Abstract
The Bacillus anthracis spore is the causative agent of the disease anthrax. The outermost structure of the B. anthracis spore, the exosporium, is a shell composed of approximately 20 proteins. The function of the exosporium remains poorly understood and is an area of active investigation. In this study, we analyzed the previously identified but uncharacterized exosporium protein ExsK. We found that, in contrast to other exosporium proteins, ExsK is present in at least two distinct locations, i.e., the spore surface as well as a more interior location underneath the exosporium. In spores that lack the exosporium basal layer protein ExsFA/BxpB, ExsK fails to encircle the spore and instead is present at only one spore pole, indicating that ExsK assembly to the spore is partially dependent on ExsFA/BxpB. In spores lacking the exosporium surface protein BclA, ExsK fails to mature into high-molecular-mass species observed in wild-type spores. These data suggest that the assembly and maturation of ExsK within the exosporium are dependent on ExsFA/BxpB and BclA. We also found that ExsK is not required for virulence in murine and guinea pig models but that it does inhibit germination. Based on these data, we propose a revised model of exosporium maturation and assembly and suggest a novel role for the exosporium in germination.
During starvation, bacteria of the genus Bacillus differentiate into dormant, highly robust cell types called spores, thereby preserving their genomes during stressful and nutrient-poor conditions (10). Spores can withstand extremely harsh environmental insults, including toxic chemicals, UV radiation, and heat (31). When conditions again become favorable for cell survival, spores can return to vegetative cell growth through a process called germination (17, 18, 31, 49). Spores are formed in an approximately 8-h process during which the developing spore first forms as a compartment (the forespore) contained within the surrounding cell (the mother cell) (34). Ultimately, the mother cell envelope lyses, releasing the mature spore into the environment.
Spores from all Bacillus species have similar architectures. At the spore interior is the core, which houses the spore chromosome. Surrounding the core is an inner membrane encased in a specialized peptidoglycan called the cortex and finally a series of outer layers that vary significantly among species (10). In some species, including Bacillus subtilis, the outermost structure is a protective layer called the coat, which guards the spore against reactive small molecules, degradative enzymes, and predation by other microbes (11, 17, 20, 38). Spores of other species, including the pathogens Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis and the nonpathogenic bacteria Bacillus megaterium and Bacillus odysseyi, have an additional structure called the exosporium, which surrounds the coat (24, 32, 47). The exosporium is composed of two structural units: the basal layer, which is a shell of proteins forming a hexagonal array, and a nap of hairlike protrusions extending outward from the basal layer (2, 32). A major component of the nap (and of the spore surface) is the collagen-like protein BclA (40, 43). The proteins that comprise the outer structures (the coat and exosporium) are synthesized in the mother cell cytoplasm, from which location they assemble onto the spore surface to form their respective structures (11).
The function of the exosporium is poorly understood. Previous studies have implicated its contribution to germination, resistance to host cells and other stresses, adhesion to inert surfaces, and interactions with epithelial cells and macrophages (1, 6, 7, 13, 33, 41, 48; G. Chen, A. Driks, K. Tawfiq, M. Mallozzi, and S. Patil, submitted for publication). In most cases, however, the roles of individual exosporium proteins in each of these functions remain unclear, in part because the location of each protein within the exosporium is largely unknown.
Interestingly, it appears that the exosporium is not essential for virulence of B. anthracis in several animal models (5, 7, 12, 13). Nonetheless, it is possible that in natural infections the exosporium plays a significant role. Because it is involved in attachment, the exosporium is also likely to have a significant impact on the persistence of B. anthracis spores in the environment.
To gain insight into the molecular basis of exosporium assembly and function, we studied a previously identified but otherwise uncharacterized exosporium protein, ExsK. Using immunofluorescence microscopy (IFM), we found that ExsK is asymmetrically distributed on the surfaces of mature spores and is also present beneath the exosporium. In the absence of ExsFA/BxpB, ExsK was restricted to one spore pole, suggesting that the encirclement of the spore by ExsK depends on ExsFA/BxpB. Western blot analysis indicated that in mature spores ExsK is present in high-molecular-mass complexes, the formation of which is BclA dependent. Although ExsK is not required for several spore resistance properties or virulence, we found that it is required for normal germination. Our results provide a deeper understanding of the composition, function, and assembly of the B. anthracis exosporium and show that proteins comprising outer-spore structures can have multiple locations.
