Abstract
Cyanobacteria perform oxygenic photosynthesis, which gives rise to the continuous production of reactive oxygen species, such as superoxide anion radicals and hydrogen peroxide, particularly under unfavorable growth conditions. Peroxiredoxins, which are present in both chloroplasts and cyanobacteria, constitute a class of thiol-dependent peroxidases capable of reducing hydrogen peroxide as well as alkyl hydroperoxides. Chloroplast peroxiredoxins have been studied extensively and have been found to use a variety of endogenous electron donors, such as thioredoxins, glutaredoxins, or cyclophilin, to sustain their activities. To date, however, the endogenous reduction systems for cyanobacterial peroxiredoxins have not been systematically studied. We have expressed and purified all five Synechocystis sp. strain PCC 6803 peroxiredoxins, which belong to the classes 1-Cys Prx, 2-Cys Prx, type II Prx (PrxII), and Prx Q, and we have examined their capacities to interact with and receive electrons from the m-, x-, and y-type thioredoxins from the same organism, which are called TrxA, TrxB, and TrxQ, respectively. Assays for peroxidase activity demonstrated that all five enzymes could use thioredoxins as electron donors, whereas glutathione and Synechocystis sp. strain PCC 6803 glutaredoxins were inefficient. The highest catalytic efficiency was obtained for the couple consisting of PrxII and TrxQ thioredoxin. Studies of transcript levels for the peroxiredoxins and thioredoxins under different stress conditions highlighted the similarity between the PrxII and TrxQ thioredoxin expression patterns.
In oxygenic photosynthetic organisms, the high oxidizing potential created in photosystem II during light-driven water splitting and the concomitant oxygen evolution, which takes place in the vicinity of electron transport reactions, exacerbates the problem of reactive oxygen species (ROS) production posed by normal aerobic metabolism (3, 37). Unleashed, these reactive compounds may oxidize and irreversibly inactivate components of the photosynthetic apparatus. Such inactivation, referred to as photoinhibition, is particularly apparent under conditions of high light intensity, nutrient deprivation, or cold, when the rate of ROS production is high due to the absorption of excess light energy (66). Therefore, chloroplasts and cyanobacteria, which are phylogenetically related, must be equipped with antioxidant enzymes, such as superoxide dismutases and peroxidases, that restrict the levels of ROS produced during photosynthesis. Despite their common origin and the similar nature of the processes leading to the formation of ROS in cyanobacteria and chloroplasts, current knowledge implies that the strategies they use to control the levels of, e.g., peroxides may be quite different (5).
In chloroplasts, ascorbate peroxidases are principal scavengers of H2O2, and ascorbate concentrations are as high as 12 to 25 mM (15). In contrast, ascorbate concentrations in cyanobacteria are much lower, ranging from 20 to 100 μM (67), and genes encoding ascorbate peroxidases are missing from the sequenced cyanobacterial genomes, including that of Synechocystis sp. strain PCC 6803. Glutathione concentrations in cyanobacteria range from 2 to 4 mM, but no glutathione-dependent peroxidase activity has been detected in any species tested from this phylum (67, 68). Catalases of plants and algae are located mainly in the peroxisomes but not in plastids. However, catalase activities have been found in all cyanobacterial species examined (38, 39, 43). A Synechocystis mutant devoid of its only catalase-peroxidase (katG) provided a useful tool for studying the nature of the remaining peroxidase activities (69). This mutant revealed the significance of a light-dependent thiol-specific peroxidase activity, raising the possibility that peroxiredoxins (Prx), which could benefit from thioredoxins (Trx) as electron donors, might be responsible for part of the peroxide decomposition in this organism (69).
Trx constitute a family of redox-active enzymes, which in cyanobacteria and chloroplasts may receive reducing equivalents directly from the linear photosynthetic electron flow by means of ferredoxin and ferredoxin Trx reductase (7, 14, 35, 59). Subsequently, Trx catalyze the reduction of a variety of target enzymes, including Prx, by disulfide/dithiol exchange (7). Prokaryotic phylogenetic groups of Trx include the m, x, and y types, all of which have homologues in the chloroplast of plants and algae (35). The completely sequenced genome of Synechocystis encodes one Trx from each of these families. TrxA (open reading frame [ORF] slr0623) is of the m type (40); TrxB (slr1139) is of the x type; and TrxQ (slr0233) is of the y type (14, 48). Our previous studies of Synechocystis deletion mutants lacking either TrxB or TrxQ have shown that the absence of TrxQ, but not that of TrxB, confers hypersensitivity to hydrogen peroxide, suggesting a role for TrxQ in peroxide tolerance in this organism (47). TrxA has been found to be essential in Synechocystis, and no deletion mutants could be obtained (42).
Prx comprise a class of peroxidases that have been extensively studied in Saccharomyces cerevisiae, mammals, and nonphotosynthetic bacteria (53, 72), as well as in plants (11, 12, 58). Based on their phylogeny and catalytic mechanisms, Prx are classified into four groups: 1-Cys Prx, 2-Cys Prx, type II Prx (PrxII), and Prx Q (11, 58, 72). The genome of the cyanobacterium Synechocystis encodes five Prx (64), which belong to each of the established classes 2-Cys Prx (sll0755), 1-Cys Prx (slr1198), PrxII (sll1621), and Prx Q (slr0242, sll0221). Studies of cyanobacterial Prx mutant strains suggest that these enzymes function in adaptation to growth at elevated light intensities, though the mechanism may not always involve simply peroxide detoxification. A cyanobacterial 2-Cys Prx homologue, highly similar to chloroplast BAS1 (4), was found to be required for optimal growth under high light conditions in Synechocystis (26) as well as in a Synechococcus sp. strain PCC 7942 (46), although it did not affect the ability to survive high concentrations of H2O2 added to cell cultures (46). The Synechocystis KatG 2-Cys Prx double mutant was impaired in translation of the photosystem II D1 protein during the repair process following high-light-induced damage (42). A Synechocystis PrxII disruptant strain showed a severely reduced growth rate relative to that of the wild type strain under normal light (23, 27), whereas a Synechocystis 1-Cys Prx disruption mutant grew somewhat more slowly than the wild-type strain but was not particularly sensitive to H2O2 or methyl viologen (23). In contrast, an Anabaena sp. strain PCC 7120 mutant failing to express one of its four Prx Q was hypersensitive to methyl viologen and grew poorly at moderate light intensities (29).
Numerous studies have explored the sources of reducing equivalents for Prx. For example, the 2-Cys Prx AhpC from some nonphotosynthetic bacteria is reduced by the flavoprotein AhpF (51), an NADPH disulfide oxidoreductase, which is absent from cyanobacteria and plants. The plant chloroplast 2-Cys Prx has instead been found to receive electrons from CDSP32, a protein containing two Trx domains in tandem (6), and NTRC, which consists of an NADPH Trx reductase (NTR) and a Trx in a single polypeptide chain (49). However, no homologue of CDSP32 can be found in cyanobacteria, and only some cyanobacterial species, such as Anabaena sp. strain PCC 7120, but not Synechocystis, harbor homologues of NTRC (14). Plastid 2-Cys Prx and Prx Q have also been reported to receive reducing equivalents from presumably ferredoxin Trx reductase-dependent, simple-module Trx of prokaryotic origin, such as Trx x (9) and Trx y (10). Even a cyclophilin (13) and glutaredoxin (Grx) (57) have been suggested as potential plant Prx electron donors. The 10 Prx of the plant Arabidopsis thaliana and its many Trx and Trx-like proteins are distributed among different cellular compartments (11, 12), whereas the five Synechocystis Prx, with the possible exception of PrxQ2, might be found in the same compartment as the Trx. Therefore, a priori, until further localization has been demonstrated, any of the Synechocystis Trx, or perhaps Grx or glutathione, as suggested in reference 23, could act as electron donors for these Prx. The Synechocystis x-type Trx has been reported to be a poor electron donor for Synechocystis 1-Cys Prx and PrxII (23). Synechocystis 2-Cys Prx (73) and five of the Synechococcus elongatus PCC 7942 Prx (63) have been shown to catalyze the decomposition of peroxides in vitro only by using NTR and Trx from Escherichia coli as sources of reducing equivalents. Until now, no systematic study of the endogenous reduction systems for cyanobacterial Prx has been performed, and none of the existent reports on cyanobacterial Prx activity compare the whole set of Prx in one cyanobacterium.
To better understand the mechanisms for peroxide decomposition in cyanobacteria, we aimed at clarifying the nature of the cyanobacterial Prx reduction systems. Therefore, we have examined the interactions of all five Synechocystis Prx with the three different Trx from this organism with regard to affinity and catalytic efficiency. Furthermore, a role for the Synechocystis Grx and glutathione as electron donors for these Prx was considered. Finally, we analyzed the expression of the prx and trx genes under various conditions (e.g., after a shift to high light intensity, addition of hydrogen peroxide, moderate heat shock, or nitrogen deprivation) in order to search for possible correlations between their distinct expression patterns.
