Abstract
The disaccharide trehalose is a well-known osmoprotectant, and trehalose accumulation through de novo biosynthesis is a common response of bacteria to abiotic stress. In this study, we have investigated the role of endogenous trehalose synthesis in the osmotolerance of Sinorhizobium meliloti. Genes coding for three possible trehalose synthesis pathways are present in the genome of S. meliloti 1021: OtsA, TreYZ, and TreS. Among these, OtsA has a major role in trehalose accumulation under all of the conditions tested and is the main system involved in osmoadaptation. Nevertheless, the other two systems are also important for growth in hyperosmotic medium. Genes for the three pathways are transcriptionally responsive to osmotic stress. The presence of at least one functional trehalose biosynthesis pathway is required for optimal competitiveness of S. meliloti to nodulate alfalfa roots.
Rhizobia are gram-negative soil bacteria that are able to establish nitrogen-fixing symbioses with legume plants under conditions of nitrogen deprivation. During this process, an exchange of molecular signals occurs between the two partners, leading to the formation of the root nodule, where biological nitrogen fixation takes place (12). It has been observed that rhizobium mutants affected in adaptation to high salinity present deficiencies in their symbiotic capacity (27, 7, 25). These results emphasize the importance of studying the adaptation mechanisms of rhizobia to osmotically unbalanced environmental conditions.
Response and adaptation to environmental stresses are probably complex phenomena involving many physiological and biochemical processes that likely reflect changes in gene expression and in the activity of enzymes and transport proteins (6, 43). Rhizobia may use distinct mechanisms in response to hyperosmotic conditions, such as stimulation of potassium uptake (7), changes in cell morphology and size, or modifications in the pattern of extracellular polysaccharides (18, 35, 42). Nevertheless, one of the most general responses for long-term adaptation to osmotic stress is the intracellular accumulation of osmoprotective compounds (24, 45). Trehalose is one of the main endogenous osmolytes synthesized by Sinorhizobium meliloti after an osmotic upshift (24).
As many as three different pathways for trehalose biosynthesis (OtsAB, TreYZ, and TreS) are present in some bacteria, like Corynebacterium glutamicum, Mycobacterium smegmatis, or M. bovis (4, 44). The OtsAB pathway represents the most common route in bacteria, fungi, and plants. This two-step pathway catalyzes the synthesis of trehalose from UDP-glucose and glucose 6-phosphate (16). Trehalose synthase (TreS) catalyzes the reversible conversion of maltose and trehalose. Finally, the two-step TreYZ pathway acts in the production of trehalose from a linear maltodextrin (e.g., glycogen) (5, 17). The existence of these three trehalose synthesis pathways has also been demonstrated in some rhizobia (37). These authors determined the enzymatic activity of the three routes in Bradyrhizobium and observed that TreS activity was prevalent in nodules while the OtsAB pathway appeared as the most important one under free-living conditions.
Trehalose accumulation has been described in rhizobia in response to several stressful conditions such as high external osmolarity, low oxygen concentrations, or desiccation (26, 45, 3, 22). Trehalose also acts as an osmoprotectant when it is exogenously supplied to S. meliloti and Rhizobium leguminosarum, but through a mechanism whereby it is not accumulated (14). On the other hand, this compound is a common disaccharide in the root nodules of legumes and is present in bacteroids at the onset of nitrogen fixation (36), which could reflect an important role for this molecule in symbiosis.
Despite the interest in studying the osmotic adaptation mechanisms in rhizobia, the characterization of the various trehalose biosynthetic pathways and their contribution to the osmotic stress tolerance of these bacteria has not been systematically approached at the genetic level. In a recent work, R. leguminosarum bv. trifolii mutants affected in trehalose biosynthesis have been described (22). In this bacterium, a double mutant lacking the OtsA and TreY pathways was more sensitive to desiccation stress. Furthermore, an R. etli otsA gene mutant, although still capable of accumulating trehalose to a certain extent, was affected in its ability to establish an efficient symbiosis with Phaseolus vulgaris and showed an increased sensitivity to osmotic stress (38). In S. meliloti strain 1021, the induction of several genes related to trehalose biosynthesis was detected in response to a sudden increase in external osmolarity (8).
In this work, we have addressed the genetic and functional characterization of three trehalose biosynthesis systems in S. meliloti 1021 and their role in nodulation of alfalfa roots.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. meliloti strains were grown at 30°C in TY complex medium (1) or in minimal medium (MM) containing glutamate (7.5 mM), mannitol (55 mM), and mineral salts (K2HPO4 at 1.3 mM, KH2PO4 · 3H2O at 2.2 mM, MgSO4 · 7H2O at 0.6 mM, CaCl2 · 2H2O at 0.34 mM, FeCl3 · 6H2O at 0.022 mM, and NaCl at 0.86 mM) (30). When a specific pH was required, the MM was buffered with Tris/morpholineethanesulfonic acid (MES) at 5 mM. Escherichia coli strains were propagated in Luria-Bertani medium (31). When required, antibiotics were added at the following final concentrations: for E. coli, streptomycin (Sm) at 50 μg/ml, spectinomycin (Sp) at 50 μg/ml, kanamycin (Km) at 50 μg/ml, gentamicin (Gm) at 20 μg/ml, tetracycline (Tc) at 10 μg/ml, and ampicillin (Ap) at 200 μg/ml; for S. meliloti, Sm at 200 μg/ml, Sp at 100 μg/ml, Km at 200 μg/ml, Gm at 15 μg/ml, and Tc at 0.1 μg/ml for chromosomal insertions or 5 μg/ml for plasmid selection.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or gene | Relevant characteristics or primer sequencesa | Reference |