MATERIALS AND METHODS
General methods.
Escherichia coli strains were grown in Luria-Bertani (LB) broth, and vegetative cultures of the Sterne strain and its derivatives (Table 1) were grown either on LB agar or in brain heart infusion broth (Becton Dickinson) plus 0.5% glycerol to suppress sporulation. When appropriate, cultures were supplemented with the following antibiotics: ampicillin at 50 to 100 μg/ml (E. coli), kanamycin at 100 μg/ml (B. anthracis) or 30 to 40 μg/ml (E. coli), and erythromycin at 3 μg/ml (B. anthracis) or 300 μg/ml (E. coli).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| B. anthracis Sterne and derivatives | ||
| Sterne | Parent; 34F2; pXO1+ pXO2− | P. Jackson |
| RG1 | cotEΔ::kan | 12 |
| RG124 | exsFA::kan | 13 |
| JAB13 | bclA::kan | 13 |
| KMS1 | exsKΩpKMS1 | This study |
| KMS2 | exsKΔ::kan | This study |
| ADL2415 | tgl::kan | 30a |
| B. anthracis Ames and derivative | ||
| Ames | Parent; fully virulent; pXO1+ pXO2+ | 26 |
| Ames exsK | exsKΔ::kan | This study |
| E. coli | ||
| DH10β | Cloning host | Laboratory collection |
| DH5α | Cloning host | Laboratory collection |
| TG1 | Cloning host | 39 |
| GM1684 | dam; B. anthracis transformation | T. Koehler |
| GM2163 | dam dcm; B. anthracis transformation | New England Biolabs |
| BL21(DE3)/pLysS | Expression host | Novagen |
| Plasmids | ||
| pKS1 | Temperature-sensitive vector in B. anthracis | 39 |
| pMGM6 | pKS1-GFP | This study |
| pKMS1 | exsK-gfp fusion vector | This study |
| pKMS2 | exsK deletion vector | This study |
| pGEM-T | Cloning vector | Promega |
| pGEM-T Easy | Cloning vector | Promega |
| pET24b | Overexpression vector | Novagen |
Spores were generated by exhaustion as previously described (15). Unless otherwise indicated, spores were purified by Renografin gradient centrifugation (Bracco Diagnostics) (16), with the following modifications. Spores from a 25-ml sporulation culture were washed three times with 25 ml of cold water and resuspended in 400 μl for strains Sterne (wild type), KMS2 (exsKΔ::kan), RG124 (exsFA bxpB::kan), and ADL2415 (tgl::kan) or in 1 ml for strains RG1 (cotEΔ::kan) and JAB13 (bclA::kan) of 20% Renografin. The resuspension (400 μl) was overlaid on 50% Renografin (800 μl) and centrifuged at 14,000 × g for 10 min in a Beckman tabletop microcentrifuge. Gradient-purified spores were washed again three times with 1 ml water and stored at 4°C.
Plasmid DNA was isolated using a Wizard DNA purification system (Promega), and genomic DNA was isolated as described by Harwood and Cutting (15).
Strain construction.
As indicated below, B. anthracis Sterne (harboring pXO1 but lacking pXO2) and B. anthracis Ames (harboring both pXO1 and pXO2) were used as parent strains for the construction of the following strains. To generate strain KMS2 (exsKΔ::kan in the Sterne background), we first amplified genomic DNA (∼500 bp) upstream and downstream of exsK by PCR using the following primers (restriction sites are underlined and indicated in parentheses at the end of each sequence): BA2554 Fwd Up SacII, CCGCGGAAGTATTTGCCTCAACAGGCGAGC (SacII); BA2554 Rev Up PstI, CTGCAGTACACCAATCGCGGCAGCTTCTAT (PstI); BA2554 Fwd Downstream, AAGCTTTGGTGATACGAGAATGAGAGGACG (HindIII); and BA2554 Rev Downstream, CTCGAGTGGTGTGAGTACCCATTCATCA (XhoI). Each PCR product was cloned into pGEM-T (Promega), and each construct was confirmed by DNA sequence analysis. We then liberated fragments from each plasmid by digestion with SacII and PstI or HindIII and XhoI. We used standard recombinant DNA manipulations to place each insert into pKS1 (39) at SacII/PstI and HindIII/XhoI sites, thereby generating pKMS2. Those operations were performed with E. coli strain TG1 at 37°C, the nonpermissive temperature for the pKS1 replicon (39). pKMS2 was isolated from TG1 and passaged through E. coli strain GM1684 to demethylate the plasmid and then used to transform the Sterne strain of B. anthracis by electroporation (39). To achieve the required integration of pKMS2 into the chromosome by marker replacement, we first determined whether the plasmid had integrated into the chromosome by a single reciprocal (Campbell-type) recombination, by testing whether candidate recombinant cells had the expected Kanr Ermr antibiotic resistance phenotype at the nonpermissive temperature (37°C). After continued growth at 37°C for 6 h in the presence of antibiotics, we then screened for cells in which the integrated DNA had undergone a reverse-reciprocal excision (thereby creating a marker replacement event), as evidenced by Kanr Erms cells after growth at 37°C. Deletions were confirmed by PCR analysis.
An exsK mutant (exsKΔ::kan) strain in the Ames background was generated by generalized phage transduction, using CP-51ts grown on Sterne strain KMS2 (exsKΔ::kan) to infect wild-type Ames (13). Transductants were confirmed by PCR analysis.
To build strain KMS1 (exsKΩpKMS1, in the Sterne background), we first used PCR to generate a DNA fragment containing the genomic sequence beginning 600 bp upstream of exsK (in the BA2553 gene) and ending at the end of exsK, using the primers TTAAATGGGGTTCACGGTGACTGGGT and AGATATCTGTTAACAATGCTTCAAT (where the underlined region corresponds to an EcoRV site added to the primer sequence). The PCR product was cloned into pGEMT-Easy (Promega) and analyzed by DNA sequence analysis. A DNA fragment was excised by digestion with NotI and EcoRV. Next, pMGM6 was generated by ligation of a DNA fragment bearing the green fluorescent protein (GFP) open reading frame (produced by digestion of pAS5 [28] with BamHI and HindIII) with BamHI/HindIII-digested pKS1. We then ligated the exsK-bearing DNA fragment with NotI- and PmeI-digested plasmid pMGM6 to generate plasmid pKMS1. This manipulation was performed with TG1 cells at 37°C, since pMGM6 is a derivative of pKS1. The resulting plasmid was passaged through E. coli strain GM2163 (New England Biolabs) and used to transform B. anthracis Sterne (39). Integration of pKMS1 and disruption of exsK were confirmed by PCR analysis.
Generation of anti-ExsK antibodies.
Recombinant ExsK was generated and purified using the pET expression system (Novagen) according to the manufacturer's suggestions. The exsK open reading frame was PCR amplified from genomic DNA using primers OS-2554-SacI, GAGCTCCGGATCTCGTTATAGTAAT (SacI), and OA-2554-XhoI, CTCGAGTGTTAACAATGCTTCAAT (XhoI), and cloned into pET24b such that it was in-frame with T7 and six-His tag sequences. Protein synthesis was induced in BL21(DE3)/pLysS E. coli (Novagen) with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C, and ExsK was purified from the insoluble fraction by use of Ni-nitrilotriacetic acid His-Bind resin as described by the manufacturer (Novagen). Following elution from the column, 1 M dithiothreitol was added to a final concentration of 6 mM and the protein was dialyzed for stepwise removal of guanidine HCl (from 5 M to 1 M), followed by stepwise removal of guanidine HCl and dithiothreitol. To prevent protein precipitation, 0.4 M l-arginine was added to 1× phosphate-buffered saline (PBS) for the final dialysis step. Purified ExsK was stored at −80°C until use. ExsK purity was determined by use of a Western blot probed with T7 tag-horseradish peroxidase (HRP) (Novagen) and Coomassie blue-stained polyacrylamide gels; concentration was determined by the Bradford protein assay (Sigma).
Mice (4 to 12 weeks of age) were immunized with 10 to 50 μg of ExsK mixed 1:1 with TiterMax Gold adjuvant (Sigma) and boosted at days 12 and 20 with 25 to 50 μg of ExsK/TiterMax. Antigen-specific antibody titers were monitored by use of an enzyme-linked immunosorbent assay, using recombinant ExsK-coated plates and donkey anti-mouse immunoglobulin G heavy plus light chains [IgG(H+L)]-HRP (Jackson Immunoresearch) as the secondary antibody.
Phase-contrast microscopy and IFM.
Sporulating cells were harvested at various time points during sporulation, stained with Hoechst 33352 (10 μg/ml) to visualize DNA, and viewed on a Leica DM IRB fluorescence microscope equipped with a MagnaFire charge-coupled-device camera. Entry into sporulation (t0) was defined as the exit from exponential growth (15).
IFM was performed as described previously (13, 28). The primary antibodies used for staining were mouse anti-ExsK serum (used at a dilution of 1:200) and biotinylated mouse anti-BclA antibody (5 μg/ml) (BA-MAB 5; Critical Reagents Program, Department of Defense). We detected primary antibody staining using Alexa-Fluor 568-goat anti-mouse IgG (Invitrogen), Cy2-goat Fab anti-mouse IgG (Jackson Immunoresearch), or streptavidin-Cy3 (Jackson Immunoresearch), each used at a dilution of 1:200. Image processing was performed using Microsoft Powerpoint picture tools.
Flow cytometry.
Spores were stained with mouse anti-ExsK serum (1:200) followed by Dylight 649-goat Fab anti-mouse IgG (Jackson Immunoresearch) (1:200) or biotinylated mouse anti-BclA antibody (BA-MAB 5; Critical Reagents Program, Department of Defense) (5 μg/ml) followed by streptavidin-phycoerythrin (Invitrogen) (1:200). Samples were analyzed using FACSCanto or FACSCantoII (Becton Dickinson) at the Loyola University Medical Center FACS Core Facility. Data were analyzed using FlowJo analysis software (Tree Star, Inc.).
Germination.
Germination of Ames and exsK mutant B. anthracis spores was assessed by fluorescent-dye uptake, in which germination is measured by the accessibility of spore DNA to a dye following core hydration during early to intermediate stages of germination (37, 50). Softmax Pro 5.2 LS software was used to record fluorescence (485/530, excitation/emission) readings in relative fluorescence units (RFU) every minute for 60 min. The mean RFU and standard deviation values of triplicate germinations from a single sporulation were determined using Excel (Microsoft). GraphPad Prism v.5 software was used to analyze the data by nonlinear regression analysis, and the statistical significance of differences between samples was determined by Kruskal-Wallis nonparametric analysis of variance with Dunn's multiple-comparison posttests. The four germinants used were (i) medium containing l-alanine, adenosine, and Casamino acids (AAC), prepared as described previously (50); (ii) 10 mM Na phosphate buffer, 0.25 mM l-alanine, 1 mM inosine, and 0.1 M NaCl, pH 7.2 (AI); (iii) 0.5 mM l-alanine and 1 mM serine (AS); and (iv) 50 mM l-alanine prepared in the same phosphate buffer as the AI germinant.
Germination of Sterne wild-type and exsK mutant (KMS2) spores was assessed by monitoring a loss of optical density (OD) as previously described (13). This assay measures early and intermediate stages of germination (37). The OD at 550 nm (OD550) of three independent sporulations was measured every minute for 30 min after the addition of two germinants, (i) AI and (ii) 1 mM inosine and 1 mM l-serine in PBS, to spores resuspended in PBS at an OD550 of 1.0.
Western blots.
Spore proteins were extracted as previously described (4), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide) (36), and transferred to nitrocellulose membranes (0.2 μm pore size; Bio-Rad). The nitrocellulose blots were probed with mouse anti-ExsK serum (1:2,000) followed by donkey anti-mouse IgG(H+L)-HRP (Jackson Immunoresearch) (1:2,500) and an ECL substrate (Thermo).
Murine challenge experiments.
Female BALB/c mice (approximately 6 to 8 weeks of age) were obtained from the National Cancer Institute, Fort Detrick (Frederick, MD). For intraperitoneal (i.p.) experiments, the spore challenges (either wild-type Ames or exsK mutant strains) were administered i.p. as 200 μl of heat-activated spores in sterile water at a dose of approximately 1,500 CFU. For intranasal (i.n.) experiments, mice were anesthetized with 100 μl of ketamine, acepromazine, and xylazine injected intramuscularly (i.m.) and then challenged with approximately 1.3 × 106 spores of either the wild-type or the exsK mutant strain. The total volume of inoculum instilled was 35 μl. Challenged mice were monitored several times each day, and mortality rates were recorded for 14 days.
Guinea pig i.m. challenge experiments.
Female Hartley guinea pigs (350 to 400 g) were obtained from Charles River Laboratories (Wilmington, MA). Competitive challenges were performed with guinea pigs injected i.m., as described previously (5, 9), with spores of approximately equal numbers of both the wild-type Ames strain and the exsK mutant (approximately 1,000 spores total). Spleens were removed from moribund animals after euthanasia and homogenized in sterile PBS with Dounce homogenizers. The bacterial loads of the homogenized spleens were titrated on LB agar plates to determine the total numbers of bacteria and on LB agar plates containing kanamycin, on which only the exsK mutant cells can grow. The relative recovery of the wild-type Ames and exsK mutant cells was determined by dividing the percentage of the corresponding strain recovered from the spleen of each animal after infection by the percentage present in the challenge dose.
RESULTS
Timing of ExsK synthesis and deposition to the developing spore.
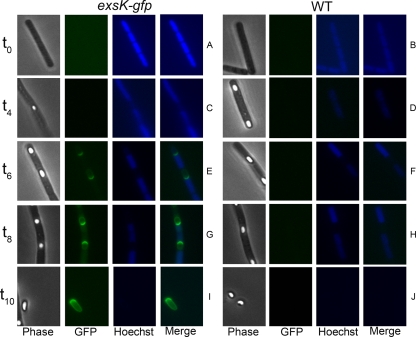
ExsK was previously identified as an exosporium protein by proteomic analyses (27, 35, 46). To analyze its assembly and role in exosporium function, we first studied the kinetics of ExsK synthesis and localization to the spore. We generated a B. anthracis strain, in the Sterne background, bearing exsK-gfp at the exsK locus (strain KMS1 [exsKΩpKMS1]) to facilitate monitoring the appearance and localization of ExsK-GFP during sporulation. By epifluorescence microscopy, we first detected GFP fluorescence 6 h after the onset of sporulation (t6) (Fig. 1E). This timing is approximately 3 h after the developing spore forms a protoplast within the sporulating cell and approximately 2 h after spores become heat resistant (based on the appearance of refractility by phase-contrast microscopy). At t6, GFP fluorescence appeared as a caplike structure at the pole of the spore adjacent to the mother cell (Fig. 1E). At t8, GFP fluorescence at the mother-cell-proximal pole increased, and GFP fluorescence was also distributed around the entire spore (Fig. 1G). GFP fluorescence remained asymmetrically distributed around the spore for the remainder of sporulation and in spores released from the mother cell (Fig. 1I). As a negative control, no GFP fluorescence was observed in wild-type spores (Fig. 1B, D, F, H, and J). These data suggest that ExsK assembles preferentially at the mother cell pole and, secondarily, elsewhere around the spore. The synthesis of ExsK-GFP only late in sporulation is consistent with the possibility that exsK is transcribed under the control of the late-acting sporulation transcription factor σK (23, 27). Nonuniform assembly of exosporium proteins has been seen previously in the cases of BclA and Alr (13, 42).
FIG. 1.
Fluorescence microscopy of B. anthracis (Sterne) sporangia. Phase-contrast (Phase) and fluorescence (GFP, Hoechst, and Merge) images of B. anthracis exsK-gfp (A, C, E, G, and I) or wild-type (WT) (B, D, F, H, and J) sporangia harvested at the indicated time points (in hours) during sporulation (t0 to t10) and visualized for GFP fluorescence and DNA staining with Hoechst 33352.
ExsK is located on the exosporium surface.
The relatively late synthesis of ExsK-GFP is consistent with its location on the spore surface. To test this directly, we stained spores with polyclonal anti-ExsK antiserum and visualized staining by IFM and flow cytometry. Anti-ExsK antibodies bound to the surfaces of wild-type spores (Fig. 2A). This binding is specific because no binding to exsK mutant spores was observed (Fig. 2B and K). The pattern of fluorescence obtained by staining spores with anti-ExsK antibodies was similar to that observed with strains bearing the ExsK-GFP fusion, indicating that GFP did not detectably interfere with ExsK assembly (Fig. 1). Surprisingly, when we stained bclA mutant spores (from strain JAB13 [bclA::kan]), which lack the well-characterized spore surface protein BclA (40, 43), we found more-intense fluorescence staining than with wild-type spores (Fig. 2A, C, and K). These data indicate that in the absence of BclA, ExsK is more accessible to antibody binding, suggesting that although ExsK is present at the spore surface, it is partially occluded by BclA and is likely present on the outside of the exosporium basal layer.
FIG. 2.
Immunofluorescence of B. anthracis (Sterne) spores stained with anti-ExsK. (A to J) Phase-contrast (top) and fluorescence (bottom) images of wild-type (WT) (A and F), exsK (B and G), bclA (C and H), exsFA bxpB (D and I), and cotE (E and J) spores stained with anti-ExsK (A to E) or preimmune (F to J) sera. (K) Flow cytometric plots of WT, exsK, bclA, exsFA bxpB, and cotE spores. Top, FSC versus SSC dot plots; bottom, staining with anti-ExsK (white histograms) or preimmune (shaded histograms) sera, gated on the indicated FSC versus SSC population. Max, maximum.
Because ExsK is present on the outside of the exosporium basal layer, we investigated whether the previously characterized basal layer protein ExsFA/BxpB (41, 44), which affects the localization of BclA in spores (13), also plays a role in the deposition or localization of ExsK. We stained exsFA (bxpB) mutant spores in the Sterne background (from strain RG124 [exsFA::kan]) with anti-ExsK antibodies and analyzed binding by IFM and flow cytometry. We readily detected fluorescence on the surfaces of exsFA (bxpB) mutant spores, but the fluorescence was detected largely at only one pole of the spore (Fig. 2D and K). This fluorescence pattern is reminiscent of the pattern of BclA localization in exsFA (bxpB) mutant spores (13). We interpret these data as evidence that ExsFA (BxpB) partially controls ExsK assembly. Therefore, ExsFA (BxpB) partially controls the assembly of at least two proteins, BclA and ExsK, and, as measured by IFM, affects the localization patterns of both proteins in similar manners.
Multiple locations of ExsK in spores.
To determine whether ExsK is present in locations of the spore other than on the exosporium, we took advantage of previous work showing that the exosporium is absent from most spores bearing a mutation in the coat protein gene cotE (12). We analyzed wild-type and cotE mutant spores (from strain RG1 [cotEΔ::kan]) by IFM and flow cytometry. We found that anti-ExsK antibodies bound to nearly all cotE spores, (Fig. 2E and K), suggesting that ExsK is also present on the surfaces of spores lacking the exosporium. To investigate this observation further, we performed two-color flow cytometry, using anti-ExsK and anti-BclA (as a marker of the exosporium) antibodies (40, 43). We observed that approximately 60% of cotE mutant spores lacked the exosporium, as evidenced by the absence of anti-BclA antibody binding (Fig. 3). Interestingly, we detected ExsK staining in both the BclA-negative and the BclA-positive population of cotE mutant spores (Fig. 3). Taken together, these data suggest that, in addition to its location on the exosporium surface, ExsK is located within the spore, beneath the exosporium.
FIG. 3.
Two-color flow cytometric analysis of B. anthracis (Sterne) spores stained with anti-ExsK and anti-BclA antibodies. Wild-type (WT), exsK, bclA, and cotE spores were stained with anti-ExsK serum (allophycocyanin [APC]) and anti-BclA (phycoerythrin [PE]) antibodies. Top, FSC versus SSC dot plots. Middle, isotype control (mouse IgG1 [MsIgG1] and preimmune serum) dot plots. Bottom, anti-ExsK and anti-BclA antibody dot plots. Gates in each FSC versus SSC plot indicate the population used for isotype and antibody stain analyses.
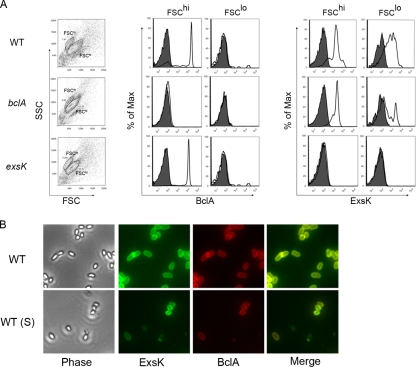
To determine whether the presence of ExsK underneath the exosporium was an artifact due to the absence of CotE, we performed two-color flow cytometry on wild-type spores after the exosporium was mechanically removed using sonication (35). Following sonication, we observed two populations of spores that differed in their forward and side light scatters (FSC and SSC, respectively), which differ depending on size and morphological characteristics of cells, respectively, and also in the staining patterns observed with anti-BclA antibodies (Fig. 4A). The wild-type spores with a lower forward scatter and higher side scatter (FSClo) did not stain with anti-BclA antibodies, indicating that they lack exosporia, whereas spores with a higher forward scatter and a lower side scatter (FSChi) did stain positive for BclA, indicating that they retained exosporia. We found that a large proportion of the BclA-positive as well as the BclA-negative sonicated wild-type spores stained positive with anti-ExsK serum (Fig. 4A). We confirmed this finding by staining wild-type sonicated B. anthracis spores simultaneously with anti-ExsK and anti-BclA antibodies and analyzing them by epifluorescence microscopy. Consistent with the flow cytometric analysis, we found that BclA-positive and -negative spores stained positive with anti-ExsK antiserum (Fig. 4B). Taken together, these data indicate that ExsK is present on the surfaces of spores bearing, as well as lacking, an exosporium and suggest that ExsK is present in multiple spore compartments.
FIG. 4.
Flow cytometric and IFM analyses of sonicated B. anthracis (Sterne) spores stained with anti-ExsK and anti-BclA antibodies. (A) Sonicated wild-type (WT) (top row), bclA (middle row), and exsK (bottom row) spores were analyzed based on FSC versus SSC plots (first column) and staining with anti-BclA antibody and mouse IgG1 (MsIgG1) isotype control (middle columns) or anti-ExsK or preimmune (right columns) sera. Gates in FSC versus SSC plots (FSClo and FSChi) indicate the populations used for BclA and ExsK analyses. Shaded histograms, MsIgG1 and preimmune serum staining; white histograms, anti-BclA antibody or anti-ExsK serum staining. Max, maximum. (B) Phase-contrast (Phase) and fluorescence (ExsK, BclA, and Merge) images of unsonicated WT (top row) and sonicated (S) WT (bottom row) spores stained with anti-ExsK serum (Cy2) and anti-BclA antibody (Cy3).
ExsK forms high-molecular-mass protein complexes.
Exosporium and coat proteins have the capacity to be cross-linked into high-molecular-mass species (19, 21, 22, 30, 40, 43, 44, 52, 53). We used SDS-PAGE and Western blot analysis to analyze the electrophoretic patterns of ExsK-containing species in B. anthracis (Sterne) spores. In wild-type spores, we detected the apparent monomeric form of ExsK, at 12 kDa (35), as well as several higher-molecular-mass species, ranging up to 250 kDa (Fig. 5). In both cotE and bclA mutant spores, we detected bands migrating as 12-kDa and 25-kDa species, but each of these bands was less intense than in wild-type spores, and spores from these two mutants lacked higher-molecular-mass complexes. Because bclA and cotE mutant spores both lack BclA, the simplest interpretation of our data is that the ExsK bands absent (or reduced in intensity) in either the bclA or the cotE mutant spores depend on BclA for their presence in wild-type spores and that ExsK assembly may be at least partially dependent on BclA. These data suggest that ExsK is present in the spore in covalently cross-linked complexes whose abundance depends on BclA.
FIG. 5.
Western blots of B. anthracis (Sterne) spores probed with anti-ExsK. Lysates (coat preparations) of wild-type (WT), cotE, exsK, bclA, exsFA (bxpB), and tgl mutant spores were probed with anti-ExsK antiserum. Sizes in kilodaltons are given on the left.
exsFA (bxpB) mutant spores assemble less BclA than wild-type spores and at only one pole of the spore (13, 41). Because BclA is abnormally localized in exsFA (bxpB) mutant spores, we used spores from this mutant to ask whether the formation of ExsK complexes depends on properly localized BclA. Using SDS-PAGE and Western blot analysis, we found that exsFA (bxpB) mutant spores possessed an essentially normal electrophoretic pattern of ExsK complexes (Fig. 5). Therefore, proper deposition of ExsK does not necessarily require that BclA be at wild-type levels or in its normal location.
We do not know the biochemical event responsible for ExsK complex formation. It is unlikely, however, to be due solely to disulfide cross-linkages or the activity of the sporulation-specific transglutaminase gene, because neither electrophoresis under reducing conditions nor mutation of the B. anthracis tgl gene disturbed the electrophoretic pattern of ExsK high-molecular-mass species (Fig. 5).
Roles of ExsK in spore functions and structure.
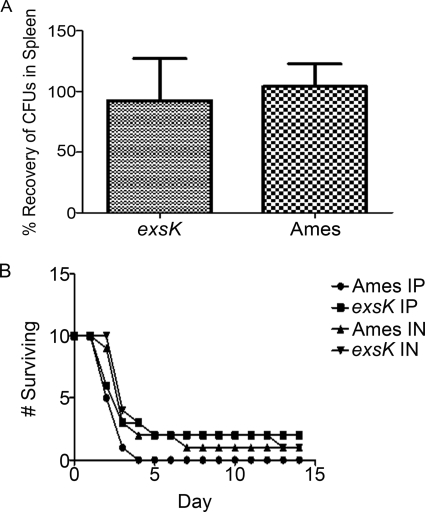
The B. anthracis exosporium is not essential for virulence in at least some animal models (12). To address the role of ExsK in disease, we inoculated guinea pigs i.m. and mice i.n. or i.p. with wild-type or exsK mutant spores in the virulent Ames background (5, 9). We observed no difference in the numbers of wild-type or exsK mutant cells recovered from guinea pig spleens in a competitive challenge assay (Fig. 6A) or in the times of death of mice inoculated i.p. or i.n. with either wild-type or exsK mutant spores (Fig. 6B). These data indicate that ExsK is not required for virulence in these animal models. We also did not observe significant structural differences between wild-type and exsK mutant spores in the Sterne background by thin-section electron microscopy (data not shown).
FIG. 6.
In vivo challenge assays using B. anthracis Ames spores and exsK mutant Ames spores. (A) