Abstract
The MnmE-MnmG complex is involved in tRNA modification. We have determined the crystal structure of Escherichia coli MnmG at 2.4-Å resolution, mutated highly conserved residues with putative roles in flavin adenine dinucleotide (FAD) or tRNA binding and MnmE interaction, and analyzed the effects of these mutations in vivo and in vitro. Limited trypsinolysis of MnmG suggests significant conformational changes upon FAD binding.
tRNA contains modified nucleosides that are posttranscriptionally generated by the activity of specific enzymes (http://modomics.genesilico.pl/sequence?seqtype=tRNA; 2). Many of these modifications frequently appear in the anticodon wobble position and are pivotal in the decoding process by stabilizing correct codon-anticodon interactions (1, 9). In Escherichia coli, the enzymes MnmE and MnmG carry out the GTP- and flavin adenine dinucleotide (FAD)-dependent incorporation of the cmnm (CH2-NH-CH2-COOH) group at position 5 of the wobble uridine in several tRNAs, a reaction whose specific steps remain to be elucidated (4, 7, 8). MnmG and MnmE form a heterotetrameric α2β2 complex in vitro (8). MnmG is a highly conserved FAD-binding protein (8) with 49% sequence identity to human MTO1.
We cloned E. coli MnmG (MnmGEc; NCBI gi:2367273), MnmGEc(1-550), and MnmE (gi:12518545) into a modified pET15b vector (Novagen). The proteins were expressed in E. coli BL21(DE3) (Novagen). Cells were grown in LB medium, induced with 100 μM isopropyl-1-thio-β-d-galactopyranoside, and incubated for ∼16 h at 16°C. Proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen, Mississauga, Ontario, Canada) and concentrated to ∼8 mg/ml in 20 mM Tris-HCl (pH 8.0), 0.8 M NaCl, 5% (vol/vol) glycerol, 5 mM dithiothreitol (DTT). MnmGEc was crystallized by the hanging-drop method, equilibrating the protein drop against reservoir solution containing 100 mM Tris-HCl (pH 7.5), 100 mM sodium formate, 6.5% (wt/vol) polyethylene glycolPEG 8000, 6% (vol/vol) ethylene glycol. The crystals belong to space group P21, with a = 85.9 Å, b = 144.1 Å, c = 147.6 Å, and β = 106.8°, with four molecules in the asymmetric unit (Vm = 3.03 Å3 Da−1) (6). They differ from the previously reported crystals and diffract to significantly higher resolution (7). Selenomethionine-labeled protein [expressed in strain DL41(DE3)] crystallized under the same conditions. Crystals of MnmGEc(1-550) were obtained at 20°C by equilibrating protein (7 mg/ml) supplemented with the anticodon stem-loop fragment of tRNAGlu (Dharmacon) (molar ratio, 1:1.2) against a reservoir solution of 1.3 M Li2SO4, 0.1 M Tris-HCl (pH 8.0). They belong to space group P3121, with a = 144 Å.6 and c = 271.0 Å, with two molecules in the asymmetric unit (Vm = 6.39 Å3 Da−1) and a solvent content of 80.8%.
The synchrotron data collected at the X12B beamline (NSLS, BNL) and at the SER-CAT beamline (APS, ANL) were used to solve the structures. The data were processed with HKL2000 (9). The structure of MnmGEc was solved at a 2.4-Å resolution by the single-wavelength anomalous diffraction method using the program SHARP/autoSHARP (2), the model was built with the help of RESOLVE (10), and the structure was refined with the program REFMAC5 (8) (see the supplemental material). The structure of MnmGEc(1-550) was determined by molecular replacement (MolRep [12]) and refined to a 3.5-Å resolution (Table 1). MnmGEc is composed of three well-ordered domains (Fig. 1a; see also Fig. S1 in the supplemental material) and a C-terminal region (residues 551 to 629) that is not visible in either crystal form (5) but is crucial for MnmE interaction (see Fig. S2 in the supplemental material). The one-dimensional nuclear magnetic resonance spectrum of MnmGEc suggested that the C-terminal domain is not unfolded but rather forms a molten globule or is loosely connected to the rest of MnmGEc.
