Abstract
Chlamydiae are unusual obligately intracellular bacteria that do not synthesize detectable peptidoglycan. However, they possess genes that appear to encode products with peptidoglycan biosynthetic activity. Bioinformatic analysis predicts that chlamydial MurE possesses UDP-MurNAc-l-Ala-d-Glu:meso-diaminopimelic acid (UDP-MurNAc-l-Ala-d-Glu:meso-A2pm) ligase activity. Nevertheless, there are no experimental data to confirm this hypothesis. In this paper we demonstrate that the murE gene from Chlamydia trachomatis is capable of complementing a conditional Escherichia coli mutant impaired in UDP-MurNAc-l-Ala-d-Glu:meso-A2pm ligase activity. Recombinant MurE from C. trachomatis (MurECt) was overproduced in and purified from E. coli in order to investigate its kinetic parameters in vitro. By use of UDP-MurNAc-l-Ala-d-Glu as the nucleotide substrate, MurECt demonstrated ATP-dependent meso-A2pm ligase activity with pH and magnesium ion optima of 8.6 and 30 mM, respectively. Other amino acids (meso-lanthionine, the ll and dd isomers of A2pm, d-lysine) were also recognized by MurECt. However, the activities for these amino acid substrates were weaker than that for meso-A2pm. The specificity of MurECt for three possible C. trachomatis peptidoglycan nucleotide substrates was also determined in order to deduce which amino acid might be present at the first position of the UDP-MurNAc-pentapeptide. Relative kcat/Km ratios for UDP-MurNAc-l-Ala-d-Glu, UDP-MurNAc-l-Ser-d-Glu, and UDP-MurNAc-Gly-d-Glu were 100, 115, and 27, respectively. Our results are consistent with the synthesis in chlamydiae of a UDP-MurNAc-pentapeptide in which the third amino acid is meso-A2pm. However, due to the lack of specificity of MurECt for nucleotide substrates in vitro, it is not obvious which amino acid is present at the first position of the pentapeptide.
Chlamydiae cause serious respiratory tract and genital infections in humans (9). They are obligately intracellular gram-negative bacteria, with a unique biphasic development cycle. Elementary bodies (EBs) are the infectious form of the organism and invade susceptible host cells. Once internalized, EBs differentiate into reticulate bodies (RBs), which have the capacity to divide (39, 40). The RBs are fragile and pleomorphic, whereas EBs are comparatively rigid and stable (19, 39). After repeated cycles of binary fission, the RBs differentiate into EBs, and the host cell lyses, releasing infectious EBs (1).
In contrast to the vast majority of eubacteria, chlamydiae lack detectable amounts of peptidoglycan (PG), an essential polymer. PG is a giant macromolecule composed of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues cross-linked by short peptides. It determines the shape of bacteria, protects the cell from lysis due to internal osmotic pressure, and also plays a role in cell division. However, PG has not been detected in EBs (14, 20) or RBs (5).
Although chlamydiae appear to lack PG, they contain penicillin-binding proteins and are sensitive to antibiotics that inhibit PG synthesis (5, 40). The Chlamydia trachomatis genome contains most of the genes coding for proteins involved in, or associated with, PG synthesis (54). Chlamydial MurA, MurC-Ddl, and the MurC domain of the latter fusion protein are active in vitro and complement Escherichia coli mutants deficient in the respective enzymes (23, 32, 33). Furthermore, proteomic analysis reveals that the murE gene product, which was assigned as UDP-MurNAc-l-Ala-d-Glu:meso-diaminopimelic acid ligase (UDP-MurNAc-l-Ala-d-Glu:meso-A2pm ligase), is expressed in RBs (52).
MurE ligases catalyze the addition of the third amino acid residue to the peptide chain of PG. This residue, generally a diamino acid, is usually meso-A2pm for gram-negative bacteria and bacilli, and l-lysine for gram-positive bacteria, although other amino acids (for example, l-ornithine, meso-lanthionine, ll-A2pm, l-diaminobutyric acid, or l-homoserine) occur in certain species (6, 50, 57). In many organisms, the third residue of the peptide chain participates in PG cross-linking; consequently, the MurE enzyme is highly specific for the relevant amino acid so as to avoid incorporation of incorrect amino acids into the macromolecule, which could result in deleterious morphological changes and cell lysis (35). Crystallization of MurE from E. coli (MurEEc) has permitted analysis of the structural basis for this high specificity (22). Sequence alignments of different MurE orthologues have also revealed the specific consensus sequences DNPR and D(D/N)P(N/A) located in the binding pockets for meso-A2pm and l-Lys, respectively (11, 17). Chlamydia trachomatis MurE (MurECt) possesses the DNPR motif, which suggests that it adds meso-A2pm (17). However, there are no experimental data to confirm this prediction.
In this paper we report for the first time the overproduction and purification of MurECt, as well as a detailed investigation of its in vivo and in vitro biochemical properties. These studies contribute to our understanding of the nature and properties of the PG biosynthetic enzymes in chlamydiae and do indeed suggest that MurECt has meso-A2pm ligase activity.
MATERIALS AND METHODS
Materials.
DNA restriction enzymes and synthetic oligonucleotides were purchased from New England Biolabs and Eurofins-MWG, respectively. UDP-MurNAc-l-Ala-d-Glu and UDP-MurNAc-l-Ala-d-[14C]Glu were prepared according to published procedures (4, 37). dd-A2pm, ll-A2pm, and meso-A2pm were separated from a commercially available mixture of the three isomers by high-performance liquid chromatography (HPLC) on a Chirobiotic T column (250 by 10 mm; Advanced Separation Technologies, Inc.), with 75% ethanol as the mobile phase, at 2.6 ml·min−1 (25). Tritiated A2pm (a mixture of all isomers; 2 TBq·mmol−1) was purchased from American Radiolabeled Chemicals, Inc.
