhood: act → hdh-thr → thrB → ask. This organization is intriguing because of the inclusion of a gene encoding a protein with a free-standing ACT domain. The act → hdh-thr → thrB → ask operon may be quite prevalent in Clostridiales, since a recent scan of new genomes (available after the cutoff date for inclusion in our collection) revealed organisms representing three additional families that have either the same gene organization or very similar ones (variant operons without thrB or variant operons with thrC preceding thrB). These are Anaerocellum thermophilum (no family rank assigned), Ruminococcus obeum (family Ruminococcaceae), and Heliobacterium modesticaldum (family Heliobacteriaceae). The Carboxydothermus hydrogenoformans ask orphan possesses an act → hdh-thr → thrC → thrB → ask operon, in which the orphan ask gene resides. In addition to C. thermocellum in Fig. 14, Clostridium phytofermentans and Clostridium nexile (Clostridiaceae) possess one of these operon variations. Thus, the essential act → hdh-thr → ask gene arrangement is distributed among at least five families of Clostridiales. It is striking that ACT, Hdh-thr, and Ask are all proteins that contain one or more ACT domains. Since the free-standing ACT domain is an amino acid binding module, and since ACT domains are known to interact with one another in complex ways (50), it seems quite plausible that lysine might be an effector for the free-standing ACT domain that somehow delivers appropriate regulation in a fashion that involves the other gene products of the operon.
In “Evolution of the ASKα assemblage” above, the recruitment of a free-standing ACT domain in Archaea was discussed. It was suggested that during periods of insufficiency of lysine and aromatic amino acids, Hdh might essentially be sequestered by association with ACT, creating an inactive state of Hdh that thereby blocks substrate flow to methionine and threonine. During lysine and aromatic amino acid sufficiency, these amino acids might bind to ACT and promote dissociation of ACT and Hdh. Dissociated Hdh would be free to complex with Ask. This would then accommodate substrate flow to methionine and threonine. A similar mechanism could prevail in the Clostridiales, with the simplification that aromatic amino acids are not relevant.
It was discussed above how the two-Ask system of some Clostridia (such as members of the family Peptococcaceae) can be equated with the three-Ask system of B. subtilis. The question addressed has boiled down to the matter of how, in the absence of an ask specialist for lysine, the need for lysine in Clostridia might be sensed at the level of Ask during vegetative growth. An even more challenging question is how the newly evolved threonine/methionine specialist Ask enzymes of Syntrophomonas wolfei and Caldicellulosiruptor saccharolyticus are sufficient alone as a one-Ask system for coordination of both their sporulation programs and the primary ASK network of biosynthesis. If the explanation of how lysine biosynthesis is regulated via the ACT-Hdh mechanism in organisms like the Peptococcaceae is accepted, the question is how S. wolfei and C. saccharolyticus accommodate the sporulation process without a CG-24 ask gene. S. wolfei is in the same family as C. saccharolyticus (Fig. 14). Both of these organisms have a single ask gene with a threonine/methionine gene context. Lysine and dipicolinate synthesis genes are clustered elsewhere. The main dilemma appears to be how to prevent threonine from disrupting the sporulation process by shutting off the precursor supply to dipicolinate and DAP. The gene context suggests that threonine controls transcription. The Swol_Aa enzyme is likely feedback inhibited by threonine (Fig. 10). The C. saccharolyticus enzyme might indeed be resistant to feedback inhibition by threonine, since the D25 residue (Fig. 10), which appears to be essential for threonine inhibition, is absent (see the individual CG-45 alignment in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG45_aln.html). S. wolfei and C. saccharolyticus differ from one another in several respects. S. wolfei lacks the ACT domain gene that is adjacent to hdh in C. saccharolyticus, but the gene may be elsewhere in the genome. S. wolfei possesses a single gene cluster to accommodate lysine and dipicolinate synthesis, whereas C. saccharolyticus separates them into two clusters. In S. wolfei, this is an intact six-gene equivalent of the dipicolinate-DAP gene cluster present in some Clostridia, except that the CG-24 ask gene that is usually embedded in the operon is missing. In C. saccharolyticus, the loss of the CG-24 ask gene from the ancestral six-gene cluster has been accompanied by dispersal of the genes encoding the alpha and beta subunits of dipicolinate synthase to a position adjacent to the gene encoding the cell division protein FtsK.
DAP/dipicolinate branch specialization.
In the Firmicutes, DAP is especially important in its role as a contributor to peptidoglycan because the peptidoglycan layer in cell walls is significantly thicker than in gram-negative bacteria. For those Firmicutes that form endospores, there is a developmental stage of sporulation accompanied by high input of peptidoglycan into the spore cortex. In addition, dipicolinate can comprise up to 10% of endospore dry weight (12). B. subtilis (12) and Clostridium perfringens (49) have been shown to possess an Ask species that is sensitive to feedback inhibition by DAP. Both of them belong to a large cohesion group (CG-24) that is quite distinctive. It is one of the few ASKβ groups that appear to lack an internal translational start site (Fig. 12). The strong cd identifiers for the twin ACT domains (cd04914-cd04937) are absolutely unique to CG-24, and the high sequence conservation seems to suggest that all members of CG-24 might prove to be sensitive to feedback inhibition by DAP. If so, however, it is an open question how a few organisms having only a CG-24 Ask enzyme could manage to support the entire ASK network without interference by DAP (as discussed below). In the class Bacilli of Firmicutes, a CG-24 member is always present in organisms that have retained the ability to make endospores. The loss of endospore capability is usually accompanied by loss of the CG-24 ask gene, but occasionally this ask gene has been retained (undoubtedly with some functional adjustments). On the other hand, most members of the class Clostridia of Firmicutes in our collection (except for T. tengcongensis) make endospores, but there are a number of examples where this is accomplished without a DAP-regulated CG-24 Ask.
The B. subtilis member of CG-24 has been shown (12) to possess a complex operon, dpaA → dpaB → asd → ask → dapA, from which multiple transcripts are made. Here, this is denoted the “five-gene operon.” The two upstream genes (encoding subunits of dipicolinate synthase and originally named spoVFA and spoVFB) are expressed only during sporulation (stage V), whereas the remaining three (which encode the first three steps of DAP biosynthesis) are expressed during both sporulation and vegetative growth. This five-gene cluster of B. subtilis is conserved at the taxon level of family (Bacillaceae), which in our genome collection includes the genera Bacillus, Geobacillus, and Oceanobacillus. Interestingly, a number of the members of the class Clostridia possess nearly identical gene organizations, differing only in the possession of an additional DAP pathway gene (encoding dihydrodipicolinate reductase) upstream of dpaA. This operon in the Clostridia is referred to as the “six-gene operon.” Figure 14 shows the lineages that have retained these ancestral operons. One genome, S. wolfei subsp. wolfei, possesses a gene cluster that is intact except for the absence of the CG-24 ask gene. The ask members of CG-24 that are not part of a five-gene cluster (class Bacilli) or of a six-gene cluster (class Clostridia) are genes associated with cell wall biosynthesis. The Clostridiaceae organisms always exhibit an ask gene located near a stage III sporulation protein that is involved in cell division. The genera Lactobacillus and Listeria possess CG-24 ask genes that are within gene neighborhoods heavily populated with various mur and fts genes of cell wall biosynthesis.
The five-gene operon within Bacilli and the six-gene operon within Clostridia present a striking aspect of incongruence in that each has closer relatives in their class that do not possess these operons. One might think that this incongruence might be explained by LGT between the two. If so, we would expect the CG-24 Ask enzymes from Bacilli having the five-gene operon and the CG-24 Ask enzymes from Clostridia with the six-gene operon to group together on the phylogenetic tree to the exclusion of CG-24 Ask proteins from other Bacilli and Clostridia. However, when the Ask tree of the CG-24 section of the overall master tree was examined, the Ask enzymes of Clostridia and Bacilli clustered separately and cohesively in congruence with phylogenetic expectations, regardless of the gene neighborhood patterns. This might indicate that the gene clustering in the two classes of the Firmicutes emerged independently. On the other hand, an ancient operon might have been widespread in the ancestor of the Bacilli and Clostridia, and operon disintegration might have occurred independently in the two classes. Consistent with the latter possibility is the fact that the portion of Bacilli that lack the five-gene operon have lost the ability to make endospores and therefore lack the now unneeded genes for dipicolinate synthase. Loss of the genes encoding the two subunits of dipicolinate synthase would disrupt the ancestral gene cluster. As far as we know, only T. tengcongensis (Tten in Fig. 14) is deficient in sporulation in the class Clostridia (67). Elsewhere in the class Clostridia, loss of the two genes encoding dipicolinate synthase also seems to explain the disruption of the six-gene operon in the family Clostridiaceae (Fig. 14). This is quite surprising, because the family Clostridiaceae is known to make endospores and dipicolinate. This includes C. botulinum, Clostridium tetani, Clostridium novyi, Clostridium acetobutylicum, and C. perfringens. Only Clostridium difficile has the established dipicolinate synthase genes, and they are remote from the CG-24 ask gene and from other dap genes. It seems that the Clostridiaceae must have an alternative pathway leading to dipicolinate! The absence of dpaA and dpaB in Clostridiaceae was previously noted by Onyenwoke et al. (67).
In summary, it appears that the five-gene or six-gene operon existed in the ancestor of endospore-forming Bacilli and Clostridia. In Bacilli, the five-gene operon was disrupted by the abandonment of the dpaA and dpaB genes in lineages where endospore capability was lost. In Clostridia, an independent abandonment of dpaA and dpaB disrupted the six-gene cluster due to replacement of dpaA and dpaB by unknown genes.
Lysine branch specialization.
There are three small-membership cohesion groups that can be considered to be coordinated specifically with lysine biosynthesis. CG-33 is the best known, because it contains the B. subtilis ask species characterized as a lysine-sensitive enzyme (13). It presumably could increase the supply of precursor under conditions where DAP withdrawal for peptidoglycan biosynthesis decreases the availability for lysine biosynthesis. CG-33 members are limited to the families Bacillaceae and Listeriaceae (Table 2 and Fig. 14). Members of CG-30 are thus far restricted to the genus Staphylococcus (Table 2 and Fig. 14). Since Staphylococcus spp. possess an ask homolog in CG-03 that resides within a threonine/methionine operon and since they lack a member of CG-24, it seems straightforward that ask members of CG-30 must be lysine specialized, a conclusion supported by colocalization of CG-30 genes with genes for lysine biosynthesis and by the indication of lysine allostery in Fig. 10. Finally, the ask members of CG-28 are specialized for lysine biosynthesis because they are closely linked to the gene encoding LysA, the only gene that is unique to lysine biosynthesis. In addition, they fit the pattern for lysine allostery in Fig. 10.
In Fig. 14, C. botulinum has a three-Ask functional profile that resembles the putative ancestor of the class Clostridia, compared to B. subtilis, which has a three-Ask profile that resembles the putative ancestor of the class Bacilli. C. botulinum has three Ask enzymes that distribute into CG-03 (threonine/methionine specialized), CG-24 (DAP/dipicolinate specialized), and CG-28 (lysine specialized). Thus, the overall functional interplay may be roughly similar to that in B. subtilis (with the CG-33 enzyme of B. subtilis and the CG-28 enzyme of C. botulinum being functionally equivalent). Even though both B. subtilis and C. botulinum possess the DAP-inhibited CG-24 enzyme, however, they differ in that B. subtilis maintains the five-gene operon, whereas C. botulinum lacks the six-gene operon entirely, owing to replacement of dpaA-dpaB by unknown functional counterparts. In addition, the C. botulinum CG-24 ask gene is no longer in a gene neighborhood context with DAP genes.
The overall evolutionary scenario in the vertical genealogy.
The scheme shown in Fig. 14 summarizes the evolutionary progression of specialized Ask enzymes in the Firmicutes. At the center of the diagram, the classes Bacilli and Clostridia are shown to have diverged from a common ancestor already in possession of the three specialized ask homologs that are familiar to us in B. subtilis. For convenience, the CG-03 Ask is denoted as having a functional specialization for biosynthesis of threonine, the most dominant end product regulator. This is an oversimplification, because it is also tuned to methionine biosynthesis, and it probably is feedback inhibited synergistically by the Thr-plus-Lys combination most of the time. The lysine-specialized ask of the Firmicutes ancestor diverged into the memberships of CG-33 and CG-30 in the Bacilli and diverged into the membership of CG-28 in the Clostridia. Genes encoding Ask members of CG-03 and CG-24 have been the most persistently retained. On the other hand, lysine specialization appears to have been abandoned quite often, especially in the class Clostridia. The lysine-specialized Ask (CG-33) that is present throughout most of the Bacillales has diverged in the family Staphylococcaceae to become a member of CG-30. In one interesting case (Fig. 14, lower right), gene duplication in the common ancestor of two Lactobacillus spp. has resulted in the emergence of a new CG-03 paralog that has shifted its specialization to lysine biosynthesis.
Figure 14 shows the trace of the lineages that have retained the five-gene operon (class Bacilli) or the six-gene operon (class Clostridia). The key common genes in these operons are the CG-24 ask genes and the two genes (dpaA and dpaB) encoding dipicolinate synthase. This ancient operon is tightly associated with endospore formation, as elucidated in B. subtilis (12). In various lineages, this elegant system has been disrupted by (i) loss of competence for endospore formation, and thus loss of dpaA-dpaB, as shown in the Bacilli; (ii) acquisition of a pathway to dipicolinate outside of the ASK network, and thus loss of dpaA-dpaB, as seen in the Clostridiaceae; and (iii) loss of the the CG-24 ask gene, as seen in S. wolfei and C. saccharolyticus from the Syntrophomonadaceae.
