Abstract
Lipoic acid is an essential cofactor required for the function of key metabolic pathways in most organisms. We report the characterization of a Bacillus subtilis mutant obtained by disruption of the lipA (yutB) gene, which encodes lipoyl synthase (LipA), the enzyme that catalyzes the final step in the de novo biosynthesis of this cofactor. The function of lipA was inferred from the results of genetic and physiological experiments, and this study investigated its role in B. subtilis fatty acid metabolism. Interrupting lipoate-dependent reactions strongly inhibits growth in minimal medium, impairing the generation of branched-chain fatty acids and leading to accumulation of copious amounts of straight-chain saturated fatty acids in B. subtilis membranes. Although depletion of LipA induces the expression of the Δ5 desaturase, controlled by a two-component system that senses changes in membrane properties, the synthesis of unsaturated fatty acids is insufficient to support growth in the absence of precursors for branched-chain fatty acids. However, unsaturated fatty acids generated by deregulated overexpression of the Δ5 desaturase functionally replaces lipoic acid-dependent synthesis of branched-chain fatty acids. Furthermore, we show that the cold-sensitive phenotype of a B. subtilis strain deficient in Δ5 desaturase is suppressed by isoleucine only if LipA is present.
Lipoic acid (LA; 6,8-thioctic acid or 1,2-dithiolane-3-pentanoic acid) is a sulfur-containing cofactor required for the function of several key enzymes involved in oxidative and single-carbon metabolism, including pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, branched-chain 2-oxoacid dehydrogenase (BCKADH), acetoin dehydrogenase, and the glycine cleavage system (10). Lipoate-requiring complexes typically contain three protein subunits, E1, E2, and E3. LA is linked through an amide bond to lysine residues in the E2 subunits (42) and acts as a swinging arm, transferring covalently attached reaction intermediates among the active sites of the enzyme complexes (40).
Although the general role of LA as a bound cofactor has been known for decades, the mechanisms by which LA is synthesized and becomes linked to its cognate proteins in different organisms continue to be elucidated. The reactions whereby LA-modified proteins are produced are best understood in Escherichia coli. In this organism, lipoylation is mediated by two separate enzymes, lipoyl protein ligase A (LplA) and octanoyl-acyl carrier protein-protein transferase (LipB) (30, 31). While LplA uses exogenous LA, LipB transfers endogenous octanoic acid to the target proteins (19). These octanoylated domains are then converted into lipoylated derivatives by the S-adenosyl-l-methionine-dependent enzyme lipoyl synthase (LipA), which catalyzes the insertion of sulfur atoms into the carbon-6 and -8 positions of the corresponding fatty acids (29). This process bypasses the requirement for an exogenous supply of LA.
In contrast to the wealth of knowledge available on LA synthesis and utilization in E. coli, the existing information about these pathways in gram-positive bacteria is scarce. It has been found that Listeria monocytogenes mutants defective in proteins homologous to the E. coli LplA enzymes are unable to scavenge exogenous LA for modification of lipoyl domains (22, 23, 38). However, L. monocytogenes is a natural lipoate auxotroph since it does not encode the enzymes necessary for lipoate biosynthesis (15, 55). Bacillus subtilis synthesizes LA, but the biosynthesis, attachment, and function of this essential nutrient in this model gram-positive organism have not yet been studied in detail (50). Analysis of the genome sequence of B. subtilis (25) revealed that it contains an open reading frame, yutB, encoding a protein with a high degree of homology to E. coli LipA and two open reading frames encoding proteins slightly similar to LplA, while no LipB homolog was detected.
LA is a critical cofactor of BCKADH, the enzyme involved in the formation of the primer carbons for the initiation of branched-chain fatty acid (BCFA) synthesis (21). Early work indicated that a bfmB mutant of B. subtilis, defective in both BCKADH and pyruvate dehydrogenase, requires short-branched-chain carboxilic acids for growth (56). However, in our hands, this mutant presented a high percentage of reversion, precluding its use in the study of lipid metabolism. Since BCFAs are the dominant acyl chains found in membrane phospholipids of B. subtilis, the goal of this study was to employ a genetic approach to investigate the role of yutB in the physiology of this organism, in particular in fatty acid metabolism. In addition, we provide compelling evidence showing that Δ5 unsaturated fatty acids (UFA), the products of the B. subtilis desaturase, can fully replace the function of BCFAs. Furthermore, we demonstrate that UFA are essential to provide cryoprotective properties in strains depleted of LipA. This work reports the first characterization of a gram-positive mutant deficient in LA synthesis and its use to study the interplay between BCFAs and UFA metabolism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in the present study are listed in Table 1. E. coli and B. subtilis strains were routinely grown in Luria-Bertani (LB) broth (44). Spizizen salts (51), supplemented with 0.5% glucose, 0.01% each tryptophan and phenylalanine, and trace elements (16), were used as the minimal medium for B. subtilis. This medium was designated MM. Different supplements, including 0.5 mM dl-α-LA, 10 mM sodium acetate, 10 mM succinic acid, and 0.1 mM (each) BCFA precursors (isobutyric acid, isovaleric acid, and 2-methylbutiric acid), were added as needed. MM supplemented with the combination of succinate and acetate was designated MM-SA, and when MM-SA was supplemented with BCFA precursors, it was named MM-SAB. For the experiments involving lipA expression under the control of the xylose-inducible promoter (PxylA), 0.5% glycerol was used as a carbon source instead of glucose. Xylose (0.01%) was added when required. Antibiotics were added to media at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; kanamycin, 5 μg ml−1; and spectinomycin, 50 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description and/or relevant characteristicsa | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| AKP3 | JH642 amyE::Pdes-lacZ | 2 |
| AM100 | JH642 desK::Kmr PKm-desR | This study |
| EL104 | JH642 lipA::PxilA-lipA Cmr | This study |
| JH642 | trpC2 pheA1 | Laboratory stock |
| LC7 | JH642 des::Kmr | 4 |
| NM03 | JH642 lipA::PxilA-lipA Spr | This study |
| NM04 | NM03 amyE::Pdes-lacZ | This study |
| NM07 | EL104 desK::Kmr PKm-desR | This study |
| NM47 | EL104 des::Kmr | This study |
| E. coli strains | ||
| DH5α | supE44 thi-1 ΔlacU169 (φ80lacZΔM15) endA1 recA1 hsdR17 gyrA96 relA1 trp-6 cysT329::lac λinmpI(209) | Laboratory stock |
| TM131 | rpsL lipA150::Tn1000dKn lplA148::Tn10dTc Kmr Tcr | 30 |
| TM136 | rpsL lipB182::Tn1000dKn lplA148::Tn10dTc Kmr Tcr | 30 |
| Plasmids | ||
| pJM116 | Integrative vector to construct transcriptional fusions to lacZ; integrates at the amyE locus of B. subtilis; Cmr | 39 |
| pBluescript II SK(+) | E. coli cloning vector; Ampr | Stratagene |
| pGES49 | Integrative vector containing PxylA and xylR | 45 |
| pAR11 | Contains Pdes cloned into the EcoRI-BamHI sites of pJM116 | 2 |
| pdes913KAN | des::Kmr cloned into pBluescript II SK(+) | 2 |
| pEL11 | lipA cloned under control of PxylA in pGES49 | This study |
| pEL10 | lipA cloned into the HindIII-HincII sites of pBluescript II SK(+) | This study |
| pCm::Sp | Antibiotic switching vector | 52 |
Cmr, Kmr, Spr, Tcr, and Ampr denote resistance to chloramphenicol, kanamycin, spectinomycin, tetracycline, and ampicillin, respectively.