MATERIALS AND METHODS
Growth of Synechocystis sp. strain PCC 6803 cultures.
Cells were grown photoautotrophically at 30°C in BG11 medium (54) supplemented with 1 g/liter NaHCO3 and were bubbled with a stream of 1% (vol/vol) CO2 in air under continuous illumination at a light intensity of 50 to 70 μE·m−2·s−1. All treatments were performed using exponentially growing Synechocystis sp. strain PCC 6803 cultures, typically displaying a chlorophyll concentration of 3 to 4 μg/ml. For high light conditions, cultures were illuminated with white light at an intensity of 500 μE·m−2·s−1, and the temperature was kept at 30°C by applying a 5-cm-thick water filter. For heat shock conditions, cells were grown in a water bath at 43°C. For oxidative stress conditions, 1 mM hydrogen peroxide was added. For nitrate starvation, cells were harvested by centrifugation at 9,000 × g for 10 min, washed three times with BG11 medium without nitrate, and resuspended in the same medium.
Sequences of genes and proteins.
Gene and protein sequences were obtained from the CyanoBase (http://genome.kazusa.or.jp/cyanobase). The trx genes were named trxA (ORF slr0623), trxB (slr1139), and trxQ (slr0233). The prx genes were 2-cys prx (sll0755), prxII (sll1621), 1-cys prx (slr1198), prxQ2 (sll0221), and prxQ1 (slr0242).
RNA isolation and Northern blot analysis.
Total RNA was isolated from 40-ml samples of Synechocystis sp. strain PCC 6803 cultures subjected to the treatments described above and at the times indicated. Cells were harvested by centrifugation and were broken using acid-washed baked glass beads (Sigma-Aldrich) in the presence of phenol and chloroform as previously described (19). For Northern blot analysis of mRNA levels, 10 μg of total RNA from each sample was electrophoresed on denaturing 1.5% agarose gels. Transfer of the RNA to nylon membranes (Millipore), prehybridization, hybridization, and washes were performed as recommended by the manufacturer. Probes for all genes were obtained by PCR, where 5 μg DNA of each trx, prx, and control gene was labeled with 32P using a Ready-To-Go kit (GE Healthcare) according to the manufacturer's instructions. As a control, filters were reprobed with a 580-bp HindIII and BamHI fragment from plasmid pAV1100 containing the constitutively expressed RNase P RNA gene (rnpB) from Synechocystis sp. strain PCC 6803 (70). The isiA, hspA, and pgr5 genes, used as indicators of the different stress conditions, were amplified using the primers listed in Table 1. The glnN gene was used as a control for nitrogen deprivation (52). Relative transcript levels were determined with a Cyclone Plus Storage Phosphor system (Perkin-Elmer).
TABLE 1.
Oligonucleotides used for amplifying thioredoxin, peroxiredoxin, glutaredoxin, and control genes used as probes
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| TrxAF | TTCCAGTATGAGTGCTACCC |
| TrxAR | AAGCCAGCGCTTAAAGATAT |
| TrxBF | GACCATTCATATGAGTTTAC |
| TrxBR | CAAATAGCTCGAGTCAGTTTAGGG |
| TrxQF | ACTGGATTAATATCTTTGCG |
| TrxQR | CTAAAGGTTTGTTGTCG |
| 2CysF | GCTACATATGACAGAGGTATTAAGGGTAG |
| 2CysR | GCTACTCGAGCTAAGGTTCCGCCACTGTCTC |
| PrxIIF | GAGGTACATATGACCCCCGAACG |
| PrxIIR | GGTTTCACTCGAGTAATCAC |
| 1CysF | GCTACATATGGCCTTACAACTCGGTGATG |
| 1CysR | GCTACTCGAGCTTATTGGGTTGGGGGGTC |
| PrxQ2F | CCTGGGGCATATGCAACCAGAGTTG |
| PrxQ2R | CGGGATTATTTACGTCGACGGGGAG |
| PrxQ1F | GAATTTTTTTCATATGGCCACTGCCTTAG |
| PrxQ1R | CCAAGGCTCGAGGATTTGCTTGC |
| GrxAF | GGCGTAGAGATATTGCACGGTC |
| GrxAR | GTTGGATCCGCTGTCTCGGCAAAAATTG |
| GrxBF | CTGGGGAATTCACGGGCAC |
| GrxBR | GGTGATCATATGGCTAATTTGTTC |
| Pgr5F | GGAGTCACTCATATGTTCGCCC |
| Pgr5R | CTCAGTTTCTCGAGAATTATTG |
| HspAF | CCACACATCAGGAGTTAACAT |
| HspAR | TTGATCATCTAGGGTCAGGAGC |
| IsiAF | CATAGGTCTCGGGTGGAC |
| IsiAR | TAAAGCTGATGGCTAATG |
Cloning, expression, and purification of recombinant proteins.
DNA fragments corresponding to the entire coding region of each prx gene (sll0755, sll1621, slr1198, sll0221, and slr0242) were amplified by PCR using appropriate forward and reverse primers, described in Table 1, and the High Expand Fidelity kit (Roche). All PCR products were cloned into the pET28a vector (Novagen), except slr1198, which was cloned into the pET24a vector, in order to produce recombinant proteins with N-terminal histidine tags. The resulting plasmids were named p2Cys, pPrxII, p1Cys, pPrxQ2, and pPrxQ1 and encoded the 2-cys prx, prxII, 1-cys prx, prxQ2, and prxQ1 genes, respectively. For expression of Prx, Escherichia coli (strain BL21) cells were transformed and grown in LB medium supplemented with kanamycin. Expression was induced by adding 1 mM isopropyl-l-d-thiogalactose (IPTG) at an optical density at 600 nm of about 0.4. The recombinant proteins were purified by nickel affinity chromatography using a HiTrap chelating HP column (GE Healthcare), and thereafter, gel filtration was performed on a Superdex 75 Hiload 16/60 column (GE Healthcare).
The wild-type version of TrxA was purified as described previously (41). For wild type TrxB and TrxQ, DNA fragments corresponding to the coding regions of trxB (slr1139) and trxQ (slr0233) were amplified by PCR using primers TrxBF and TrxBR or primers TrxQF and TrxQR, respectively (Table 1). PCR products were cloned into the pET28a vector, and recombinant TrxB and TrxQ were purified by nickel affinity chromatography and gel filtration as described for the Prx. His-tagged cysteine-to-serine mutants of TrxA, TrxB, and TrxQ, designated TrxA(C35S), TrxB(C34S), and TrxQ(C33S), respectively, were purified as described previously (32, 48).
The grxA (ssr2061) and grxB (slr1562) genes were amplified by PCR using primer pairs GrxA4-GrxA5 and GrxB-F2-GrxB-R2, respectively (Table 1). Fragments were cloned into pET28a, and the expressed GrxA and GrxB proteins were purified as described for the Prx.
Protein concentrations were determined according to reference 33.
SDS-PAGE and Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 10 or 12% acrylamide concentration and were stained with Coomassie brilliant blue (Sigma-Aldrich). For solubilization under reducing conditions, the loading buffer included 100 mM dithiothreitol (DTT) and 5% (vol/vol) β-mercaptoethanol. For nonreducing conditions, DTT and β-mercaptoethanol were omitted from the loading buffer, and 50 mM iodoacetamide was included. All samples were boiled for 5 min prior to electrophoresis. For Western blot analysis, proteins were resolved on SDS-PAGE (15% acrylamide) gels, transferred to nitrocellulose membranes (Bio-Rad), and probed with anti-TrxA (1:1,000), anti-TrxB (1:10,000), or anti-TrxQ (1:5,000) antibodies as described previously (14). Signals were detected using an anti-rabbit secondary antibody (Sigma-Aldrich) and the ECL-Plus immunoblotting detection system (GE Healthcare).
Peroxidase assay.
Peroxide decomposition catalyzed by Prx was measured using the ferrous ion oxidation (FOX) assay in the presence of xylenol orange as described in reference 71. The reduction of peroxides, such as hydrogen peroxide (H2O2), tert-butyl hydroperoxide (t-BOOH), or cumene hydroperoxide (CHP), was measured kinetically with several time points during the initial linear rate of catalysis. The assay mixture typically contained 50 mM HEPES-NaOH (pH 7.0), 0.1 or 0.2 μg/μl Prx, DTT at 0.2, 2, or 5 mM as indicated, and either 100 μM H2O2, 100 μM t-BOOH, or 50 μM CHP. Trx or Grx was included as an electron donor at the concentrations specified for each experiment and was kept reduced by addition of 0.2 mM DTT (Sigma Aldrich). To measure glutathione-dependent peroxidase activity, a reaction mixture containing 100 μM reduced glutathione (GSH) (Sigma-Aldrich) was used. When the GSH-Grx system was tested, 100 μM GSH, 0.15 U glutathione reductase from yeast (Sigma-Aldrich), 10 mM NADPH, and 4 μM GrxA or GrxB were added to the reaction mixtures.