Endospore biosynthesis probably evolved once in a Firmicutes ancestor, judging from the extremely large number of genes engaged in this developmental process (26). Although the particular gene-enzyme relationships that underpin endospore biosynthesis appear to be more variable than previously assumed (67), substantial evolutionary divergences in individual lineages could account for this. In Fig. 14, various points are shown in the Bacilli lineage where capability for endospore synthesis is asserted to have been lost. In most cases, there is a correlated loss of the DAP-regulated ask (CG-24), e.g., in Staphylococcaceae and four of the five families of the Lactobacillales. The only exception to the loss of CG-24 ask correlated with the loss of endospore formation is in the family Listeriaceae, where gene members of CG-24 are retained in close linkage with the gene encoding peptidoglycan synthetase (ftsI). Most of the genomes shown (Fig. 14, top) in the class Clostridia can produce endospores, but a few of them nevertheless have not retained a CG-24 ask. This includes the two genomic members of the Syntrophomonadaceae. In a separate loss event, C. thermocellum has lost the CG-24 ask, leaving behind the CG-03 ask to support the entire network. It seems that the CG-03 Ask must differ from the B. subtilis CG-03 Ask, which is unstable during sporulation physiology and therefore would not be able to supply precursor for DAP and dipicolinate. The C. thermocellum ask gene is of further interest in that it exists in an act → hdh-met → ask operon. This gene organization is similar to that discussed above for the novel CG-45 members and four orphans, which appear to have replaced CG-03 enzymes as threonine specialists. If the significance of this gene organization is reflective of a mechanism for lysine control, as proposed, it would be appropriate, since no lysine-specialized ask is present in the genome. In this vein, it is of further interest to consider a number of genomes that have retained only a CG-24 ask gene. Recalling that high sensitivity to feedback inhibition by DAP prevents the B. subtilis enzyme from supporting threonine, methionine, and lysine biosynthesis (95), how can a single CG-24 Ask enzyme support the overall ASK network in C. perfringens and T. tengcongensis (Fig. 14, top), as well as Geobacillus thermodenitrificans and Lactobacillus salivarius (Fig. 14, bottom)? Either there are regulatory mechanisms to keep the DAP pool sizes very small during vegetative growth or the feedback sensitivity to DAP is highly relaxed. In the case of C. perfringens, there would only need to be enough slippage to accommodate lysine biosynthesis, since the threonine/methionine branches have been lost.
The prominent threonine/methionine specialist in the Firmicutes is the CG-03 species, which belongs to the ASKα subdivision. In the class Clostridia, the losses of the CG-03 ask gene have been followed by the emergence of a new threonine/methionine specialist belonging to the ASKβ subdivision. This has apparently happened on two independent occasions: once in the lineage leading to the Thermoanaerobacteriaceae and once in a common ancestor of Peptococcaceae and Syntrophomonadaceae. In each case, there is a perfect correlation of the disappearance of the lysine Ask specialist belonging to CG-28 and the appearance of threonine specialists in the form of the four orphans in the bottom section of Table 2, as well as the three members of CG-45. It is therefore suggested that the ancestral CG-28 lysine specialist diverged in conjunction with the acquisition of a new threonine/methionine gene context, thus switching its functional role. The lysine allostery of CG-28 members was additionally transformed into threonine allostery (Fig. 10). This scenario is consistent with the close proximity of the four orphans, CG-45, and CG-28 to one another in the tree in Fig. 5. No ask genes of the novel ASKβ type, specialized for threonine/methionine biosynthesis and present in some Clostridia, are present in the Bacilli.
Bacteroidetes-Chlorobi Superphylum
The overall evolutionary scenario in the vertical genealogy.
The phyla Bacteroidetes and Chlorobi possess ACTα Ask paralogs. A summary of evolutionary events in the vertical genealogy is given in Fig. 13, left. Although the phylum Chlorobi may appear to have been less dynamic in evolutionary change, only five genomes in a single family (Chlorobiaceae) are available. The five genomes are listed in Fig. 9. In contrast, genomes representing the phylum Bacteroidetes are drawn from five families.
A ubiquitous Ask of the superphylum is an Ask-Hdh fusion that belongs to CG-05. The gene encoding this highly successful bifunctional enzyme is strongly conserved but has diverged sufficiently to yield another cohesion group (CG-07), which, however, exhibits no obvious functional changes. The gene has undergone duplication a number of times to yield paralogs with different functional specializations, as described below. Finally, the two catalytic domains of some duplicates have become separated in a reversal of the original fusion and have taken on individual roles in at least two cases. The cd04892-cd04892 pattern of the twin ACT regulatory region and the strong conformation with the QXXSE…QXXSE motif (Fig. 8A) indicates that both the Ask and homoserine dehydrogenase domains are allosterically inhibited by threonine. In the phylum Bacteroidetes, the gene encoding the CG-05 Ask is located within a threonine/methionine pathway operon. In the phylum Chlorobi, the Ask gene is adjacent to two extraneous genes that separate it from thrB and thrC (which are also divergently transcribed with respect to ask).
A second paralog of Ask is almost as prominent throughout the superphylum. This has diverged to form CG-10 (in Chlorobi) and CG-11 (in Bacteroidetes). Members of both CG-10 and CG-11 possess a cd04890-cd04892 pattern in the twin ACT regulatory region and exhibit strong conformation (Fig. 8A) with the residues known to be associated with lysine allostery (Fig. 13, lower left). The fundamental picture of the ASK network in the superphylum is thus one in which two ancient paralog types are deployed, one (CG-05 and CG-07) specialized to control threonine and methionine biosynthesis and the other (CG-10 and CG-11) tuned to the cellular demand for lysine. While all Chlorobi (so far) use these two enzyme paralogs, the phylum Bacteroidetes exhibits a larger paralog repertoire (Fig. 13). In this phylum, only Cytophaga hutchinsonii is limited to the two foregoing paralog types. In one case of reductive evolution, P. gingivalis has lost the threonine/methionine branches and has accordingly also lost the CG-05 ask gene. It has retained a single Ask in CG-11, which, as would be expected, is regulated by lysine. In most cases, the evolutionary direction appears to be one in which an additional paralog is generated in order to allow a more refined responsivity to demands of the methionine branch. Phylogenetic changes along these lines occurred independently in each of three Bacteroidetes classes, as discussed below.
The classes Flavobacteria and Bacteroidetes have generated an additional species of Ask by two completely different mechanisms, the first derived from a CG-05 gene and the second from a CG-11 gene. In the proposed step D of Fig. 13, the bifunctional ask-hdh gene was duplicated and the catalytic domains of one copy were separated to yield genes encoding a monofunctional Ask (belonging to CG-06) and Hdh-min. The selective advantage may primarily have been to obtain a second hdh gene, which then was inserted into a methionine operon. The Hdh-min gene product would not be feedback inhibited by threonine, unlike the parental Hdh domain of the bifunctional ask-hdh gene in CG-05. The monofunctional Ask of CG-06 has a disrupted twin ACT domain and is probably insensitive to allostery. It is color coded as an unregulated “basal” Ask activity in Fig. 13. The foregoing is descriptive of Gramella, but other genera in the family Flavobacteriaceae had a common ancestor that underwent the additional steps E and F. Here, the CG-05 ask-hdh gene was duplicated, followed by substantial disruption of the QXXSE…QXXSE motif in one copy, as illustrated in the case of F. johnsonii at the bottom of the alignment portion of Fig. 9. The Fjoh_Ab paralog has deviated markedly (with respect to the twin ACT region) from the Fjoh_Ac paralog, as well as from all other members of the cohesion group. (Note that sequences from Robiginitalea, Tenacibaculum, Kordia, and Polaribacter were not included in the dynamic table or Fig. 9. Had they been, there would have been paralog sets from each organism, which would have paralleled Fjoh_Ac and Fjoh_Ab). The altered paralog copy, Fjoh_Ab (as well as its counterparts in the genomes of the other four sister genera), was inserted into the methionine operon. Clustered methionine branch genes are compared for the Gramella forsetii and F. johnsonii genomes at the bottom of Fig. 9. Thus, the methionine operons of all Flavobacteriaceae had the hdh-min gene (which was generated by gene scission in step D) inserted into the methionine operon. At a later time, following the divergence of Gramella from the common ancestor of at least five other genera, an ask-hdh gene duplicate that generated a disrupted twin ACT region was additionally appended to the methionine operon. The gene context of the CG-06 Ask gene is not informative, but since the CG-11 Ask has disappeared from the cluster of five or more genera, the CG-06 Ask appears to be the default candidate for lysine specialization and is so color coded in Fig. 13. The CG-06 Ask enzymes are derived from the cd04892-cd04892 combination of ACT domains, which are expected to be threonine regulated. In Fig. 8A, the CG-06 twin ACT region groups with “disrupted twin ACT” regions. Perhaps this disrupted region represents an alternative mode of lysine binding that has not yet been experimentally demonstrated.
In the case of the family Bacteroidaceae, a paralog of the CG-11 Ask arose by gene duplication. The motif signature for lysine allostery has been totally disrupted in one copy, and important residues for catalysis have been altered, including the K(F/Y/I)GG motif. This is color coded as a basal activity in Fig. 13, but it is likely a pseudogene.
The most dynamic evolutionary progression is seen in the class Sphingobacteria, and it should be interesting eventually to examine a greater sampling of genomes. Salinibacter possesses four Ask genes that are clearly tuned to different ASK branches. Differences from the common ancestor, which is retained in the contemporary C. hutchinsonii, occurred via phenomena of gene fusion, gene scission, and gene duplication as follows. Srub_Ac, a member of CG-07, diverged from CG-05. However, it has retained its features of bifunctionality, threonine allostery, and theonine gene context. Second, Srub_Ad (CG-09) diverged from a CG-11 antecedant after fusing with the gene encoding LysA. The genes encoding Ask and LysA were probably adjacent prior to fusion, since phylogenetic neighbors, such as Bacteroides fragilis currently exhibit these gene neighbors. The fused protein, a member of CG-09, has retained a strong signature for lysine allostery in the twin ACT region (Fig. 8A). Third, a duplicated copy of ask-hdh was cleaved in such a way as to generate hdh-min and a gene encoding an Ask lacking the ACT domains altogether (Srub_Ab). The genes encoding this allosteric-incompetent Ask belonging to CG-47 and the hdh-min remain adjacent and have become incorporated into S. ruber's methionine operon. Fourth, Srub_Ab was duplicated, and one copy (encoding Srub_Aa, a second member of CG-47) was translocated to a gene context (genes encoding dihydrodipicolinate reductase and d-alanine d-alanine carboxypeptidase) that implies functional specialization for peptidoglycan/cell wall synthesis.
Proteobacteria
The overall evolutionary scenario in the vertical genealogy.
At the phylum level, two ancestral Ask enzymes can be traced in the vertical genealogy of the Proteobacteria. One is ubiquitous throughout the phylum and has diverged into the many cohesion groups and orphans, as indicated in Fig. 7, right. It belongs to the ASKβ subhomology division and has CDD aspartate kinase domain cd04246, the latter almost always being cd04261 at a more shallow hierarchical level (Fig. 7). The second ancestral Ask of the phylum belongs to the ASKα subhomology division, and its members are tightly gathered into a single cohesion group (CG-02). These ectoine-specialized Ask enzymes (Ask_ect) are broadly distributed within the phylum, but their occurrence is quite erratic, as shown in the far-right column of Fig. 11. At the level of any class, only a minority of genomes possess Ask_ect. Even at the level of the order, only a fraction of genomes are represented by Ask_ect, the best represented being those of Vibrionales, where four of five genomes have Ask-ect. The distribution of ask_ect within Proteobacteria could perhaps be explained by frequent LGT exchanges within the phylum. However, the explanation that frequent independent losses occurred in different sublineages seems more probable in view of the overall instability of the ect genes (gene shuffling, inversions, transpositions, and deletions) that comprise the ask_ect operon.
Figure 11 lists the classes of Proteobacteria in the middle of the diagram, with the distribution of cohesion groups and orphans that persist within the vertical genealogy shown on the right. Epsilonproteobacteria (6 genomes) and Betaproteobacteria (33 genomes) are the most straightforward classes in that they (i) have a single ancestral Ask per genome, (ii) lack any CG-02 Ask, and (iii) lack any additional Ask enzymes originating from LGT. Ask enzymes from the entire class Epsilonproteobacteria belong to a single cohesion group (CG-35), and those from the entire class Betaproteobacteria belong to CG-40.
The overall pictures of Ask distribution in both Deltaproteobacteria and Alphaproteobacteria are similar in that most genomes are represented by a single ancestral Ask, but a few genomes additionally possess Ask_ect (Fig. 11, far right column) or an Ask of LGT origin (Fig. 11, left). The ancestral Ask orthologs of the 16 genomes of Deltaproteobacteria have diverged to yield three cohesion groups and two orphans. Three of these genomes possess an Ask_ect Ask. Two of these appear to be remnants that have survived a deletion of all ect genes previously comprising a presumed ask_ect operon. One of the latter genomes (D. psychrophila) has also lost the ancestral Ask, leaving ask_ect as a remnant Ask that now supports the entire ASK network. Only two other Deltaproteobacteria genomes possess additional Ask enzymes of LGT origin (see below and Table 3 in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/LGT.html). Reminiscent of the Deltaproteobacteria, the 48 genomes of Alphaproteobacteria possess Ask homologs that have diverged to yield three cohesion groups and one orphan. The overwhelming majority of ancestral Ask enzymes belong to CG-37. Four genomes of Alphaproteobacteria additionally possess a specialized Ask_ect Ask belonging to CG-02, and two other genomes possess additional Ask enzymes of LGT origin.
TABLE 3.
LGT genes for aspartokinase
| Organism | Dynamic table linka | Cohesion groupb | Near-donor lineagec | Ancient-donor lineagec | Cotransferred domains/genesd |
|---|---|---|---|---|---|
| Escherichia coli K-12 | Ecol_Aa | 21 (39) | Bacteroidetes | ||
| Ecol_Ac | 04 (41) | Sphingobacteriales | hdh/thrB/thrC | ||
| Ecol_Ab | 08 (29) | Sphingobacteriales | hdh | ||
| Bdellovibrio bacteriovorus HD100 | Bbac_Ab | 21 (39) | Lower Gammaproteobacteria | Bacteroidetes | |
| Francisella tularensis subsp. novicida U112 | Ftul_Aa | 21 (39) | Lower Gammaproteobacteria | Bacteroidetes | |
| Ftul_Ab | 07 (6) | Sphingobacteriales | hdh/thrB/thrC | ||
| Maricaulis maris MCS10 | Mmar_Ab | 09 (7) | Sphingobacteriales | lysA | |
| Mmar_Ac | 52 (2) | Sphingobacteriales | hdh/thrB/thrC | ||
| Methanopyrus kandleri AV19 | Mkan_Aa | 19 (1) | Desulfurococcales | ||
| Myococcus xanthis | Mxan_Ac | Orphan | Lower Gammaproteobacteria | Sphingobacteriales | hdh |
| Mxan_Ad | Orphan | Flavobacteriaceae | hdh | ||
| Thermus thermophilus HB27 | Tthe_Aa | Orphan | ? |
In the full dynamic version of this table in the supplemental files, the AroPath acronyms in this column are hyperlinked to the dynamic table, facilitating access to all available gene detail.
The numbers in parentheses indicate the number of xenolog sequences in the cohesion group, all of which are included in the full dynamic version of Table 3 in the supplemental files.
In the three examples of piggyback LGT shown, the most recent donor lineage is indicated in the “Near-donor lineage” column, with the donor itself having been obtained via an earlier LGT event in which the donor is referred to as the “Ancient-donor lineage.”
Operonic LGT transfers include the fused C-terminal domains (hdh or lysA) attached to ask, as well as thrB and thrC in some cases.
Within the Alphaproteobacteria, the divergence of Ask enzymes to the “Candidatus Pelagibacter ubique” orphan (Pubi_Aa) and to memberships in CG-37 or CG-38 is strikingly parallel to the divergence exhibited by Alphaproteobacteria in the genome tree published by Wu and Eisen (88). A large group of nonpathogenic Alphaproteobacteria genomes possess CG-37 Ask enzymes, whereas a smaller group of pathogenic or endosymbiotic genomes possess CG-38 Ask enzymes. The smaller assemblage of genomes, including “Candidatus Pelagibacter ubique” as the outlying genome branch on the tree, corresponds to the order Rickettsiales. The genome of “Candidatus Pelagibacter ubique” is the only one in this order that possesses the threonine/methionine branches of the ASK network. Thus, after the divergence of “Candidatus Pelagibacter ubique,” the loss of the Thr/Met branches occurred in the common ancestor of all of the remaining Rickettsiales. At about the same time, the gene encoding LysA of the lysine pathway was discarded, leaving an intact pathway to DAP but loss of competence for lysine biosynthesis. This includes the genera Rickettsia, Orientia, Wolbachia, and Anaplasma. This simple ASK network consisting of a linear path to DAP is reminiscent of what is present in Coxiella burnetii and Dichelobacter nodosum and in the Chlamydiae. The only genus in the Rickettsiales that possesses LysA, and therefore competence for lysine biosynthesis, is Ehrlichia. If the recently diverged position of Ehrlichia in the genome tree (88) is accepted, either lysA was lost independently a number of times or lysA was reacquired recently in a common ancestor of Ehrlichia spp. by LGT. If lysA in Ehrlichia were a gene retained in the vertical genealogy, it should be more similar to that of “Candidatus Pelagibacter ubique,” which is a member of the same order (Rickettsiales). Since it is more similar to lysA genes of other orders of Alphaproteobacteria, LGT from another Alphaproteobacteria organism is a strong possibility.
A greater level of detail is shown in Fig. 11 for the class Gammaproteobacteria, which is subdivided into the “superorders” that represent upper Gammaproteobacteria and lower Gammaproteobacteria (Fig. 3). Throughout the Gammaproteobacteria, the gene encoding the ancestral Ask (shown in Fig. 11) is conspicuous because it is located within a gene neighborhood context that is highly conserved throughout the class. This cluster of genes is shown on the bottom right of the column. Ask genes that are associated with this gene cluster are designated ask_alaS. Members of the order Xanthomonadales in the upper Gammaproteobacteria, as well as members of the order Enterobacterales in the lower Gammaproteobacteria, have lost ask_alaS. It is interesting that genomes in these two orders still retain the conserved gene cluster (including alaS), except that ask_alaS has either been deleted (Enterobacterales) or deleted in concert with replacement by another gene (Xanthomonadales). It is ironic that the best-studied organism, E. coli, belongs to an order that is the least typical of its phylum, since it has no Ask remnants of its vertical genealogy. The Enterobacterales (and other lower Gammaproteobacteria) have experienced a massive and complex replacement of Ask genes, as shown in Fig. 11, left, and detailed in “LGT” below.
Reductive losses occurring in Ask networks originally having multiple Ask enzymes.
Endosymbionts and pathogens often evolve reductively, with the loss of some or all of the branches of the ASK network. In endosymbiotic or pathogenic derivatives of enteric bacteria (which coordinate three specialized Ask paralogs in tune to the differential demands for threonine, methionine, and lysine end products), one might expect loss of a given pathway branch to be accompanied by loss of the corresponding paralog. As outlined below, recent events of reductive evolution do not always follow simplistic expectations.
The set of differentially regulated Ask enzymes (CG-04, CG-08, and CG-21) encoded by lysC, metL, and thrA, respectively, are generally present in those lower Gammaproteobacteria that have a complete ASK amino acid network. However, a variety of endosymbionts and pathogens rely upon their hosts as sources of lysine, methionine, and/or threonine. Alternatively, endosymbionts are known to be important sources of essential amino acids for their insect hosts (60). Buchnera aphidicola lacks metL, which is in accord with expectations raised by the absence of the methionine pathway branch. Since an intact lysine pathway is present, one might have expected LysC to have been retained. However, only ThrA has survived reductive evolution and thus must support both threonine and lysine biosynthesis. If the regulatory competence of ThrA is fully intact in B. aphidicola, it could tend to interfere with lysine provisioning to the host. Within the CG-04 cohesion group, the QXXSE…QXXSE motif of B. aphidicola is unusual in being imperfect (see individual CG alignments in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/CG04_aln.html), and it seems quite possible that threonine inhibition of Ask and especially homoserine dehydrogenase is nil or much weakened. Similar considerations apply to several Blochmannia spp. that have completely intact ASK pathways for the three amino acids but have nevertheless lost lysC and metL. The regulation of the remaining ThrA Ask by threonine appears to have been completely obliterated, judging from the substantial disruption of the QXXSE…QXXSE motif. Thus, the ThrA paralog has been selected for its relatively high expression level, and its allosteric control has been eliminated in order to feed all three branches for excess end product production in order to provision the carpenter ant host with these essential amino acids.
Wigglesworthia glossinidia, another endosymbiont living in tsetse fly hosts, represents an instance where both metL and lysC have been lost. A single amino acid branch has survived, and one might expect it to be the threonine branch. However, in this case, only the lysine pathway remains, and lysine biosynthesis therefore depends upon the thrA-encoded paralog of Ask. The Hdh domain of this bifunctional ThrA is still present, but in view of the threonine/methionine auxotrophy of W. glossinidia, the Hdh domain seemingly has no functional role to exercise. Indeed, a multiple alignment with various other homoserine dehydrogenases and the S. cerevisiae enzyme, whose structure has been documented by X-ray crystal studies (20), showed that nearly all of the highly conserved active-site residues and residues important for metal coordination have undergone mutational change (data not shown). Within the Ask domain, recognizable ACT domains are no longer present in the CDD, although sequence has been retained in that region. Thus, what was originally a threonine-regulated bifunctional Ask-Hdh has been conscripted to function as an unregulated monofunctional Ask for lysine production. Endosymbionts are well known to overproduce certain amino acids for the host organism. Significant overproduction of lysine probably occurs due to lack of feedback inhibition of ThrA by threonine, and ThrA might have been further selected (instead of LysC, the apparently logical choice) because ThrA is usually the most highly expressed of the three paralog Ask enzymes.
Comparison of different species of Haemophilus pathogens reveals surprising variations of reductive evolution. Haemophilus ducreyi exhibits an expected correlation of pathway loss and Ask homolog loss. It has retained only the lysine-sensitive Ask member of CG-21 and, accordingly, has retained only the lysine pathway. Haemophilus influenzae and Haemophilus somnus, on the other hand, have retained apparently intact ASK pathway branches to threonine, methionine, and lysine but have nevertheless lost both lysC and metL. If the allosteric regulation of ThrA is intact (as seems to be the case, judging from the similarity of the twin ACT regulatory region to other members of CG-04 known to be sensitive to feedback inhibition by threonine), threonine may interfere substantially with lysine and methionine biosynthesis. This may not matter if these species are in the process of losing the lysine and methionine branches anyway, similar to what has happened in H. ducreyi.
Reductive losses occurring in ASK networks supported by a single Ask enzyme.
In most Proteobacteria, a single Ask supports the entire ASK network, and regulatory mechanisms that can successfully sense and respond appropriately to multiple end products have been discussed. In these organisms, the loss of the only Ask species available is, of course, not an option in response to loss of one or more branches of the network. It is interesting to consider in these cases what adjustments might be made to the loss of any network branches.
Upper Gammaproteobacteria typically have a single Ask subject to concerted feedback inhibition by the threonine-lysine combination. C. burnetii and Dichelobacter nodosus possess single orphan Ask_AlaS enzymes that must be relevant only to DAP biosynthesis, since these organisms lack a capability for methionine, threonine, and lysine biosynthesis. The lysine auxotrophy is due to lack of a LysA connection from DAP (Fig. 1). One might expect the Ask enzymes from these two organisms to be feedback inhibited by DAP. Indeed, the Cbur_Aa enzyme is in the second group from the top in Fig. 10, where the DAP-inhibited enzymes of CG-24 are located. However, the Dnod_Aa enzyme is in the third group in Fig. 10, a group for which no pattern of feedback inhibition is predicted. Divergence of the Cbur_Aa and Dnod_Aa sequences to orphan status may be associated with a change to specialization for DAP biosynthesis. The C. burnetii Ask exhibits an apparent lack of an internal start site (Fig. 12). L. pneumophila, which belongs to the same order as C. burnetii (Fig. 11), is also auxotrophic for methionine and threonine but does support lysine (DAP) biosynthesis. In this case, ask_alaS has been lost and replaced via LGT (see below) with an Ask gene fused to the gene encoding LysA, the last gene of lysine biosynthesis. The Legionella Ask sequence belongs to CG-09 and has the signature motif for lysine allostery (Fig. 8A).
Provided that a number of LGT events are excluded, the overall divergence of Alphaproteobacteria Ask enzymes into a few cohesion groups and an orphan exactly parallels the organismal tree of Wu and Eisen (88). Most Alphaproteobacteria genomes possess a single Ask belonging to CG-37 that supports a complete ASK network to threonine, methionine, and lysine. All Ask proteins of the Alphaproteobacteria (except for a few LGT arrivals and a few CG-02 Ask_ect Ask enzymes) have the cd signatures that typify the ASKβ division of Ask enzymes: cd04261 for the aspartate kinase domain and cd04913-cd04936 for the tandem ACT domains. This cd signature in almost all of the Alphaproteobacteria within CG-37 may be correlated with a generally present allosteric pattern whereby a single Ask is synergistically inhibited by the combination Thr plus Lys. Although the enzyme of Rhodobacter sphaeroides has instead been found to be inhibited only by aspartate semialdehyde, both threonine and lysine were found to bind the Ask enzyme as a protectant (19). Consistent with a generally present Thr-plus-Lys pattern of concerted feedback inhibition in all CG-37 enzymes is the conformation of all 34 members of CG-37 with the pattern of color-coded residues illustrated in Fig. 10, top. It is interesting that this is true even of Bartonella quintana, a human pathogen that is reductively derived from Bartonella henselae (1). B. quintana, alone among all of the Alphaproteobacteria having a CG-37 Ask, has lost the threonine/methionine section of the ASK network. It should be sufficient for the B. quintana Ask to be feedback inhibited by lysine, since Ask no longer has a role in threonine biosynthesis. There may be little or no selection against a more complex pattern of feedback inhibition than is necessary, and loss of threonine binding by Ask may be essentially neutral. Probably the reductive loss of threonine/methionine biosynthesis has occurred so recently that loss of threonine binding has yet to occur.
Those bacteria that belong to the families Anaplasmatacea and Rickettsiaceae within the order Rickettsiales of the Alphaproteobacteria are represented by 11 sequences that belong to CG-38. These organisms have lost the methionine and threonine branches of the ASK network. Thus, there is a correlation between divergence to CG-38 and reductive evolution to loss of threonine and methionine biosynthesis. In addition, all organisms having Ask enzymes in CG-38 have lost competence for lysine biosynthesis, since LysA is absent (except in the three Ehrlichia spp.). As in the cases discussed above, it seems likely that the solitary Ask of CG-38 organisms may have altered allostery so that DAP alone is an adequate allosteric effector (or perhaps lysine alone in the case of Ehrlichia). This is consistent with the observation that the ACT domains of many of the CG-38 sequences are not recognized in the CDD as specific hits. “Candidatus Pelagibacter ubique” is unique among the Rickettsiales in having a single orphan sequence that does not belong to CG-38. Since “Candidatus Pelagibacter ubique” is exceptional among the Rickettsiales in having an intact threonine/methionine/lysine network, its single orphan Ask probably reflects ancestral properties that were otherwise altered by rapid reductive evolution in the pathogenic CG-38 clade. “Candidatus Pelagibacter ubique” also differs from all of the other Alphaproteobacteria in having an Ask that appears to lack an internal translation start site (Fig. 12).
LGT
The Cohesion Group Approach to LGT Detection
Since membership in a cohesion group is constrained by a limited range of divergence, the protein members of the group are expected to be phylogenetically congruent orthologs (and perhaps one or more recent sets of paralogs). The phylogenetic breadth of the organisms associated with a given cohesion group varies considerably depending upon the pace of evolutionary change. The presence in the cohesion group of any sequence(s) that is incongruent with phylogenetic expections indicates that the organismal host of the sequence received it via LGT from a donor organism that belongs to the lineage that is defined by the non-LGT members of the cohesion group.
Below, ask genes obtained by LGT at various levels of certainty are discussed at length. ask genes that we consider to be of certain LGT origin are enumerated fully in Table 3 in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/LGT.html. This sortable table is linked to the dynamic table (http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html) through clickable links in the “Acronyms” column. This allows rapid and convenient navigation to any bioinformatics information desired for a given ask xenolog. To illustrate the functionality of the online Table 3, a highly condensed version is included in Table 3 here. It includes one example from each cohesion group that contains xenolog intruders, as well as the three orphan intruders.