|---|---|---|
| S. meliloti | ||
| 1021 | SU47 str-21 (wild type) Smr | 23 |
| 10OtSS | 1021 (ΔotsA::Sm/Sp) Smr Spr | This work |
| 10MOTK | 1021 (ΔtreY::Km) Smr Kmr | This work |
| 10treS | 1021 (treS::pSUP202Pol4) Smr Tcr | This work |
| 10OtM | 10OtSS (ΔtreY::Km) Smr Spr Kmr | This work |
| 10treSY | 10treS (ΔtreY::Km) Smr Tcr Kmr | This work |
| 10trOt | 10treS (ΔotsA::Sm/Sp) Smr Tcr Spr | This work |
| 10SYOt | 10treSY (ΔotsA::Sm/Sp) Smr Kmr Tcr Spr | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 φ80 lacZΔM recA1 endA1 gyrA96 thi-1 relA1 5hsdR171 | Bethesda Research Laboratories |
| S17-1 | thi pro recA hsdR hsdM Rp4Tc::Mu Km::Tn7 Tpr Smr Spr | 33 |
| Plasmids | ||
| pBSKS(+) | Cloning vector; Apr | Stratagene |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pHP45Ω | Plasmid containing Sm/Sp cassette; Apr Smr Spr | 29 |
| pHP45 Ω-Km | Plasmid containing Km cassette; Apr Kmr | 9 |
| pK18mobsacB | Suicide plasmid; Kmr | 32 |
| pSUP202Pol4 | Suicide plasmid; Tcr | 11 |
| pGUS3 | Plasmid containing nfeD::gus fusion; Kmr | 13 |
| p53Gus | Plasmid containing uidA gene; Gmr | L. Girard (CCG, Mexico) |
| p53otsA | p53Gus derivative containing a otsA::gus fusion; Gmr | This work |
| p53treY | p53Gus derivative containing a treY::gus fusion; Gmr | This work |
| p53treS | P53Gus derivative containing a treS::gus fusion; Gmr | This work |
| pJB3Tc19 | Plasmid RK2 derivative; Tcr Apr | 2 |
| pJB3otsA | pJB3Tc19 derivative containing a PCR-amplified fragment with the otsA gene from S. meliloti 1021; Tcr | Our laboratory |
| pJB3treY | pJB3Tc19 derivative containing a PCR-amplified fragment with the treY gene from S. meliloti 1021; Tcr | Our laboratory |
| pJB3treS | pJB3Tc19 derivative containing a PCR-amplified fragment with the treS gene from S. meliloti 1021; Tcr | Our laboratory |
| Genes | ||
| otsA (SMa0322) | TTATCTAGAGCAGGTCCATGGTTTCGATA/TAATCTAGAGAAATCTAGTCACTCTGGACACb | This work |
| treY (SMb20574) | TAAGGATCCGTCGAAGACGTGCTTGAAGA/TAAGGTACCACGCTCGAATGGCTGATCCT | This work |
| treS (SMb20099) | TAATCTAGACGTCTGCTTTTTCGCTACAT/TAATCTAGACCGACATTGTGGAGGTAGAT | This work |
Smr, streptomycin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Gmr, gentamicin resistance; Tpr, trimethoprim resistance; Apr, ampicillin resistance; Tcr, tetracycline resistance.
Forward/reverse primer sequences (5′-3′) are shown. Underlined are the endonuclease XbaI action sites used to clone the mutated gene versions into the shuttle vector.
For the different experiments, MM was inoculated from cultures grown in TY with antibiotics to stationary phase. During the experiments, antibiotics were only added to avoid the possibility of plasmid loss in the case of transcriptional studies. When osmotic stress conditions were required, the osmolarity of the medium was increased by addition of appropriate amounts of NaCl from a 4 M stock solution (made in MM) or sucrose. These conditions were either imposed from the beginning of the experiment or, in the case of experiments involving an osmotic shock, when the culture reached mid-exponential phase. Growth was followed by measuring the optical density at 600 nm in cultures incubated in a rotary shaker or by comparative growth on solid medium.
Genetic procedures and construction of S. meliloti mutants.
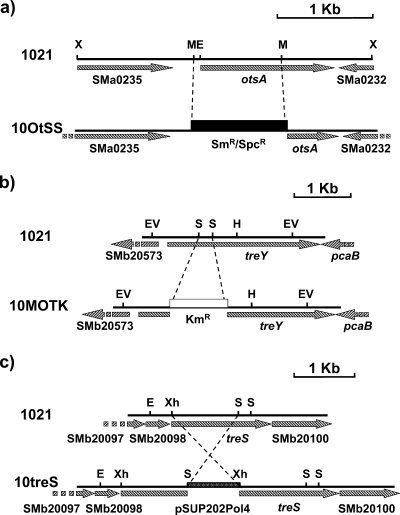
Standard techniques were used for genetic manipulations (31). To construct plasmids for the disruption of the different genes, we first amplified the corresponding regions of the S. meliloti genome by PCR with custom-synthesized oligonucleotides (Table 1). After cloning the amplicons into appropriate vectors, we deleted part of the gene sequence by endonuclease digestion and inserted antibiotic resistance cassettes to obtain OtsA and TreY mutant constructs in vitro. The constructs were then subcloned into the shuttle vectors, and the plasmids obtained were transferred by conjugation to S. meliloti 1021. In the case of the TreS system, an internal fragment from treS was directly cloned into the shuttle vector. Transconjugants were selected on medium containing the appropriate antibiotics. In the case of pK18mobsacB, the use of sucrose addition during selection as described by Schäfer and associates (32) was avoided since its osmotic action could affect the growth of osmosensitive mutants. Double and triple mutants were obtained by transduction with bacteriophage φM12 as described by Finan and associates (10). Disruption of the genes of interest was confirmed by Southern hybridization with specific gene probes.
Determination of the trehalose contents of cells.
For the analysis of trehalose content, four aliquots of the bacterial culture were collected by centrifugation. Cells were washed with water and then extracted in 75% (vol/vol) ethanol at room temperature for 24 h. The supernatants were collected, and the pellet was extracted once more. The resulting supernatants were combined and dried in a SpeedVac (SPD111V; ThermoSavant). The solids were dissolved in 250 μl of 135 mM citrate buffer, pH 5.7, and then preheated to 37°C, and three of the aliquots were treated with 0.008 U trehalase (Sigma). After 1 h at 37°C, the reaction was stopped by adding 250 μl of 500 mM Tris buffer, pH 7.5, and the glucose derived from the trehalose present in the sample was determined with the glucose (GO) assay kit (Sigma) using the non-trehalase-treated aliquot as blank. Mean values were calculated from the three replicates, and differences between samples were assessed by using a single-factor analysis of variance (ANOVA) test with five degrees of freedom (P < 0.01). The experiments presented were performed at least twice.