Peroxidase activity in the presence of 100 μM GSH, 0.15 U gluthatione reductase, and 10 mM NADPH was also measured indirectly by monitoring the rate of NADPH oxidation as described previously (23).
One unit of enzyme activity was defined as the amount of protein required to catalyze the reduction of 1 μmol of H2O2, t-BOOH, or CHP per min.
RESULTS
Synechocystis Prx-Trx protein-protein interactions.
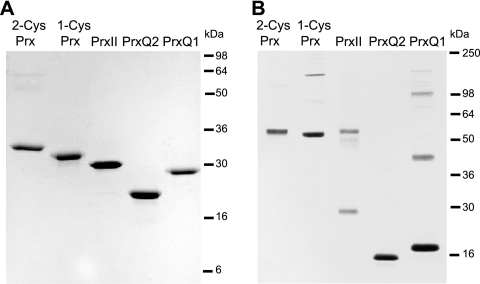
In order to assess the interactions between the Prx and Trx in Synechocystis sp. strain PCC 6803, the His-tagged versions of 2-Cys Prx (sll0755), 1-Cys Prx (slr1198), PrxII (sll1621), and Prx Q (slr0242 and sll0221) were expressed in Escherichia coli and purified to homogeneity (Fig. 1A). The PrxQ protein encoded by ORF sll0221, here designated PrxQ2, was expressed without the 34-amino-acid N-terminal extension, which is predicted to be a signal peptide according to the SignalP, version 3.0 (http://www.cbs.dtu.dk/services/SignalP/), and TargetP, version 1.1 (http://www.cbs.dtu.dk/services/TargetP/), programs. Electrophoresis under denaturing conditions in the absence of a reductant showed that 2-Cys Prx and 1-Cys Prx migrate mainly as dimers; PrxQ2 and PrxQ1 migrate for the most part as monomers; and PrxII is distributed between dimeric and monomeric forms, though minor amounts of a few high-molecular-mass adducts could also be observed (Fig. 1B). Notably, the monomeric forms of PrxII, PrxQ2, and PrxQ1 migrated more rapidly under nonreducing than under reducing conditions (Fig. 1A and B). This would be consistent with the presence of intramolecular disulfides, which render the proteins more compact (74).
FIG. 1.
Synechocystis sp. strain PCC 6803 peroxiredoxins. Four micrograms of each purified recombinant peroxiredoxin was incubated in the presence (A) or in the absence (B) of 100 mM DTT for 1 h on ice. Nonreduced samples (B) were solubilized in a buffer that included 50 mM iodoacetamide, whereas reduced samples (A) were solubilized in normal sample buffer. Thereafter, proteins were separated on 12% (A) or 10% (B) SDS-PAGE gels and stained with Coomassie brilliant blue.
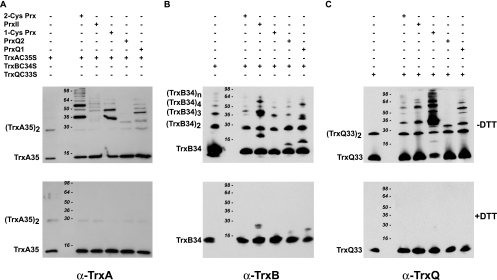
Our previous screening of the Synechocystis cytosolic compartment for Trx-interacting proteins using a TrxA(C35S) site-directed mutant as the bait resulted in the identification of 1-Cys Prx and PrxII among the target proteins (32). The subsequent study, in which we compared the target proteomes of the three Synechocystis Trx, showed that 1-Cys Prx and PrxII, in addition to being targets for TrxA, are also targets for TrxQ and, to a lesser extent, for TrxB (48). In this study, we tested the abilities of all five Prx to interact with each of the three Trx. To this end, the recombinant Prx were incubated with the site-directed mutated versions of each Trx—TrxA(C35S), TrxB(C34S), and TrxQ(C33S)—thus obtaining stable mixed disulfides between the respective Trx and Prx when interactions did occur. Mixed disulfides were detected by Western blot analysis using antibodies raised against the respective Trx following electrophoretic separation under nonreducing conditions (Fig. 2). Since we have observed previously that Synechocystis Trx carrying site-directed mutations of their second active-site cysteines tend to form disulfide-bridged dimers and, in the case of TrxB(C34S), oligomers (48), the Trx were also incubated in the absence of Prx as a control in order to distinguish Trx dimers or oligomers from Prx/Trx adducts (Fig. 2). Incubation of the Prx with TrxA(C35S) followed by Western blotting using antibodies specific for TrxA showed that mixed disulfides were formed mainly between this Trx and 2-Cys Prx, 1-Cys Prx, and PrxQ1, though smaller amounts of mixed disulfides formed with PrxII and PrxQ2 were also detected (Fig. 2A, upper panel). As expected, electrophoresis under reducing conditions abolished the observed Prx/Trx adducts, as detected by Western blotting (Fig. 2A, lower panel). The formation of disulfide-linked trimers and tetramers of TrxB(C34S) (Fig. 2B) due to the presence of a non-active-site cysteine in the amino acid sequence (48) made interpretation more difficult. However, bands with different migrations, indicating mixed disulfides, did appear for PrxII and PrxQ1 (Fig. 2B). It should be noted that the antibody raised against TrxB showed weak cross-reactivity against PrxII, PrxQ2, and PrxQ1 (Fig. 2B, lower panel). TrxQ(C33S) formed mixed disulfides with all Prx, particularly 1-Cys Prx, which in fact bound most of the added Trx (Fig. 2C). From this experiment it may be concluded that 2-Cys Prx is capable of interacting with TrxA and TrxQ; PrxII interacts with all Trx; 1-Cys Prx interacts strongly with TrxQ but also with TrxA; PrxQ2 interacts with TrxQ and weakly with TrxA; and PrxQ1 interacts moderately with all Trx.
FIG. 2.
Analysis of the thioredoxin-peroxiredoxin interaction. Twenty (A) or 11.25 (B and C) μg of each peroxiredoxin was incubated with 8 μg TrxA(C35S) (A), 4 μg TrxB(C34S) (B), or 4 μg TrxQ(C33S) (C). Thereafter, proteins were resolved on 12% SDS-PAGE gels under reducing (lower panels) or nonreducing (upper panels) conditions. Thioredoxins were detected by Western blotting using specific antibodies at the following dilutions: anti-TrxA (α-TrxA), 1:1,000; α-TrxB, 1:10,000; α-TrxQ, 1:5,000.
Peroxidase activities of the Synechocystis Prx using the Trx as electron donors.
Next, we explored the ability of the Synechocystis Prx to receive electrons and protons from each of the Trx in order to sustain peroxidase activity. To this end, equal amounts of the Prx were incubated with 4 μM each Trx reduced with a low (0.2 mM) concentration of DTT, and decomposition of H2O2 was measured (Fig. 3). DTT at this concentration did not produce appreciable rates of Prx activity in the absence of Trx (Fig. 3). Except for the couple TrxQ and PrxQ1, the Synechocystis Trx could indeed serve as electron donors for the entire complement of Prx from the same organism. The finding that nearly all Prx were able to receive electrons from all Trx prompted a detailed analysis of the kinetic parameters in order to establish the possible preferences of each Prx for a particular Trx as the electron donor. To this end, Prx activities were measured at constant H2O2 concentrations (100 μM), and the Trx concentrations were varied in the micromolar range for determinations of the apparent Km values. The kinetic parameters presented in Table 2 allowed some conclusions to be drawn about catalytic efficiencies. First, TrxQ was the most efficient electron donor for 2-Cys Prx, PrxII, and PrxQ2, mainly due to the low apparent Km values observed. Second, TrxB produced the highest reaction rates (kcat) for 2-Cys Prx and PrxQ2, but these kcat were counteracted by high apparent Km values, and the catalytic efficiencies (kcat/Km) were fourfold lower than when TrxQ was used as the electron donor. Third, 1-Cys Prx and PrxQ1 displayed very low kcat and kcat/Km irrespective of the Trx used to donate reducing equivalents; however, TrxA proved a more efficient electron donor for 1-Cys Prx than the other Trx, due to a low apparent Km (Table 2).
FIG. 3.