General DNA techniques and E. coli cell transformation.
The construction of the pET2160 vector from pET21d (Novagen) was described previously (13). Plasmid vector pTrc99A (2) was obtained from Amersham Biosciences, and plasmid pTrcHis60 has been described previously (45). Plasmid purification kits were purchased from Macherey-Nagel. Standard procedures for molecular biology were used (49). E. coli cells were made competent and transformed with plasmid DNA according to the method of Dagert and Ehrlich (16), or by electroporation.
Enzymatic synthesis of UDP-MurNAc-Gly-d-Glu and UDP-MurNAc-l-Ser-d-Glu.
The reaction mixtures (5 ml) contained 100 mM Tris-HCl (pH 8.6), 2 mM 2-mercaptoethanol, 10 mM MgCl2, 5 mM ATP, 1 mM UDP-MurNAc, 4 mM glycine or l-serine, 5 mM d-glutamate, and pure MurC (50 μg) and MurD (20 μg) enzymes from E. coli. The reactions were followed by analytical HPLC on a Vydac 218TP54 column (250 by 4.6 mm; Agilent France) using 50 mM ammonium phosphate, pH 3.5, as the mobile phase, at 0.6 ml·min−1. After 20 h at 37°C, the reaction mixtures were lyophilized, and each nucleotide product was purified by filtration on a Sephadex G-25 column (115 by 2 cm) in water (4). The identity and purity of the products were verified by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry. The [M − H]− ions of UDP-MurNAc-Gly-d-Glu and UDP-MurNAc-l-Ser-d-Glu were at m/z 864.18 (calculated, 864.16) and 894.18 (calculated, 894.17), respectively.
Bacterial strains and growth conditions.
E. coli strains DH5α (Invitrogen), C43(DE3) (Avidis), and TKL-11 (27, 28) were used as hosts for plasmids, overproduction of the MurECt enzyme, and complementation assays, respectively. 2YT and LB media (38) were used to grow cells, and growth was monitored at 600 nm with a Shimadzu UV-1601 spectrophotometer. When required, ampicillin was added at a concentration of 100 μg·ml−1.
Construction of plasmids.
The C. trachomatis murE gene was amplified from the L2 serotype by PCR using primers 5′-GCGCCCATGGCGCATTTAGACCAACTTCTTCAGAATATTCCGGC-3′ and 5′-GCGCAAGCTTAGTGATGGTGATGGTGATGACAAAGGGCTGCCAAGGCTTCACACACAAC-3′. The primer upstream of the start codon (underlined) contained an engineered NcoI site, shown in boldface. The primer 3′ to the gene contained an engineered HindIII site, shown in boldface, and a His6 tag. The amplified product was first digested by NcoI and HindIII and then inserted between the same sites in vector pET2160 (T7 promoter), generating a plasmid that encodes MurECt with a His6 C-terminal extension. The construction was verified by DNA sequencing (Eurofins-MWG). Plasmid pET2160::murECt was transformed into E. coli C43(DE3) for expression experiments. The C. trachomatis murE gene (with its His6 tag) was also cloned into the pTrc99A expression vector, generating plasmid pTrc99A::murECt. The pMLD117 plasmid (pTrc99A::murEEc), which expresses the E. coli murE gene (22), was used as a control in functional complementation assays.
Functional complementation assays.
The E. coli murE temperature-sensitive lysis mutant TKL-11 (27, 28) was transformed with plasmid pTrc99A, pTrc99A::murEEc, or pTrc99A::murECt. Transformants were grown in low-salt LB medium (0.2% NaCl instead of 0.5% NaCl) supplemented with thymine (100 μg·ml−1) at the permissive temperature (30°C) before transfer to the restrictive temperature (42°C). Cell growth was followed by monitoring the optical density at 600 nm, and complementation was judged by restoration of the ability of the E. coli mutant to grow at the restrictive temperature.
Overproduction and purification of the MurECt protein.
An overnight preculture of E. coli C43(DE3) (pET2160::murECt) was used to inoculate 1 liter of 2YT supplemented with ampicillin. The culture was incubated at 37°C with shaking until the optical density at 600 nm reached 0.8. Then the temperature was reduced to 15°C; isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 0.75 mM; and incubation was continued for 18 h at 15°C with shaking. The cells were harvested at 4°C, and the pellet was washed with cold 20 mM phosphate buffer (pH 7.2) containing 1 mM dithiothreitol (DTT) (buffer A). Bacteria were resuspended in buffer A (6 ml) and disrupted by sonication in the cold using a Bioblock Vibracell 72412 sonicator. The resulting suspension was centrifuged at 4°C for 30 min at 200,000 × g with a Beckman TL100 apparatus, and the pellet was discarded. The supernatant was kept at −20°C.
The His6-tagged MurECt protein was purified on Ni2+-nitrilotriacetate (Ni2+-NTA) agarose according to the manufacturer's recommendations (Qiagen). All procedures were performed at 4°C. The supernatant was mixed for 1 h with the polymer, which had previously been washed with buffer A containing 0.3 M KCl and 10 mM imidazole. The washing and elution steps were performed with a discontinuous gradient of imidazole (20 to 400 mM) in buffer A containing 0.3 M KCl. Eluted protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and relevant fractions were pooled and dialyzed against buffer A. Glycerol (10% final concentration) was added for storage of the protein at −20°C. Protein concentrations were determined by quantitative amino acid analysis with a Hitachi L8800 analyzer (ScienceTec) after hydrolysis of samples in 6 M HCl at 105°C for 24 h.
MurE activity assays.