Gene expression assays.
Three different fusions to the β-glucuronidase gene uidA were constructed by cloning the promoter regions from otsA, treY, and treS in the appropriate direction into plasmid p53Gus. The promoter regions from otsA and treY were obtained by endonuclease digestion (XbaI-EcoRI for otsA and EcoRI-XhoI for treY) of the PCR fragments used for mutagenesis. For the treS fusion, primers PTreS-1 (5′-TTTTATCTAGACAGGACGAGATTAGAAGGTT-3′) and PTreS-2 (5′-TTTAATCTAGAGATATCGATTCCATAGATGACCGAGC-3′) were used to amplify a 4,040-bp DNA fragment containing part of treS and the upstream genes and promoter region of the putative operon in which treS is included. The PCR fragment was cloned as an XbaI fragment into p53Gus (restriction enzyme sites underlined in the primer sequences), and the correct direction was selected to obtain the treS::uidA fusion. The p53Gus derivatives were then introduced into S. meliloti 1021 by conjugation. To quantify β-glucuronidase activity, three aliquots of the bacterial culture were collected by centrifugation, washed, and resuspended in assay buffer (dithiothreitol, 5 mM; EDTA, 1 mM; Na2HPO4-NaH2PO4, 50 mM). A 200-μl portion of each aliquot was used to determine the β-glucuronidase activity by mixing with 740 μl assay buffer, 50 μl 0.1% sodium dodecyl sulfate, and 100 μl chloroform and vortexing for 15 s twice. Samples were incubated at 37°C for 10 min, and 10 μl of 100 mM 4-nitrophenyl-β-d-glucuronide, preheated at 37°C, was then added. When the mixture turned yellow, the reactions were ended by adding 200 μl of 1 M Na2CO3. Samples were centrifuged for 5 min (12,000 rpm), and the absorbance of the upper phase was measured at 405 nm. β-Glucuronidase activity was expressed in Miller units. Mean values were calculated from three different replicates, and the experiments presented were performed at least twice.
Plant assays.
Alfalfa (Medicago sativa L. cv. Aragon) seeds were sterilized, germinated in the dark, and then transferred to glass tubes or Leonard jars for growth (28).
To test the infectivity of the rhizobial strains, 24 individual plants were inoculated with each rhizobial suspension (106 CFU/plant). After inoculation, the number of nodulated plants and the number of nodules per plant were recorded daily. When needed, osmotic stress conditions were imposed by addition of NaCl to the nutrient solution at the time of inoculation to a final concentration of 75 mM.
For nodulation competitiveness tests, 12 plants were individually inoculated with 1:1 mixtures of strain 1021(pGUS3) and each of the strains tested. Nodule occupancy was determined as described previously (7). All of the nodules from the 12 inoculated plants were screened (approximately 100 nodules). The percentage of nodules occupied by the strain tested was recorded for each plant. The results for any given strain were compared to those obtained with wild-type strain 1021 in the same experiment by using a single-factor ANOVA test with 23 degrees of freedom (P < 0.05).
To study the symbiotic effectiveness of the strains, 32 individual plants grown in Leonard jars (8 plants per jar) were inoculated with rhizobial suspensions (109 CFU/plant). Seven weeks after inoculation, the plants were collected and the extent of nitrogen fixation was assessed by measurement of the shoot dry weight and its nitrogen content. When required, osmotic stress conditions were imposed 3 weeks after inoculation by addition of NaCl to the nutrient solution to a final concentration of 75 mM, the highest salt concentration tested that did not compromise the growth and development of alfalfa.
RESULTS
Genes for trehalose biosynthesis in S. meliloti 1021.
An in silico analysis of the S. meliloti 1021 genome revealed the presence of putative genes coding for three trehalose biosynthesis systems: OtsA, TreYZ, and TreS. A gene annotated as otsA (SMa0233) is located in plasmid pSymA, and its product presents high sequence similarity to trehalose 6-phosphate synthases from other rhizobia (46% to 94% sequence identity). The OtsAB pathway involves a second step, which is catalyzed by the OtsB protein. We could not find any open reading frame in the S. meliloti 1021 genome encoding an OtsB-like product. Nevertheless, the activity of this biosynthetic pathway has been previously reported (37).
The S. meliloti gene SMb20574, located in plasmid pSymB, is annotated as a putative maltooligosyl-trehalose synthase, and in fact its product presents 32 to 34% identity with TreY from Mycobacterium or Corynebacterium and up to 47% identity with TreY from R. leguminosarum bv. trifolii (4, 44, 22). Although no treZ gene has been annotated in the S. meliloti genome, a gene homologous to treZ could be identified. The product of gene SMb21447, annotated as glgB2 (encodes a putative 1,4α-glucan branching enzyme), presents significant sequence similarity to TreZ from Arthrobacter sp. strain Q36 (38% identity) or Rhizobium sp. strain M11 (37% identity) (20, 21). Although both glgB and treZ code for α-amylases, GlgB is well conserved among distantly related bacteria and the product of the S. meliloti open reading frame SMc03922, annotated as glgB1, presents as much as 53% identity with E. coli GlgB, which in turn shows only 27% identity with the N-terminal part of the SMb21447 product. Due to its high homology to treZ genes and its low similarity to glgB genes, we concluded that SMb21447 is misannotated in the S. meliloti 1021 genome and should be renamed treZ.
Finally, the gene SMb20099, located in the S. meliloti 1021 plasmid pSymB, is annotated as a maltose α-d-glucosyltransferase, an alternative name for trehalose synthase. The product of the SMb20099 gene presents significant sequence similarity (35 to 40% identity) to known trehalose synthases from M. tuberculosis, C. glutamicum, or Pimelobacter sp. strain R48 (41, 4, 44). treS-like genes are commonly found in rhizobial genomes, and some may carry more than one copy. In S. meliloti, treS is the fifth gene of a putative operon of six genes (SMb20095 to SMb20100). None of the other five genes has a specific function assigned.