Thioredoxin-dependent peroxidase activities of 2-Cys Prx, PrxII, 1-Cys Prx, PrxQ1, and PrxQ2. Reaction mixtures contained 50 mM HEPES-NaOH (pH 7.0), 0.2 mM DTT, 4 μM Trx, 100 μM H2O2, and 25 μg Prx in a final volume of 250 μl. This concentration corresponds to 5 μM for 2-Cys Prx, 1-Cys Prx, and PrxII and to 5.5 μM for PrxQ1 and PrxQ2. The concentration of H2O2 was measured at 0, 10, 20, and 30 min after hydrogen peroxide addition. As a control, for each peroxiredoxin an assay was performed in the absence of thioredoxins but in the presence of DTT (dashed lines). The values at each time point are averages from at least three independent experiments.
TABLE 2.
Kinetic parameters of the five different Synechocystis peroxiredoxins using the three different thioredoxins as electron donorsa
| Peroxiredoxin and thioredoxin | Kmapp (μM) | Vmax (nmol min−1 mg Prx−1) | kcat (s−1) (103) | kcat/Km (103) |
|---|---|---|---|---|
| 2-Cys Prx | ||||
| TrxA | 9.6 ± 1.8 | 237 ± 38 | 92 ± 1 | 10 |
| TrxB | 21.8 ± 6.5 | 547 ± 29 | 212 ± 12 | 9.7 |
| TrxQ | 2.1 ± 0.2 | 205 ± 27 | 79 ± 10 | 39 |
| PrxII | ||||
| TrxA | 4.6 ± 0.8 | 273 ± 34 | 99 ± 10 | 22 |
| TrxB | 20.8 ± 0.1 | 124 ± 5 | 45 ± 2 | 2 |
| TrxQ | 0.74 ± 0.1 | 170 ± 10 | 63 ± 4 | 85 |
| 1-Cys Prx | ||||
| TrxA | 0.9 ± 0 | 25 ± 1 | 10 ± 0 | 11 |
| TrxB | 2.9 ± 1.4 | 23 ± 3 | 9 ± 1 | 3 |
| TrxQ | 2.8 ± 0.2 | 24 ± 1 | 10 ± 1 | 4 |
| PrxQ2 | ||||
| TrxA | 12.2 ± 3 | 269 ± 65 | 78 ± 10 | 6 |
| TrxB | 17.3 ± 2.5 | 776 ± 57 | 226 ± 16 | 13 |
| TrxQ | 2.6 ± 0.5 | 82 ± 7 | 140 ± 40 | 54 |
| PrxQ1 | ||||
| TrxA | 6.6 ± 3 | 48 ± 14 | 15 ± 4 | 2 |
| TrxB | 2.6 ± 0.2 | 23 ± 6 | 72 ± 2 | 3 |
| TrxQ |
Peroxidase activities were measured using 100 μM hydrogen peroxide as the substrate. Each peroxiredoxin was added at a final concentration of 10 μM, except for PrxQ1 and PrxQ2, which were added at 11.5 μM. Thioredoxin concentrations were varied from 0.1 to 20 μM for determination of apparent Km (Km app) values. The data are means of the values obtained from three or four independent assays.
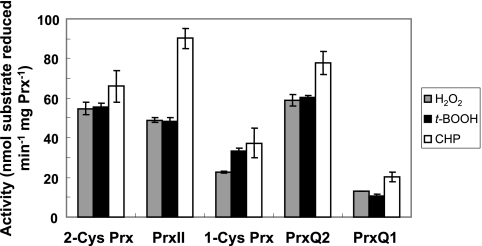
The specificities of the Synechocystis Prx for peroxide substrates were examined by comparing the decomposition rates of hydrogen peroxide, t-BOOH, and CHP. Generally, the rates of decomposition of hydrogen peroxide and alkyl hydroperoxides were similar, except for PrxII, which decomposed CHP twice as fast as the other peroxides (Fig. 4).
FIG. 4.
Substrate specificities of 2-Cys Prx, PrxII, 1-Cys Prx, PrxQ1, and PrxQ2. Reaction mixtures contained 50 mM HEPES-NaOH (pH 7.0), 0.2 mM DTT, 4 μM TrxA, 25 μg Prx, and either 100 μM H2O2, 100 μM t-BOOH, or 50 μM CHP in a final volume of 250 μl. The reduction of peroxides was measured as described in Materials and Methods. The values at each time point are means from at least three independent experiments. Error bars, standard deviations.
Peroxidase activity of the Synechocystis Prx using GSH and Grx's as electron donors.
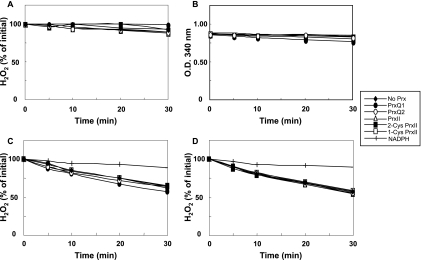
To compare the efficiency of GSH as an electron donor for the five Synechocystis Prx, assays for peroxidase activity were performed using H2O2 as the substrate (Fig. 5A). Surprisingly, none of the Prx displayed peroxidase activity, as measured by the disappearance of peroxide, when supplied with GSH as the sole electron source (Fig. 5A). Assays for peroxidase activity were also performed using a system to regenerate GSH, consisting of glutathione reductase and NADPH, and the absorbance at 340 nm was monitored. However, no oxidation of NADPH above the background level occurred in the presence of Prx, demonstrating that GSH was not consumed by the Prx (Fig. 5B). Since the cytosolic poplar PrxII has been shown to use Grx as an electron donor (56, 57), the His-tagged versions of Synechocystis GrxA and GrxB, encoded by the ssr2061 and slr1562 ORFs, respectively, were expressed, purified, and examined for activity as electron donors for the five Synechocystis Prx (Fig. 5C and D). These experiments showed that both GrxA and GrxB possess some intrinsic peroxidase activity but are not able to donate electrons to any of the Prx (Fig. 5C and D). The peroxidase activities of GrxA and GrxB were found to depend on the presence of either GSH or DTT (data not shown). Since our results were partially in disagreement with those of Hosoya-Matsuda et al. (2005) concerning PrxII, which was reported to use GSH rather than Trx as the electron donor (23), we also tested a peroxide concentration (250 μM) higher than that used in their study. However, this only increased the nonenzymatic GSH-dependent background activity, and the presence of PrxII did not accelerate NADPH oxidation (see Fig. S1 in the supplemental material).
FIG. 5.
Peroxidase activities of 2-Cys Prx, PrxII, 1-Cys Prx, PrxQ1, and PrxQ2 using GSH or Synechocystis glutaredoxins as electron donors. Each reaction mixture contained 75 mM potassium phosphate (pH 7.6), 100 μM GSH, 100 μM H2O2, and 25 μg Prx in a final volume of 250 μl. This concentration corresponds to 5 μM for 2-Cys Prx, 1-Cys Prx, and PrxII and to 5.5 μM for PrxQ1 and PrxQ2. (A) Reduction of hydrogen peroxide by peroxiredoxins by using GSH alone as the electron donor. Concentrations of hydrogen peroxide were determined by the FOX assay. (B) NADPH oxidation coupled to peroxiredoxin activity using GSH as the electron donor and glutathione reductase-dependent regeneration. To this end, reaction mixtures were supplemented with 0.15 U glutathione reductase from yeast and 10 mM NADPH. NADPH oxidation was measured spectrophotometrically at 340 nm. (C and D) Decomposition of hydrogen peroxide using a GSH-glutaredoxin system as the electron donor for the peroxiredoxins. Reaction mixtures were supplemented with 0.15 U glutathione reductase from yeast, 10 mM NADPH for regeneration, and either 4 μM GrxA (C) or 4 μM GrxB (D). Concentrations of hydrogen peroxide were determined by the FOX assay. Controls were performed with reaction mixtures containing no Prx but still containing the respective Grx (No Prx). Controls were also performed in the presence of NADPH, glutathione reductase, and GSH but without Grx and Prx (NADPH). The values at each time point are averages for at least three independent experiments.
Expression of the Synechocystis prx and trx genes.
Previous studies have shown that the expression of prx genes may differ in response to changes in growth conditions in plants as well as in cyanobacteria (8, 22, 63, 64). Little is known about trx gene expression in cyanobacteria, except that Synechocystis trxA transcription depends on the operation of photosynthetic or respiratory electron transport and that prolonged exposure of cultures to high light intensity results in a decrease in trxA transcript levels (41). Considering a possible correlation between the expression patterns of the prx and trx genes, we examined the transcript levels of all five Synechocystis prx genes and the three trx genes under various conditions. To this end, Northern blot analyses were performed on RNA isolated from Synechocystis cultures following transfer to high light intensity, addition of H2O2 (Fig. 6), a temperature increase, or the removal of a nitrogen source from the medium (Fig. 7), and the relative mRNA levels were quantified.
FIG. 6.