The standard MurE activity assay measured the formation of UDP-MurNAc-l-Ala-γ-d-[14C]Glu-meso-A2pm in a mixture (final volume, 40 μl) containing 100 mM Tris-HCl (pH 8.6), 30 mM MgCl2, 5 mM DTT, 5 mM ATP, 100 μM UDP-MurNAc-l-Ala-d-[14C]Glu (400 Bq), 100 μM meso-A2pm, and the enzyme (20 μl of an appropriate dilution in buffer A). After 30 min at 37°C, the reaction was terminated by the addition of glacial acetic acid (10 μl), followed by lyophilization. The radioactive substrate and product were separated on a Nucleosil 100 C18 5U column (150 by 4.6 mm; Alltech France) using 50 mM ammonium formate, pH 3.9, at a flow rate of 0.6 ml·min−1. Radioactivity was detected with a flow detector (model LB506-C1; Berthold) using the Quicksafe Flow 2 scintillator (Zinsser Analytic) at 0.6 ml·min−1. Quantification was performed with Radiostar software (Berthold).
Identical assay conditions were used when amino acids other than meso-A2pm were tested as substrates. However, the buffer and pH used for the separation of the radiolabeled substrate and product varied, depending on the substrate used. For meso-A2pm, ll-A2pm, dd-A2pm, and meso-lanthionine, 50 mM ammonium formate (pH 3.9) was used; for l-ornithine, d-lysine, l-lysine, and l-homoserine, 50 mM sodium phosphate (pH 2.5) containing 7.2 mM sodium hexanesulfonate-acetonitrile (98.5:1.5, vol/vol) (11) were used. When different nucleotide substrates (UDP-MurNAc-l-Ala-d-Glu, UDP-MurNAc-Gly-d-Glu, and UDP-MurNAc-l-Ser-d-Glu) were tested, [3H]A2pm was used as the labeled substrate and HPLC analysis was conducted using 50 mM ammonium formate, pH 3.2, as the mobile phase.
For determination of kinetic constants, the same assay was used with various concentrations of one substrate and fixed concentrations of the others. In all cases, the enzyme concentration was chosen so that substrate consumption was <20%, the linearity being ensured within this interval even at the lowest substrate concentration. Data were fitted to the equation v = VmaxS/(Km + S + [S2]/Ki) by the Levenberg-Marquardt method (47), where v is the initial velocity and S is the substrate concentration, and values ± standard deviations at 95% confidence were calculated. The MDFitt software developed by M. Desmadril (UMR 8619, CNRS, Orsay, France) was used for this purpose.
RESULTS
Overproduction and purification of MurE ligase from C. trachomatis.
The murE gene encoding MurECt was cloned into plasmid pET2160, allowing expression of the protein with a C-terminal His6 tag. Preliminary assays of overexpression in strain C43 using classical conditions (induction for 3 h at 37°C with 0.75 mM IPTG) revealed that the MurE protein was produced but was recovered almost exclusively as inclusion bodies in the particulate fraction (data not shown). To overcome this problem and to increase the solubility of the protein, different induction temperatures (15, 22, 30, and 37°C) and different E. coli host strains (BL21, C43, and Rosetta) were tested (data not shown). Under all conditions, MurECt formed inclusion bodies in induced cells, but in some cases, a significant proportion of the protein was also found in the soluble fraction. The best results were obtained by using overnight expression in C43(DE3) at 15°C with 0.75 mM IPTG. Under these conditions, about 50% of the protein was recovered in the soluble fraction, and this protein was subsequently purified by affinity chromatography on Ni2+-NTA agarose. As judged by SDS-PAGE (Fig. 1), MurECt exhibited a molecular mass in agreement with the calculated value (54,093 Da, taking the His6 tag into account). The yield was approximately 7 mg of purified protein per liter of culture.
FIG. 1.
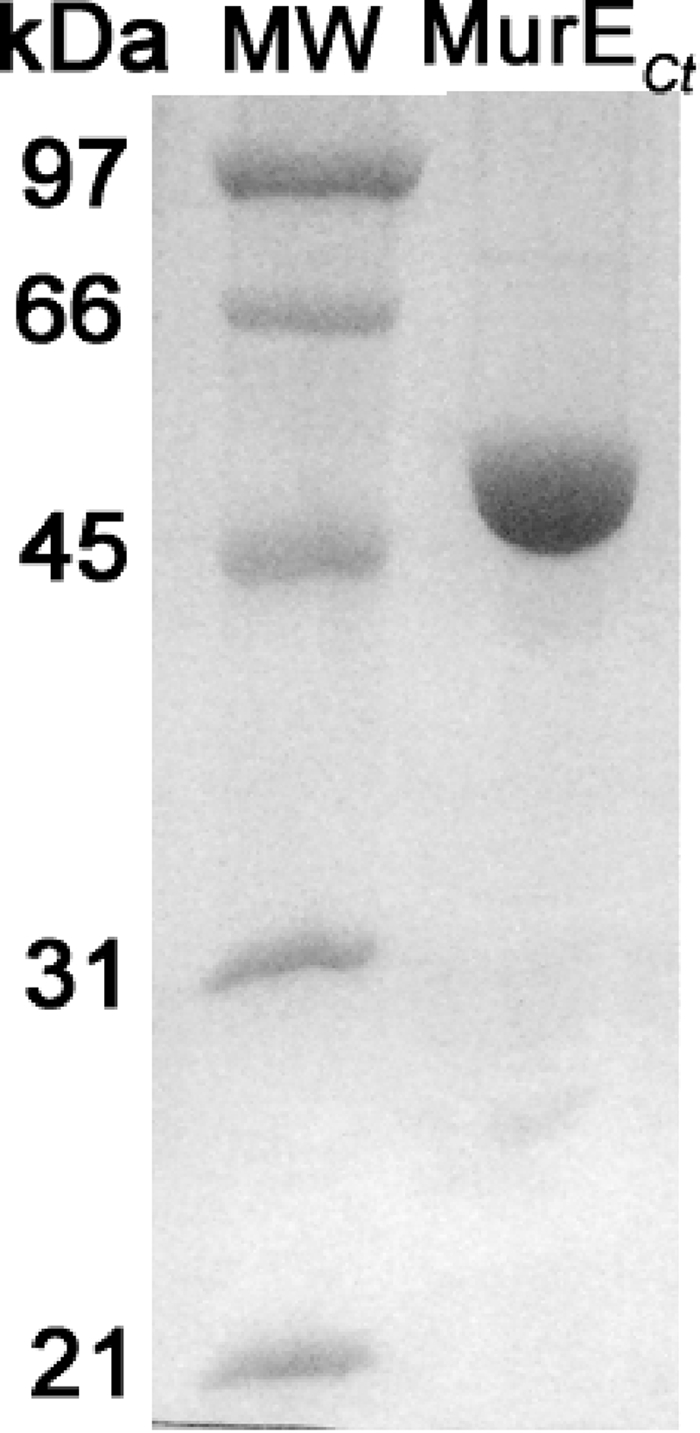
SDS-PAGE of purified C. trachomatis MurE. The MurECt protein was overproduced in E. coli cells in the His6-tagged form. Purification on Ni2+-NTA agarose was performed as described in the text. Lanes: MW, molecular mass standards; MurECt, purified MurECt. Staining was performed with Coomassie brilliant blue R250.