To study the importance of each of the three possible trehalose biosynthesis systems identified in the S. meliloti 1021 genome, we constructed strains lacking each of them by allelic exchange with interrupted versions of the genes otsA, treY, and treS (Fig. 1). We also obtained double mutants retaining only one of the systems described and a triple mutant devoid of all three systems.
FIG. 1.
Construction of S. meliloti 1021 otsA (a), treY (b), and treS (c) mutants. The wild-type version of the amplified region obtained by PCR is represented, and the mutated version is shown below. E, EcoRI; M, MluI; EV, EcoRV; S, SmaI; H, HindIII; Xh, XhoI; KmR, kanamycin resistance cassette; SmR/SpcR, streptomycin/spectinomycin resistance cassette.
Role of trehalose biosynthesis systems in S. meliloti osmotolerance.
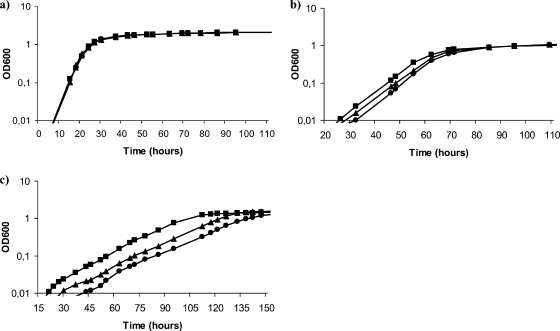
To check whether any mutation could cause growth defects in S. meliloti, the different mutants were tested in TY complex medium and in defined MM. In both media, all mutants displayed wild-type growth (data not shown). To determine the involvement of each trehalose biosynthesis system in osmotolerance, growth curves of all of the mutants in liquid MM containing different concentrations of NaCl (0.3 M, 0.4 M, or 0.5 M) or sucrose (0.5 M or 0.7 M) were obtained. Among all of the mutants tested, only 10trOt (TreS− OtsA−) and 10SYOt (TreS− TreY− OtsA−), lacking both the OtsA and TreS systems, presented a significant growth delay in medium containing NaCl or sucrose. For both mutants, the growth delay was only detectable at the highest concentrations of osmolytes tested and was more evident in medium with NaCl than with sucrose addition (Fig. 2). Thus, in medium containing 0.5 M NaCl, the lag phase was about 10 h longer in the double mutant 10trOt (TreS− OtsA−) than in the parental strain. A stronger growth delay was observed in the triple mutant, which presented a lag phase approximately 25 h longer than that of the parental strain, thus revealing the involvement of TreY in the osmotolerance of S. meliloti in the absence of OtsA and TreS.
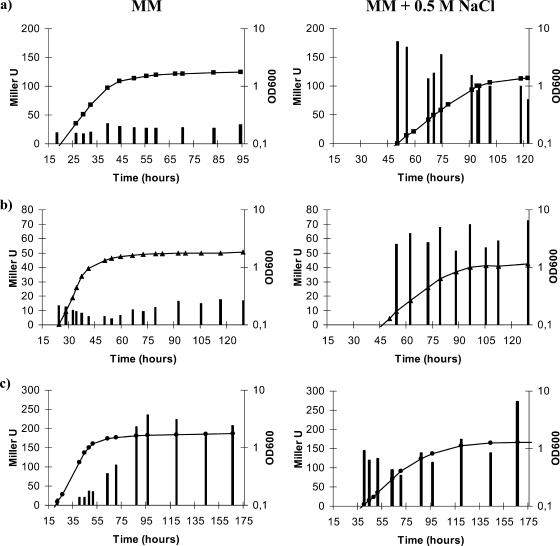
FIG. 2.
Growth curves of S. meliloti 1021-derived strains in MM (a), MM supplemented with 0.7 M sucrose (b), and MM supplemented with 0.5 M NaCl (c). Only the growth curves of mutants showing significant differences with the wild-type strain are shown. Wild-type strain 1021, squares; TreS− OtsA− mutant 10trOt, triangles; TreS− TreY− OtsA− mutant 10SYOt, circles. The data shown are representative of at least two independent experiments. OD600, optical density at 600 nm.
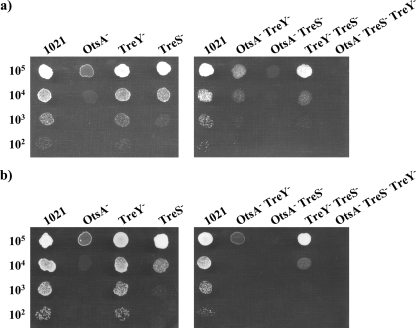
On the other hand, the study of bacterial growth on solid MM revealed interesting differences among the mutants. For these experiments, we used buffered MM plates at different pHs (6, 6.5, and 7) and containing different concentrations of NaCl (0.3 M, 0.4 M, or 0.5 M) or sucrose (0.5 M, 0.6 M, or 0.7 M). Wild-type strain 1021 was unable to grow at the lowest pH combined with the highest concentrations of NaCl or sucrose. The lowest concentrations of osmolytes used (0.3 M NaCl and 0.5 M sucrose) had no differential effect on the growth of the different mutants and the wild type (data not shown). However, at higher solute concentrations, we could observe a reduction in the tolerance of all of the mutants except 10MOTK (TreY−) (an illustrative example is presented in Fig. 3). In these experiments, the triple mutant 10SYOt (TreS− TreY− OtsA−) again displayed the lower levels of osmotolerance, followed by the double mutant 10trOt (TreS− OtsA−), mutants 10OtM (OtsA− TreY−) and 10OtSS (OtsA−), and finally mutant 10treS (TreS−). Thus, the observed negative effects were solute dose dependent and stronger at lower pHs and, as in liquid medium, were more apparent with NaCl than sucrose addition (data not shown).
FIG. 3.
Ability of 1021-derived mutants to grow in MM with 0.5 M NaCl added at pH 7 (a) and pH 6.5 (b). In each row, drops contain approximately the number of CFU indicated at the left. A representative example of at least two experiments is shown. Pictures were taken after 6 to 8 days of incubation under the indicated conditions.