TABLE 1.
X-ray data and refinement statistics
| Statistics | Value(s) or data for: |
||
|---|---|---|---|
| Se-Meta | MnmGEc | MnmGEc(1-550) | |
| Space group | P21 | P21 | P3121 |
| Unit cell parameters | |||
| a, b, c (Å) | 85.9, 144.1, 147.6 | 85.9, 144.3, 147.7 | 144.6, 144.6, 271.0 |
| α, β, γ (°) | 90, 106.8, 90 | 90, 106.6, 90 | 90, 90, 120 |
| Wavelength (Å) | 0.9793 | 0.9798 | 0.9793 |
| Resolution rangeb (Å) | 50-3.00 (3.11-3.00) | 50-2.41 (2.50-2.4) | 50-3.49 (3.61-3.49) |
| No. of observed reflections | 415,264 | 480,442 | 331,255 |
| No. of unique reflections | 133,967 (unmerged) | 129,269 | 42,430 |
| Completeness of data (%) | 99.6 (99.5) | 97.5 (81.6) | 99.8 (98.5) |
| Rsymc | 0.115 (0.497) | 0.065 (0.356) | 0.108 (0.640) |
| I/σ(I) | 11.6 (3.0) | 13.1 (2.6) | 9.0 (2.2) |
| Rworkd (no. of reflections) | 0.201 (122,715) | 0.227 (40,204) | |
| Rfree (no. of reflections) | 0.238 (6,511) | 0.265 (2,137) | |
| B-factor (no. of atoms) for: | |||
| Protein | 54.4 (15,648) | 83.3 (8,136) | |
| Solvent/ligand | 39.6 (325) | 79.0 (20) | |
| Ramachandran plot—residues in: | |||
| Allowed regions (%) | 98.5 | 95.0 | |
| Generously allowed regions (%) | 0.8 | 4.2 | |
| Disallowed regions (%) | 0.7 | 0.8 | |
| RMSDe | |||
| Bond length (Å) | 0.013 | 0.016 | |
| Bond angle (°) | 1.5 | 2.3 | |
| PDB code | 3CES | 3G05 | |
Se-Met, selenomethionyl (data obtained by single-wavelength anomalous diffraction).
The first range is for all reflections, and the range in parentheses is for the last resolution shell.
Rsym = (Σ Iobs − Iavg)/ΣIavg, where obs and avg indicate observed and average values of I. Values in parentheses are Rsym for the last resolution shell.
Rwork = (Σ Fobs − Fcalc)/ΣFobs, where obs and calc indicate observed and calculated F values.
RMSD, root mean square deviation.
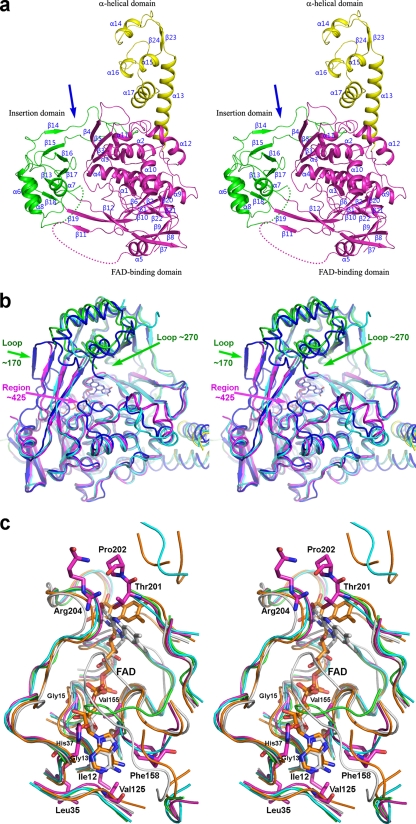
FIG. 1.