Genetic techniques.
E. coli competent cells were transformed with supercoiled plasmid DNA by the calcium chloride procedure (44). Transformation of B. subtilis was carried out by the method of Dubnau and Davidoff-Abelson (13). The amy phenotype was assayed, with colonies grown for 48 h on LB starch plates, by flooding the plates with 1% I2-KI solution (46). amy+ colonies produced a clear halo, while amy colonies gave no halo.
Plasmids and strain construction.
In all cases, DNA fragments were obtained by PCR using the oligonucleotides listed in Table 2. Chromosomal DNA from strain JH642 was used as the template. The yutB conditional mutant was constructed as follows. A 430-bp fragment corresponding to the 5′ region of the yutB gene without its promoter was obtained by PCR amplification using the oligonucleotides yutBup and yutBdw (Table 2). The fragment was cloned into the HindIII and SalI sites of vector pGES49 (45), which allows expression of the yutB gene under the control of PxylA. The resulting plasmid was named pEL11. This construct was then integrated by a single-crossover event at the yutB locus of JH642, yielding strain EL104. For E. coli complementation analysis, a 954-bp fragment containing the yutB gene of B. subtilis was PCR amplified with oligonucleotides yutBup and yutBfill (Table 2). The fragment was cloned into the HindIII and HincII sites of pBluescriptII SK(+). The resulting plasmid was named pEL10 and was used to transform the E. coli lipA lplA strain TM131 (30). Strain EL104 was transformed with vector pCm::Sp (52) to exchange the chloramphenicol resistance cassette present in the yutB locus for a spectinomycin resistance cassette. The transformed strain was named NM03. To construct a yutB conditional mutant containing a transcriptional fusion of lacZ to the promoter region of the desaturase gene (Pdes-lacZ), strain NM03 was transformed with pAR11 linearized with ScaI (2), yielding strain NM04. Strain AM100 was constructed by transformation of JH642 with chromosomal DNA of AKP20 (2) and selection of the amy+ transformants. A derivative of strain EL104 that overexpresses the desaturase gene constitutively was obtained by transformation with chromosomal DNA of strain AM100. This strain was named NM07.
TABLE 2.
Oligonucleotide primers
| Primer name | Sequence (5′-3′)a |
|---|---|
| yutBup | TTGAGaAgcTTGGATATTGGAGATGATG |
| yutBdw | TGCGagTCGacCGGACAGTTTCC |
| yutHind | CGTGTaAGCTTCGATTTTATGATATAATTC |
| yutBam | ATTAGGaTccATTCGGGCTTTCTGAG |
| yutBfill | cgCCAGtTaAcCTGGCGTTTTGG |
Lowercase letters indicate variations with respect to the wild-type B. subtilis sequence. Restriction sites are underlined.
Strain NM47 was obtained by transformation of strain EL104 with plasmid pdes913KAN (2), previously linearized with ScaI.
β-Galactosidase assays.
B. subtilis strains harboring the Pdes-lacZ fusion were grown overnight in MM-SAB. The cells were collected by centrifugation and diluted in MM-SA, with or without the addition of 0.01 mM 2-methylbutiric acid. Samples were taken at 1-h intervals after resuspension and assayed for β-galactosidase activity as described previously (27). The specific activity was expressed in Miller units (28). The results shown are the averages of results from three independent experiments.
Fatty acid analysis.
To determine the fatty acid composition, cells were grown at 37°C in MM-SA with the addition of each BCFA precursor or LA. The cultures were harvested in stationary phase. Total cellular fatty acids were extracted by the method of Bligh and Dyer (6). The fatty acid methyl esters were prepared by transesterification of glycerolipids with 0.5 M sodium methoxide in methanol (9) and then analyzed in a PerkinElmer TurboMass gas chromatograph-mass spectrometer with a capillary column (30 m by 0.25 mm in diameter; Varian) of 100% dimethyl polysiloxane (PE-1; PerkinElmer). Helium at 1 ml min−1 was used as the carrier gas, and the column temperature was programmed to rise by 4°C min−1 from 165 to 275°C. The saturated fatty acids (SFA) and UFA used as reference compounds were obtained from Sigma Chemical Co.
To determine the in vivo desaturase activities of B. subtilis strains, cells were grown overnight at 37°C in MM-SAB and then resuspended in MM-SA supplemented with isovalerate, with the addition of 2-methylbutyrate where indicated. Two-milliliter samples were labeled with 0.2 μCi of [14C]palmitate for 6 h. Following incubation, the labeled fatty acids were extracted and methyl esters were prepared as described above. Each sample was fractionated by argentation-thin-layer chromatography, as described previously (1). The spots of the different fatty acids were quantified by ImageQuant 5.2.
Immunoblot analysis.
Strains JH642 (yutB+) and EL104 (yutB) were grown overnight in MM-SAB at 37°C. Cells were resuspended in fresh MM-SAB or MM with the addition of 0.01% xylose or 0.5 mM LA. A 1-ml aliquot of each culture was harvested after 22 h of growth. The samples were centrifuged, and the pellets were washed with buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl). The pellets were then resuspended in 180 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 10 μM phenylmethylsulfonyl fluoride) per unit of optical density at 600 nm (OD600). Resuspended cells were disrupted by incubation with lysozyme (100 μg/ml) for 15 min at 37°C, followed by 5 min of boiling in the presence of loading buffer. Each sample was fractionated by sodium dodecyl sulfate gel electrophoresis in an 8% acrylamide gel. Proteins were electroeluted onto a nitrocellulose membrane and detected using anti-LA rabbit antibody and a secondary anti-rabbit immunoglobulin G antibody conjugated to alkaline phosphatase. The blots were scanned, and the intensity of the bands was quantified by ImageQuant 5.2.
RESULTS
Depletion of YutB.
A BLAST search of the SubtiList database (32) using the amino acid sequence of the E. coli LA synthase (LipA) detected a gene of B. subtilis, yutB, encoding a homologous polypeptide of 298 amino acid residues with a molecular mass of 33,766 Da, classified as a possible LA synthase. Comparison of the deduced amino acid sequence of the B. subtilis YutB protein with the sequence of E. coli LipA rendered values of 52% identity and 67% similarity. Several stretches of conserved residues that may be important for the function of the YutB enzyme were found. A conserved Cys motif, CXXXCXXC, was found at positions 67 to 74 in the YutB protein. This motif is common among radical S-adenosyl-l-methionine enzymes and is thought to bind an Fe-S center involved in 5′-deoxyadenosyl radical generation (17, 29, 43). A second Cys motif, CXXXXCXXXXXC, which is unique to LA synthases and is involved in the coordination of the second [4Fe-4S] cluster present in these enzymes (7), was located at positions 40 to 51 in YutB.
The yutB gene has not been considered to be essential based on the results of systematic genomewide inactivation of B. subtilis genes (24) in a study performed using a standard laboratory rich medium, LB medium. As LA auxotrophy would be in evidence only in minimal medium, to perform this study we constructed a conditional mutant, EL104, which expressed yutB under the control of PxylA. This mutant was able to grow in MM in the presence of xylose, and removal of the inducer resulted in no growth, indicating that yutB was indeed essential (Fig. 1). Moreover, EL104 grew as well as wild-type B. subtilis in MM containing LA (Fig. 1), demonstrating that it is able to transfer the exogenously provided cofactor to the apoproteins, although it would be unable to synthesize it.
FIG. 1.
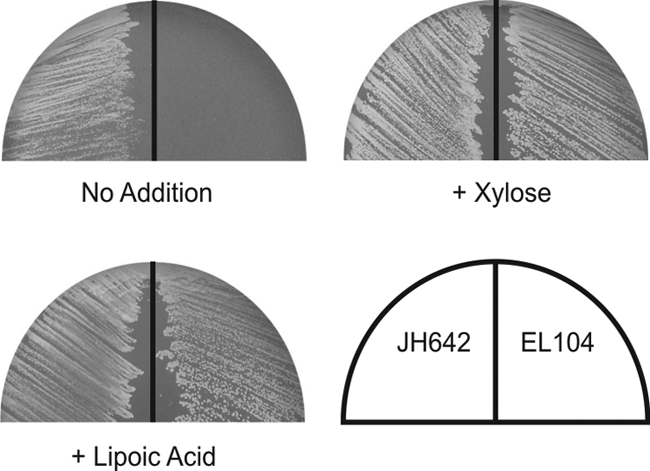
Growth of a yutB mutant on minimal medium. Strains JH642 (yutB+) and EL104 (yutB) were streaked onto MM plates with the supplements indicated in the figure. The plates were incubated for 24 h at 37°C.
During vegetative growth, B. subtilis expresses at least three E2 apoproteins that require lipoylation for activity, namely, pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, and BCKADH. If indeed YutB is involved in LA synthesis, the growth of EL104 in MM should also be rescued by the addition of the metabolites that yield the products of the lipoylated enzymes, i.e., acetate, succinate, and BCFA precursors, bypassing the lipoylation requirement. As shown in Fig. 2, EL104 needs the addition of both acetate and BCFA precursors to MM to restore growth, indicating that pyruvate dehydrogenase and BCKADH are not functional in this strain. It should be noted that the addition of succinate is not required to complement the growth of EL104 in MM, in agreement with previous reports that succinate is dispensable for the growth of B. subtilis in minimal medium supplemented with glucose (14).
FIG. 2.
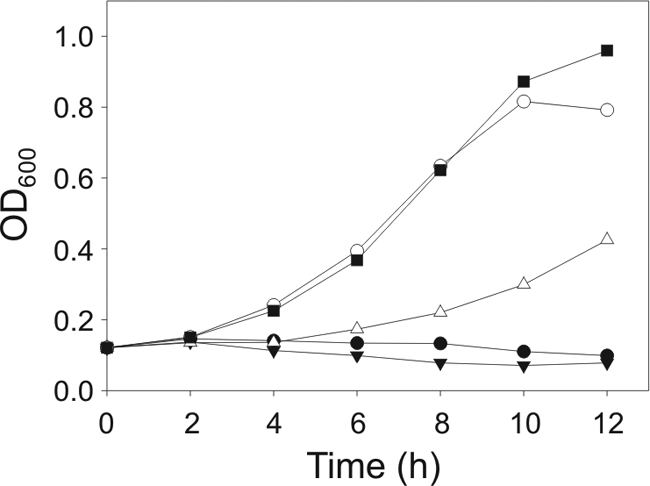
Effects of the addition of different supplements on the growth of the yutB mutant. Strain EL104 (yutB) was grown overnight in MM-SAB. Cells were resuspended in MM alone (closed circles); MM with the addition of acetate and succinate (closed triangles), acetate and BCFA precursors (open circles), or succinate and BCFA precursors (open triangles); or MM-SAB (closed squares). Samples were taken every 2 h, and the OD600 was measured.
Lipoylation of proteins in a YutB-depleted strain.
To determine if the E2 apoproteins of the dehydrogenase complexes have been lipoylated in the yutB mutant EL104, we analyzed cell extracts using antiserum specific for lipoylated proteins. By Western blotting, two major bands with apparent masses of 60 and 52 kDa were detected in samples from the wild-type strain, JH642, growing in three different media (Fig. 3, lanes 1 to 3). The same pattern was observed when EL104 was grown in MM supplemented with xylose (Fig. 3, lane 5) or LA (Fig. 3, lane 6). However, almost no reaction was observed when cell extracts of EL104 were obtained from cultures grown in MM-SAB (Fig. 3, lane 4). These results indicate that the growth defect of this strain in MM is due to the absence of lipoylation of essential apoproteins.
FIG. 3.
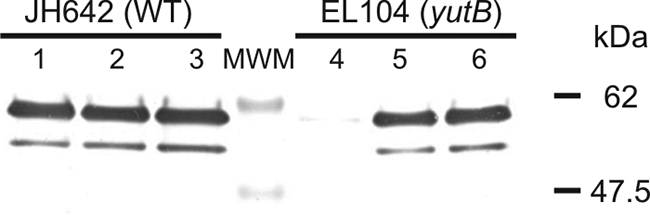
Immunoblot with anti-LA antibody. JH642 (yutB+) and EL104 (yutB) cells were grown overnight in MM-SAB. Cells were resuspended in fresh MM-SAB (lanes 1 and 4) or MM with the addition of xylose (lanes 2 and 5) or LA (lanes 3 and 6). Cells were harvested after 22 h of growth and treated as described in Materials and Methods. The upper band corresponds to the pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase E2 subunits. The lower band presumably corresponds to the E2 subunit of BCKADH. WT, wild type; MWM, molecular size marker.
The B. subtilis yutB gene encodes a functional lipoyl synthase enzyme.
Complementation experiments were used to confirm the prediction that yutB encodes a functional LipA-like lipoyl synthase. E. coli strains harboring both a lipA mutation and a lplA mutation fail to synthesize LA and are unable to grow even in the presence of exogenous lipoate since it cannot be attached to the apoproteins (30). Therefore, growth of a lipA lplA double mutant in M9 minimal medium can only be rescued by supplementation with acetate and succinate (30) (Fig. 4). Transformation of strain TM131 (lipA lplA) or TM136 (lipB lplA) with plasmid pEL10, expressing yutB, showed that E. coli lipA lplA was complemented by yutB expression (Fig. 4) but that the lipB lplA strain was not (data not shown). Thus, based on the molecular characteristics of the yutB gene, the growth phenotype and protein lipoylation pattern of the yutB mutant, and the ability of yutB to complement the lipA but not the lipB defect, we have designated this gene lipA in concordance with E. coli nomenclature.
FIG. 4.
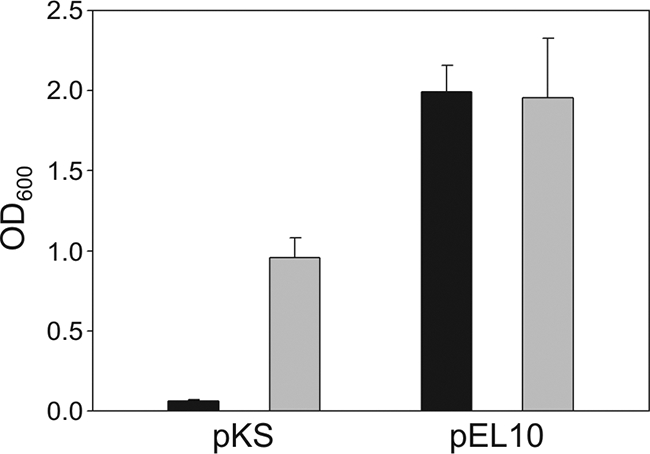
B. subtilis yutB gene complementation of an E. coli strain deficient in lipoate synthesis and utilization. E. coli TM131 (lipA lplA) was transformed with either the empty vector bearing the lacZ inducible promoter (pKS) or the same vector expressing B. subtilis yutB under the control of PlacZ (pEL10). The strains were inoculated into M9 minimal medium containing IPTG (isopropyl-β-d-thiogalactopyranoside; black bars) or the same medium supplemented with acetate and succinate (gray bars). The OD600 values of the cultures were determined after 24 h of growth at 37°C. The results shown are the averages of results from three independent experiments.
Membrane fatty acid alterations in the lipA mutant.
Iso- and anteiso-BCFAs are the dominant components of B. subtilis membranes (21). The primer carbons for the synthesis of these fatty acids are isobutyryl coenzyme A (isobutyryl-CoA), isovaleryl-CoA, and 2-methylbutyryl-CoA, which in turn are derived from the branched-chain amino acids valine, leucine, and isoleucine, respectively (20, 33). The enzyme that catalyzes the formation of acyl-CoA primers from the branched-chain α-keto acids is the BCKADH complex, which requires LA as a coenzyme (34).
Due to the lipoate requirement of the BCKADH complex, we expected the fatty acid composition of EL104 to be altered by depletion of LipA. In fact, at 37°C, even-numbered straight-chain SFA largely predominated in the lipA mutant while BCFAs were the major fatty acid species in the wild-type strain (Table 3; see also Table S1 in the supplemental material). In agreement with the findings of previous studies (56, 57), we observed that the addition of each of the short-BCFA precursors increased the proportion of the corresponding BCFA. However, the total fatty acid composition of EL104 membranes became similar to that of wild-type membranes only when the medium was supplemented with LA, indicating that lipoate-dependent metabolism is essential for normal BCFA biosynthesis. Despite these membrane alterations, analysis of bacterial growth in MM-SA supplemented with any one of the BCFA precursors showed that all of them were able to support the growth of the lipA strain. However, 2-methylbutyrate, the precursor of anteiso-BCFAs, was slightly more effective in stimulating the growth of lipA mutants than isobutyrate or isovalerate, which are used as primers for iso-BCFA biosynthesis (data not shown).
TABLE 3.
Fatty acid compositions of total membrane lipid extracts from B. subtilis strains JH642 (lipA+) and EL104 (lipA)
| Strain | Supplementa | % of total fatty acid in the form of: |
Iso-BCFA/anteiso-BCFA ratio | (Iso-BCFA + straight-chain fatty acid)/anteiso-BCFA ratio | |||
|---|---|---|---|---|---|---|---|
| Straight-chain fatty acids | Even-numbered iso-BCFAs | Odd-numbered iso-BCFAs | Anteiso-BCFAs | ||||
| JH642 | IBU | 12.95 | 28.84 | 23.87 | 33.65 | 1.57 | 1.95 |
| IVAL | 14.84 | 7.17 | 40.01 | 37.20 | 1.27 | 1.67 | |
| 2-MB | 23.88 | 3.12 | 12.51 | 60.10 | 0.26 | 0.66 | |
| LA | 19.57 | 14.96 | 23.41 | 41.65 | 0.92 | 1.39 | |
| EL104 | IBU | 63.32 | 14.84 | 10.47 | 11.04 | 2.29 | 8.03 |
| IVAL | 60.39 | 6.18 | 23.01 | 10.16 | 2.87 | 8.82 | |
| 2-MB | 59.55 | 7.15 | 9.25 | 23.69 | 0.69 | 3.20 | |
| LA | 7.41 | 10.72 | 35.18 | 46.62 | 0.98 | 1.14 | |
Cells were grown at 37°C in MM-SA supplemented with isobutyrate (IBU), isovalerate (IVAL), 2-methylbutyrate (2-MB), or LA.
Expression and activity of the lipid desaturase in a lipA mutant.