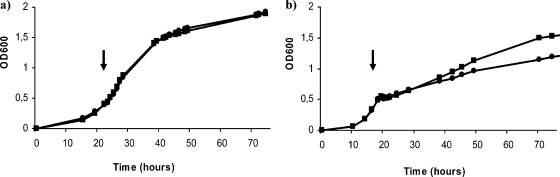
The behavior of the different mutants pointed out the importance of trehalose biosynthesis systems for S. meliloti adaptation to hyperosmotic conditions. To determine whether the corresponding genes were only involved in long-term adaptation or also important for adaptation to osmotic shock, growth experiments using the triple mutant 10SYOt (TreS− TreY− OtsA−) were conducted in which osmotic stress was imposed on exponentially growing cultures by addition of NaCl to a final concentration of 0.4 M, 0.5 M, or 0.6 M. Again, the addition of the highest NaCl concentration had a negative effect on the mutant's growth ability compared to that of the wild-type strain (Fig. 4), suggesting an involvement of trehalose biosynthesis systems in the early stages of the adaptation to hyperosmotic conditions. Nevertheless, the behavior of the mutant after such a sudden osmotic shock differed from that observed when the osmotic stress was imposed from the beginning of the culture (compare Fig. 4 and 3). The lag phase after the NaCl shock was not particularly extended in the mutant, but once it resumed, growth was slower in the mutant, which was unable to reach the optical density values of the wild-type strain.
FIG. 4.
Growth curves of strains 1021 (wild type; squares) and 10SYOt (TreS− TreY− OtsA− mutant; circles) in MM with NaCl added at the times indicated by the arrows to a final concentration of 0 M (a) or 0.6 M (b). The data shown are representative of at least two independent experiments. OD600, optical density at 600 nm.
Expression of trehalose biosynthesis genes in S. meliloti 1021.
The expression of the otsA, treY, and treS genes in S. meliloti was assayed throughout growth in MM. All of the genes presented low levels of expression during exponential growth on MM, but induction of treS transcription could be detected upon entry into stationary phase, whereas otsA and treY expression remained virtually unchanged throughout the growth curve (Fig. 5). However, the expression of otsA and treY was significantly increased during growth on MM with NaCl added (Fig. 5). Induction of treS expression could also be detected under these conditions but to a lower extent than that of otsA and treY, and the effect was masked during early stationary phase due to the induction of the gene at this stage. Gene expression at different NaCl concentrations (0.4 M and 0.5 M) was also studied, and dose-dependent induction could be assessed (data not shown). For instance, the expression of otsA at mid-exponential phase in MM was around 20 Miller units, 40 in MM with 0.4 M NaCl added, and 130 when the NaCl concentration was 0.5 M. These observations were corroborated by studying gene expression after the addition of different concentrations of NaCl (0.5 M and 0.6 M) to exponentially growing cultures (NaCl shock). The addition of the highest salt concentration also resulted in the highest expression levels for all three genes (data not shown).
FIG. 5.
Expression of the transcriptional fusions otsA::uidA (a), treY::uidA (b), and treS::uidA (c) during growth in MM (left) or MM plus 0.5 M NaCl (right). The data shown are representative of two independent experiments. OD600, optical density at 600 nm; Miller U, β-glucuronidase activity expressed as Miller units.
Trehalose accumulation.
The accumulation of trehalose in R. leguminosarum bv. trifolii has been reported to reach a peak at the early stationary phase, declining afterwards (22). To establish how the disaccharide accumulates in S. meliloti, the levels of trehalose were determined in the wild-type strain throughout the growth curve. We observed that in S. meliloti 1021, trehalose accumulated during exponential growth in MM and its levels continued to increase in the stationary phase, reaching a maximum at the end of the growth curve (Fig. 6a). To determine the involvement of the different biosynthetic systems in trehalose accumulation, the amounts of trehalose present in different mutants were measured at the late stationary phase of cultures. We observed that the mutants lacking OtsA or TreY (10OtSS and 10MOTK) presented reduced levels of trehalose with respect to the parental strain, whereas mutants lacking both systems (10OtM and 10SYOt) showed a further reduction of the accumulated trehalose, suggesting that accumulation of trehalose in the stationary phase is mediated by the OtsA and TreY systems (Fig. 6b).
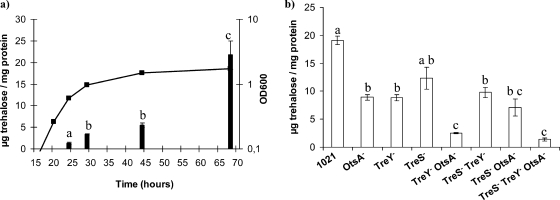
FIG. 6.
Trehalose accumulation by S. meliloti 1021 during growth in MM (a) and by 1021-derived mutants at the late stationary phase of growth (70 h after inoculation) (b). OD600, optical density at 600 nm. Bars correspond to standard errors, and different letters indicate significant differences according to an ANOVA test (P < 0.01). The data are representative of two independent experiments.
The ability of the parental strain S. meliloti 1021 to accumulate trehalose upon NaCl addition to exponentially growing cultures was also tested by using different concentrations of the stressing compound. We found that trehalose accumulation was part of the response to osmotic stress, but in contrast to the dose-dependent induction of the biosynthetic genes, the amount of trehalose accumulated was lower with the highest concentration of NaCl tested (Fig. 7a).
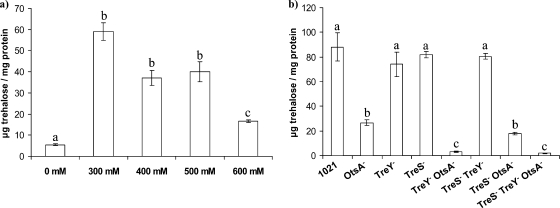
FIG. 7.
Trehalose accumulation by S. meliloti 1021 after an osmotic shock caused by the addition of different NaCl concentrations to mid-exponential-phase cultures growing in MM (a) and by 1021-derived mutants after an osmotic shock with 0.3 M NaCl (b). Cells were harvested 24 h after the saline shock. Bars correspond to standard errors, and different letters indicate significant differences according to an ANOVA test (P < 0.01). The data shown are representative of two independent experiments.
We also determined the trehalose accumulated by each of the mutants after a 0.3 M NaCl osmotic shock. The results indicated that the main system involved in trehalose accumulation under these conditions was OtsA, although in the absence of OtsA, the involvement of TreY became apparent, so that the lack of both systems caused a further reduction in trehalose accumulated in 10OtM (OtsA− TreY−) and 10SYOt (TreS− TreY− OtsA−) (Fig. 7b). Trehalose accumulation by the triple mutant 10SYOt (TreS− TreY− OtsA−) recovered to the levels of wild-type strain 1021 upon complementation with the otsA gene alone (pJB3otsA), while complementation with the treY gene (pJB3treY) restored trehalose accumulation in the triple mutant to the levels of the double mutant 10trOt (TreS− OtsA−), still showing the effect of the missing OtsA pathway (data not shown). The triple mutant 10SYOt (TreS− TreY− OtsA−), the triple mutant complemented with the treS gene (pJB3treS), and the double mutant 10OtM (OtsA− TreY−) accumulated comparably low amounts of trehalose under these conditions, showing the lack of involvement of the TreS system in the accumulation process (data not shown).
Importance of trehalose biosynthesis systems for symbiosis.
All of the mutants obtained were able to induce nitrogen-fixing root nodules on alfalfa plants and displayed nodulation kinetics comparable to that of wild-type strain 1021 (data not shown). Furthermore, the nodulation kinetics of the triple mutant in a solution containing 75 mM NaCl was identical to that of the wild-type strain (data not shown). The symbiotic effectiveness of the triple mutant, measured as the dry weight or nitrogen content of the shoots of inoculated plants was, as well, comparable to that of the wild-type strain both in the absence and in the presence of 75 mM NaCl (data not shown).
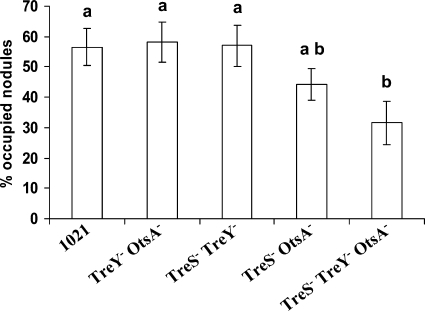
Nevertheless, the triple mutant 10SYOt (TreS− TreY− OtsA−) showed a significant reduction in competitive nodule occupancy (Fig. 8). Although the double mutant 10trOt (TreS− OtsA−) presented a tendency to be less competitive than the wild type or the other double mutants, the difference was not statistically significant. These results underline the importance of trehalose biosynthesis by S. meliloti for optimal nodulation competitiveness.
FIG. 8.
Nodule competition assays. The data shown represent the percentages of nodules occupied by the indicated strain after coinoculation (ratio 1:1) with strain 1021 (pGUS3). Bars correspond to standard errors, and different letters indicate significant differences according to an ANOVA test (P < 0.05). The data shown are representative of two independent experiments.
DISCUSSION
Many rhizobia accumulate trehalose in response to different stresses (26, 3). The biosynthesis of this disaccharide occurs in most organisms through the OtsAB pathway, although the coexistence of up to three different trehalose biosynthetic systems in some bacteria has been previously reported (4, 44, 19). The present study demonstrates that S. meliloti 1021 carries genes for three likely trehalose biosynthetic pathways (OtsA, TreYZ, and TreS) and that all are involved in the osmoadaptive response.
The relevance of trehalose biosynthesis for osmotic tolerance could only be revealed at very high osmolyte concentrations. Trehalose has been previously proposed to act in the osmotic adaptation of S. meliloti only under conditions of harsh stress, since the accumulation of other compounds like glutamate or N-acetyl glutaminyl glutamine amide during the exponential growth phase could protect the cell at lower stress levels (34). We assessed differences in the relative importance of each of the three systems for osmoadaptation, the OtsA system being the most relevant one in this process. The OtsAB pathway has been reported to be active in S. meliloti 1021 under free-living conditions and is known to be involved in the osmotic tolerance of other rhizobia (37, 38). Nevertheless, we could also observe a role for treS in the osmotolerance of S. meliloti 1021, since the absence of both OtsA and TreS caused a reduction of the growth ability of the strain at high solute concentrations compared to otsA single mutants. The possibility of a polar effect of the treS mutation on the downstream gene (Smb20100) cannot be ruled out, and in fact, the complete operon (SMb20095 to SMb20100) could be involved in osmoadaptation since all of the genes are upregulated after an NaCl shock (9). On the other hand, this is the first report of the possible involvement of TreS in the osmoadaptation ability of rhizobia, although this system is known to be active in Bradyrhizobium japonicum and its activity increases during symbiosis to become the dominant trehalose biosynthetic pathway in the bacteroids of this species (37). The minor relevance of the TreYZ system during the osmotic adaptation of S. meliloti 1021 observed in our studies contrasts with the reported involvement of both TreYZ and OtsAB in the desiccation tolerance of R. leguminosarum bv. trifolii (22). Nevertheless, we observed that the TreYZ pathway is involved in trehalose accumulation under various growth conditions, although its importance during osmotic adaptation was clearly lower than that of the OtsA system.
It has been reported that the accumulation of trehalose in S. meliloti in response to osmotic stress is maximal at the end of the exponential growth phase and the beginning of the stationary growth phase (39). A similar result was obtained for R. leguminosarum bv. trifolii during growth under nonstress conditions (22). However, our studies show that S. meliloti 1021 accumulates trehalose during growth, reaching maximum amounts at late stationary phase. Furthermore, this accumulation depends mainly on the activity of both the OtsA and TreYZ systems. On the other hand, gene expression studies showed that, as observed in R. leguminosarum bv. trifolii (22), the S. meliloti 1021 otsA and treY genes display low constitutive expression levels during growth in osmotically balanced medium. Only treS expression was enhanced upon entry into stationary phase; however, TreS was apparently not involved in trehalose accumulation. These results suggest that trehalose accumulation during the stationary phase could be controlled at the posttranscriptional level or through control of trehalose breakdown rates. Our studies also revealed that in S. meliloti 1021, the response to osmotic stress includes induction of the otsA, treY, and treS genes, in a dose-dependent manner, both after an osmotic shock and in response to prolonged growth on saline medium. These results confirm our previous microarray data showing upregulation of the three genes after an osmotic upshift (8).
It was unexpected to observe that the osmolyte concentration yielding the highest gene expression levels in S. meliloti 1021 did not cause maximal trehalose accumulation. While the expression levels of the three genes positively correlated with increasing NaCl concentrations, the amount of accumulated trehalose decreased at very high NaCl concentrations (Fig. 7). Such a paradoxical situation is not without precedents. In C. glutamicum, otsA and treS expression increase significantly in response to osmotic stress, whereas treY transcription remains unchanged despite TreY being the main system involved in the osmoregulated synthesis of trehalose in this bacterium (44). Likewise, the transcriptional profile of the trehalose biosynthetic genes in S. meliloti may not necessarily correlate with their relative importance during osmotic adaptation, which would again suggest the existence of other control mechanisms in addition to transcriptional regulation. Nevertheless, the lower amounts of trehalose accumulated at the highest osmolarities could reflect a metabolic compromise caused by the severe stress conditions imposed rather than differential regulation. Thus, the inability of the metabolic systems to keep high trehalose biosynthesis rates under conditions of harsh stress would cause the observed reduction in disaccharide accumulation.
None of the trehalose biosynthesis systems studied in S. meliloti 1021 was essential for nodulation or nitrogen fixation, even under osmotic stress conditions. Nevertheless, the lack of the corresponding genes caused a significant reduction in the competitive ability of the triple mutant, which is in accordance with the observation by McIntyre and associates (22) that an R. leguminosarum bv. trifolii mutant unable to accumulate trehalose was less competitive than the parental strain for nodule occupancy but capable of nodulation and nitrogen fixation. It has also been reported that S. meliloti 1021 mutants defective in trehalose catabolism present higher rates of nodule occupancy than their parental strain but a similar nitrogen fixation ability (15). As suggested by these authors, the ability to accumulate trehalose (either by increasing biosynthesis or by reducing catabolism) could help bacteria to tolerate infection-related stresses during early stages of the interaction.
The situation in S. meliloti and R. leguminosarum seems different from that in R. etli, where an otsA mutant still able to accumulate trehalose to a certain extent displayed reduced nodulation and lower nitrogenase activity in symbiosis with Phaseolus vulgaris and consequently reduced plant biomass (38). In contrast, S. meliloti (this study) or R. leguminosarum (22) strains virtually unable to accumulate trehalose seem to display a more discrete symbiotic phenotype. Suárez and associates (38) suggested that the reduction in symbiotic effectiveness could be related to a putative role for trehalose as a signal molecule coordinating nitrogen and carbon metabolism and responses to stress. Taken together, these results suggest that the activity of rhizobial trehalose as a protectant compound and/or as a signal molecule may depend on the nature of the symbiotic partners. Our results show that in S. meliloti 1021, trehalose biosynthesis becomes important during the response to harsh stress conditions. The presence within the plant of molecules such as proline betaine, able to act as osmoprotectants for S. meliloti, has been reported in alfalfa (40). Such osmoprotectants would make trehalose biosynthesis unnecessary for stress adaptation during nodule development, although its production could still be an advantage during the early stages of the symbiotic interaction, as proposed by Jensen and associates (15).
Acknowledgments
This work was supported by grants AP2000-3118 and I3P-postgrado-2007 to A.D.F. and BOS2002-04182-C02-01, BIO2005-08089-CO2-01, and BIO2008-02447 to J.S.
The assistance of Socorro Muñoz and help by Salwa Moussaid are gratefully acknowledged.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 2.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cytryn, E. J., D. P. Cangurdekar, J. G. Streeter, W. L. Franck, W. S. Chang, G. Stacey, D. W. Emerich, T. Joshi, D. Xu, and M. J. Sadowsky. 2007. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 189:6751-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smet, K. A., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 5.Di Lernia, I., A. Morana, A. Ottombrino, S. Fusco, M. Rossi, and M. De Rosa. 1998. Enzymes from Sulfolobus shibatae for the production of trehalose and glucose from starch. Extremophiles 2:409-416. [DOI] [PubMed] [Google Scholar]
- 6.Djordjevic, M. A., H. C. Chen, S. Natera, G. Van Noorden, C. Menzel, S. Taylor, C. Renard, O. Geiger, the Sinorhizobium DNA Sequencing Consortium, and G. F. Weiller. 2003. A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant-Microbe Interact. 16:508-524. [DOI] [PubMed] [Google Scholar]
- 7.Domínguez-Ferreras, A., S. Muñoz, J. Olivares, M. J. Soto, and J. Sanjuán. 2009. Role of potassium uptake systems in Sinorhizobium meliloti osmoadaptation and symbiotic performance. J. Bacteriol. 191:2133-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domínguez-Ferreras, A., R. Pérez-Arnedo, A. Becker, J. Olivares, M. J. Soto, and J. Sanjuán. 2006. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 188:7617-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 10.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, H. M., M. Babst, T. Kaspar, G. Acuña, F. Arigoni, and H. Hennecke. 1993. One member of a groESL-like chaperonin multigene family of Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12:2901-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Rodríguez, F. M., and N. Toro. 2000. Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation. Mol. Plant-Microbe Interact. 13:583-591. [DOI] [PubMed] [Google Scholar]