MALDI-TOF mass spectrometry confirmed the purity and integrity of the MurECt preparation. Peaks of m/z 53,956 and 26,981, corresponding to the [M + H]+ and [M + 2H]2+ ions, respectively, were observed (data not shown). These results are in perfect agreement with the molecular mass of the protein that has lost its N-terminal methionine (53,961 Da), indicating that the latter residue had been eliminated during in vivo heterologous expression of the recombinant protein, in contrast to chlamydial MurC (MurCCt) (23).
Complementation analysis of MurECt in a temperature-sensitive E. coli murE mutant.
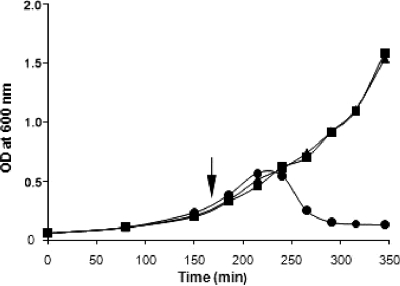
The murECt gene was also subcloned into the IPTG-inducible expression vector pTrc99A, generating pTrc99A::murECt. The pTrc99A, pTrc99A::murEEc, and pTrc99A::murECt plasmids were transformed separately into the temperature-sensitive E. coli strain TKL-11, which harbors a defective murE gene. The aim was to determine whether the chlamydial murE gene could complement the temperature-sensitive defect of the E. coli murE mutant, which would demonstrate that murECt was functional in vivo. Growth of the control TKL-11(pTrc99A) strain at 30°C to early-exponential phase, followed by a shift to 42°C, led to rapid lysis of the culture (Fig. 2). In contrast, the TKL-1 (pTrc99A::murEEc) and TKL-11(pTrc99A::murECt) strains continued to grow at 42°C (Fig. 2). Thus, expression of the chlamydial MurE ligase encoded by pTrc99A::murECt complemented the specific defect in the UDP-MurNAc-l-Ala-d-Glu:meso-A2pm ligase (MurE) activity of the E. coli mutant. Complementation of the mutation did not require IPTG induction of murECt expression, indicating that basal expression of the enzyme from the plasmid was sufficient to restore cell viability.
FIG. 2.
Growth of E. coli TKL-11 carrying either plasmid pTrc99A (circles), pTrc99A::murEEc (squares), or pTrc99A::murECt (triangles). Cells were grown at 30°C, and at the time indicated by the arrow, the cultures were shifted to 42°C.
Enzymatic properties and kinetic parameters of MurECt.
The kinetic parameters of MurECt were investigated in detail. As complementation studies suggested that chlamydial MurE possesses meso-A2pm-adding activity, assays were first performed with meso-A2pm as a substrate. The activity of MurECt as a function of pH and of Mg2+ and DTT concentrations was determined. Profiles displayed a typical bell shape with optimal values of pH 8.6, 30 mM, and 5 mM, respectively (data not shown).
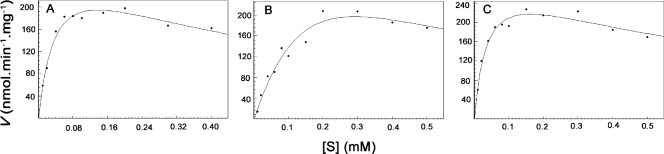
With meso-A2pm and UDP-MurNAc-l-Ala-d-Glu as the amino acid and nucleotide substrates, the kcat of the enzyme was ca. 17 min−1 (Vmax, ca. 320 nmol·min−1·mg−1) (Table 1). Inhibition by an excess of the UDP-MurNAc-dipeptide substrate was observed (Fig. 3A). To make a compromise between enzyme saturation and substrate inhibition and to determine the kinetic parameters for ATP and meso-A2pm, the UDP-MurNAc-dipeptide concentration was fixed at 0.1 mM. The kinetic parameters for the main substrates are reported in Table 1.
TABLE 1.
Kinetic parameters of MurECt
| Substratea | Apparent Km (μM) | Apparent kcat (min−1) | Apparent kcat/Km ratio (min−1·μM−1) |
|---|---|---|---|
| ATP | 300 ± 110 | 12 ± 1 | 0.040 ± 0.015 |
| UDP-MurNAc-l-Ala-d-Glub | 42 ± 12 | 17 ± 1 | 0.40 ± 0.12 |
| UDP-MurNAc-Gly-d-Glub | 300 ± 100 | 33 ± 5 | 0.11 ± 0.04 |
| UDP-MurNAc-l-Ser-d-Glub | 37 ± 12 | 17 ± 2 | 0.46 ± 0.16 |
| meso-A2pm | 23 ± 10 | 14 ± 2 | 0.61 ± 0.28 |
The kinetic parameters were determined as described in Materials and Methods. The concentrations of the fixed substrates were 5 mM for ATP, 0.1 mM for UDP-MurNAc-l-Ala-d-Glu, and 0.1 mM for meso-A2pm. The concentration ranges for the varied substrates were 0.025 to 4 mM for ATP, 10 to 400 μM for UDP-MurNAc-l-Ala-d-Glu, 10 to 500 μM for UDP-MurNAc-Gly-d-Glu and UDP-MurNAc-l-Ser-d-Glu, and 7.5 to 300 μM for meso-A2pm.
The kinetic parameters for this substrate were determined by fitting the data to the equation v = VmaxS/(Km + S + [S2]/Ki). The Ki values found were 450 ± 180 μM (UDP-MurNAc-l-Ala-d-Glu), 280 ± 140 μM (UDP-MurNAc-Gly-d-Glu), and 700 ± 320 μM (UDP-MurNAc-l-Ser-d-Glu).
FIG. 3.
Graphic representation of the kinetic parameters of MurECt for UDP-MurNAc-l-Ala-d-Glu (A), UDP-MurNAc-Gly-d-Glu (B), and UDP-MurNAc-l-Ser-d-Glu (C). MurE assays were performed as described in Materials and Methods, and the data were fitted to the equation v = VmaxS/(Km + S + [S2]/Ki).
Other amino acids were tested as substrates (Table 2). Significant addition of meso-lanthionine was observed (30% velocity with respect to meso-A2pm). The two other isomers of A2pm (the ll and dd isomers) and d-lysine were also incorporated, but approximately 25-fold (ll-A2pm), 3,700-fold (dd-A2pm), and 3,200-fold (d-lysine) less rapidly than meso-A2pm. In contrast, we failed to detect any significant incorporation of l-ornithine, l-lysine, or l-homoserine under our assay conditions, even with prolonged incubation times and large amounts of enzyme.
TABLE 2.
Specificity of MurECt for the amino acid substrate
| Substrate | Enzymatic activity (nmol·min−1·mg−1)a |
|---|---|
| meso-A2pm | 260 |
| meso-Lanthionine | 80 |
| ll-A2pm | 11 |
| dd-A2pm | 0.07 |
| d-Lysine | 0.08 |
| l-Lysine | NDb |
| l-Ornithine | ND |
| l-Homoserine | ND |
Determined as described in Materials and Methods with fixed concentrations of ATP (5 mM), UDP-MurNAc-l-Ala-d-Glu (0.1 mM), and amino acid (0.1 mM).
ND, no activity detected after overnight incubation with 10 μg of the enzyme.
With regard to the specificity for the nucleotide substrate, the purified MurECt enzyme also exhibited activity with UDP-MurNAc-Gly-d-Glu and UDP-MurNAc-l-Ser-d-Glu (Table 1; Fig. 3B and C). Comparable Km and kcat values for UDP-MurNAc-l-Ala-d-Glu and UDP-MurNAc-l-Ser-d-Glu were determined. While the kcat value for UDP-MurNAc-Gly-d-Glu was approximately twofold higher, a sevenfold-higher Km value was observed for this substrate.
DISCUSSION
The murE gene product from C. trachomatis shares 36% amino acid identity with its E. coli counterpart. Examination of its sequence shows the presence of a meso-A2pm-binding motif (see Introduction), thereby predicting that MurECt is a UDP-MurNAc-l-Ala-d-Glu:meso-A2pm ligase. Chlamydiae possess a functional, although unusual, pathway for the biosynthesis of meso-A2pm (30), indicating that this amino acid may indeed be available for incorporation in the PG biosynthesis pathway.
In the present work, we have cloned the murECt gene and purified the corresponding protein. Two observations support the presence of meso-A2pm at the third position of the peptide in chlamydial PG: (i) complementation of the E. coli murE-defective strain and (ii) the specificity of the purified MurECt protein for the amino acid meso-A2pm.
The behavior of MurECt toward its substrates resembles that of MurEEc and, possibly, that of MurE ligases from most gram-negative bacteria. The preference for meso-A2pm mentioned above is shared by MurEEc, which possesses only very weak activities for addition of l- and d-lysine and of ll- and dd-A2pm, and which exhibits no l-ornithine-adding activity (10, 11, 34, 36). On the other hand, very close analogues of meso-A2pm, such as meso-lanthionine, can be added by MurECt, though with a lower efficiency. Sulfur analogues of meso-A2pm, such as meso-lanthionine and l-allo-cystathionine, can be incorporated by MurEEc and can totally replace meso-A2pm in the PG of genetically engineered E. coli cells (3, 34, 36). The maximum velocity of MurECt reported here (320 nmol·min−1·mg−1) differs those of MurE enzymes in other gram-negative bacteria, with 1.4 μmol·min−1·mg−1 (10) and 2.6 μmol·min−1·mg−1 (42) being reported for MurEEc and MurE from Pseudomonas aeruginosa, respectively. A low catalytic activity (ca. 75 nmol·min−1·mg−1) had also been observed for MurCCt (23). Such low values may be sufficient for the requirements of these slow-growing, primarily intracellular bacteria.