Effects of hydrogen peroxide and high-light (HL) treatments on the expression of thioredoxin and peroxiredoxin genes. Synechocystis cells growing photoautotrophically were either treated with hydrogen peroxide (0.5 mM) (A and C) or exposed to a high light intensity (500 μE·m−2 s−1) (B and D). At the indicated times, samples were collected, and total RNA was obtained and hybridized with specific probes for each gene. Relative mRNA levels were determined and normalized to rnpB expression levels.
FIG. 7.
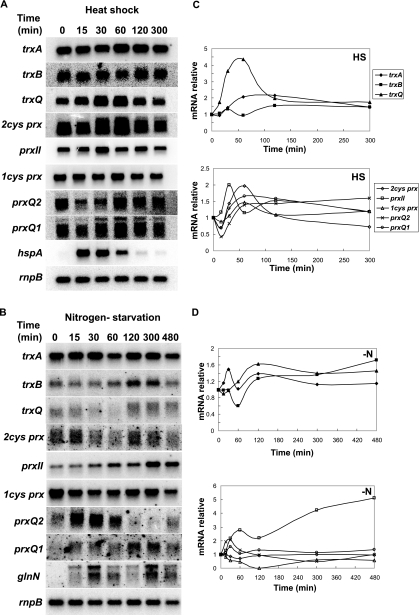
Effects of heat shock (HS) and nitrogen deprivation (−N) on the expression of thioredoxin and peroxiredoxin genes. Either Synechocystis cells growing photoautotrophically were subjected to a heat shock treatment (43°C) (A and C) or the culture was transferred to a medium lacking a nitrogen source (B and D). At the indicated times, samples were collected, and total RNA was obtained and hybridized with the specific probes for each gene. Relative mRNA levels were determined and normalized to rnpB expression levels.
Following the addition of 1 mM hydrogen peroxide to Synechocystis cultures, the transcript levels of the trxA, trxB, 2-cys prx, 1-cys prx, and prxQ2 genes decreased during the first 15 min and thereafter increased to the initial levels (Fig. 6A and C). In contrast, the transcript levels of trxQ, prxII, and prxQ1 showed a pronounced increase during the first 30 min; thereafter, they were reduced to the initial levels for trxQ and prxII but not for prxQ1, which remained upregulated (Fig. 6A and C). The expression of the isiA gene, which encodes an iron deficiency-related chlorophyll-binding protein (36), was monitored under the same conditions as a control, since expression of this gene is known to be induced by hydrogen peroxide (30, 62). The isiA gene was found to respond with a pattern similar to that of trxQ, prxII, and prxQ1 (Fig. 6A).
Exposure to high light intensity resulted in enhanced transcription of several of the prx genes, and the most conspicuous response was observed for the prxII transcript, the level of which displayed a 10-fold increase (Fig. 6B and D). The expression of 1-cys prx appeared to be the least affected by high light conditions. Concerning trx expression, the trxA transcript and, to a greater extent, the trxB transcript accumulated continuously throughout the experiment, whereas the trxQ transcript level showed a transient increase during the first 30 min of high light (Fig. 6B and D). The trxB transcript exhibited the most dramatic change, with an increase of almost sixfold during the 5-h experiment (Fig. 6D). As a control, the pgr5 transcript level was determined (Fig. 6B), since expression of this gene (ssr2016) has been shown to be enhanced following a shift from low to high light intensity (2).
Heat shock treatment produced no considerable changes in the expression of any of the prx genes, trxA, or trxB. In contrast, the trxQ transcript level increased about fourfold during the first hour of exposure to 43°C and thereafter dropped again (Fig. 7A and C). The expression of the hspA gene, which encodes the 16.6-kDa small heat shock protein (65), was examined as a positive control and was found to be induced more than 100-fold during the first 30 min (Fig. 7A).
Nitrogen starvation did not affect the expression of the trx genes significantly (Fig. 7B and D). In contrast to the other prx genes, the transcript level of the prxII gene increased throughout the experiment, whereas that of the 1-cys prx gene decreased sharply after 60 min. As expected, expression of the glnN gene, which encodes a glutamine synthetase (52), was rapidly induced by nitrogen starvation (Fig. 7B and D).
Thus, similar transcription patterns were observed for the Synechocystis prxII, prxQ1, and trxQ genes with respect to induction in response to H2O2. Common patterns were also detected for the 1-cys prx and trxA genes with respect to repression of transcription in response to H2O2. The 2-cys prx and trxB genes shared the common features of induction in response to high light intensity but not to H2O2.
DISCUSSION
Interactions between Synechocystis Prx and Trx.
In this study an effort was made to provide an overview of the entire Prx complement from one oxygenic photosynthetic prokaryote, the cyanobacterium Synechocystis, with particular emphasis on the role of the Trx from this organism as electron donors to sustain Prx activity. To establish which Trx may interact with each Prx is of special relevance for a prokaryotic organism, where all these enzymes might reside in the same compartment. The only exception to the presumed cytosolic location is PrxQ2, which belongs to a phylogenetic subgroup of the cyanobacterial Prx Q homologues referred to as the GCT4 subcluster (8). Sequence analysis showed that all cyanobacterial Prx Q homologues from the GCT4 subcluster possess an N-terminal extension that is predicted to be a signal peptide with a probability close to unity (our unpublished data). Interestingly, the only Arabidopsis PrxQ was recently shown to be associated with chloroplast thylakoid membranes (28) and was further demonstrated to reside within the thylakoid lumen (50).
Since Prx enzymatic activity depends on the redox state of cysteines, resolution of Prx by protein gel electrophoresis is commonly performed both with and without disulfide reductants, such as DTT and β-mercaptoethanol. Under nonreducing conditions, the Synechocystis 2-Cys Prx migrated, as expected, as a dimer. This is a result of the disulfide formed during catalysis between the N-terminal peroxidatic cysteine from one subunit and the C-terminal resolving cysteine from another subunit, which is characteristic for typical 2-Cys Prx (72). Plant PrxII and Prx Q have been classified as atypical 2-Cys Prx, which means that the disulfides formed between peroxidatic and resolving cysteines are intramolecular (13, 55). Nevertheless, a fraction of Synechocystis PrxII, which conserves both cysteines with respect to plant PrxII isoforms, migrates as a dimer under nonreducing conditions (Fig. 1B). PrxQ2 migrated exclusively as a monomer in the absence of a reductant. This is in agreement with the study of Cha et al. (2007), which showed that all four Prx Q from Anabaena sp. strain PCC 7120 form intramolecular disulfides (8). Synechocystis PrxQ1 existed principally in the monomeric form under nonreducing conditions, but a minor fraction could also be observed as a dimer. This finding is somewhat unexpected, since the PrxQ1 homologue from Anabaena clearly is a monomeric enzyme (8). It is probably due to the fact that PrxQ1 contains a second cysteine 70 amino acids downstream from the peroxidatic cysteine, which apparently permits disulfide formation with another PrxQ1 molecule. Whether this disulfide is relevant for catalysis remains to be elucidated. The catalytic mechanism for 1-Cys Prx is still unknown (13), but the migration of Synechocystis 1-Cys Prx under nonreducing conditions (Fig. 1B) indicates that either of its two additional cysteines, apart from the conserved peroxidatic cysteine, may participate in intermolecular disulfide formation (23).
Plant Prx have been reported to be relatively slow peroxidases compared to ascorbate peroxidases and catalases (58). The peroxide decomposition rates measured for the Synechocystis Prx, using any of the three Synechocystis Trx as electron donors, lie within a kcat range of 0.01 to 0.2 s−1 (Table 2). These values are compatible with those observed for Arabidopsis 2-Cys Prx and Prx Q, which were reported to range from 0.08 to 0.6 s−1 when various chloroplast simple-module Trx were used as electron donors with DTT as the ultimate reductant of these Trx (9, 10). Notably, 100-fold-higher turnover numbers have been determined for the Salmonella enterica serovar Typhimurium 2-Cys Prx (AhpC) by using an assay that included a highly efficient reductant consisting of the S. Typhimurium AhpF N-terminal domain carrying an S128W substitution (44, 45). In the present study, however, we aimed at comparing the efficiencies and limitations of the endogenous Synechocystis Trx as electron donors for the Prx, and therefore, the assay developed in references 44 and 45 was not applicable.
Hosoya-Matsuda et al. (2005) have determined the peroxidase activities of the Synechocystis 1-Cys Prx and PrxII using the Synechocystis x-type Trx, which in our study is referred to as TrxB, as the electron donor (23). Using 0.5 mM H2O2 as the substrate, the measured maximal activities were 2.7 and 0.2 nmol per mg of protein per min for PrxII and 1-Cys Prx, respectively, considerably lower than the activities of all Synechocystis Prx determined in the present study, which range from 22 to 776 nmol per mg of protein per min (Table 2). However, Hosoya-Matsuda et al. (2005) used an Arabidopsis NTR as the electron donor for TrxB (23). Notably, the cyanobacterial x-type Trx was previously found to be unable to receive electrons from E. coli NTR (1), unlike the cyanobacterial m-type Trx (20, 31). Therefore, a possible explanation for the discrepancy between the activities observed in the present study (Fig. 3 and Table 2) and those observed in reference 23 is the choice of reductant of the Trx, since electron transfer between the NTR and the Trx may be rate limiting.
Previous biochemical characterization of plant chloroplast 2-Cys Prx and Prx Q, including assays of various chloroplast Trx together with DTT as electron donors (9, 10), allows a comparison with the data obtained from this study of their cyanobacterial homologues. It should be noted that in our assays we used 100 μM H2O2, whereas in references 9 and 10, 400 μM t-BOOH was used as the peroxide substrate. However, in all three studies, one could consider the peroxide concentration to be well above saturation. The Arabidopsis 2-Cys Prx was reduced in vitro by plastid simple-module Trx of the m, f, and x types, and the highest catalytic efficiency was obtained for Trx x, with a kcat of 0.18 s−1 and a Km of 25 μM (9). These values are strikingly similar to those measured for Synechocystis 2-Cys Prx using the x-type Trx, TrxB, as the electron donor, which yielded a kcat of 0.21 s−1 and a Km of 22 μM (Table 2). However, Synechocystis 2-Cys Prx could be considered most efficiently reduced by the y-type Trx, TrxQ, since the apparent Km was as low as 2 μM (Table 2). The plastid Trx y isoforms had not yet been discovered at the time of the study by Collin et al. (2003) (9). Later, Arabidopsis Prx Q activity was found to be sustained in vitro by Arabidopsis Trx y1, Trx y2, Trx x, and Trx m4, and the highest catalytic efficiencies were measured for Trx y1 and Trx y2, which produced kcat values of 0.29 and 0.56 s−1 and Km values of 1 and 2.6 μM, respectively (10). The Synechocystis y-type Trx, TrxQ, is also the best electron donor for PrxQ2, with a kcat of 0.14 s−1 and a Km of 2.6 μM. However, it should be kept in mind that these activities may be physiologically irrelevant if the plant Prx Q is located in the thylakoid lumen (50) and the Synechocystis PrxQ2 is indeed luminal or periplasmic. Surprisingly, Synechocystis TrxQ proved completely inefficient toward PrxQ1, emphasizing that Prx Q represents a heterogeneous family in cyanobacteria (8; also this study).
The next issue to be addressed was whether there is a role for GSH or Grx as sources of reducing equivalents for cyanobacterial Prx, since, for example, a cytosolic poplar PrxII is able to use Grx as an electron donor (55-57). In contrast to the results in reference 23, where very high activities for Synechocystis PrxII were obtained using GSH with or without Grx, we observed no activity for any Synechocystis Prx by using GSH or Grx as the reducing agent. It should be noted that Arabidopsis chloroplast PrxII-E, one of the closest plant relatives of Synechocystis PrxII, could be reduced neither by GSH nor by chloroplast Grx (13). Furthermore, Tichy and Vermaas (1999) demonstrated that Synechocystis possesses no GSH-dependent peroxidase activity in vivo (69), which is also in agreement with the finding that the two glutathione peroxidase-like proteins encoded by the Synechocystis ORFs slr1171 and slr1992 are able to use NADPH, but not GSH, as an electron donor for the reduction of lipid hydroperoxides (16, 17). Concerning peroxide substrate specificity, we detected no significant preferences of any of the Synechocystis Prx, except for PrxII, which was markedly more active with the aromatic peroxide CHP. PrxII proteins from other photosynthetic organisms have not been studied previously in this regard, but poplar Prx Q decomposes H2O2, t-BOOH, and CHP, with a higher efficiency toward CHP (55), and Arabidopsis Prx Q reduces H2O2 and CHP at compatible rates but is less efficient toward t-BOOH and completely inefficient toward lipid hydroperoxides (28).
Expression of prx and trx genes under different stress conditions.
Enhanced transcription of the Synechocystis prxII gene under high light conditions and in the presence of H2O2 is well documented. Synechocystis cDNA microarray analyses revealed that expression of the prxII gene was strongly upregulated by H2O2 (30) and methyl viologen, particularly under high light conditions (27). This was also observed in the study reported in reference 64, which confirmed the transient increase in Synechocystis prxII mRNA levels following treatment with either H2O2 or methyl viologen. Here we show a similar expression pattern for the Synechocystis trxQ gene following the addition of H2O2, which is consistent with the fourfold increase observed in a recent cDNA microarray analysis (25). The initial decrease in transcription of the trxA and 1-cys prx genes during the first 30 min of peroxide treatment in our study is also in agreement with the values obtained from a cDNA microarray analysis (30), where these genes are reported to be repressed 2.1-fold and 2.0-fold, respectively, 30 min after addition of H2O2. Interestingly, global expression profiling of Synechocystis cells exposed to UV-B radiation displayed twofold and fourfold increases in the trxQ and prxII mRNA levels, respectively, whereas the trxA and 1-cys prx transcript levels decreased threefold and twofold, respectively (24). Thus, there is a consistent pattern of upregulation of expression of the prxII and trxQ genes and downregulation of expression of the 1-cys prx and trxA genes in response to H2O2 as well as to UV-B radiation.
In contrast to the transient nature of the responses to H2O2 addition, we found that nitrogen starvation triggered continuous induction of prxII transcription. Since nitrogen-containing macromolecules, such as proteins and DNA, represent major sinks for the primary products of photosynthesis, nitrogen deprivation may lead to enhanced production of ROS (60), due to absorption of excess excitation energy even at moderate light intensities. Thus, it is likely that nitrogen starvation initiates continuous intracellular release of ROS, in contrast to the addition of exogenous H2O2 or methyl viologen, which results in a brief burst of intracellular ROS. In this context, it is worth mentioning that 1 mM H2O2 added to a culture of Synechococcus sp. strain PCC 7942 cells was decomposed within 15 min (46). Therefore, it is not surprising that no differences in the PrxII and 1-Cys Prx protein levels were detected during the hours following the addition of 0.5 mM H2O2 to Synechocystis cultures (23).
Unlike prxII and trxQ, the Synechocystis 2-cys prx gene did not respond appreciably to the addition of H2O2, though its transcription was induced by an increase in light intensity, suggesting that it is subject to a different control mechanism, independent of ROS. Previous studies have shown that illumination with high light intensity enhances transcription of both the Synechocystis 2-cys prx gene (64) and the Synechococcus sp. strain PCC 7942 2-cys prx gene (46). Among the Synechocystis trx genes, trxB represents the clearest example of induction under high light conditions but not in response to the addition of H2O2.
The Synechocystis prxQ genes were both induced by increased light intensity but responded differently to the addition of H2O2 and to nitrogen deficiency: prxQ1 was transcribed more under the former condition and prxQ2 was transcribed more under the latter. Stork et al. (2005) failed to detect any transcript of the Synechocystis prxQ2 gene, referred to as prxQ-B2 in their study, under any conditions (64). In the present study we clearly detected the prxQ2 transcript in all Northern blot analyses, confirming that this gene is indeed expressed. In comparison, in plants the genes encoding the chloroplast PrxQ and PrxII proteins appear to be transcribed at higher levels under conditions that stimulate ROS production (50). Concerning the protein levels of the Synechocystis Prx, under standard growth conditions 1-Cys Prx and PrxII are highly abundant proteins, which are readily detectable on Coomassie-stained 2-dimensional electrophoresis gels (61) and are invariably found in the Synechocystis Trx target proteome (32, 34, 48). The 2-Cys Prx, PrxQ2, and PrxQ1 proteins have not been detected in proteomic studies (18, 61). As for Trx abundance, we have determined previously by quantitative Western blotting that the TrxA, TrxB, and TrxQ proteins constitute 0.25, 0.016, and 0.0046% of the total Synechocystis cellular protein, respectively (14), and in a different study, TrxA and TrxB were reported to constitute 1 and 0.25% of the total Synechocystis soluble protein, respectively (21). From experimental values of total cellular protein concentrations in Synechocystis cultures and volumes of cell pellets after centrifugation (data not shown), we estimate the intracellular protein concentration to be about 358 mg/ml. This would yield concentrations of 76, 4.7, and 1.4 μM for TrxA, TrxB, and TrxQ, respectively. Taking into account the kinetic parameters determined in this study, TrxA might be present at saturating concentrations for all Prx, whereas TrxB is saturating only for 1-Cys Prx and PrxQ1, and TrxQ is saturating only for PrxII.
We conclude that all Synechocystis Prx are expressed and are genuine Trx-dependent peroxidases and that GSH and Grx appear not to play roles in peroxide decomposition in this organism. Taken together, our in vitro analyses of protein-protein interactions and enzymatic activities show that the Synechocystis Prx in general are able to interact with and receive electrons from the different Trx from the same organism. Hence, it is likely that in vivo, there is a high degree of functional overlap between the Trx with respect to reduction of these Prx. Here we show that the Synechocystis TrxQ is at least fourfold as efficient as an electron donor for PrxII, PrxQ2, and 2-Cys Prx as the other Trx and that, in contrast, TrxQ is completely inefficient toward PrxQ1. However, the 50-fold-higher abundance of TrxA (76 μM) than of TrxQ (1.4 μM) indicates that TrxA could be of relative importance as an electron donor to Synechocystis Prx, despite the superior catalytic efficiency of TrxQ. Since PrxII is one of the most abundant and, in addition, one of the most active Synechocystis Prx, it seems plausible that PrxII accounts for most of the Prx activity in this organism in vivo. As a result of the attempt to establish connections between individual Trx and Prx, we found that TrxQ and PrxII constitute the most remarkable couple, which stands out for its catalytic efficiency as measured by apparent kcat/Km, pronounced affinity, and similar expression patterns. The second couple worth noting is TrxA and 1-Cys Prx: they coincide in the unusual expression pattern of reduced transcription under conditions of elevated ROS concentrations, and TrxA is also the most efficient electron donor for Synechocystis 1-Cys Prx.
Supplementary Material
Acknowledgments
This work was supported by grant BFU2007-603007/BMC) from Ministerio de Ciencia e Innovación (MICINN) and a grant from Junta de Andalucia (group BIO-284 and CVI-099).
A.M.-C. and A.M.S.-R. are recipients of fellowships from the Ministerio de Educación y Ciencia of Spain (M.E.C.).
Footnotes
Published ahead of print on 9 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alam, J., S. Curtis, F. K. Gleason, M. Gerami-Nejad, and J. A. Fuchs. 1989. Isolation, sequence, and expression in Escherichia coli of an unusual thioredoxin gene from the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 171:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allakhverdiev, S. I., Y. Nishiyama, S. Miyairi, H. Yamamoto, N. Inagaki, Y. Kanesaki, and N. Murata. 2002. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiol. 130:1443-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apel, K., and H. Hirt. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55:373-399. [DOI] [PubMed] [Google Scholar]
- 4.Baier, M., and K. J. Dietz. 1997. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 12:179-190. [DOI] [PubMed] [Google Scholar]
- 5.Bernroitner, M., M. Zamocky, P. G. Furtmuller, G. A. Peschek, and C. Obinger. 2009. Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J. Exp. Bot. 60:423-440. [DOI] [PubMed] [Google Scholar]
- 6.Broin, M., S. Cuine, F. Eymery, and P. Rey. 2002. The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 14:1417-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan, B. B., and Y. Balmer. 2005. Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56:187-220. [DOI] [PubMed] [Google Scholar]
- 8.Cha, M. K., S. K. Hong, and I. H. Kim. 2007. Four thiol peroxidases contain a conserved GCT catalytic motif and act as a versatile array of lipid peroxidases in Anabaena sp. PCC7120. Free Radic. Biol. Med. 42:1736-1748. [DOI] [PubMed] [Google Scholar]
- 9.Collin, V., E. Issakidis-Bourguet, C. Marchand, M. Hirasawa, J. M. Lancelin, D. B. Knaff, and M. Miginiac-Maslow. 2003. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J. Biol. Chem. 278:23747-23752. [DOI] [PubMed] [Google Scholar]
- 10.Collin, V., P. Lamkemeyer, M. Miginiac-Maslow, M. Hirasawa, D. B. Knaff, K.-J. Dietz, and E. Issakidis-Bourguet. 2004. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol. 136:4088-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietz, K. J. 2003. Plant peroxiredoxins. Annu. Rev. Plant Biol. 54:93-107. [DOI] [PubMed] [Google Scholar]
- 12.Dietz, K. J. 2008. Redox signal integration: from stimulus to networks and genes. Physiol. Plant. 133:459-468. [DOI] [PubMed] [Google Scholar]
- 13.Dietz, K. J., S. Jacob, M. L. Oelze, M. Laxa, V. Tognetti, S. M. de Miranda, M. Baier, and I. Finkemeier. 2006. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57:1697-1709. [DOI] [PubMed] [Google Scholar]
- 14.Florencio, F. J., M. E. Pérez-Pérez, L. Lopez-Maury, A. Mata-Cabana, and M. Lindahl. 2006. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 89:157-171. [DOI] [PubMed] [Google Scholar]
- 15.Foyer, C. H., and G. D. Noctor. 2009. Redox regulation in photosynthetic organisms: signaling, acclimation and practical implications. Antioxid. Redox Signal. 11:861-905. [DOI] [PubMed] [Google Scholar]
- 16.Gaber, A., M. Tamoi, T. Takeda, Y. Nakano, and S. Shigeoka. 2001. NADPH-dependent glutathione peroxidase-like proteins (Gpx-1, Gpx-2) reduce unsaturated fatty acid hydroperoxides in Synechocystis PCC 6803. FEBS Lett. 499:32-36. [DOI] [PubMed] [Google Scholar]
- 17.Gaber, A., K. Yoshimura, M. Tamoi, T. Takeda, Y. Nakano, and S. Shigeoka. 2004. Induction and functional analysis of two reduced nicotinamide adenine dinucleotide phosphate-dependent glutathione peroxidase-like proteins in Synechocystis PCC 6803 during the progression of oxidative stress. Plant Physiol. 136:2855-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan, C. S., K. F. Reardon, and P. C. Wright. 2005. Comparison of protein and peptide prefractionation methods for the shotgun proteomic analysis of Synechocystis sp. PCC 6803. Proteomics 5:2468-2478. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Dominguez, M., and F. J. Florencio. 1997. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 35:723-734. [DOI] [PubMed] [Google Scholar]
- 20.Gleason, F. K., and A. Holmgren. 1981. Isolation and characterization of thioredoxin from the cyanobacterium, Anabaena sp. J. Biol. Chem. 256:8306-8309. [PubMed] [Google Scholar]
- 21.Hishiya, S., W. Hatakeyama, Y. Mizota, N. Hosoya-Matsuda, K. Motohashi, M. Ikeuchi, and T. Hisabori. 2008. Binary reducing equivalent pathways using NADPH-thioredoxin reductase and ferredoxin-thioredoxin reductase in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 49:11-18. [DOI] [PubMed] [Google Scholar]
- 22.Horling, F., P. Lamkemeyer, J. Konig, I. Finkemeier, A. Kandlbinder, M. Baier, and K. J. Dietz. 2003. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosoya-Matsuda, N., K. Motohashi, H. Yoshimura, A. Nozaki, K. Inoue, M. Ohmori, and T. Hisabori. 2005. Anti-oxidative stress system in cyanobacteria. Significance of type II peroxiredoxin and the role of 1-Cys peroxiredoxin in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 280:840-846. [DOI] [PubMed] [Google Scholar]
- 24.Huang, L., M. P. McCluskey, H. Ni, and R. A. LaRossa. 2002. Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 184:6845-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanesaki, Y., H. Yamamoto, K. Paithoonrangsarid, M. Shoumskaya, I. Suzuki, H. Hayashi, and N. Murata. 2007. Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J. 49:313-324. [DOI] [PubMed] [Google Scholar]
- 26.Klughammer, B., M. Baier, and K. J. Dietz. 1998. Inactivation by gene disruption of 2-cysteine-peroxiredoxin in Synechocystis sp. PCC 6803 leads to increased stress sensitivity. Physiol. Plant. 104:699-706. [Google Scholar]
- 27.Kobayashi, M., T. Ishizuka, M. Katayama, M. Kanehisa, M. Bhattacharyya-Pakrasi, H. B. Pakrasi, and M. Ikeuchi. 2004. Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 45:290-299. [DOI] [PubMed] [Google Scholar]
- 28.Lamkemeyer, P., M. Laxa, V. Collin, W. Li, I. Finkemeier, M. A. Schottler, V. Holtkamp, V. B. Tognetti, E. Issakidis-Bourguet, A. Kandlbinder, E. Weis, M. Miginiac-Maslow, and K. J. Dietz. 2006. Peroxiredoxin Q of Arabidopsis thaliana is attached to the thylakoids and functions in context of photosynthesis. Plant J. 45:968-981. [DOI] [PubMed] [Google Scholar]
- 29.Latifi, A., M. Ruiz, R. Jeanjean, and C. C. Zhang. 2007. PrxQ-A, a member of the peroxiredoxin Q family, plays a major role in defense against oxidative stress in the cyanobacterium Anabaena sp. strain PCC7120. Free Radic. Biol. Med. 42:424-431. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., A. K. Singh, L. M. McIntyre, and L. A. Sherman. 2004. Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186:3331-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim, C. J., F. K. Gleason, and J. A. Fuchs. 1986. Cloning, expression, and characterization of the Anabaena thioredoxin gene in Escherichia coli. J. Bacteriol. 168:1258-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindahl, M., and F. J. Florencio. 2003. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. USA 100:16107-16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 34.Mata-Cabana, A., F. J. Florencio, and M. Lindahl. 2007. Membrane proteins from the cyanobacterium Synechocystis sp. PCC 6803 interacting with thioredoxin. Proteomics 7:3953-3963. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, Y., J. P. Reichheld, and F. Vignols. 2005. Thioredoxins in Arabidopsis and other plants. Photosynth. Res. 86:419-433. [DOI] [PubMed] [Google Scholar]
- 36.Michel, K. P., and E. K. Pistorius. 2004. Adaptation of the photosynthetic electron transport chain in cyanobacteria to iron deficiency: the function of IdiA and IsiA. Physiol. Plant. 120:36-50. [DOI] [PubMed] [Google Scholar]
- 37.Mittler, R., S. Vanderauwera, M. Gollery, and F. Van Breusegem. 2004. Reactive oxygen gene network of plants. Trends Plant Sci. 9:490-498. [DOI] [PubMed] [Google Scholar]
- 38.Miyake, C., F. Michihata, and K. Asada. 1991. Scavenging of hydrogen-peroxide in prokaryotic and eukaryotic algae—acquisition of ascorbate peroxidase during the evolution of cyanobacteria. Plant Cell Physiol. 32:33-43. [Google Scholar]
- 39.Mutsuda, M., T. Ishikawa, T. Takeda, and S. Shigeoka. 1996. The catalase-peroxidase of Synechococcus PCC 7942: purification, nucleotide sequence analysis and expression in Escherichia coli. Biochem. J. 316:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro, F., and F. J. Florencio. 1996. The cyanobacterial thioredoxin gene is required for both photoautotrophic and heterotrophic growth. Plant Physiol. 111:1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navarro, F., E. Martin-Figueroa, and F. J. Florencio. 2000. Electron transport controls transcription of the thioredoxin gene (trxA) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 43:23-32. [DOI] [PubMed] [Google Scholar]
- 42.Nishiyama, Y., H. Yamamoto, S. I. Allakhverdiev, M. Inaba, A. Yokota, and N. Murata. 2001. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 20:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obinger, C., G. Regelsberger, G. Strasser, U. Burner, and G. A. Peschek. 1997. Purification and characterization of a homodimeric catalase-peroxidase from the cyanobacterium Anacystis nidulans. Biochem. Biophys. Res. Commun. 235:545-552. [DOI] [PubMed] [Google Scholar]
- 44.Parsonage, D., P. A. Karplus, and L. B. Poole. 2008. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. USA 105:8209-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsonage, D., D. S. Youngblood, G. N. Sarma, Z. A. Wood, P. A. Karplus, and L. B. Poole. 2005. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44:10583-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perelman, A., A. Uzan, D. Hacohen, and R. Schwarz. 2003. Oxidative stress in Synechococcus sp. strain PCC 7942: various mechanisms for H2O2 detoxification with different physiological roles. J. Bacteriol. 185:3654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Pérez, M., E. Martin-Figueroa, and F. J. Florencio. 2009. Photosynthetic regulation of the cyanobacterium Synechocystis sp. PCC 6803 thioredoxin system and functional analysis of TrxB (Trx x) and TrxQ (Trx y) thioredoxins. Mol. Plant 2:270-283. [DOI] [PubMed] [Google Scholar]
- 48.Pérez-Pérez, M. E., F. J. Florencio, and M. Lindahl. 2006. Selecting thioredoxins for disulphide proteomics: target proteomes of three thioredoxins from the cyanobacterium Synechocystis sp. PCC 6803. Proteomics 6(Suppl. 1):S186-S195. [DOI] [PubMed] [Google Scholar]
- 49.Pérez-Ruiz, J. M., M. C. Spinola, K. Kirchsteiger, J. Moreno, M. Sahrawy, and F. J. Cejudo. 2006. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18:2356-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersson, U. A., T. Kieselbach, J. G. Garcia-Cerdan, and W. P. Schroder. 2006. The Prx Q protein of Arabidopsis thaliana is a member of the luminal chloroplast proteome. FEBS Lett. 580:6055-6061. [DOI] [PubMed] [Google Scholar]
- 51.Poole, L. B. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433:240-254. [DOI] [PubMed] [Google Scholar]
- 52.Reyes, J. C., and F. J. Florencio. 1994. A new type of glutamine synthetase in cyanobacteria: the protein encoded by the glnN gene supports nitrogen assimilation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 176:1260-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhee, S. G., H. Z. Chae, and K. Kim. 2005. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 38:1543-1552. [DOI] [PubMed] [Google Scholar]
- 54.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignment, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 55.Rouhier, N., E. Gelhaye, J. M. Gualberto, M. N. Jordy, E. De Fay, M. Hirasawa, S. Duplessis, S. D. Lemaire, P. Frey, F. Martin, W. Manieri, D. B. Knaff, and J. P. Jacquot. 2004. Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol. 134:1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouhier, N., E. Gelhaye, and J. P. Jacquot. 2002. Glutaredoxin-dependent peroxiredoxin from poplar: protein-protein interaction and catalytic mechanism. J. Biol. Chem. 277:13609-13614. [DOI] [PubMed] [Google Scholar]
- 57.Rouhier, N., E. Gelhaye, P. E. Sautiere, A. Brun, P. Laurent, D. Tagu, J. Gerard, E. de Fay, Y. Meyer, and J. P. Jacquot. 2001. Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol. 127:1299-1309. [PMC free article] [PubMed] [Google Scholar]
- 58.Rouhier, N., and J. P. Jacquot. 2005. The plant multigenic family of thiol peroxidases. Free Radic. Biol. Med. 38:1413-1421. [DOI] [PubMed] [Google Scholar]
- 59.Schürmann, P., and B. B. Buchanan. 2008. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10:1235-1274. [DOI] [PubMed] [Google Scholar]
- 60.Schwarz, R., and K. Forchhammer. 2005. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151:2503-2514. [DOI] [PubMed] [Google Scholar]
- 61.Simon, W. J., J. J. Hall, I. Suzuki, N. Murata, and A. R. Slabas. 2002. Proteomic study of the soluble proteins from the unicellular cyanobacterium Synechocystis sp. PCC6803 using automated matrix-assisted laser desorption/ionization-time of flight peptide mass fingerprinting. Proteomics 2:1735-1742. [DOI] [PubMed] [Google Scholar]
- 62.Singh, A. K., and L. A. Sherman. 2007. Reflections on the function of IsiA, a cyanobacterial stress-inducible, Chl-binding protein. Photosynth. Res. 93:17-25. [DOI] [PubMed] [Google Scholar]
- 63.Stork, T., M. Laxa, M. S. Dietz, and K. J. Dietz. 2009. Functional characterisation of the peroxiredoxin gene family members of Synechococcus elongatus PCC 7942. Arch. Microbiol. 191:141-151. [DOI] [PubMed] [Google Scholar]
- 64.Stork, T., K. P. Michel, E. K. Pistorius, and K. J. Dietz. 2005. Bioinformatic analysis of the genomes of the cyanobacteria Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 for the presence of peroxiredoxins and their transcript regulation under stress. J. Exp. Bot. 56:3193-3206. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki, I., W. J. Simon, and A. R. Slabas. 2006. The heat shock response of Synechocystis sp. PCC 6803 analysed by transcriptomics and proteomics. J. Exp. Bot. 57:1573-1578. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi, S., and N. Murata. 2008. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 13:178-182. [DOI] [PubMed] [Google Scholar]
- 67.Tel-Or, E., M. Huflejt, and L. Packer. 1985. The role of glutathione and ascorbate in hydroperoxide removal in cyanobacteria. Biochem. Biophys. Res. Commun. 132:533-539. [DOI] [PubMed] [Google Scholar]
- 68.Tel-Or, E., M. E. Huflejt, and L. Packer. 1986. Hydroperoxide metabolism in cyanobacteria. Arch. Biochem. Biophys. 246:396-402. [DOI] [PubMed] [Google Scholar]
- 69.Tichy, M., and W. Vermaas. 1999. In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 181:1875-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolff, S. P. 1994. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233:182-189. [Google Scholar]
- 72.Wood, Z. A., E. Schroder, J. Robin Harris, and L. B. Poole. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32-40. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto, H., C. Miyake, K. J. Dietz, K. Tomizawa, N. Murata, and A. Yokota. 1999. Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 447:269-273. [DOI] [PubMed] [Google Scholar]
- 74.Yano, H., S. Kuroda, and B. B. Buchanan. 2002. Disulfide proteome in the analysis of protein function and structure. Proteomics 2:1090-1096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.