Crystal structure of MnmGEc. (a) MnmG monomer. The N-terminal FAD-binding domain is colored magenta, the insertion domain is green, and the α-helical domain is yellow. The locations of disordered loops (residues 167 to 179 and 263 to 277) are schematically marked by dotted lines. Arrows indicate a deep cleft between the FAD-binding and insertion domains. (b) Stereoview looking down at the cleft between the insertion and FAD-binding domains. The insertion domain is at the top, and the FAD-binding domain is at the bottom. Several MnmG molecules are superimposed, showing conformational flexibility of this putative tRNA-binding cleft. MnmGEc from our structure is colored as in panel a (magenta, green, and yellow), MnmGEc with Protein Data Bank (PDB) code 3CP2 is in cyan, and MnmGCt (PDB code 3CP8) is in blue (5). (c) Superposition of the FAD-binding sites from our form of MnmGEc (magenta), MnmGEc with PDB code 3CP2 (cyan), MnmGCt (PBD code 3CP8; orange), MnmGEc(1-550) (green), and GidAr (PDB code 2CUL; gray) (3). The FAD molecules from MnmGCt and GidAr are shown in stick mode. No bound FAD molecules were found in the crystal structures of MnmGEc. Some of the residues of MnmGEc discussed in the text are shown explicitly. The major structural deviation in this region is limited to the loop 153LTVGTFLDG161. This figure was prepared with PyMOL (http://pymol.org/).
MnmGEc and MnmGEc(1-550) form dimers in the crystals and in solution. The N-terminal, FAD-binding domain consists of segments comprising residues 1 to 201 and 342 to 458. Residues 202 to 341 form an insertion domain. Residues 459 to 550 form an α-helical domain, protruding outward from the N-terminal domain (Fig. 1a).
Comparison of the available MnmG structures identified three highly mobile segments that might be functionally important (loops encompassing approximately residues 165 to 180 and 245 to 295 and the region consisting of approximately residues 420 to 435 [MnmGEc numbering]) (Fig. 1b). The segment of residues 155 to 160, adjoining the first loop and partially disordered in various structures, forms part of the FAD's adenosine-binding site, and its conformation varies among MnmGs (Fig. 1c). All conformations observed in MnmGEcs are incompatible with FAD binding (Fig. 1c). The intrinsic flexibility of this region likely contributes to the rather low affinity of MnmGEc for FAD (see below), with a Kd (dissociation constant) of 3.0 ± 0.4 μM as measured by isothermal titration calorimetry (ITC) (see Fig. S3 in the supplemental material). The second segment, Thr245 to Ile295, is also disordered to a different extent in various MnmGs and adopts different conformations in ordered parts (Fig. 1b). The largest deviation occurs for residues D289RNQH293 at the top of this segment and residues Thr247 to Asn248 in the next β-strand. The region from Thr419 to Leu437 (Fig. 1b) is slightly longer in Chlorobium tepidum MnmG (MnmGCt), and the conformation of this region differs from that in MnmGEc. Specifically, the N-terminal end of MnmGCt lies close to the FAD molecule, with Arg423 contacting the flavin rings. The analogous Arg427 in MnmGEc is within sequence T422KEPYR427MFT430, identical to the corresponding sequence in MnmGCt and highly conserved among MnmGs (6). However, Arg427 is directed toward the solvent, over 13 Å away from its counterpart in MnmGCt. This structural flexibility is likely biologically relevant, providing a partner-induced fit mechanism.
MnmGEc is an FAD-binding protein with the tendency to lose its FAD cofactor (7, 14). In fact, it has not been possible to obtain crystals of MnmGEc with FAD, as far as we know. Here, the binding of FAD to MnmGEc was determined by ITC in a Microcal VP-ITC calorimeter (MicroCal, Northampton, MA). Experiments consisted of the titration of a 225 μM solution of nucleotide to a 15 μM MnmG solution at 25°C in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5 mM MgCl2. Binding was observed for FAD with an affinity of 3.0 ± 0.4 μM (see Fig. S4 in the supplemental material).
Based on findings from structural comparisons, we mutated highly conserved MnmG residues to alanines and explored the abilities of the resulting proteins to modify tRNA in vivo and to be protected by FAD against trypsinolysis in vitro. We analyzed the nucleoside compositions of tRNAs extracted from strain IC5241 (mnmG::Tn10) and IC5241 carrying pIC1180 or its derivative plasmids (see Table S1 in the supplemental material), which express, under the control of PBAD, wild-type or mutant MnmG protein. To this end, strains were grown in LB until the optical density at 600 nm reached 0.4. The culture was then divided into two equal parts, arabinose (0.2%) was added to one of them, and incubation was continued until the optical density at 600 nm reached 1.0. Cells were subsequently collected and processed as described previously for nucleoside analysis (5). In the absence of MnmG, the accumulation of s2U (defective conversion of wobble s2U into mnm5s2U) was observed (Table 2; see also Fig. S4a in the supplemental material), whereas expression of Flag-MnmG (wild type) reduced the fraction of s2U to a level close to that observed in a wild-type strain, even in the absence of an inducer (arabinose), indicating a leakiness of the pIC1180 expression system. In fact, wild-type levels of Flag-MnmG were observed without the inducer (see Fig. S4b in the supplemental material). The mutation of most of the selected residues impaired the tRNA-modifying function of MnmG (as summarized in Table 2). In some cases, the phenotype was rescued by overexpression of the mutant protein.
TABLE 2.
Properties of the MnmG mutant proteins
| Straina/plasmidb | MnmG protein | Mutated domainc | s2U/s4U ratiod |
FAD protectione | Putative function of mutated residue(s)f | |
|---|---|---|---|---|---|---|
| −ara | +ara | |||||
| MG1655 | Wild type | 0.000* | 0.000* | |||
| IC5241 | None | 0.024 | 0.024 | |||
| IC5241/pBAD22 | None | 0.026 | 0.028 | |||
| IC5241/pIC1180 | Flag-MnmG | 0.001 | 0.000* | + | ||
| IC5241/pIC1181 | G13A | FAD | 0.015 | 0.002 | − | FAD (Nuc) binding |
| IC5241/pIC1182 | G15A | FAD | 0.015 | 0.004 | − | FAD (Nuc) binding |
| IC5241/pIC1183 | G13A/G15A | FAD | 0.024 | 0.024 | FAD (Nuc) binding | |
| IC5241/pIC1387 | N48A/P49A | FAD | 0.015 | 0.007 | + | Catalysis or tRNA binding |
| IC5241/pIC1326 | G52A/G53A | FAD | 0.025 | 0.028 | + | Catalysis or tRNA binding |
| IC5241/pIC1382 | K56A | FAD | 0.014 | 0.005 | + | Catalysis or tRNA binding |
| IC5241/pIC1327 | G67A/G68A | FAD | 0.026 | 0.020 | Change of local conformation | |
| IC5241/pIC1386 | K88A | FAD | 0.002 | 0.000* | tRNA binding | |
| IC5241/pIC1388 | G89A/P90A | FAD | 0.016 | 0.004 | + | Change of local conformation |
| IC5241/pIC1365 | G156A/T157A | FAD | 0.023 | 0.030 | − | FAD (Nuc) binding |
| IC5241/pIC1368 | F158A | FAD | 0.000* | 0.000* | FAD (Nuc) binding | |
| IC5241/pIC1366 | R174A | FAD | 0.011 | 0.004 | − | FAD (Nuc) binding |
| IC5241/pIC1391 | R196A | FAD | 0.010 | 0.000* | tRNA biding | |
| IC5241/pIC1392 | T199A/G200A | FAD | 0.021 | 0.024 | FAD (IAM) interaction or conformational change | |
| IC5241/pIC1390 | T201A/P202A | FAD | 0.020 | 0.018 | − | FAD (IAM) binding or conformational change |
| IC5241/pIC1389 | R204A | INS | 0.015 | 0.000* | − | FAD (IAM) binding or conformational change |
| IC5241/pIC1383 | K283A | INS | 0.018 | 0.017 | + | tRNA biding |
| IC5241/pIC1384 | R286A | INS | 0.012 | 0.000* | + | tRNA binding |
| IC5241/pIC1364 | Y377A | FAD | 0.015 | 0.013 | + | Catalysis or tRNA binding |
| IC5241/pIC1385 | R427A | FAD | 0.018 | 0.004 | + | tRNA binding |
| IC5241/pIC1442 | R436A | FAD | 0.023 | 0.018 | tRNA binding | |
| IC5241/pIC1443 | R440A | FAD | 0.019 | 0.000* | tRNA binding | |
| IC5241/pIC1444 | R447A | FAD | 0.017 | 0.000* | tRNA binding | |
| IC5241/pIC1440 | E585K | C-term | 0.019 | 0.004 | MnmE interaction? | |
| IC5241/pIC1480 | E585A | C-term | 0.007 | 0.000* | MnmE interaction? | |
| IC5241/pIC1283 | MnmG1-535 | C-term | 0.024 | 0.026 | MnmE interaction | |
IC5241 is MG1655 mnmG::Tn10 (8).