Although the presence of meso-A2pm at the third position of the peptide of chlamydial PG is supported by the present results, the nature of the amino acid at position 1 is still unknown. In most bacteria, this amino acid is l-alanine, but in rare cases, glycine or l-serine occurs (6, 50, 57). In E. coli, where the amino acid at position 1 is l-alanine, the Vmax/Km ratios of MurC are (expressed in relative values) 100, 2.3, and 0.8 for l-alanine, l-serine, and glycine, respectively (26), emphasizing that the preferred amino acid substrate is the one found in the polymer. In contrast, we found that these three amino acids were added by MurCCt with similar efficiencies: the relative Vmax/Km values were 100, 53, and 30, respectively (23). Consequently, it was impossible to deduce which UDP-MurNAc:amino acid ligase activity was catalyzed by chlamydial MurC in vivo. A way of solving this problem could be to study the behavior of the subsequent enzymes toward the nucleotide precursors containing these amino acids. An example is provided by Corynebacterium poinsettiae, where glycine is found at position 1 of PG. Although glycine is the preferred MurC substrate (ca. 70-fold-higher velocity with glycine than with l-alanine), a further discrimination occurs two steps ahead: the velocity of MurE with UDP-MurNAc-Gly-d-Glu is 38-fold higher than that with UDP-MurNAc-l-Ala-d-Glu (58). Therefore, we tried to purify chlamydial MurD. However, despite numerous attempts, we did not succeed in overproducing the enzyme in a soluble, active form (data not shown). Having subsequently purified active MurECt, we logically tested it on the three nucleotides UDP-MurNAc-l-Ala-d-Glu, UDP-MurNAc-l-Ser-d-Glu, and UDP-MurNAc-Gly-d-Glu: the relative kcat/Km values found are 100, 115 and 27, respectively (Table 1). Here again, no straightforward discrimination was found, although the glycine-containing nucleotide is slightly disfavored, as was glycine by MurC. Since chlamydiae may scavenge their amino acids from the host, the amino acid added in vivo may depend on the concentration of available amino acids and the growth conditions, as described for Mycobacterium leprae (29). It is therefore not obvious which amino acid is present at the first position of the pentapeptide chain in chlamydial PG.
MurECt displays maximum activity at pH 8.6. The optimal pH values for the chlamydial enzymes of PG synthesis studied so far are somewhat diverse: 8 to 8.5 for MurC (23), 6 to 8 for Ddl (32), and 6 for MurA (33). In E. coli, the Mur ligases have optimal pH values between 8 and 9.2 (15, 26, 37, 46). Chlamydial MurC and MurE fall within this range. E. coli DdlB and MurA are active over a wide pH range: 6 to 10 for DdlB and 6 to 9 for MurA (24, 43). The pH range found for chlamydial Ddl is within this range. The general preference of these enzyme activities for a slightly alkaline pH is not observed with chlamydial MurA, which is active at a slightly acidic pH and whose activity dramatically drops at pH values higher than 7 (33). An important characteristic of MurACt is the change of the active-site cysteine (position 115 in the E. coli numbering), found in most bacteria, to aspartic acid. Introduction of a C115D mutation in E. coli MurA brings about a shift in the pH dependency (24), which becomes similar to that of MurACt, thereby providing an explanation for the particular pH optimum of the chlamydial enzyme.
Although our understanding of the hypothetical structure of PG in chlamydiae is becoming clearer, the function of this macromolecule remains unknown. A role for chlamydial PG in cell division has been suggested, since treatment with antibiotics targeting enzymes involved in cell wall biosynthesis in other organisms arrests the division of RBs, which become large and aberrant. Chlamydiae lack an identifiable ftsZ homologue (48, 55). In other bacteria, FtsZ is required for cell septation, and in chlamydiae, PG may act as an FtsZ substitute in the assembly of a septum (21). This is supported by the observation that an antigen that may have affinity for a structure similar to PG localizes in ring structures within or between RBs (12).
Using reverse transcription-PCR, McCoy et al. (33) detected murA expression early in the chlamydial cell cycle and murB expression approximately 6 h later. The enzymes involved in the synthesis of meso-A2pm could be detected 8 h postinfection (30). However, microarray analysis demonstrates that murABCDEF transcripts are not present before 16 h postinfection, with the highest levels of expression observed 40 h postinfection (7). This is in agreement with the findings of Nicholson et al. (41), who reported an upregulation of murA 24 to 36 h postinfection. In a separate study, murG was detected 12 and 20 h postinfection using reverse transcription-PCR (51). For C. trachomatis serovar L2, RB production reaches maximal levels at 18 h postinfection, at which point reorganization into EBs is initiated (41). This general lack of correlation between the initiation of RB division and the expression of the enzymes involved in PG biosynthesis may indicate either that the levels of expression are very low and therefore undetectable or that chlamydial PG is not intimately involved in RB cell division. Alternatively, chlamydial PG may be involved in modulating the host immune response and inducing a state of persistence rather than active growth of RBs (31), a possibility that certainly warrants further investigation.
The recent genetic manipulation of Chlamydia psittaci by electroporation with recombinant DNA (8) may provide opportunities to directly manipulate enzymes involved in PG biosynthesis in chlamydiae. Also, the increasing effort to identify novel specific inhibitors of these essential and conserved enzyme targets (18, 42, 44, 53, 56) may help advance our understanding of the role of PG in this important human pathogen.
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique (PICS 2950) and the European Union EUR-INTAFAR project (LSHM-CT-2004512138).
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Abdelrahman, Y. M., and R. J. Belland. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 3.Auger, G., J. van Heijenoort, J. C. Vederas, and D. Blanot. 1996. Effect of analogues of diaminopimelic acid on the meso-diaminopimelate-adding enzyme from Escherichia coli. FEBS Lett. 391:171-174. [DOI] [PubMed] [Google Scholar]
- 4.Babič, A., D. Patin, A. Boniface, M. Hervé, D. Mengin-Lecreulx, S. Pečar, S. Gobec, and D. Blanot. 2007. Chemoenzymatic synthesis of the nucleotide substrates of the Mur ligases, p. 1-4. In D. Kikelj (ed.), 5th Joint Meeting on Medicinal Chemistry, 17 to 21 June, Portorož, Slovenia. Medimond Srl, Bologna, Italy.
- 5.Barbour, A. G., K. Amano, T. Hackstadt, L. Perry, and H. D. Caldwell. 1982. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barreteau, H., A. Kovač, A. Boniface, M. Sova, S. Gobec, and D. Blanot. 2008. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:168-207. [DOI] [PubMed] [Google Scholar]
- 7.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binet, R., and A. T. Maurelli. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc. Natl. Acad. Sci. USA 106:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard, T. J., and D. C. Mabey. 1994. Chlamydial infections. Br. J. Clin. Pract. 48:201-205. [PubMed] [Google Scholar]
- 10.Boniface, A. 2007. Etude des relations structure-activité au sein de la famille des Mur synthétases, enzymes de la voie de biosynthèse du peptidoglycane. Ph.D. thesis. Université Paris-Sud, Orsay, France.
- 11.Boniface, A., A. Bouhss, D. Mengin-Lecreulx, and D. Blanot. 2006. The MurE synthetase from Thermotoga maritima is endowed with an unusual d-lysine adding activity. J. Biol. Chem. 281:15680-15686. [DOI] [PubMed] [Google Scholar]
- 12.Brown, W. J., and D. D. Rockey. 2000. Identification of an antigen localized to an apparent septum within dividing chlamydiae. Infect. Immun. 68:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caravano, A., D. Mengin-Lecreulx, J. M. Brondello, S. P. Vincent, and P. Sinaÿ. 2003. Synthesis and inhibition properties of conformational probes for the mutase-catalyzed UDP-galactopyranose/furanose interconversion. Chem. Eur. J. 9:5888-5898. [DOI] [PubMed] [Google Scholar]
- 14.Chopra, I., C. Storey, T. J. Falla, and J. H. Pearce. 1998. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology 144:2673-2678. [DOI] [PubMed] [Google Scholar]
- 15.Comb, D. G. 1962. The enzymatic addition of d-alanyl-d-alanine to a uridine nucleotide peptide. J. Biol. Chem. 237:1601-1604. [PubMed] [Google Scholar]
- 16.Dagert, M., and S. D. Ehrlich. 1979. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23-28. [DOI] [PubMed] [Google Scholar]
- 17.Dementin, S. 2001. Etude du mécanisme réactionnel des Mur synthétases, enzymes impliquées dans la biosynthèse du peptidoglycane bactérien. Ph.D. thesis. Université Paris-Sud, Orsay, France.
- 18.Dunsmore, C. J., K. Miller, K. L. Blake, S. G. Patching, P. J. F. Henderson, J. A. Garnett, W. J. Stubbings, S. E. V. Phillips, D. J. Palestrant, J. De Los Angeles, J. A. Leeds, I. Chopra, and C. W. G. Fishwick. 2008. 2-Aminotetralones: novel inhibitors of MurA and MurZ. Bioorg. Med. Chem. Lett. 18:1730-1734. [DOI] [PubMed] [Google Scholar]
- 19.Everett, K. D., and T. P. Hatch. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J. Bacteriol. 177:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox, A., J. C. Rogers, J. Gilbart, S. Morgan, C. H. Davis, S. Knight, and P. B. Wyrick. 1990. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect. Immun. 58:835-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghuysen, J. M., and C. Goffin. 1999. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob. Agents Chemother. 43:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon, E., B. Flouret, L. Chantalat, J. van Heijenoort, D. Mengin-Lecreulx, and O. Dideberg. 2001. Crystal structure of UDP-N-acetylmuramoyl-l-alanyl-d-glutamate: meso-diaminopimelate ligase from Escherichia coli. J. Biol. Chem. 276:10999-11006. [DOI] [PubMed] [Google Scholar]
- 23.Hesse, L., J. Bostock, S. Dementin, D. Blanot, D. Mengin-Lecreulx, and I. Chopra. 2003. Functional and biochemical analysis of Chlamydia trachomatis MurC, an enzyme displaying UDP-N-acetylmuramate:amino acid ligase activity. J. Bacteriol. 185:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, D. H., W. J. Lees, K. E. Kempsell, W. S. Lane, K. Duncan, and C. T. Walsh. 1996. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35:4923-4928. [DOI] [PubMed] [Google Scholar]
- 25.Koo, C. W., and J. S. Blanchard. 1999. Chemical mechanism of Haemophilus influenzae diaminopimelate epimerase. Biochemistry 38:4416-4422. [DOI] [PubMed] [Google Scholar]
- 26.Liger, D., A. Masson, D. Blanot, J. van Heijenoort, and C. Parquet. 1995. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Eur. J. Biochem. 230:80-87. [DOI] [PubMed] [Google Scholar]
- 27.Lugtenberg, E. J. J., L. De Haas-Menger, and W. H. M. Ruyters. 1972. Murein synthesis and identification of cell wall precursors of temperature-sensitive lysis mutants of Escherichia coli. J. Bacteriol. 109:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lugtenberg, E. J. J., and A. van Schijndel-van Dam. 1972. Temperature-sensitive mutants of Escherichia coli K-12 with low activity of the diaminopimelic acid adding enzyme. J. Bacteriol. 110:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahapatra, S., D. C. Crick, and P. J. Brennan. 2000. Comparison of the UDP-N-acetylmuramate:l-alanine ligase enzymes from Mycobacterium tuberculosis and Mycobacterium leprae. J. Bacteriol. 182:6827-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy, A. J., N. E. Adams, A. O. Hudson, C. Gilvarg, T. Leustek, and A. T. Maurelli. 2006. l,l-Diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. USA 103:17909-17914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy, A. J., and A. T. Maurelli. 2006. Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 14:70-77. [DOI] [PubMed] [Google Scholar]
- 32.McCoy, A. J., and A. T. Maurelli. 2005. Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting d-alanyl-d-alanine ligase activity involved in peptidoglycan synthesis and d-cycloserine sensitivity. Mol. Microbiol. 57:41-52. [DOI] [PubMed] [Google Scholar]
- 33.McCoy, A. J., R. C. Sandlin, and A. T. Maurelli. 2003. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J. Bacteriol. 185:1218-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mengin-Lecreulx, D., D. Blanot, and J. van Heijenoort. 1994. Replacement of diaminopimelic acid by cystathionine or lanthionine in the peptidoglycan of Escherichia coli. J. Bacteriol. 176:4321-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mengin-Lecreulx, D., T. Falla, D. Blanot, J. van Heijenoort, D. J. Adams, and I. Chopra. 1999. Expression of the Staphylococcus aureus UDP-N-acetylmuramoyl-l-alanyl-d-glutamate:l-lysine ligase in Escherichia coli and effects on peptidoglycan biosynthesis and cell growth. J. Bacteriol. 181:5909-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mengin-Lecreulx, D., C. Michaud, C. Richaud, D. Blanot, and J. van Heijenoort. 1988. Incorporation of ll-diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diaminopimelate epimerase encoded by dapF. J. Bacteriol. 170:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaud, C., D. Mengin-Lecreulx, J. van Heijenoort, and D. Blanot. 1990. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramoyl-l-alanyl-d-glutamate:meso-2,6-diaminopimelate ligase from Escherichia coli. Eur. J. Biochem. 194:853-861. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics, p. 431-435. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moulder, J. W. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect. Agents Dis. 2:87-99. [PubMed] [Google Scholar]
- 41.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paradis-Bleau, C., A. Lloyd, F. Sanschagrin, H. Maaroufi, T. Clarke, A. Blewett, C. Dowson, D. I. Roper, T. D. Bugg, and R. C. Levesque. 2009. Pseudomonas aeruginosa MurE amide ligase: enzyme kinetics and peptide inhibitor. Biochem. J. 421:263-272. [DOI] [PubMed] [Google Scholar]
- 43.Park, I. S., C. H. Lin, and C. T. Walsh. 1996. Gain of d-alanyl-d-lactate or d-lactyl-d-alanine synthetase activities in three active-site mutants of the Escherichia coli d-alanyl-d-alanine ligase B. Biochemistry 35:10464-10471. [DOI] [PubMed] [Google Scholar]
- 44.Perdih, A., A. Kovač, G. Wolber, D. Blanot, S. Gobec, and T. Solmajer. 2009. Discovery of novel benzene 1,3-dicarboxylic acid inhibitors of bacterial MurD and MurE ligases by structure-based virtual screening approach. Bioorg. Med. Chem. Lett. 19:2668-2673. [DOI] [PubMed] [Google Scholar]
- 45.Pompeo, F., J. van Heijenoort, and D. Mengin-Lecreulx. 1998. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J. Bacteriol. 180:4799-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pratviel-Sosa, F., D. Mengin-Lecreulx, and J. van Heijenoort. 1991. Over-production, purification and properties of the uridine diphosphate N-acetylmuramoyl-l-alanine:d-glutamate ligase from Escherichia coli. Eur. J. Biochem. 202:1169-1176. [DOI] [PubMed] [Google Scholar]
- 47.Press, W. H., B. P. Flannery, S. A. Teukolsky, and W. T. Vetterling. 1986. Numerical recipes: the art of scientific computing. Cambridge University Press, Cambridge, United Kingdom.
- 48.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913-925. [DOI] [PubMed] [Google Scholar]
- 52.Skipp, P., J. Robinson, C. D. O'Connor, and I. N. Clarke. 2005. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics 5:1558-1573. [DOI] [PubMed] [Google Scholar]
- 53.Sova, M., G. Čadež, S. Turk, V. Majce, S. Polanc, S. Batson, A. J. Lloyd, D. I. Roper, C. W. G. Fishwick, and S. Gobec. 2009. Design and synthesis of new hydroxyethylamines as inhibitors of d-alanyl-d-lactate ligase (VanA) and d-alanyl-d-alanine ligase (DdlB). Bioorg. Med. Chem. Lett. 19:1376-1379. [DOI] [PubMed] [Google Scholar]
- 54.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 55.Thomson, N. R., M. T. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Connor, I. Goodhead, H. Norbertzcak, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turk, S., A. Kovač, A. Boniface, J. M. Bostock, I. Chopra, D. Blanot, and S. Gobec. 2009. Discovery of new inhibitors of the bacterial peptidoglycan biosynthesis enzymes MurD and MurF by structure-based virtual screening. Bioorg. Med. Chem. 17:1884-1889. [DOI] [PubMed] [Google Scholar]
- 57.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149-167. [DOI] [PubMed] [Google Scholar]
- 58.Wyke, A. W., and H. R. Perkins. 1975. The specificity of enzymes adding amino acids in the synthesis of the peptidoglycan precursors of Corynebacterium poinsettiae and Corynebacterium insidiosum. J. Gen. Microbiol. 88:159-168. [DOI] [PubMed] [Google Scholar]