High Density and Even Spacing of Genome Representation Assist Evolutionary Deduction
If phylogenetic space is represented with a well-spaced continuity of genomes, then ideal information is available to trace both vertical and lateral evolutionary progressions. Density of genome representation becomes increasingly useful in cases where a dynamic pace of evolutionary change has occurred. Two phylogenetic groupings of particular interest because of frequent LGT exchanges have been the beneficiaries of sequencing choices that have resulted in the availability of a relatively high density of sequenced genomes. Figure 13 shows a detailed analysis of evolutionary events that are proposed for Ask divergence in the superphylum that includes the phyla Bacteroidetes and Chlorobi. For the second grouping, Fig. 11 shows a comparable analysis of evolutionary events that are proposed for Ask divergence in the phylum Proteobacteria. Although both Fig. 11 and 13 were discussed above in the context of vertical evolutionary progressions, these figures also summarize LGT events. Instances where members of the superphylum Bacteroidetes-Chlorobi were LGT donors are included in Fig. 13, whereas occasions on which Proteobacteria were LGT recipients are included in Fig. 11. These companion figures overlap to the extent that the phylum Bacteroidetes was a conspicuous LGT donor for the Proteobacteria, particularly for the lower-Gammaproteobacteria component.
Sorting the Mix between LGT Acquisitions and Evolutionary Events in the Vertical Genealogy of Proteobacteria
An effort is made in Fig. 11 to summarize the entire history of Ask evolution in the Proteobacteria. Evolution in the vertical genealogy is represented in the two right-hand columns, whereas LGT is shown on the left. The visual effect may give the impression that LGT is more frequent than it is, because each individual LGT event is shown. For example, 48 genomes of Alphaproteobacteria are included in our analysis, but only 2 closely related genomes exhibit LGT for ask genes (Fig. 11, third row). The common ancestor of O. alexandrii and M. maris received an ask-hdh fusion (belonging to CG-52) and an ask-lysA fusion (belonging to CG-09), possibly as a single LGT event (see below). These two organisms have retained a third Ask, which is the monofunctional ancestral enzyme (belonging to CG-51). An overwhelming majority (42) of the Alphaproteobacteria genomes possess only the single ancestral Ask. Four additional genomes possess the ancestral ask gene as well, but an additional specialized ask_ect gene is also present. The ask_ect genes are displayed in the vertical genealogy in Fig. 12 as a conservative choice, but the uncertain possibility that CG-02 Ask enzymes originated in Proteobacteria via LGT is considered below. If so, such an origin was sufficiently ancient that they have since endured within the vertical genealogy for most of the history of the Proteobacteria. Note that CG-08 Ask enzymes on the lower left are displayed with vertical hatching. This is technically incorrect, because the original CG-04 and CG-08 genes had exactly the same history in their common ancestor following LGT acquisition until the time of gene duplication that produced two paralogs. In Fig. 11, the vertical hatching is intended to convey the fact that MetL evolved rapidly in the vertical genealogy in acquiring a new functional specialization, whereas the ThrA paralog retained its previous functional role in threonine biosynthesis.
LGT can potentially present substantial hurdles to the goal of tracing the vertical genealogy of Ask enzymes. Two orders in the Gammaproteobacteria have lost the ancestral Ask entirely in favor of LGT replacement (Fig. 11), and therefore, the record of the ask vertical genealogy has been obliterated in these 21 genomes. First, within the upper Gammaproteobacteria, Xanthomonadales (four genomes) possess ask-hdh and ask-lysA fusions of LGT origin belonging to CG-07 and CG-09, respectively. Second, within the lower Gammaproteobacteria, the 17 genomes of Enterobacterales typically possess a threonine-specialized ask-hdh fusion obtained by LGT from a CG-07 source (which subsequently diverged in the recipient lineage to become CG-04 members), they possess a lysine-specialized ask obtained via LGT from a CG-11 source (which subsequently diverged in the recipient lineage to become CG-21 members), and they possess a methionine-specialized paralog (CG-08 members) derived from the CG-04 xenolog following gene duplication, as detailed below.
Given the adequate breadth of genome sampling available for Proteobacteria, the ask replacements in Xanthomonadales and Enterobacterales can be appreciated as intriguing and important events without masking the correct overall scenario. Thus, the general persistence of the primary ancestral Ask throughout the Proteobacteria is quite apparent when the entire phylum is examined en bloc, as summarized in Fig. 11. It is clear that in the upper Gammaproteobacteria, all of the various orders, other than Xanthomonadales, possess a single ancestral Ask with the cd04246 signature (and occasionally also possess a second Ask_ect Ask). Even in the lower Gammaproteobacteria, where every order typically possesses the LGT-derived trio of ASKα Ask enzymes (described above for the Enterobacterales) that are otherwise alien to the Proteobacteria, the ancestral Ask is quite apparent in a larger context. Thus, the three other orders of lower Gammaproteobacteria have so far retained the ancestral ASKβ Ask gene, in addition to the newcomer ASKα genes.
Epic LGT acquisition of three specialized ASKα genes in lower Gammaproteobacteria.
Members of the Epsilonproteobacteria, Deltaproteobacteria, Alphaproteobacteria, Betaproteobacteria, and the upper Gammaproteobacteria usually have solitary ASKβ Ask enzymes with the cd04246 signature, and it is only the occasional genome or small clade of organisms that has received one or more xenolog ask genes. In contrast, the lower-Gammaproteobacteria taxon is distinctive in having undergone a relatively ancient importation via LGT of two genes encoding an ASKα type of Ask, followed by duplication of one of them. This must have occurred near the time of divergence of lower Gammaproteobacteria from upper Gammaproteobacteria, since all lower Gammaproteobacteria possess the full alien gene set, except for occasional cases of subsequent gene losses in endosymbionts or pathogens.
ThrA of lower Gammaproteobacteria is a bifunctional Ask-Hdh belonging to CG-04. CG-04 Ask enzymes originated in lower Gammaproteobacteria from an LGT donor related to S. ruber (which so far can be pinpointed only somewhere in the order Sphingobacteriales). The latter possesses a bifunctional Ask-Hdh protein belonging to CG-07. Members of CG-04, CG-07, and CG-05 are all very similar bifunctional proteins that are specialized for threonine biosynthesis based on the criteria of allostery and gene context (Fig. 8A and B). The relationship between these cohesion groups is that CG-05 is the ancestral and generally present threonine-regulated Ask of the superphylum Bacteroidetes-Chlorobi (Fig. 13). CG-07 contains an Ask member from S. ruber that has diverged from those in CG-05 at a time that, due to lack of sufficient genome representation, can only be stated to be sometime after the divergence of the two different families to which C. hutchinsonii and S. ruber belong (Fig. 13). Members of CG-04 are most closely related to members of CG-07, and the LGT ancestor of CG-04 Ask enzymes diverged to form a new cohesion group in adjustment to the new host lineage.
The second ASKα Ask in lower Gammaproteobacteria is MetL, a bifunctional Ask-Hdh that is specialized for methionine biosynthesis and members of which belong to CG-08. This enzyme has quite reasonably been considered to have originated by gene duplication of the bifunctional thrA (28). We now can appreciate a slight variation of this in that the original incoming ask-hdh xenolog from the phylum Bacteroidetes underwent a gene duplication, with one copy remaining true to its various properties of threonine specialization whereas the other copy diverged dramatically with subsequent alteration of allostery, migration away from the threonine gene context, and acquisition of a new gene context near a methionine repressor gene. Thus, the first paralog is ThrA, and the second paralog is MetL. Lack of inhibition of MetL is accompanied by lack of a cd signature assignment for the ACT domain region (although sequence is retained, undoubtedly for structural reasons).
The third alien ASKα (LysC) of lower Gammaproteobacteria is a monofunctional Ask belonging to CG-21. Members of CG-21, CG-10, and CG-11 comprise a set of closely related ASKα Ask enzymes that are lysine regulated based on the criteria of allostery and gene context (Fig. 8A and B). The relationship between these cohesion groups is that members of CG-10 and CG-11 diverged from a common ancestor at the time of divergence of the phyla Bacteroidetes and Chlorobi (Fig. 13). Members of CG-21 originated in a common ancestor of lower Gammaproteobacteria via LGT from a donor member of CG-11. In ameliorative adaptation to the new host lineage, the ancestor of CG-21 diverged to form a new cohesion group.
As is the case for many gene-enzyme relationships, E. coli has been one of the major organismal subjects in the literature for the study of Ask enzymes. Since it belongs to the order Enterobacterales, it is striking to consider that the three Ask genes (thrA, metL, and lysC), received via LGT, do not reflect the properties that generally progressed in the Proteobacteria lineage. Rather, they are reflective of properties that originally evolved in the phylum Bacteroidetes. Cassan et al. (9) were correct in their conclusion many years ago that the three isofunctional Ask enzymes of E. coli had a common ancestor. However, the common ancestor with respect to all three was in the phylum Bacteroidetes, rather than in the Gammaproteobacteria phylum. On the other hand, the most recent common ancestor with respect to the thrA metL pair was within the lower Gammaproteobacteria lineage, since the gene duplication generating them occurred there. All organisms are mosaics in that the history of some fraction of the genes is not the same as the history of the organism. Compared to the prevailing outlook prior to genome sequencing, it is quite incredible to consider that this history can be disambiguated.
(i) Paralog, ortholog, and xenolog relationships summarized.
The functional counterparts of E. coli thrA and lysC (thrAEC and lysCEC) in the superphylum Bacteroidetes-Chlorobi (thrAB/C and lysCB/C) were very ancient paralogs from which ortholog pairs were successfully maintained throughout the lineage. The relationship between thrAEC and metLEC is one of relatively recent paralogy, whereas each of them is an ancient paralog of lysCEC. Put another way, thrAEC and metLEC can be stated to be coparalogs of lysCEC. thrAEC and metLEC were equivalent at the time of duplicative origin, but their subsequent evolutionary progressions were quite different. thrAEC has remained true to its functional role, gene context, and allostery, whereas metLEC acquired a new gene context, new regulation, and a new functional role. lysCEC and lysCB/C are xenologs; thrAEC and metLEC are coxenologs of thrAB/C. The overall outcome is a three-member set of ask orthologs of xenologous origin that is distributed throughout the lower Gammaproteobacteria. These homology relationships are diagrammed in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/figure-2S.html (Fig. 2S), and a succinct presentation of the Fitch homology rules given by Jensen (40) might be helpful.
Miscellaneous LGT events elsewhere in the Proteobacteria. (i) F. tularensis.
The order Thiotrichales of the upper Gammaproteobacteria currently is represented by only two genomes. That of Thiomicrospira crunogena is straightforward, having the ancestral genes encoding Ask_alaS and Ask_ect. In contrast, the genome of F. tularensis has not only lost the ancestral Ask(s) but maintains two ask genes obtained by LGT import (Fig. 11). One of them is a bifunctional threonine-regulated Ask-Hdh and belongs to CG-07, the LGT donor being within the lineage of S. ruber (Sphingobacteriales). The second is a lysine-regulated monofunctional Ask and belongs to CG-21, the LGT donor being a member of the lower Gammaproteobacteria. The latter xenolog appears to have arrived very recently in the common ancestor of F. tularensis and Francisella philomiragia (ATCC 25017). A presumed gene duplication in F. philomiragia has generated a second paralog, which no longer has recognizable ACT domains. Perhaps this paralog has become specialized for methionine responsivity. F. tularensis appears to be very unstable with respect to the gene encoding the CG-21 Ask. Although the strain selected (F. tularensis subsp. novicida U112) in our dynamic table (http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html) has an intact CG-21 Ask, seven other strains with complete genomes in the database have discarded this gene.
(ii) B. bacteriovorus.
Bdellovibrio bacteriovorus, a member of the Deltaproteobacteria, also received an ask xenolog from a lower Gammaproteobacteria donor belonging to CG-21 (Fig. 11). B. bacteriovorus lacks intact pathways for methionine and threonine. By default, it seems likely that the imported xenolog has retained the responsivity to regulation by lysine that is typical of CG-21 enzymes. In addition, B. bacteriovorus has thus far retained its ancestral cd04246 ask gene, one that encodes an Ask enzyme belonging to CG-27 (populated by two other Ask enzymes from Deltaproteobacteria). The twin ACT domain region of CG-27 Ask enzymes yields no recognizable cd hits from the CDD.
(iii) Xanthomonadales.
Xanthomonadales organisms have replaced their ancestral ask genes with two bifunctional ask genes (ask-hdh and ask-lysA) from a Bacteroidetes donor, as discussed fully below.
(iv) Hyphomonadaceae.
A common ancestor of M. maris and O. alexandrii (belonging to genera within the Hyphomonadaceae of Alphaproteobacteria) received exactly the same two xenolog genes (ask-hdh and ask-lysA), one diverging from members of CG-07 to CG-52 and the other remaining in CG-09 (Fig. 11). In this case, the ancestral ask genes in CG-51 have been retained.
(v) M. xanthus.
Myxococcus xanthus (Deltaproteobacteria) possesses two ask-hdh gene fusions, which, like all ask-hdh fusions, belong to the ASKα subhomology grouping. Mxan_Ac and Mxan_Ad are orphan sequences that are proposed in Fig. 11 to have originated by LGT. Mxan_Ac is related to the Ask-Hdh fusions, which comprise CG-04 from the lower Gammaproteobacteria. It possesses the Q…Q motif in the twin ACT region, which indicates that it is subject to feedback inhibition by threonine. Unlike members of CG-04, however, it is not adjacent to genes encoding other threonine branch enzymes. Mxan_Ad has a disrupted Q…Q motif in the twin ACT region (Fig. 8A), which indicates it probably is not feedback inhibited by threonine. Indeed, specialization for methionine biosynthesis is indicated because the encoding gene colocalizes with genes of methionine biosynthesis. The LGT donor of Mxan_Ad is from CG-05 of the Flavobacteria (Fig. 11). The membership of CG-05 usually has a robust Q…Q motif region indicating threonine specialization. However, five of six genera within the Flavobacteriaceae have a common ancestral gene duplication that generated two CG-05 paralogs, one of which evolved a disrupted Q…Q region in combination with its translocation to a methionine gene context (Fig. 13). One of these paralog pairs from F. johnsoniae is included in the alignment in Fig. 9, where Fjoh_Ac is the canonical threonine-regulated member and Fjoh_Ab is the newly evolved methionine-specialized Ask member. A priori, Mxan_Ac and Mxan_Ad could be paralogs comparable to Fjoh_Ac and Fjoh_Ab, i.e., a gene duplication occurring in Myxococcus with Mxan_Ac remaining true to the ancestral threonine allostery and Mxan_Ad diverging to methionine specialization. However, Mxan_Ad clearly exhibits sequence similarity to the methionine-specialized Ask enzymes present in most Flavobacteriaceae and was therefore obtained from that lineage by LGT. In this connection, it is interesting that Fjoh_Ab is the outlying member of CG-05 and the Mxan_Ad orphan is most closely related to Fjoh_Ab. This relationship can be seen in Fig. 4B, left. (Note that Ask sequences from F. johnsoniae and G. forsetii were the only representatives of the family Flavobacteriaceae in our initial assemblage of genomes screened. However, close counterparts of Fjoh_Ab [namely, a newly generated methionine-specialized paralog] are found in other genera in the family, such as Polaribacter, Kordia, Tenacibaculum, and Robiginitalea.)