- 14.Gouffi, K., N. Pica, V. Pichereau, and C. Blanco. 1999. Disaccharides as a new class of nonaccumulated osmoprotectants for Sinorhizobium meliloti. Appl. Environ. Microbiol. 65:1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, J. B., O. Y. Ampomah, R. Darrah, N. K. Peters, and T. V. Bhuvaneswari. 2005. Role of trehalose transport and utilization in Sinorhizobium meliloti-alfalfa interactions. Mol. Plant-Microbe Interact. 18:694-702. [DOI] [PubMed] [Google Scholar]
- 16.Kaasen, I., P. Falkenberg, O. B. Styrvold, and A. R. Strøm. 1992. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by KatF (AppR). J. Bacteriol. 174:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, K., M. Kato, Y. Miura, M. Kettoku, T. Komeda, and A. Iwamatsu. 1996. Gene cloning and expression of new trehalose-producing enzymes from the hyperthermophilic archaeum Sulfolobus solfataricus KM1. Biosci. Biotech. Biochem. 60:1882-1885. [DOI] [PubMed] [Google Scholar]
- 18.Lloret, J., B. B. H. Wulff, J. M. Rubio, J. A. Downie, I. Bonilla, and R. Rivilla. 1998. Exopolysaccharide II production is regulated by salt in the halotolerant strain Rhizobium meliloti EFB1. Appl. Environ. Microbiol. 64:1024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makihara, F., M. Tsuzuki, K. Sato, S. Masuda, K. V. Nagashima, M. Abo, and A. Okubo. 2005. Role of trehalose synthesis pathways in salt tolerance mechanism of Rhodobacter sphaeroides f. sp. denitrificans IL106. Arch. Microbiol. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 20.Maruta, K., H. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Arthrobacter sp. Q36. Biochim. Biophys. Acta 1289:10-13. [DOI] [PubMed] [Google Scholar]
- 21.Maruta, K., H. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Rhizobium sp. M-11. Biosci. Biotech. Biochem. 60:717-720. [DOI] [PubMed] [Google Scholar]
- 22.McIntyre, H. J., H. Davies, D. T. Hore, S. H. Miller, J.-P. Dufour, and C. W. Ronson. 2007. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl. Environ. Microbiol. 73:3984-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meade, H. M., and E. R. Signer. 1977. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 74:2076-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50:101-136. [DOI] [PubMed] [Google Scholar]
- 25.Miller-Williams, M., P. C. Loewen, and I. J. Oresnik. 2006. Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiology 152:2049-2059. [DOI] [PubMed] [Google Scholar]
- 26.Müller, J., T. Boller, and A. Wiemken. 2001. Trehalose becomes the most abundant non-structural carbohydrate during senescence of soybean nodules. J. Exp. Bot. 52:943-947. [DOI] [PubMed] [Google Scholar]
- 27.Nogales, J., R. Campos, H. BenAbdelkhalek, J. Olivares, C. Lluch, and J. Sanjuan. 2002. Rhizobium tropici genes involved in free-living salt tolerance are required for the establishment of efficient nitrogen-fixing symbiosis with Phaseolus vulgaris. Mol. Plant-Microbe Interact. 15:225-232. [DOI] [PubMed] [Google Scholar]
- 28.Olivares, J., J. Casadesús, and E. J. Bedmar. 1980. Method for testing degree of infectivity of Rhizobium meliloti strains. Appl. Environ. Microbiol. 39:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-312. [DOI] [PubMed] [Google Scholar]
- 30.Robertsen, B. K., P. Aiman, A. G. Darwill, M. McNeil, and P. Albersheim. 1981. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 67:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor NY.
- 32.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 34.Smith, L. T., G. M. Smith, M. D'Souza, J.-A. Pocard, D. LeRudulier, and M. A. Madkour. 1994. Osmoregulation in Rhizobium meliloti: mechanism and control by other environmental signals. J. Exp. Zool. 268:162-165. [Google Scholar]
- 35.Soussi, M., M. Santamaría, A. Ocaña, and C. Lluch. 2001. Effects of salinity on protein and lipopolysaccharide pattern in a salt-tolerant strain of Mesorhizobium ciceri. J. Appl. Microbiol. 90:476-481. [DOI] [PubMed] [Google Scholar]
- 36.Streeter, J. G. 1985. Accumulation of α,α-trehalose by Rhizobium bacteria and bacteroids. J. Bacteriol. 164:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streeter, J. G., and M. L. Gomez. 2006. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and bacteroids from soybean nodules. Appl. Environ. Microbiol. 72:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suárez, R., A. Wong, M. Ramírez, A. Barraza, M. del Carmen Orozco, M. A. Cevallos, M. Lara, G. Hernández, and G. Iturriaga. 2008. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol. Plant-Microbe Interact. 21:958-966. [DOI] [PubMed] [Google Scholar]
- 39.Talibart, R., M. Jebbar, K. Gouffi, V. Pichereau, G. Gouesbet, C. Blanco, T. Bernard, and J. Pocard. 1997. Transient accumulation of glycine betaine and dynamics of endogenous osmolytes in salt-stressed cultures of Sinorhizobium meliloti. Appl. Environ. Microbiol. 63:4657-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trinchant, J. C., A. Boscari, G. Spennato, G. Van de Sype, and D. Le Rudulier. 2004. Proline betaine accumulation and metabolism in alfalfa plants under sodium chloride stress. Exploring its compartmentalization in nodules. Plant Physiol. 135:1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsusaki, K., T. Nishimoto, T. Nakada, M. Kubota, H. Chaen, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose synthase gene from Pimelobacter sp. R48. Biochim. Biophys. Acta 1290:1-3. [DOI] [PubMed] [Google Scholar]
- 42.Vriezen, J. A., F. J. De Bruijn, and K. Nüsslein. 2007. Responses of rhizobia to desiccation in relation to osmotic stress, oxygen, and temperature. Appl. Environ. Microbiol. 73:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, W., J. Jiang, W. Li, L. Wang, and S. S. Yang. 2004. Isolation of salt-sensitive mutants from Sinorhizobium meliloti and characterization of genes involved in salt tolerance. Lett. Appl. Microbiol. 39:278-283. [DOI] [PubMed] [Google Scholar]
- 44.Wolf, A., R. Krämer, and S. Morbach. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC 13032 and their significance in response to osmotic stress. Mol. Microbiol. 49:1119-1134. [DOI] [PubMed] [Google Scholar]
- 45.Zahran, H. H. 1999. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 63:968-989. [DOI] [PMC free article] [PubMed] [Google Scholar]