All plasmids expressing mutant MnmG proteins are pIC1180 derivatives.
FAD, FAD-binding domain; INS, insertion domain (equivalent to I2 domain described in reference 5). The C-terminal region (C-term) of MnmG carries a point mutation and is deleted in the E585K mutant and MnmG1-535, respectively.
Levels of nucleoside s2U detected in the high-performance liquid chromatography analysis of tRNA purified from the indicated strain. −ara and +ara indicate the absence and presence of the inducer arabinose in the growth medium. The numbers are calculated as the absorbance of s2U relative to the absorbance of s4U at 314 nm and are the means of results from at least two independent experiments. The asterisks indicate that s2U was undetectable.
+ or − indicates that the protein is protected or not protected by the cofactor against trypsinolysis.
Putative function assigned to the residue(s) on the basis of structural, tRNA modification, and proteolysis data. Nuc, nucleotide moiety of FAD; IAM, isoalloxazine moiety of FAD.
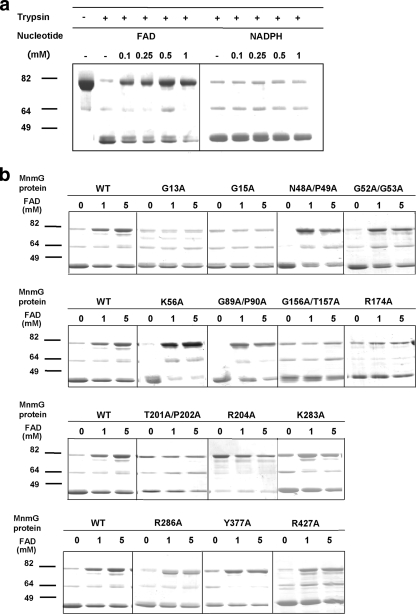
Limited proteolysis was used to explore the role of MnmG residues in FAD binding. FLAG-MnmG proteins (5 μg) were incubated with the indicated nucleotide concentrations in 25 μl of digestion buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 5 mM DTT) for 2 h at room temperature. Proteolysis reactions were started by adding TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (Sigma) at an MnmG-trypsin ratio of 300:1 (wt/wt) at 30°C. After 10 min incubation, reactions were stopped with the addition of phenylmethylsulfonyl fluoride (2 mM final concentration) and 4× Laemmli sample buffer, and the products were heated for 8 min at 95°C and subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. FAD protected MnmGEc from trypsinolysis, suggesting binding-induced conformational changes (Fig. 2a). The mutation of residues located in the vicinity of FAD (G13A, G15A, G156A/T157A, R174A, T201A/P202A, and R204A) impaired the protection (Fig. 1c and 2b), supporting evidence for the interaction of these residues with FAD or their role in the FAD-induced conformational change. These mutations also affect the tRNA modification function of MnmG (Table 2), in agreement with the idea that this function is FAD dependent (8). Mutation F158A does not impair the tRNA modification (Table 2), indicating that the aromatic ring of phenylalanine 158 is not critical for FAD binding.
FIG. 2.
(a) Sodium dodecyl sulfate-polyacrylamide gels showing the patterns of MnmGEc digestion by limited trypsinolysis in the presence of increasing concentrations (0 to 1 mM) of FAD and NADPH (negative control). Proteins were visualized by Coomassie staining. Positions of mass markers and their sizes, in kilodaltons, are indicated to the left. Full-length MnmGEc, at the top of each gel, migrates as a protein of 70 kDa. Bands below the full-length protein result from proteolysis. −, absent; +, present. (b) Sodium dodecyl sulfate-polyacrylamide gels showing the patterns of digestion of wild-type (WT) MnmGEc and various mutants (indicated by the corresponding mutations) by limited trypsinolysis in the presence of increasing concentrations (0 to 5 mM) of FAD. Molecular mass markers are shown as described above.
Residues Ser46 to Pro49, Gly52 to Gly53, Lys56, Ile343, Thr375, and Tyr377 are presumed to be in the vicinity of the FAD isoalloxazine ring. However, N48A/P49A, G52A/G53A, K56A, and Y377A mutants, while displaying partial or complete loss of function, are protected by FAD against trypsinolysis, which suggests that the roles of the mutated residues in FAD binding are minimal and that these residues are involved instead in maintaining the active-site architecture.
A double mutation, G67A/G68A, leads to a complete loss of function. Although according to our structural model, Gly67 and Gly68 are not directly involved in FAD binding, they are located at the turn between two α-helices involved in FAD binding and affect the local structure.
A distinguishing feature of MnmG proteins is a large cleft formed between the FAD-binding domain and the insertion domain (Fig. 1b). The cleft is lined with a series of conserved basic residues, including Lys56, Arg82, Lys88, Arg96, Lys198, Arg427, and Arg432, suitable for interaction with a negatively charged tRNA molecule. These basic residues are situated in the FAD-binding domain on one side of the cleft, while Lys283, Arg286, and Arg290 from the insertion domain are on the opposite side of the cleft. The presence of this cluster of basic residues gives the cleft a highly positive electrostatic potential. The FAD cofactor-binding pocket is located just below this cleft, with the riboflavin moiety partially exposed to solvent within the cleft. Interestingly, some of the above-mentioned residues are located in the mobile loops consisting of approximately residues 245 to 295 and 420 to 435 within the cleft. Mutations K56A, K283A, R286A, and R427A, while affecting tRNA modification, do not impair the FAD-induced protection against trypsinolysis (Table 2 and Fig. 2b), which is in line with a putative role of the corresponding residues in tRNA, but not FAD, binding. ITC measurements (see Fig. S3 in the supplemental material) confirmed that K283A has no effect on FAD binding (Kd for the K283A mutant, 3.7 ± 0.5 μM) and that G13A (affecting the first Gly residue of the consensus dinucleotide binding motif GXGXXG) reduces the affinity by 10-fold (Kd for the G13A mutant, 27 ± 4 μM). The ITC data for both mutants are in agreement with the results obtained from the proteolysis analysis, supporting the validity of this approach to gain information on the capabilities of MnmG mutants to bind FAD or undergo conformational changes associated with FAD binding. Note that other mutations affecting basic residues neighboring the mobile region from approximately residue 420 to residue 435 (R436A, R440A, and R447A) also impair the tRNA modification function, whereas the K88A mutant exhibits a nearly wild-type phenotype (Table 2).
Mutation G89A/P90A causes partial loss of the tRNA modification function, but it does not affect the FAD-induced resistance to trypsinolysis (Table 2 and Fig. 2b), suggesting that the corresponding residues, while affecting activity, are not associated with binding to the cofactor. Gly89 and Pro90 are situated in the turn between β4 and β5 (Fig. 1a; see also Fig. S1 in the supplemental material), and their change to alanine may affect the putative tRNA-binding site.
ITC and surface plasmon resonance experiments were conducted to quantify the binding of MnmEEc to MnmGEc (see Fig. S5a and S6 in the supplemental material). In the ITC experiments, a 150 μM solution of MnmE was titrated into a 10 μM solution of the MnmG or MnmG1-550 protein. Surface plasmon resonance studies were carried out at 25°C with a Biacore T100 instrument (Biacore AB, Uppsala, Sweden). Anti-FLAG M2 monoclonal antibody from Sigma (100 ng/μl in 10 mM sodium acetate, pH 4.5) was immobilized onto a CM-5 sensor chip (Biacore AB) at 10,000 to 12,000 resonance units using an amine coupling kit (Biacore AB). FLAG-MnmG at 1 μM in TBS buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, 0.005% Tween 20) was immobilized by capture, giving about 1,500 relative units. MnmE in TBS buffer at various concentrations was passed over the sensor chip at a flow rate of 30 μl/min, and the interactions were monitored for 1 min. The sensor surface was washed with TBS buffer to detect dissociation and then regenerated with a pulse of 5 mM NaOH (15 s at 60 μl/min). T100 Biacore evaluation software (Biacore AB) was used to evaluate the data. We obtained a Kd value of 3.3 μM by ITC and a value of 2.0 μM by SPR (see Fig. S5a and S6 in the supplemental material). In agreement with results from size exclusion chromatography (see Fig. S2 in the supplemental material), no binding of MnmEEc to MnmGEc(1-550) was observed by using ITC (see Fig. S5b in the supplemental material) and surface plasmon resonance. The expression of MnmGEc(1-550) and MnmGEc(1-535) in appropriate mnmG::Tn10 strains was unable to restore the wild-type phenotype (Table 2 and data not shown), suggesting that the formation of the MnmE-MnmG complex mediated by the MnmG C-terminal region is crucial for tRNA modification. Curiously, this function was impaired by mutation E585K and, to a lesser extent, E585A (Table 2), but neither of these changes affected the MnmG-MnmE interaction (see Fig. S6b in the supplemental material), in contrast to the data in a previous report (5). Thus, the role of Glu585 remains to be determined.
Supplementary Material
Acknowledgments
We thank I. Ekiel for help in nuclear magnetic resonance characterization of MnmGEc; Z. Dauter for help in data collection at the Southeast Regional Collaborative Access Team (SER-CAT) beamline facilities at Advanced Photon Source, Argonne National Laboratory; and M. Medina for valuable advice. X-ray diffraction data for this study were obtained by using beamline X12B of the National Synchrotron Light Source, Brookhaven National Laboratory, the Lilly Research Laboratory Collaborative Access Team beamline (LRL-CAT), and the SER-CAT beamline at Advanced Photon Source, Argonne National Laboratory. Use of the LRL-CAT beamline facilities in sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company, which operates the facility.
This is National Research Council of Canada publication no. 49586.
This work was supported by grant GSP-48370 from the Canadian Institutes of Health Research (to M.C. and A.M.) and grant BFU2007-66509 from the Ministerio de Ciencia e Innovación (to M.-E.A.).
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Björk, G. R. 1994. Biosynthesis and function of modified nucleosides, p. 165-205. In D. Soll and U. L. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. American Society for Microbiology, Washington, DC.
- 2.Czerwoniec, A., S. Dunin-Horkawicz, E. Purta, K. H. Kaminska, J. M. Kasprzak, J. M. Bujnicki, H. Grosjean, and K. Rother. 2009. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37:D118-D121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki, W., H. Miyatake, and K. Miki. 2005. Crystal structure of the small form of glucose-inhibited division protein A from Thermus thermophilus HB8. Proteins 61:1121-1126. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Vicente, M., L. Yim, M. Villarroya, M. Mellado, E. Perez-Paya, G. R. Bjork, and M. E. Armengod. 2005. Effects of mutagenesis in the switch I region and conserved arginines of Escherichia coli MnmE protein, a GTPase involved in tRNA modification. J. Biol. Chem. 280:30660-30670. [DOI] [PubMed] [Google Scholar]
- 5.Meyer, S., A. Scrima, W. Versées, and A. Wittinghofer. 2008. Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate. J. Mol. Biol. 380:532-547. [DOI] [PubMed] [Google Scholar]
- 6.Urbonavicius, J., S. Skouloubris, H. Myllykallio, and H. Grosjean. 2005. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria—evolutionary implications. Nucleic Acids Res. 33:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yim, L., M. Martinez-Vicente, M. Villarroya, C. Aguado, E. Knecht, and M. E. Armengod. 2003. The GTPase activity and C-terminal cysteine of the Escherichia coli MnmE protein are essential for its tRNA modifying function. J. Biol. Chem. 278:28378-28387. [DOI] [PubMed] [Google Scholar]
- 8.Yim, L., I. Moukadiri, G. R. Björk, and M. E. Armengod. 2006. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 34:5892-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama, S., and S. Nishimura. 1994. Modified nucleosides and codon recognition, p. 207-224. In D. Soll and U. L. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. American Society for Microbiology, Washington, DC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.