Piggyback LGT.
There are a few instances where it has been possible to deduce a recent LGT event superimposed upon a more ancient LGT event (“piggyback” LGT), i.e., where an initial recipient lineage has in turn been an LGT donor for a second recipient (Fig. 11). In the examples enumerated above, a CG-21 member of the lower Gammaproteobacteria was an LGT donor for the gene encoding the Bbac_Ab Ask of B. bacteriovorus, and a CG-21 member of the lower Gammaproteobacteria was also an LGT donor for the gene encoding the Ftul_Aa Ask of F. tularensis. Since the ancestor of the CG-21 members was itself obtained by LGT from a Bacteroidetes donor gene encoding a CG-11 Ask, Bbac_Ab and Ftul_Aa can be considered to be encoded by alien genes derived from the Bacteroidetes lineage, followed by an intermediate history in a second lineage before contributing to the mosaicism of the current host.
A CG-04 member of the lower Gammaproteobacteria was an LGT donor for the gene encoding the Mxan_Ac orphan Ask of M. xanthus (Deltaproteobacteria). Since the ancestor of the CG-04 members was itself obtained by LGT from a Bacteroidetes donor gene encoding a CG-07 Ask-Hdh, the Mxan_Ac orphan can be considered to be derived from the Bacteroidetes lineage.
Other Cases where the LGT Donor Can Be Deduced
Challenging instances where the CG membership is derived from mixed phylogenies. (i) Within CG-09.
Eight bifunctional proteins that contain an N-terminal Ask domain fused to a LysA domain make up the complete membership of CG-09, and this domain fusion combination is not thus far represented within any other CG grouping. The subhierarchical cd signatures for both the Ask domain and the twin ACT domains are absolutely unique to this fusion group. The Ask domain carries the cd identifier cd04259, and the tandem ACT domains have the signature cd0435-cd04920. The erratic phylogenetic relationships of the organisms possessing these eight sequences suggest LGT. Among these, the S. ruber lineage can be identified with confidence as the donor lineage for two reasons. (i) When the sequences of the two domain segments of each bifunctional protein were separated and used as queries for BLAST analysis of the monofunctional counterparts, the best hits were all monofunctional proteins from the phylum Bacteroidetes. This indicates that the fusion originated somewhere within the lineage that includes S. ruber. (ii) Additional compelling support is provided by the fact that the other phylum Bacteroidetes members, which all lack the fusion, possess monofunctional Ask enzymes (in CG-11) that are adjacent to LysA. Presumably, a near ancestor of S. ruber possessed the side-by-side genes and fused them, subsequently diverging to become a defining member of CG-09. No genomes are available that are even moderately related to that of S. ruber, the closest being at the taxon level of order. It is quite possible that when additional genomes become available, multiple taxa at the level of family might prove to possess the fusion.
The CG-09 fusion has been passed by LGT to a common ancestor of the order Xanthomonadales of the upper Gammaproteobacteria, which includes the genera Xanthomonas, Xylella, and Stenotrophomonas (Fig. 11 and Fig. 13). An independent LGT recipient was the common ancestor of Maricaulis and Oceanicaulis, the sole members found to have the fusion within the large group of Alphaproteobacteria included in our analysis. This LGT event must have been particularly recent, and it is quite possible that the donor genome was within the order Xanthomonadales rather than directly from the S. ruber lineage (in which case this would be an instance of piggyback LGT). Yet another independent and very recent LGT of the CG-09 fusion was to Legionella spp., and it is also possible that this might have been a piggyback LGT.
(ii) Within CG-07 (and CG-52).
Members of CG-07 comprise one of the cohesion groups populated by Ask-Hdh fusions. This fusion group has been suggested to have diverged in the S. ruber lineage from the Ask-Hdh fusions that are present in CG-05 in all other Bacteroidetes and Chlorobi included in this study (Fig. 13). Aside from the S. ruber sequence, other members of CG-07 are two paralogs from A. thaliana (higher plant) and from genera of the upper Gammaproteobacteria: Franciscella and the Xanthomonadales genera (Xanthomonas, Xylella, and Stenotrophomonas). Franciscella and the three Xanthomonadales genera are not typical of the upper Gammaproteobacteria, whose single ask gene in the vertical genealogy faithfully exhibits the ask_alaS gene neighborhood (Fig. 11). If the ask-hdh gene fusion originated by LGT in these upper Gammaproteobacteria, the attached Hdh domain should be distinctly different from the Hdh domains of other upper Gammaproteobacteria. Figure 15 clearly illustrates that this is so. Other than those having CG-07 fusions, upper Gammaproteobacteria possess Hdh-thr, which has a C-terminal ACT domain. Even an Hdh as distant as that of a Betaproteobacteria organism (Nitrosomonas europaea) clusters with the Hdh-thr sequences at the top of Fig. 15. The Hdh-min sequences from various organisms that have ASKα Ask enzymes cluster separately at the bottom of the figure. They include the well-studied yeast (S. cerevisiae) enzyme and the CG-05 Ask-Hdh fusions (represented by the B. fragilis enzyme). The Hdh domains of the CG-07 fusions (represented by the Xylella, Salinibacter, and Franciscella enzymes) and the CG-04 fusions (represented by the Pseudoalteromonas atlantica enzyme) are expected to cluster with CG-05 enzymes, since CG-07 enzymes diverged from a CG-05 ancestor and CG-04 enzymes are derived from a CG-07 ancestor via LGT.
FIG. 15.
Phylogenetic-tree positions of Hdh from Xylella, Xanthomonas, and Franciscella imply an LGT origin from a Bacteroidetes donor. Hdh sequences from organisms with ASKβ Ask enzymes, which include upper-Gammaproteobacteria Hdh enzymes (shown in yellow), typically are elongated (amino acid lengths are given) due to the presence of a C-terminal ACT domain, which is responsible for sensitivity to threonine-mediated inhibition. This Hdh species is denoted Hdh-thr. On a phylogenetic tree, Hdh-thr sequences form a compact cluster that includes many other Proteobacteria, e.g., the Hdh from N. europaea (Neur Hdh), which is a member of the Betaproteobacteria, is shown in the upper cluster. On the other hand, organisms with ASKα Ask enzymes have shorter Hdh sequences (Hdh-min) because they lack an allosteric domain, and these cluster separately in the bottom cluster of the tree. They include the Hdh-min species from S. cerevisiae (Scer Hdh), the Hdh domain from a CG-04 Ask-Hdh fusion, and Hdh from various Bacteroidetes (shown in red). In the bottom cluster of the tree, Hdh domains from three members of the upper Gammaproteobacteria (shown in blue) are distinctly separated from the Hdh domains of other upper Gammaproteobacteria (top) and cluster instead with those from the Bacteroidetes, the proposed LGT donor lineage. The Hdh sequences from G. forsetii (Gfor Hdh) and F. johnsoniae (Fjoh Hdh) are monofunctional enzymes that were generated by scission of a duplicated CG-05 ask-hdh to yield a monofunctional Ask and a monofunctional Hdh, as indicated by step D in Fig. 13.
As with CG-05 members, the ask-hdh gene of CG-07 is typically part of a threonine operon that includes thrB and thrC. (Arabidopsis is an exception, of course, because higher plants lack operonic gene organizations.) This raises the question of whether an entire thrABC operon might have been transferred to the bacterial recipients. Indeed, BLAST evaluations of ThrB and ThrC from the above-mentioned upper Gammaproteobacteria indicate that they are more similar to one another (and to the ThrB and ThrC enzymes of S. ruber) than they are to those from other upper Gammaproteobacteria. It is interesting that the homology group defined by ThrB and ThrC also includes the enzymes from M. maris and O. alexandrii. These two genera are so far unique among Alphaproteobacteria in having an ask-hdh fusion that occupies an operon along with thrB and thrC. Thus, it appears that the entire thrA-thrB-thrC operon has been passed via LGT from the S. ruber lineage to the common ancestor of the above-cited upper Gammaproteobacteria, as well as to the common ancestor of Maricaulis and Oceanicaulis. Unlike ThrB and ThrC, the Ask-Hdh fusion has diverged to such an extent in Maricaulis and Oceanicaulis as to be assigned to a different cohesion group (CG-52). Two striking features of variation in the two CG-52 members may account for divergence to form a new cohesion group. One is the reductive loss of the twin ACT domain region (Fig. 8A), and the other is deletion (Fig. 5) of the region referred to as the indel region in Fig. 6. The CG-07 cluster is very compact, suggesting that all of these are derived from a common source at a fairly recent time. Although thrA and thrB are unlinked in Arabidopsis and other higher plants, both thrA and thrB (but not thrC) were obtained in plants by LGT.
(iii) The extensive LGT radiation of the Bacteroidetes ask-hdh-thrB-thrC operon summarized.
The ask-hdh-thrB-thrC operon originated in the Bacteroidetes at about the time of its divergence from the phylum Chlorobi. The phylum component ask-hdh fusion must have occurred much earlier in the common ancestor of Chlorobi and Bacteroidetes, since CG-05 is populated by Ask-Hdh enzymes from both phyla. It appears that the entire ask-hdh-thrB-thrC operon of Bacteroidetes (the S. ruber genome being the closest current relative) was transferred to CG-04 members (lower Gammaproteobacteria), to CG-07 members (several orders of upper Gammaproteobacteria), and to CG-52 members (one isolated family of Alphaproteobacteria). Thus, the Ask-Hdh-ThrB-ThrC gene products of the threonine operon from E. coli (and the many other members of the lower Gammaproteobacteria), Franciscella, Xylella, Xanthomonas, Stenotrophomonas, Maricaulis, and Oceanicaulis are alien newcomers that do not parallel the organismal phylogeny. In addition, it can be deduced that Legionella spp. originally received the operon but, as a consequence of their evolved reductive auxotrophy, have retained only a thrC remnant. Although higher plants do not have operons, both ask-hdh and thrB were obtained from a Bacteroidetes donor.
(iv) Were the ask-hdh-thrB-thrC operon and ask-lysA transferred via a single LGT event?
As discussed above, the ask-hdh-thrB-thrC operon was obtained by recipient organisms having members belonging to CG-07 and CG-52, and ask-lysA was obtained by recipient organisms having members belonging to CG-09. In both cases, the LGT donor was closely related to S. ruber (Fig. 13). Exactly the same combination of genes was obtained by two LGT recipients: the common ancestor of the order Xanthomonadales and the common ancestor of the genera Maricaulis and Oceanicaulis. It seems unlikely that the same combination of LGT events would occur in both sets of organisms if the same two independent LGT events were required in each case. The most parsimonious explanation is that ask-hdh-thrB-thrC and ask-lysA were adjacent or at least near one another in the donor genome. If so, joint transfer would be easier to envision if the two gene areas were closely linked in the donor organism, which they are not in the contemporary S. ruber. In fact, even thrC has become separated from the rest of the thr operon in S. ruber. However, given the frequency of gene scrambling, it is quite possible that at the ancestral time when the common ancestor of the above-mentioned upper Gammaproteobacteria existed, the donor gene sets were at least relatively closely placed. It is also possible that thus far unsequenced relatives of S. ruber that did not experience the putative gene scrambling exist.
The above joint-transfer hypothesis must take into account a small list of organisms that have either the ask-hdh-thrB-thrC operon or the ask-lysA fusion but not both. All organisms that possess the ask-lysA fusion also possess the ask-hdh-thrB-thrC operon, except for Legionella spp. However, the ask-hdh-thrB-thrC operon could have been lost via reductive evolution in Legionella at a time after LGT acquisition, since intact branches of methionine and threonine biosynthesis are absent in this pathogen. Indeed, the latter is strongly indicated because L. pneumophila strains possess a ThrC remnant whose sequence clusters closely with the ThrC enzymes present in the organisms having the CG-07 Ask enzymes. Franciscella spp. received the ask-hdh-thrB-thrC operon, but not the ask-lysA fusion. Even here, it is possible that the ask-lysA fusion was initially obtained but then displaced by another lysine-specialized Ask that arrived via LGT from lower Gammaproteobacteria (Fig. 11). Of course, just because the ask-hdh-thrB-thrC operon and the ask-lysA fusion were adjacent, if they were, this does not necessarily mean that the entire region was successfully transferred to or fully retained by a Franciscella recipient.
(v) CG-19.
The CG-19 cohesion group has only two members. Although both ask members are produced by Archaea, the organisms are members of different phyla. It seems clear that this represents a case of LGT, but there are no absolute indicators pointing to which of the two is the xenolog intruder, since the genomic representation here is currently sparse. However, since the Archaea almost always possess a single ask gene and since M. kandleri possesses two paralogs, the most parsimonious inference is that the M. kandleri ask gene in CG-19 is a xenolog derived from a close relative of A. pernix.
Where the LGT Donor Is Uncertain
In some cases, there may be feasible evidence that points to an LGT origin, but the likely LGT donor is uncertain because sufficiently close relatives of the donor genome have not yet been sequenced. This will increasingly be rectified as large gaps in genome representation are filled via modern genome-sequencing projects. In other cases, there may be a basis to suspect LGT involvement, but it may be difficult to decide which member or group of members is the donor and which member or group of members is the recipient. Until an LGT donor is identified, the assertions of LGT listed below must be considered tentative.
CG-02.
CG-02 members are ASKα division Ask enzymes that are components of an ectoine operon, with the exception of a few cases where ectoine genes have been discarded, leaving an ask_ect remnant behind. They are specific to the phylum Proteobacteria. Since this phylum appears to have originally been populated with ASKβ Ask enzymes, CG-02 members might have arrived via LGT as a very ancient event soon after the divergence of the phylum. CG-02 members clearly arose from the lysine signature (cd04890-cd04892) group of ASKα Ask enzymes (Fig. 8A). However, at the present time, there are no genomes that possess an ask_ect gene that are not members of the Proteobacteria. Thus, no donor lineages can yet be implicated.
CG-03.
CG-03 presents exactly the same situation as does CG-02. Its members belong to the ASKα division of Ask enzymes, and they are uniquely present in the phylum Firmicutes. They are also derived from the lysine signature (cd04890-cd04892) group (Fig. 8A), and at least some members are known to be subject to allosteric control by a synergistic combination of Thr plus Lys. This phylum probably was originally populated with ASKβ Ask enzymes (which still exist) and received the CG-03 Ask gene via LGT. As with CG-02 members, there are no current members of CG-03 that are not in Firmicutes organisms, and therefore, no potential LGT donors can be implicated. It must be conceded that an alternative scenario is plausible. Perhaps the CG-03 type of Ask predated the evolution of endospore differentiation in the Firmicutes. In the absence of this complex developmental state, a single Ask species subject to allostery by Thr plus Lys would present an effective control mechanism (as indeed is the case in many contemporary organisms). With the acquisitive evolution of endospore formation, perhaps the various ACTβ genes present in contemporary Firmicutes were imported via LGT.
CG-33 and Bacillus-Listeria.
CG-33 contains a relatively small number of sequences from the Firmicutes (Table 2). They include three Listeria spp. that belong to the family Listeriaceae and six Bacillus spp. that belong to the family Bacillaceae. However, other genera of Bacillaceae (Oceanobacillus and Geobacillus) do not contribute any sequences to CG-33. This incongruence could be explained by LGT transfer between Bacillaceae and Listeraceae. It is quite possible, on the other hand, that Oceanobacillus and Geobacillus independently lost the CG-33 paralog. These losses could be explained parsimoniously as a single event if Oceanobacillus and Geobacillus share a common ancestor to the exclusion of other Bacillaceae genera.
Thermus.
T. thermophilus (and also Thermus aquaticus) possesses a single Ask orphan that belongs to the ASKβ subhomology division, whereas two Deinococcus spp. (CG-17) have Ask enzymes that belong to the ASKα subhomology grouping. These genera are members of the same phylum (Fig. 7), one in which lysine is generated via the alpha aminoadipate pathway (62). The positions of Ask proteins from the Deinococcus spp. in a protein tree meet phylogenetic expectations, whereas the positions of Ask proteins from Thermus spp. are incongruent with phylogenetic expectations (as previously observed [62]). Both Thermus and Deinococcus possess a single Hdh. In contrast to the difference observed for Ask, the two Hdh proteins are the closest BLAST hits (outside of their genus). Both Hdh enzymes lack an ACT domain (Hdh-min), as is typical of enzymes from ASKα organisms. This suggests that Thermus obtained an ask replacement from an ASKβ bacterium via LGT while retaining an Hdh species that is typical of ASKα organisms. The Ask donor is uncertain, but the closest BLAST hits point to the order Clostridiales within the phylum Firmicutes.
PERSPECTIVE
This article is cast in the spirit of the recent essay of Downs (22), which pursues the thesis that an integration of experimentally based information with bioinformatics information helps us to appreciate a metabolic network as something that is more than simply the sum of its atomistic parts. The cohesion group approach has been used to analyze Ask within the larger context of the ASK network. This can be viewed as a beginning effort from the vantage point that complementary analyses could be done using other key enzymes of the network as the focal point, e.g., with the homoserine dehydrogenase genes. Such projects could proceed with relative speed and efficiency because of the underlying groundwork provided by the Ask evaluation. Each analysis would reinforce, correct, and expand the existing concept of the ASK network. The consideration of how the network is regulated is an extremely important part of the current effort, but much more could be integrated with the current analysis, e.g., not only the evaluation of likely protein regulators, but also the progressively more sophisticated bioinformatics assessments of RNA regulation by riboswitches (65, 80) could be added to the scope of the existing workup with great benefit.
Studies of LGT have usually been carried out at global levels without much effort to evaluate which particular genes have moved from what particular donor organisms to what particular recipients. Implementation of the cohesion group approach shows how such LGT events can be cataloged in a systematic way. One can envision that multiple cohesion group studies that are focused upon different metabolic subsystems will eventually lead to a cumulative inventory of LGT genes that will describe the mosaic makeup of individual organisms and groups of organisms at various hierarchical levels.
Ask enzymes have been generally described as falling into one of two groups: either homo-oligomers or α2β2 heterotetramers. X-ray crystal studies have been carried out with several enzymes of each type, and interpretations have been advanced without benefit of the current realization that the two groups actually correspond to the ASKα and ASKβ subhomology divisions, which are quite distinct. The current analysis should greatly assist the rational selection of additional enzymes within each division in order to get a sense of what the consensus features of ASKα and ASKβ are. Ultimately, it will be possible to reconstruct the steps that led to the substantial divergence that currently exists.
Specific research initiatives that might have substantial merit have been brought into focus. They include the following. (i) Is it correct that a homoserine dehydrogenase (Hdh-met) species with a C-terminal extension that is distinct from the threonine-inhibited ACT domain of Hdh-thr is an enzyme specialized for methionine biosynthesis and perhaps subject to feedback inhibition by methionine? (ii) The indel region in ASKα Ask enzymes (which corresponds to a proposed deletion in ASKβ Ask enzymes) is persistently present and therefore must have functional importance. It contains no catalytic or substrate-binding residues. Is this a regulatory region, perhaps a region dictating protein-protein interactions with Hdh-min and/or perhaps interacting with the twin ACT regulatory domains? (iii) Is it correct that the cofactor-independent phosphoglycerate mutase homolog present in some organisms that lack thrB is an alternative ThrB as gene context analyses seem to suggest? (iv) Is the ectR gene that is divergently oriented to the ectoine operon in Gammaproteobacteria (and actually inside of the ect operon of Betaproteobacteria) a regulatory gene for ectoine biosynthesis, and if so, how does it work? (v) A free-standing ACT domain-containing protein is frequently adjacent to hdh-min in correlation with an absence of any obvious arrangements for regulation of lysine biosynthesis in two widely separated lineages. Is it correct that this ACT domain complexes with Hdh and interacts with lysine (in Clostridia) and with lysine plus at least one aromatic amino acid (in a large clade of Archaea) in a mode that blocks flux toward methionine and threonine? (vi) Is it correct that the functional roles of aroA′ and aroB′ in an organism like S. griseus are specialized for some secondary-metabolism role, such as antibiotic synthesis? (vii) What is the alternative pathway to dipicolinate that appears to exist in the Clostridiaceae?
SUPPLEMENTARY FILES
The supplementary files include (i) sequences (Fasta files partitioned into cohesion group and orphan files), (ii) trees available in Newick or PDF files (the original master Ask tree, the tree of cohesion group respresentative sequences/orphans depicted in Fig. 5, and separate trees of ASKα and ASKβ subhomology divisions that make up the Fig. 5 tree), (iii) the alignment of orphan sequences and representative members of each cohesion group used to obtain Fig. 5 and separate alignments of the individual cohesion groups, (iv) tables (links to the comprehensive dynamic table; to a scaled-down representative-sequences table; and to a dynamic, sortable, complete list of LGT genes [Table 3] that is linked to the dynamic table), (v) figures (Fig. 1S and Fig. 2S), and (vi) links to the SEED and AroPath home pages. These supplementary files can be accessed at http://www.theseed.org/Papers/MMBR-Aspartokinase/.
Acknowledgments
This article is dedicated to Georges Cohen, whose enduring scholarship initiated interest in the Ask network.
This work was supported by Contract HHSN266200400042C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services. We used the SEED environment (http://www.theseed.org/) (a comprehensive resource developed by the Fellowship for Interpretation of Genomes in collaboration with an international team of researchers, Argonne National Laboratory, and the National Microbial Pathogen Data Resource [NMPDR] [http://www.nmpdr.org/]) as our primary source of data and bioinformatics tools. Contract NIH NIDCR Y1-DE-6006-02 partially supported G. Xie and C.-C. Lo.
We thank the precocious undergraduate efforts of Emily Sayas for help with data analysis. We thank David E. Graham and Claude Parsot for a substantial input of constructive criticism. We appreciate the cooperation of Jian Song in developing some of the AroPath tools and for his various troubleshooting efforts.
Biography
 Chien-Chi Lo received a dual B.S. degree in public health and medical technology (2002) from the National Taiwan University and his M.S. degree in bioinformatics (2007) from the National Yang-Ming University, both in Taipei, Taiwan. During this time, he was engaged in research projects that required a combination of experimental and computational skills. Here, he developed a strong and continuing interest in how bioinformatics approaches can contribute insights into biological systems. He currently is a graduate research associate at Los Alamos National Laboratory (Los Alamos, NM) and a graduate student in the Department of Computer Science at the University of New Mexico (Albuquerque). His research interests are in bioinformatics, including sequence analysis, biological database development, computational biology, and programming.
Chien-Chi Lo received a dual B.S. degree in public health and medical technology (2002) from the National Taiwan University and his M.S. degree in bioinformatics (2007) from the National Yang-Ming University, both in Taipei, Taiwan. During this time, he was engaged in research projects that required a combination of experimental and computational skills. Here, he developed a strong and continuing interest in how bioinformatics approaches can contribute insights into biological systems. He currently is a graduate research associate at Los Alamos National Laboratory (Los Alamos, NM) and a graduate student in the Department of Computer Science at the University of New Mexico (Albuquerque). His research interests are in bioinformatics, including sequence analysis, biological database development, computational biology, and programming.
 Carol A. Bonner received a B.A. degree in biology at the State University of New York at Binghamton and her Ph.D. degree at the University of Florida (Gainesville). She managed a large program at the University of Florida, where she was in charge of a plant tissue culture laboratory and a microbiology laboratory. She directed laboratory studies focused upon the enzymology and regulation of aromatic amino acid biosynthesis in both higher plants and bacteria. Her present interest lies in bioinformatics analysis of biochemical pathways. She has been working as an annotator for the SEED database and is interested in helping apply the approaches described in this article to develop a semiautomated program that would facilitate rapid and efficient evaluation of the SEED subsystems.
Carol A. Bonner received a B.A. degree in biology at the State University of New York at Binghamton and her Ph.D. degree at the University of Florida (Gainesville). She managed a large program at the University of Florida, where she was in charge of a plant tissue culture laboratory and a microbiology laboratory. She directed laboratory studies focused upon the enzymology and regulation of aromatic amino acid biosynthesis in both higher plants and bacteria. Her present interest lies in bioinformatics analysis of biochemical pathways. She has been working as an annotator for the SEED database and is interested in helping apply the approaches described in this article to develop a semiautomated program that would facilitate rapid and efficient evaluation of the SEED subsystems.
 Gary Xie obtained his medical degree (B.M.) from Xianya School of Medicine, Central South University (formerly Hunan Medical University), in China (1992). He received both the M.S. (1998) and Ph.D. (2002) degrees from the University of Florida (Gainesville). His Ph.D. studies focused on the comparative genome analysis of aromatic amino acid biosynthesis. He did postdoctoral research on the human chromosome and bacterial genome annotation and analysis at JGI—Los Alamos (2002 to 2004). Subsequently, he was appointed as a Technical Staff Member of the Bioscience Division, Los Alamos National Laboratory. He currently is the Principal Investigator on a project to annotate and curate all oral pathogen genome sequences for the ORALGEN database. Most recently, he has been (i) working on a metagenome project involving sequenced environmental samples, (ii) developing bioinformatics applications for single-genome/metagenome annotation and analysis, and (iii) conducting comparative studies of human pathogens.
Gary Xie obtained his medical degree (B.M.) from Xianya School of Medicine, Central South University (formerly Hunan Medical University), in China (1992). He received both the M.S. (1998) and Ph.D. (2002) degrees from the University of Florida (Gainesville). His Ph.D. studies focused on the comparative genome analysis of aromatic amino acid biosynthesis. He did postdoctoral research on the human chromosome and bacterial genome annotation and analysis at JGI—Los Alamos (2002 to 2004). Subsequently, he was appointed as a Technical Staff Member of the Bioscience Division, Los Alamos National Laboratory. He currently is the Principal Investigator on a project to annotate and curate all oral pathogen genome sequences for the ORALGEN database. Most recently, he has been (i) working on a metagenome project involving sequenced environmental samples, (ii) developing bioinformatics applications for single-genome/metagenome annotation and analysis, and (iii) conducting comparative studies of human pathogens.
 Mark D'Souza received his B.Sc. degree from the University of Mumbai and an M.Sc. degree from the University of Poona in India. He obtained a Ph.D. degree in physics from the State University of New York at Albany and then switched fields, conducting postdoctoral research in bioinformatics at Argonne National Laboratory, where he worked on genome annotation through comparative genomics. After working as a bioinformatics scientist with the private company Integrated Genomics for some years, he is now employed as a software developer at the Computation Institute of the University of Chicago, working with the bioinformatics group in the Math and Computer Science Division at Argonne National Laboratory. He has worked on a number of genome annotation systems, including WIT2, Ergo, Puma2, and RAST, as well as the metagenome analysis system MG-RAST.
Mark D'Souza received his B.Sc. degree from the University of Mumbai and an M.Sc. degree from the University of Poona in India. He obtained a Ph.D. degree in physics from the State University of New York at Albany and then switched fields, conducting postdoctoral research in bioinformatics at Argonne National Laboratory, where he worked on genome annotation through comparative genomics. After working as a bioinformatics scientist with the private company Integrated Genomics for some years, he is now employed as a software developer at the Computation Institute of the University of Chicago, working with the bioinformatics group in the Math and Computer Science Division at Argonne National Laboratory. He has worked on a number of genome annotation systems, including WIT2, Ergo, Puma2, and RAST, as well as the metagenome analysis system MG-RAST.
 Roy A. Jensen received an undergraduate degree from Ripon College in Wisconsin and a Ph.D. degree from the University of Texas. Following postdoctoral studies at the University of Washington, he has held faculty positions at the State University of New York at Buffalo, Baylor College of Medicine, M. D. Anderson Hospital and Tumor Institute, the State University of New York at Binghamton, and the University of Florida, where he is currently Professor Emeritus. He had a strong interest in evolution before it became fashionable for biochemists and geneticists. This interest, dynamically stimulated by the advent of genome sequencing, has merged nicely in recent times with an enduring fascination with the alternative biochemical pathways that can lead to a given end product, the diversity of alternative patterns of regulation that exist to control them, and the complex relationships (metabolic interlock) between what are often considered unrelated pathways.
Roy A. Jensen received an undergraduate degree from Ripon College in Wisconsin and a Ph.D. degree from the University of Texas. Following postdoctoral studies at the University of Washington, he has held faculty positions at the State University of New York at Buffalo, Baylor College of Medicine, M. D. Anderson Hospital and Tumor Institute, the State University of New York at Binghamton, and the University of Florida, where he is currently Professor Emeritus. He had a strong interest in evolution before it became fashionable for biochemists and geneticists. This interest, dynamically stimulated by the advent of genome sequencing, has merged nicely in recent times with an enduring fascination with the alternative biochemical pathways that can lead to a given end product, the diversity of alternative patterns of regulation that exist to control them, and the complex relationships (metabolic interlock) between what are often considered unrelated pathways.
APPENDIX
ANALYSIS OF RAW PROTEIN SEQUENCE DATA
Ask amino acid sequences were collected from the SEED (http://www.theseed.org/), IMG (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi), and NCBI (http://www.ncbi.nlm.nih.gov) databases. The cutoff date for the wholesale inclusion of new entries was about 8 August 2008, although various entries deemed important to the analysis were periodically added. The original collection of sequences was examined for incorrect start sites by screening for a K(F/Y/I)GG motif, which marks the N-terminal regions of all Ask proteins. A file of trimmed sequences was created by trimming away fused domains or extensions that were obvious from inspection of an initial multiple alignment. On occasion, it was necessary to correct start sites that had been incorrectly annotated. For example, the P. stutzeri Ask_ect sequence in CG-02 was missing about 40 amino acids at the N terminus; this was obvious from inspection of the preliminary alignment, where the KIGG motif was absent. If necessary, the CDD graphic (53) was also used as a guide to help pinpoint fusion boundaries. Then, the trimmed Ask sequence file was imported into the Molecular Evolutionary Genetics Analysis (MEGA version 4) Alignment Explorer (85) in order to generate an alignment using the built-in ClustalW implementation. Manual adjustments were then made to obtain a final high-quality alignment. The alignment was used as input for generation of a phylogenetic tree using the MEGA Evolutionary Analysis feature. The neighbor-joining program with 1,000 bootstrap replications was used to obtain a distance-based tree. Trees were visualized with the MEGA Tree Explorer.
DELINEATION OF COHESION GROUPS
In the initial tree of 518 trimmed sequences from 339 genomes, nodes were collapsed at bootstrap values exceeding 75%. An arbitrarily chosen member of each collapsed group was selected as a representative sequence of that node position. The resulting set of representative sequences was used to build another tree, and node positions having bootstrap values in excess of 75% were again collapsed to generate a smaller number of representative sequences (as illustrated by Fig. 4). An additional repetition of this process resulted in a final tally of 52 cohesion groups and 31 orphans. The ultimate collapsed tree (Fig. 5) exhibited nodes with bootstrap values below 74%.
16S rRNA TREE CONSTRUCTION
16S rRNA subtrees were derived by use of the tools available at the Ribosomal Database Project site (14).
DYNAMIC TABLE
The sources and properties of Ask cohesion groups are tabulated in a large working table (“dynamic table”) which can be accessed online (http://www.theseed.org/Papers/MMBR-Aspartokinase/dynamic.html) at the SEED. In addition to the assigned cohesion group and the assigned subhomology division (ASKα or ASKβ), this table includes the entire taxonomy lineage, the gene acronyms obtained from AroPath (http://www.aropath.lanl.gov/cgi-bin/Get_absolute_acronyms.cgi), variations of the K(F/Y/I)GG motif, fused domains, internal translational start sites for ASKβ Ask enzymes, and hyperlinks to gene detail pages for other enzymes of interest (such as homoserine dehydrogenase). The online version of the dynamic table is linked to the SEED Viewer pages, the IMG gene detail pages, NCBI taxonomy, the NCBI CDD, the PDB for crystal structures, a dynamic and full version of Table 3 that lists LGT genes, and a scaled-down version of the dynamic table in which a single member of each cohesion group is used to represent the cohesion group (http://www.theseed.org/Papers/MMBR-Aspartokinase/representatives.html). Each column can be sorted by left clicking the column header. Right clicking on any column header will deliver a pop-up of check boxes for each of the columns so that any of them can be hidden by unchecking the appropriate boxes. The “Acronym” column contains highlighted entries that exemplify cases of LGT. These are hyperlinked to the appropriate section of Table 3, which is a complete, dynamic, and sortable table of LGT genes in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/LGT.html.
The dynamic table contains links that allow gene neighborhoods to be viewed at three different levels. The gene neighborhood graphic on the front page of the SEED Viewer is accessed from the SEED ID column. This has been set to display conserved gene neighborhoods over a fairly wide phylogenetic range and can readily be set to view greater numbers of gene neighborhoods. Two additional innovative gene neighborhood tools that are linked to the SEED system of genome annotation are provided (69). In the second column, clicking the triarrow icon in any given cohesion group will deliver a comparison of gene neighborhoods for every member of the cohesion group. From this SEED Viewer, built-in tools can be accessed to accomplish other tasks, e.g., to download all of the sequences in the cohesion group or to obtain a multiple alignment of the members of the cohesion group. Another column entitled “strain level gene neighborhoods” will deliver, following clicking of the triarrows, a comparison of gene neighborhoods for multiple strains in the SEED collection, one of which has been selected arbitrarily for the table proper. Here, one can screen for very recent phylogenetic changes in close relatives, as is illustrated by Fig. 1S in the supplementary files posted at http://www.theseed.org/Papers/MMBR-Aspartokinase/figure-1S.html.
The dynamic table is also linked at the upper left to a shorter version called the “representatives table.” This shows all of the Ask orphans and one “representative sequence” chosen to represent each cohesion group. The representative sequences and the orphans are the same sequences used to do an alignment and to construct the tree shown in Fig. 5. The representatives table possesses all of the same columns and functionalities as the dynamic table and is linked back to the dynamic table.
ACRONYMS
The nomenclature of genes follows the rules of nomenclature posted at AroPath (http://www.aropath.lanl.gov). The Fasta sequences extracted from the IMG database were imported into the “convert sequence files” tool at AroPath, which then generated four-letter acronyms that code organisms to the species level and two additional extension letters to define the strain and protein (or paralog). For example, E. coli (designated Ecol) strain K-12 (designated _A) has three Ask homologs that were assigned the acronym designations Ecol_Aa, Ecol_Ab, and Ecol_Ac. The system is set up so that a given gene product from a given strain will never change (absolute acronym).
THE NCBI CDD
The CDD version 2.13 was downloaded from the NCBI (ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/). The domain search was conducted locally by a RPS-BLAST program, using all sequences as queries. The RPS-BLAST results then were parsed for E values, hit positions, and description fields for further manual inspection. For the Ask domain, and for the ACT domains, the top BLAST hit was entered into the dynamic table.
ASSESSMENT OF OTHER ENZYMES
In order to understand Ask in the context of the ASK network, it proved to be helpful to evaluate the presence or absence of key enzymes in the network. For example, if homoserine dehydrogenase is absent, one knows that the genome lacks the capability to make threonine and methionine (Fig. 1). It was also quite useful to access the KEGG link from the dropdown list of the “comparative tools” tab from the SEED Viewer in order to assess which variation of a pathway branch exists in a genome or whether the branch is absent altogether.
Homoserine dedydrogenase (Hdh).
The list of genes having the COG0460 homoserine dehydrogenase (ThrA) domain was retrieved from the IMG database. Taxonomy object identifiers (taxon_oid) of retrieved sequences were used to compare with the Ask list in the dynamic table, and only those genes with the same taxon_oid were kept. There are five columns that are related to Hdh in the dynamic table. The column “HDh present?” displays yes or no. The column “Fused Domains” lists any Ask-HDh or HDh-Ask fusions. For all of the monofunctional enzymes, the presence of pfam01842 (ACT domain) was used to recognize threonine-inhibited Hdh (denoted Hdh-thr). For the remaining sequences, a sequence length cutoff of 380 amino acids was used to distinguish short unregulated sequences (denoted Hdh-min) from those having a C-terminal extension (denoted Hdh-met).
Threonine deaminase.
“Biosynthetic” and “catabolic” threonine deaminases are homologs, except that the biosynthetic ones have a unique carboxy-terminal extension that provides an allosteric module. The list of biosynthetic threonine deaminases (IlvA), was retrieved from the IMG database using genome context analysis based on two pfam domains, pfam00585 (the C-terminal regulatory domain of threonine dehydratase) and pfam00291 (a pyridoxal phosphate-dependent enzyme). Catabolic threonine deaminases lack the pfam00585 domain, as do other enzyme classes belonging to pfam00291. Thus, only biosynthetic threonine deaminases possess both pfam domains, and these were entered into the dynamic table.
Citramalate synthase.
Citramalate synthase is the key enzyme for l-isoleucine biosynthesis from pyruvate. Its gene list was retrieved from a subsystem of the SEED database, and “yes” entries were made in the dynamic table as appropriate.
REFERENCES
- 1.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arevalo-Rodriguez, M., I. L. Calderon, and S. Holmberg. 1999. Mutations that cause threonine sensitivity identify catalytic and regulatory regions of the aspartate kinase of Saccharomyces cerevisiae. Yeast 15:1331-1345. [DOI] [PubMed] [Google Scholar]
- 3.Arevalo-Rodriguez, M., X. Pan, J. D. Boeke, and J. Heitman. 2004. FKBP12 controls aspartate pathway flux in Saccharomyces cerevisiae to prevent toxic intermediate accumulation. Eukaryot. Cell 3:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bareich, D. C., and G. D. Wright. 2003. Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase. Biochem. Biophys. Res. Commun. 311:597-603. [DOI] [PubMed] [Google Scholar]
- 5.Black, S., and N. G. Wright. 1955. β-Aspartokinase and β-aspartyl phosphate. J. Biol. Chem. 213:27-38. [PubMed] [Google Scholar]
- 6.Bonner, C. A., T. Disz, K. Hwang, J. Song, V. Vonstein, R. Overbeek, and R. A. Jensen. 2008. Cohesion group approach for evolutionary analysis of TyrA, a protein family with wide-ranging substrate specificities. Microbiol. Mol. Biol. Rev. 72:13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bursy, J., A. U. Kuhlmann, M. Pittelkow, H. Hartmann, M. Jebbar, A. J. Pierik, and E. Bremer. 2008. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74:7286-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bursy, J., A. J. Pierik, N. Pica, and E. Bremer. 2007. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J. Biol. Chem. 282:31147-31155. [DOI] [PubMed] [Google Scholar]
- 9.Cassan, M., C. Parsot, G. N. Cohen, and J. C. Patte. 1986. Nucleotide sequence of lysC gene encoding the lysine-sensitive aspartokinase III of Escherichia coli K12. Evolutionary pathway leading to three isofunctional enzymes. J. Biol. Chem. 261:1052-1057. [PubMed] [Google Scholar]
- 10.Charon, N. W., R. C. Johnson, and D. Peterson. 1974. Amino acid biosynthesis in the spirochete Leptospira: evidence for a novel pathway of isoleucine biosynthesis. J. Bacteriol. 117:203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, N. Y., F. M. Hu, and H. Paulus. 1987. Nucleotide sequence of the overlapping genes for the subunits of Bacillus subtilis aspartokinase II and their control regions. J. Biol. Chem. 262:8787-8798. [PubMed] [Google Scholar]
- 12.Chen, N. Y., S. Q. Jiang, D. A. Klein, and H. Paulus. 1993. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J. Biol. Chem. 268:9448-9465. [PubMed] [Google Scholar]
- 13.Chen, N. Y., and H. Paulus. 1988. Mechanism of expression of the overlapping genes of Bacillus subtilis aspartokinase II. J. Biol. Chem. 263:9526-9532. [PubMed] [Google Scholar]
- 14.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Curien, G., V. Biou, C. Mas-Droux, M. Robert-Genthon, J. L. Ferrer, and R. Dumas. 2008. Amino acid biosynthesis: new architectures in allosteric enzymes. Plant Physiol. Biochem. 46:325-339. [DOI] [PubMed] [Google Scholar]
- 17.Daniel, R. A., and J. Errington. 1993. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J. Mol. Biol. 232:468-483. [DOI] [PubMed] [Google Scholar]
- 18.Datta, P., and H. Gest. 1964. Control of enzyme activity by concerted feedback inhibition. Proc. Natl. Acad. Sci. USA 52:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta, P., and L. Prakash. 1966. Aspartokinase of Rhodopseudomonas spheroides. Regulation of enzyme activity by aspartate beta-semialdehyde. J. Biol. Chem. 241:5827-5835. [PubMed] [Google Scholar]
- 20.DeLaBarre, B., P. R. Thompson, G. D. Wright, and A. M. Berghuis. 2000. Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases. Nat. Struct. Biol. 7:238-244. [DOI] [PubMed] [Google Scholar]
- 21.Dereppe, C., G. Bold, O. Ghisalba, E. Ebert, and H. P. Schar. 1992. Purification and characterization of dihydrodipicolinate synthase from pea. Plant Physiol. 98:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downs, D. M. 2006. Understanding microbial metabolism. Annu. Rev. Microbiol. 60:533-559. [DOI] [PubMed] [Google Scholar]
- 23.Drevland, R. M., A. Waheed, and D. E. Graham. 2007. Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J. Bacteriol. 189:4391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dungan, S. M., and P. Datta. 1973. Concerted feedback inhibition. Modifier-induced oligomerization of the Pseudomonas fluorescens aspartokinase. J. Biol. Chem. 248:8541-8546. [PubMed] [Google Scholar]
- 25.Ekechukwu, C. R., T. A. Burns, and T. Melton. 1995. Selection and characterization of aspartokinase feedback-insensitive mutants of Azotobacter vinelandii. Appl. Environ. Microbiol. 61:3189-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Follettie, M. T., O. P. Peoples, C. Agoropoulou, and A. J. Sinskey. 1993. Gene structure and expression of the Corynebacterium flavum N13 ask-asd operon. J. Bacteriol. 175:4096-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fondi, M., M. Brilli, and R. Fani. 2007. On the origin and evolution of biosynthetic pathways: integrating microarray data with structure and organization of the Common Pathway genes. BMC Bioinformatics 8(Suppl. 1):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gophna, U., E. Bapteste, W. F. Doolittle, D. Biran, and E. Z. Ron. 2005. Evolutionary plasticity of methionine biosynthesis. Gene 355:48-57. [DOI] [PubMed] [Google Scholar]
- 30.Gosset, G., C. A. Bonner, and R. A. Jensen. 2001. Microbial origin of plant-type 2-keto-3-deoxy-d-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and tryptophan-regulated enzyme from Xanthomonas campestris. J. Bacteriol. 183:4061-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham, D. E., and H. K. Huse. 2008. Methanogens with pseudomurein use diaminopimelate aminotransferase in lysine biosynthesis. FEBS Lett. 582:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant, G. A. 2006. The ACT domain: a small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 281:33825-33829. [DOI] [PubMed] [Google Scholar]
- 34.Graves, L. M., and R. L. Switzer. 1990. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J. Bacteriol. 172:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grundy, F. J., S. C. Lehman, and T. M. Henkin. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. USA 100:12057-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamano, Y., I. Nicchu, T. Shimizu, Y. Onji, J. Hiraki, and H. Takagi. 2007. ɛ-Poly-l-lysine producer, Streptomyces albulus, has feedback-inhibition resistant aspartokinase. Appl. Microbiol. Biotechnol. 76:873-882. [DOI] [PubMed] [Google Scholar]
- 37.Han, J. H., S. Batey, A. A. Nickson, S. A. Teichmann, and J. Clarke. 2007. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 8:319-330. [DOI] [PubMed] [Google Scholar]
- 38.Hudson, A. O., C. Gilvarg, and T. Leustek. 2008. Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of ll-diaminopimelate aminotransferase. J. Bacteriol. 190:3256-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson, A. O., B. K. Singh, T. Leustek, and C. Gilvarg. 2006. An ll-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 140:292-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen, R. A. 2001. Orthologs and paralogs—we need to get it right. Genome Biol. 2:INTERACTIONS1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen, R. A., and S. Ahmad. 1990. Nested gene fusions as markers of phylogenetic branchpoints in prokaryotes. Trends Ecol. Evol. 5:219-224. [DOI] [PubMed] [Google Scholar]
- 42.Jensen, R. A., and D. S. Nasser. 1968. Comparative regulation of isoenzymic 3-deoxy-d-arabino-heptulosonate 7-phosphate synthetases in microorganisms. J. Bacteriol. 95:188-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen, R. A., D. S. Nasser, and E. W. Nester. 1967. Comparative control of a branch-point enzyme in microorganisms. J. Bacteriol. 94:1582-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King, N. D., and M. R. O'Brian. 2001. Evidence for direct interaction between enzyme I (Ntr) and aspartokinase to regulate bacterial oligopeptide transport. J. Biol. Chem. 276:21311-21316. [DOI] [PubMed] [Google Scholar]
- 45.Kleeb, A. C., P. Kast, and D. Hilvert. 2006. A monofunctional and thermostable prephenate dehydratase from the archaeon Methanocaldococcus jannaschii. Biochemistry 45:14101-14110. [DOI] [PubMed] [Google Scholar]
- 46.Kobashi, N., M. Nishiyama, and H. Yamane. 2001. Characterization of aspartate kinase III of Bacillus subtilis. Biosci. Biotechnol. Biochem. 65:1391-1394. [DOI] [PubMed] [Google Scholar]
- 47.Kotaka, M., J. Ren, M. Lockyer, A. R. Hawkins, and D. K. Stammers. 2006. Structures of R- and T-state Escherichia coli aspartokinase III. Mechanisms of the allosteric transition and inhibition by lysine. J. Biol. Chem. 281:31544-31552. [DOI] [PubMed] [Google Scholar]
- 48.Kuhlmann, A. U., J. Bursy, S. Gimpel, T. Hoffmann, and E. Bremer. 2008. Synthesis of the compatible solute ectoine in Virgibacillus pantothenticus is triggered by high salinity and low growth temperature. Appl. Environ. Microbiol. 74:4560-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuramitsu, H. K., and R. M. Watson. 1973. Regulation of aspartokinase activity in Clostridium perfringens. J. Bacteriol. 115:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberles, J. S., M. Thorolfsson, and A. Martinez. 2005. Allosteric mechanisms in ACT domain containing enzymes involved in amino acid metabolism. Amino Acids 28:1-12. [DOI] [PubMed] [Google Scholar]
- 51.Liu, X., A. G. Pavlovsky, and R. E. Viola. 2008. The structural basis for allosteric inhibition of a threonine-sensitive aspartokinase. J. Biol. Chem. 283:16216-16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lysenko, T. G., M. I. Mendzhul, N. V. Koltukova, O. A. Shainskaia, and S. I. Perepelitsa. 1993. Isolation, purification and various properties of aspartate kinase from the cyanobacterium Plectonema boryanum. Ukr. Biokhim. Zh. 65:54-61. (In Russian.) [PubMed] [Google Scholar]
- 53.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35:D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mas-Droux, C., G. Curien, M. Robert-Genthon, M. Laurencin, J. L. Ferrer, and R. Dumas. 2006. A novel organization of ACT domains in allosteric enzymes revealed by the crystal structure of Arabidopsis aspartate kinase. Plant Cell 18:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarron, R. M., and Y. F. Chang. 1978. Aspartokinase of Streptococcus mutans: purification, properties, and regulation. J. Bacteriol. 134:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy, A. J., N. E. Adams, A. O. Hudson, C. Gilvarg, T. Leustek, and A. T. Maurelli. 2006. l,l-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. USA 103:17909-17914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendelovitz, S., and Y. Aharonowitz. 1982. Regulation of cephamycin C synthesis, aspartokinase, dihydrodipicolinic acid synthetase, and homoserine dehydrogenase by aspartic acid family amino acids in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 21:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merino, E., R. A. Jensen, and C. Yanofsky. 2008. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 11:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mongodin, E. F., K. E. Nelson, S. Daugherty, R. T. Deboy, J. Wister, H. Khouri, J. Weidman, D. A. Walsh, R. T. Papke, G. Sanchez Perez, A. K. Sharma, C. L. Nesbo, D. MacLeod, E. Bapteste, W. F. Doolittle, R. L. Charlebois, B. Legault, and F. Rodriguez-Valera. 2005. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. USA 102:18147-18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moran, N. A., G. R. Plague, J. P. Sandstrom, and J. L. Wilcox. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14543-14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motoyama, H., K. Maki, H. Anazawa, S. Ishino, and S. Teshiba. 1994. Cloning and nucleotide sequences of the homoserine dehydrogenase genes (hom) and the threonine synthase genes (thrC) of the gram-negative obligate methylotroph Methylobacillus glycogenes. Appl. Environ. Microbiol. 60:111-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishida, H., and I. Narumi. 2007. Phylogenetic and disruption analyses of aspartate kinase of Deinococcus radiodurans. Biosci. Biotechnol. Biochem. 71:1015-1020. [DOI] [PubMed] [Google Scholar]
- 63.Nishida, H., M. Nishiyama, N. Kobashi, T. Kosuge, T. Hoshino, and H. Yamane. 1999. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res. 9:1175-1183. [DOI] [PubMed] [Google Scholar]
- 64.Nishiyama, M., M. Kukimoto, T. Beppu, and S. Horinouchi. 1995. An operon encoding aspartokinase and purine phosphoribosyltransferase in Thermus flavus. Microbiology 141:1211-1219. [DOI] [PubMed] [Google Scholar]
- 65.Nudler, E., and A. S. Mironov. 2004. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29:11-17. [DOI] [PubMed] [Google Scholar]
- 66.Ono, H., K. Sawada, N. Khunajakr, T. Tao, M. Yamamoto, M. Hiramoto, A. Shinmyo, M. Takano, and Y. Murooka. 1999. Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic eubacterium, Halomonas elongata. J. Bacteriol. 181:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onyenwoke, R. U., J. A. Brill, K. Farahi, and J. Wiegel. 2004. Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch (Firmicutes). Arch. Microbiol. 182:182-192. [DOI] [PubMed] [Google Scholar]
- 68.Oren, A. 2008. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 96:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paris, S., C. Viemon, G. Curien, and R. Dumas. 2003. Mechanism of control of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase by threonine. J. Biol. Chem. 278:5361-5366. [DOI] [PubMed] [Google Scholar]
- 72.Parsot, C., and G. N. Cohen. 1988. Cloning and nucleotide sequence of the Bacillus subtilis hom gene coding for homoserine dehydrogenase. Structural and evolutionary relationships with Escherichia coli aspartokinases-homoserine dehydrogenases I and II. J. Biol. Chem. 263:14654-14660. [PubMed] [Google Scholar]
- 73.Pavelka, M. S., Jr. 2007. Another brick in the wall. Trends Microbiol. 15:147-149. [DOI] [PubMed] [Google Scholar]
- 74.Pearce, F. G., M. A. Perugini, H. J. McKerchar, and J. A. Gerrard. 2006. Dihydrodipicolinate synthase from Thermotoga maritima. Biochem. J. 400:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Porat, I., M. Sieprawska-Lupa, Q. Teng, F. J. Bohanon, R. H. White, and W. B. Whitman. 2006. Biochemical and genetic characterization of an early step in a novel pathway for the biosynthesis of aromatic amino acids and p-aminobenzoic acid in the archaeon Methanococcus maripaludis. Mol. Microbiol. 62:1117-1131. [DOI] [PubMed] [Google Scholar]
- 76.Raoult, D., H. Ogata, S. Audic, C. Robert, K. Suhre, M. Drancourt, and J. M. Claverie. 2003. Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res. 13:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rebello, J. L., and R. A. Jensen. 1970. Metabolic interlock. The multi-metabolite control of prephenate dehydratase activity in Bacillus subtilis. J. Biol. Chem. 245:3738-3744. [PubMed] [Google Scholar]
- 78.Robert-Gero, M., L. Le Borgne, and G. N. Cohen. 1972. Concerted feedback inhibition of the aspartokinase of Rhodospirillum tenue by threonine and methionine: a novel pattern. J. Bacteriol. 112:251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roberts, S. J., J. C. Morris, R. C. Dobson, and J. A. Gerrard. 2003. The preparation of (S)-aspartate semi-aldehyde appropriate for use in biochemical studies. Bioorg. Med. Chem. Lett. 13:265-267. [DOI] [PubMed] [Google Scholar]
- 80.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 31:6748-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saum, S. H., and V. Muller. 2008. Growth phase-dependent switch in osmolyte strategy in a moderate halophile: ectoine is a minor osmolyte but major stationary phase solute in Halobacillus halophilus. Environ. Microbiol. 10:716-726. [DOI] [PubMed] [Google Scholar]
- 82.Shiio, I., and R. Miyajima. 1969. Concerted inhibition and its reversal by end products of aspartate kinase in Brevibacterium flavum. J. Biochem. 65:849-859. [DOI] [PubMed] [Google Scholar]
- 83.Soma, A., Y. Ikeuchi, S. Kanemasa, K. Kobayashi, N. Ogasawara, T. Ote, J. Kato, K. Watanabe, Y. Sekine, and T. Suzuki. 2003. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell 12:689-698. [DOI] [PubMed] [Google Scholar]
- 84.Song, J., C. A. Bonner, M. Wolinsky, and R. A. Jensen. 2005. The TyrA family of aromatic-pathway dehydrogenases in phylogenetic context. BMC Biol. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 86.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Velasco, I., M. Arevalo-Rodriguez, P. Marina, and I. L. Calderon. 2005. A new mutation in the yeast aspartate kinase induces threonine accumulation in a temperature-regulated way. Yeast 22:99-110. [DOI] [PubMed] [Google Scholar]
- 88.Wu, M., and J. A. Eisen. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie, G., C. A. Bonner, J. Song, N. O. Keyhani, and R. A. Jensen. 2004. Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy. BMC Biol. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67:303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang, C., D. A. Rodionov, I. A. Rodionova, X. Li, and A. L. Osterman. 2008. Glycerate 2-kinase of Thermotoga maritima and genomic reconstruction of related metabolic pathways. J. Bacteriol. 190:1773-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoshida, A., T. Tomita, H. Kono, S. Fushinobu, T. Kuzuyama, and M. Nishiyama. 2009. Crystal structures of the regulatory subunit of Thr-sensitive aspartate kinase from Thermus thermophilus. FEBS J. 276:3124-3136. [DOI] [PubMed] [Google Scholar]
- 93.Yoshida, A., T. Tomita, T. Kurihara, S. Fushinobu, T. Kuzuyama, and M. Nishiyama. 2007. Structural Insight into concerted inhibition of alpha 2 beta 2-type aspartate kinase from Corynebacterium glutamicum. J. Mol. Biol. 368:521-536. [DOI] [PubMed] [Google Scholar]
- 94.Zhang, J. J., F. M. Hu, N. Y. Chen, and H. Paulus. 1990. Comparison of the three aspartokinase isozymes in Bacillus subtilis Marburg and 168. J. Bacteriol. 172:701-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang, J. J., and H. Paulus. 1990. Desensitization of Bacillus subtilis aspartokinase I to allosteric inhibition by DAP allows aspartokinase I to function in amino acid biosynthesis during exponential growth. J. Bacteriol. 172:4690-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]