Previous work has revealed that the DesK-DesR two-component system regulates the expression of the des gene coding for the Δ5-acyl-lipid desaturase from B. subtilis (2). It is believed that a decrease in membrane fluidity activates the DesK-DesR transducing cascade, which results in synthesis of the desaturase and desaturation of membrane phospholipids (26). Transcription of des is tightly regulated by temperature: although des mRNA is barely detected in cells growing in rich medium at 37°C, its production is dramatically induced upon a temperature downshift (3). However, the des gene is also derepressed at 37°C when the supply of isoleucine is limited, leading to a decrease in the synthesis of anteiso-BCFAs (11). This is probably because these fatty acids share with UFA the ability to decrease the transition temperature of membrane phospholipids to maintain the appropriate fluidity.
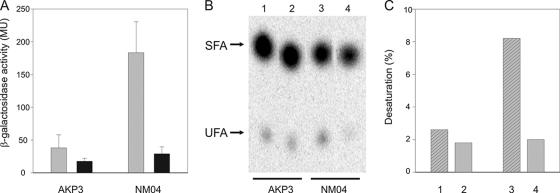
As the amount of anteiso-BCFAs is dramatically decreased in a lipA strain deprived of LA (Table 3), we wished to determine whether the lipA mutation has some effects on des transcription and on fatty acid desaturation. To this end, we used strains AKP3 (lipA+) and NM04 (lipA), which contain a fusion of the lacZ gene to the des promoter integrated ectopically at the nonessential amyE locus. These strains were grown in either MM-SA or MM-SA supplemented with 2-methylbutyrate. As shown in Fig. 5A, the activity of the des promoter in the lipA strain NM04 reaches induction levels about fourfold higher than the levels found in strain AKP3. The expression of the des-lacZ fusion was repressed in both strains after supplementation with 2-methylbutyrate (Fig. 5A), supporting previous evidence that transcription of des is regulated by the order of membrane lipids.
FIG. 5.
lipA depletion stimulates transcription and activity of the desaturase. (A) Effect of lipA mutation on des-lacZ expression. AKP3 (lipA+ Pdes-lacZ) and NM04 (lipA Pdes-lacZ) cells were grown overnight at 37°C in MM-SAB and resuspended in MM-SA (gray bars) or MM-SA with the addition of 2-methylbutyric acid (black bars). β-Galactosidase activities of samples taken after 6 h of growth are shown. The results presented are the averages of results from three independent experiments. MU, Miller units. (B) Argentation-thin-layer chromatography analysis of fatty acid methyl esters. AKP3 (lipA+) and NM04 (lipA) were grown overnight in MM-SAB at 37°C. Cells were then resuspended in MM-SA supplemented with isovalerate (lanes 1 and 3) or with the addition of isovalerate and 2-methylbutyrate (lanes 2 and 4) and exposed to 0.2 μCi of [1-14C]palmitate. After 6 h of incubation at 37°C, lipids were extracted and transesterified and the resulting methyl esters were analyzed as described in Materials and Methods. About 8,000 cpm of radioactivity was loaded into all lanes. The final positions of UFA and SFA are indicated to the left. (C) Percentages of UFA synthesis. The radioactivity levels of the spots of the UFA and SFA shown in panel B were quantified by ImageQuant 5.2. Results are expressed as percentages of the total methyl esters recovered. The numbers below the bars correspond to the lane numbers indicated in panel B.
We also tested the desaturase activities of strains NM04 and AKP3 growing at 37°C in MM-SA supplemented with isovalerate by labeling the cells with radioactive palmitate and assaying the conversion of this fatty acid to cis-hexadecenoic acid (Fig. 5B). In agreement with the results of the transcriptional experiments (Fig. 5A), the levels of desaturation of exogenously added palmitate were about threefold higher in strain NM04 than in strain AKP3 (Fig. 5C, bars 1 and 3). As expected, the addition of 2-methylbutyrate decreases [14C]palmitate desaturation in both strains (Fig. 5C, bars 2 and 4).
Although depletion of LipA induces the expression of the desaturase at 37°C (Fig. 5A), the UFA synthesized by the lipA strain did not improve the growth of this strain in the absence of BCFA precursors (Fig. 2). This may be because the amount of UFA synthesized is not sufficient to decrease the phase transition of membrane lipids or because the branching of fatty acids has an essential function other than disrupting the packing of the phospholipid acyl chains. To distinguish between these two possibilities, we constructed strain NM07, a derivative of EL104, which lacks DesK and overexpresses the transcriptional activator DesR, resulting in constitutive overproduction of the desaturase (2). At 37°C, a strain with such characteristics contains an unusually high proportion of membrane UFA, which reach values near 13% of total fatty acids, representing levels about sixfold higher than those found in the wild-type strain (4). When NM07 is grown in MM-SA, a condition under which EL104 does not grow (Fig. 2, closed triangles), it reaches an OD600 similar to that of EL104 grown in the presence of acetate, succinate, and BCFA precursors (compare Fig. 2, closed squares, with Fig. 6, closed triangles). Moreover, strain NM07 reached OD600 values about twofold higher than EL104 after 12 h of growth in MM-SAB at 37°C (compare Fig. 2 and Fig. 6, closed squares). These results clearly indicate that UFA are able to fully complement the BCFA deficiency of LipA-depleted strains. It follows that the main role of BCFAs at 37°C is to maintain optimal fluidity for normal cell growth.
FIG. 6.
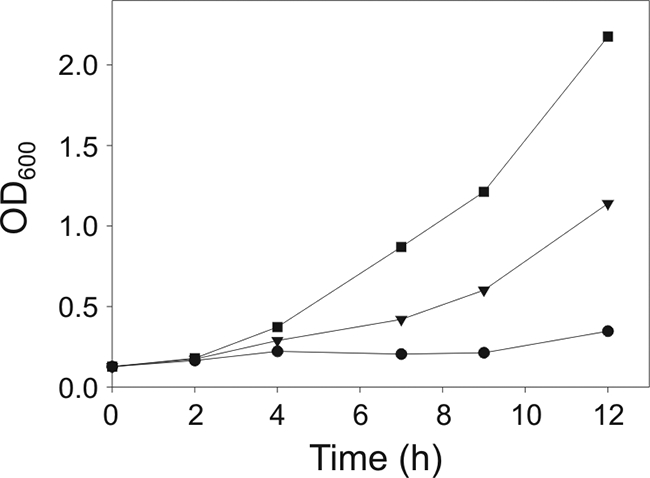
Effect of des overexpression on the growth of the lipA mutant. Strain NM07 (lipA desK::Km PKm-desR, where Km indicates a kanamycin resistance cassette) was grown overnight in MM-SAB. Cells were resuspended in fresh MM (circles), MM-SA (triangles), or MM-SAB (squares). Samples were taken at different times, and the OD600 was measured.
Cold sensitivity of des mutants in the absence of lipoylation.
It has been reported previously that isoleucine is essential for a B. subtilis des mutant to survive cold shock (54). This protective effect seems to be based on the function of this amino acid as a precursor in the biosynthesis of anteiso-BCFAs. Interestingly, branched-chain amino acids, especially isoleucine, seem to have a key regulatory role in adaptation to a wide range of nutritional environments by different mechanisms, in both gram-positive and gram-negative bacteria (for a review, see reference 49). Therefore, it was of interest to test if isoleucine suppresses the cold-sensitive phenotype of a des mutant by using a yet unknown regulatory mechanism(s) together with the well-established role of this amino acid as a precursor of anteiso-BCFAs. To asses this possibility, we constructed a des derivative of EL104, strain NM47. This strain is unable to synthesize anteiso-BCFAs due to the absence of lipoylation of the BCKADH complex and cannot desaturate fatty acids owing to des mutation (1). Therefore, if the major role of isoleucine in the B. subtilis cold shock response is to prime the synthesis of anteiso-BCFAs, a des lipA mutant should be unable to grow at 15°C even in the presence of this amino acid. However, if isoleucine has an additional role in cold adaptation, partial restoration of the growth would be observed. Thus, we analyzed the growth at 15°C of NM47 (des lipA), LC7 (des), EL104 (lipA), and JH642 (des+ lipA+) in MM-SA supplemented with 0.1 mM isovalerate, 0.1 mM isobutyrate, 0.05 mM 2-methylbutyrate, and 0.05 mM isoleucine. Growth of NM47 was almost completely inhibited at 15°C, despite the presence of isoleucine, while EL104 and LC7 were able to grow at this temperature, although at a reduced rate compared to a wild-type strain (Fig. 7). Increasing concentrations of isoleucine, ranging from 0.05 to 0.75 mM, failed to confer a cold resistance phenotype on NM47 (data not shown). These data, combined with the observation that NM47 grew normally at 37°C in the above-described medium (data not shown), allowed us to conclude that the suppressor effect of isoleucine on the cold sensitivity of des mutants is almost entirely due to its role as a precursor of anteiso-BCFAs.
FIG. 7.
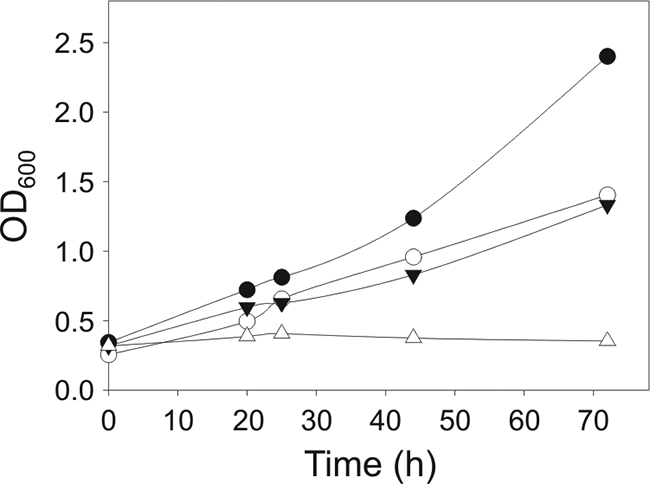
Cold sensitivity of a des lipA double mutant. Cells were grown overnight at 37°C in MM-SA supplemented with 0.1 mM isobutyric acid, 0.1 mM isovaleric acid, 0.05 mM 2-methylbutyric acid, and 0.05 mM isoleucine and were then resuspended in the same medium and grown at 37°C until early exponential phase, when cultures were transferred to 15°C. Samples were taken, and the OD600 was measured. Closed circles, JH642 (des+ lipA+); open circles, LC7 (des); closed triangles, EL104 (lipA); open triangles, NM47 (des lipA).
DISCUSSION
Although a large body of information concerning LA biosynthesis and utilization has been accumulated over the years (for a review, see reference 10), these pathways in gram-positive organisms have not been elucidated completely. A recent report suggested that LipA (YutB) of B. subtilis is involved in competence development, functioning as a transcriptional regulator of comE (36). For these reasons and due to our interest in fatty acid biosynthesis, we began the present study in which we obtained genetic and physiological data demonstrating that lipA is an essential B. subtilis gene coding for LipA, the enzyme responsible for the sulfur insertion step of LA biosynthesis. The genetic evidence is based on results from experiments showing that the B. subtilis lipA gene can complement the growth of E. coli lipA lplA double mutants but not of lipB mutants. In agreement with the complementation data, the growth of lipA strains can be restored by supplementation with either LA or the metabolites that yield the products of the lipoylated enzymes, pyruvate dehydrogenase and BCKADH.
Extensive work using B. subtilis as a model gram-positive organism has shown that the acyl-CoA esters with three to five carbons are synthesized by oxidative decarboxylation of branched-chain keto acids by the LA-dependent enzyme BCKADH (34). Three genes, bfmBAA, bfmBAB, and bfmBB, encode the E1α, E1β and E2 components of this complex, respectively (53). They form part of the bkd gene cluster (12), which also contains a gene, lpd, encoding a putative E3 subunit that is distinct from the one associated with the pyruvate dehydrogenase complex. However, an alternative pathway to generate the precursors for BCFA synthesis, involving a branched-chain α-keto acid decarboxylase, was proposed previously (37). It was suggested that this enzyme is a free form of the dehydrogenase (E1α) of the BCKADH complex (53) that would catalyze the synthesis of short carboxylic acyl-CoA esters that in turn would be used for the initiation of BCFA elongation, by condensation with malonyl-acyl carrier protein (21). The findings of the present study rule out the physiological significance of this hypothetical pathway in B. subtilis since we have demonstrated that in the absence of enzyme lipoylation, which should not affect E1α activity, the synthesis of BCFAs is dramatically decreased. More importantly, the lipA mutant was unable to grow in minimal medium unless the medium was supplemented with short-branched-chain carboxylic acids. Thus, we conclude that the lipoate-dependent BCKADH complex is the major pathway responsible for providing the precursors for BCFA biosynthesis in B. subtilis. This may not be the case for other gram-positive organisms, since in an L. monocytogenes strain deficient in protein lipoylation the synthesis of BCFAs is not completely abolished, indicating that a compensatory pathway is functioning (22).
The branched-chain acyl-CoA esters are introduced into the fatty acid biosynthetic pathway at the initiation step by two FabH isozymes, named FabHA and FabHB (8). These enzymes catalyze the condensation of the acyl-CoA esters with malonyl-acyl carrier protein to form the first β-ketoacyl intermediate in the type II fatty acid synthase (8). While the single E. coli FabH has a high degree of specificity for acetyl-CoA (18), the FabH enzymes from B. subtilis are less active with this substrate and readily utilize branched-chain acyl-CoA primers (8). Here, we found that in the absence of lipoylated metabolic enzymes, including BCKADH, the lipA strain EL104 growing in defined medium supplemented with any one of the BCFA precursors produced copious amounts of even-numbered straight-chain SFA. This result contrasts with the one observed for L. monocytogenes BCKADH mutants, which contain an increased proportion of even-numbered BCFAs only when grown in medium without the addition of BCFA precursors (5, 57). LA supplementation of the lipA mutant could substantially restore the BCFA profile observed in wild-type strains. Thus, when the availability of branched-chain acyl-CoA primers is restricted by LipA depletion, B. subtilis FabHA and/or FabHB can efficiently utilize straight-chain acyl-CoA primers, such as acetyl-CoA or butyryl-CoA, to prime fatty acid synthesis. Even though the B. subtilis genome contains an operon (mmgABCDE) encoding enzymes homologous to that involved in the synthesis of butyryl-CoA (35), it is unlikely that this compound is being used for fatty acid biosynthesis since, as demonstrated by Choi et al. (8), both FabH isoenzymes have much higher affinity for acetyl-CoA than for butyryl-CoA. In summary, our findings strongly indicate that at 37°C the supply of precursors, rather than the substrate specificity of the FabH isozymes, is the most significant factor in determining the composition of fatty acids produced by the B. subtilis fatty acid synthase.
It was shown previously that in B. subtilis a two-component system composed of a membrane-associated sensor histidine kinase, DesK, and a response regulator, DesR, controls the expression of the des gene coding for the Δ5 desaturase (2). We proposed that DesK could assume different signaling states in response to changes in membrane fluidity (26). A kinase dominant state of DesK predominates upon an increase of the packing of membrane lipids. This increase can be accomplished either under cold shock conditions or at the optimal growth temperature, 37°C, by restricting the supply of isoleucine, which reduces the amount of anteiso-BCFAs in membrane phospholipids (2, 11). Here, we show that the dramatic inhibition of fatty acid branching produced by LipA depletion from cells growing at 37°C (Table 3) leads to transcriptional induction of the des gene to levels fourfold higher than those observed in lipA+ strains growing in an isoleucine-free medium. These transcriptional data give further support to the hypothesis that DesK senses membrane fluidity, since the membrane lipids of lipA-deficient cells contain a much higher proportion of high-melting-point fatty acids than those of cells expressing LipA. As shown in Fig. 2, the UFA synthesized by lipA cells in the absence of supplementation with BCFA precursors cannot overcome BCFA deficiency in the absence of lipoylation. However, this BCFA precursor auxotrophy of lipA mutants was easily abolished by overexpression of the DesR transcription factor, which leads to overproduction of UFA. These results mean that UFA can fulfill the role of BCFAs in cell growth and that regulation of the expression of the B. subtilis desaturase by the two-component system DesK-DesR is designed to fine-tune membrane fluidity rather than to substitute for the role of fatty acid branching.
The des gene from B. subtilis is induced severalfold by cold shock, although this organism can grow without UFA at low temperatures in rich medium (1). However, a B. subtilis des deletion mutation underlies a dramatic reduction of viability during cold shock in the absence of exogenous isoleucine (54). It has been suggested that this cold sensitivity is because the desaturase provides a backup mechanism to survive at low temperatures in the absence of an isoleucine-dependent switch of the branching pattern, from iso- to anteiso-BCFAs. However, this proposal does not rule out another role of isoleucine in the survival of des mutants under cold shock conditions, since this model is based on results from experiments performed in the absence of the amino acid (54). It is well documented that branched-chain amino acids play a key regulatory role in bacterial adaptation (12, 47, 48). Interestingly, the existence of a novel regulatory small RNA in B. subtilis, BsrF, whose transcription is activated by the global regulator CodY only in the presence of GTP and branched-chain amino acids, has been reported recently. Notably, of the three branched-chain amino acids, isoleucine was found to be the most potent activator of BsrF transcription (41). Here, we show that a des lipA double mutant growing in medium supplemented with acetate and BCFA precursors is viable at 37°C but is unable to survive after a shift from 37 to 15°C, even in the presence of isoleucine. Therefore, our data provide direct evidence that the major role of exogenous isoleucine in allowing B. subtilis des mutants to withstand cold shock is to stimulate an increase in the anteiso-BCFA content of the membranes. In addition, the results of our experiments directly demonstrate that UFA are essential to provide cryoprotective properties to a B. subtilis strain defective in lipoylation of BCKADH for membrane adaptation during cold shock.
Supplementary Material
Acknowledgments
We thank J. E. Cronan, Jr., for the gift of strains TM131 and TM136.
This work was supported by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Agencia Nacional de Promoción Científica y Tecnológica (FONCYT). N. Martin is a fellow of CONICET, and S. G. Altabe, M. C. Mansilla, and D. de Mendoza are Career Investigators from the same institution. D. de Mendoza is an International Research Scholar from Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 9 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguilar, P. S., J. E. Cronan, Jr., and D. de Mendoza. 1998. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 180:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. S., A. M. Hernandez-Arriaga, L. E. Cybulski, A. C. Erazo, and D. de Mendoza. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, P. S., P. Lopez, and D. de Mendoza. 1999. Transcriptional control of the low-temperature-inducible des gene, encoding the Δ5 desaturase of Bacillus subtilis. J. Bacteriol. 181:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altabe, S. G., P. Aguilar, G. M. Caballero, and D. de Mendoza. 2003. The Bacillus subtilis acyl lipid desaturase is a Δ5 desaturase. J. Bacteriol. 185:3228-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 7.Cheek, J., and J. B. Broderick. 2001. Adenosylmethionine-dependent iron-sulfur enzymes: versatile clusters in a radical new role. J. Biol. Inorg. Chem. 6:209-226. [DOI] [PubMed] [Google Scholar]
- 8.Choi, K. H., R. J. Heath, and C. O. Rock. 2000. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, W. (ed.). 1989. Gas chromatography and lipids. The Oily Press, Ayr, Scotland.
- 10.Cronan, J. E., X. Zhao, and Y. Jiang. 2005. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 50:103-146. [DOI] [PubMed] [Google Scholar]
- 11.Cybulski, L. E., D. Albanesi, M. C. Mansilla, S. Altabe, P. S. Aguilar, and D. de Mendoza. 2002. Mechanism of membrane fluidity optimization: isothermal control of the Bacillus subtilis acyl-lipid desaturase. Mol. Microbiol. 45:1379-1388. [DOI] [PubMed] [Google Scholar]
- 12.Debarbouille, M., R. Gardan, M. Arnaud, and G. Rapoport. 1999. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, S. H., and B. Magasanik. 1984. Synthesis of oxaloacetate in Bacillus subtilis mutants lacking the 2-ketoglutarate dehydrogenase enzymatic complex. J. Bacteriol. 158:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Harwood, C. R., and S. M. Cuttings (ed.). 1990. Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England.
- 17.Hayden, M. A., I. Huang, D. E. Bussiere, and G. W. Ashley. 1992. The biosynthesis of lipoic acid. Cloning of lip, a lipoate biosynthetic locus of Escherichia coli. J. Biol. Chem. 267:9512-9515. [PubMed] [Google Scholar]
- 18.Heath, R. J., and C. O. Rock. 1996. Inhibition of β-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:10996-11000. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, S. W., and J. E. Cronan, Jr. 2003. The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J. Bacteriol. 185:1582-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneda, T. 1973. Biosynthesis of branched long-chain fatty acids from the related short-chain α-keto acid substrates by a cell-free system of Bacillus subtilis. Can. J. Microbiol. 19:87-96. [DOI] [PubMed] [Google Scholar]
- 21.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeney, K., L. Colosi, W. Weber, and M. O'Riordan. 2009. Generation of branched-chain fatty acids through lipoate-dependent metabolism facilitates intracellular growth of Listeria monocytogenes. J. Bacteriol. 191:2187-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney, K. M., J. A. Stuckey, and M. X. O'Riordan. 2007. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol. Microbiol. 66:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Mansilla, M. C., and D. de Mendoza. 2005. The Bacillus subtilis desaturase: a model to understand phospholipid modification and temperature sensing. Arch. Microbiol. 183:229-235. [DOI] [PubMed] [Google Scholar]
- 27.Mansilla, M. C., and D. de Mendoza. 1997. l-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. (ed.). 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Miller, J. R., R. W. Busby, S. W. Jordan, J. Cheek, T. F. Henshaw, G. W. Ashley, J. B. Broderick, J. E. Cronan, Jr., and M. A. Marletta. 2000. Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39:15166-15178. [DOI] [PubMed] [Google Scholar]
- 30.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 269:16091-16100. [PubMed] [Google Scholar]
- 31.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1995. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J. Bacteriol. 177:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moszer, I. 1998. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430:28-36. [DOI] [PubMed] [Google Scholar]
- 33.Naik, D. N., and T. Kaneda. 1974. Biosynthesis of branched long-chain fatty acids by species of Bacillus: relative activity of three α-keto acid substrates and factors affecting chain length. Can. J. Microbiol. 20:1701-1708. [DOI] [PubMed] [Google Scholar]
- 34.Namba, Y., K. Yoshizawa, A. Ejima, T. Hayashi, and T. Kaneda. 1969. Coenzyme A- and nicotinamide adenine dinucleotide-dependent branched chain α-keto acid dehydrogenase. I. Purification and properties of the enzyme from Bacillus subtilis. J. Biol. Chem. 244:4437-4447. [PubMed] [Google Scholar]
- 35.Nielsen, D. R., E. Leonard, S. H. Yoon, H. C. Tseng, C. Yuan, and K. L. Prather. 2009. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 11:262-273. [DOI] [PubMed] [Google Scholar]
- 36.Ogura, M., and T. Tanaka. 2009. The Bacillus subtilis late competence operon comE is transcriptionally regulated by yutB and under post-transcription initiation control by comN (yrzD). J. Bacteriol. 191:949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oku, H., and T. Kaneda. 1988. Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J. Biol. Chem. 263:18386-18396. [PubMed] [Google Scholar]
- 38.O'Riordan, M., M. A. Moors, and D. A. Portnoy. 2003. Listeria intracellular growth and virulence require host-derived lipoic acid. Science 302:462-464. [DOI] [PubMed] [Google Scholar]
- 39.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 40.Perham, R. N. 2000. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69:961-1004. [DOI] [PubMed] [Google Scholar]
- 41.Preis, H., R. A. Eckart, R. K. Gudipati, N. Heidrich, and S. Brantl. 2009. CodY activates transcription of a small RNA in Bacillus subtilis. J. Bacteriol. 191:5446-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reche, P., and R. N. Perham. 1999. Structure and selectivity in post-translational modification: attaching the biotinyl-lysine and lipoyl-lysine swinging arms in multifunctional enzymes. EMBO J. 18:2673-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed, K. E., and J. E. Cronan, Jr. 1993. Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J. Bacteriol. 175:1325-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Schujman, G. E., K. H. Choi, S. Altabe, C. O. Rock, and D. de Mendoza. 2001. Response of Bacillus subtilis to cerulenin and acquisition of resistance. J. Bacteriol. 183:3032-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekiguchi, J., N. Takada, and H. Okada. 1975. Genes affecting the productivity of α-amylase in Bacillus subtilis Marburg. J. Bacteriol. 121:688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 48.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 49.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917-927. [DOI] [PubMed] [Google Scholar]
- 50.Sonenshein, A. L., J. A. Hoch, and R. Losick (ed.). 2002. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 51.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 53.Wang, G. F., T. Kuriki, K. L. Roy, and T. Kaneda. 1993. The primary structure of branched-chain α-oxo acid dehydrogenase from Bacillus subtilis and its similarity to other α-oxo acid dehydrogenases. Eur. J. Biochem. 213:1091-1099. [DOI] [PubMed] [Google Scholar]
- 54.Weber, M. H., W. Klein, L. Muller, U. M. Niess, and M. A. Marahiel. 2001. Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol. Microbiol. 39:1321-1329. [DOI] [PubMed] [Google Scholar]
- 55.Welshimer, H. J. 1963. Vitamin requirements of Listeria monocytogenes. J. Bacteriol. 85:1156-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willecke, K., and A. B. Pardee. 1971. Fatty acid-requiring mutant of Bacillus subtilis defective in branched chain α-keto acid dehydrogenase. J. Biol. Chem. 246:5264-5272. [PubMed] [Google Scholar]
- 57.Zhu, K., D. O. Bayles, A. Xiong, R. K. Jayaswal, and B. J. Wilkinson. 2005. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain α-keto acid dehydrogenase. Microbiology 151:615-623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.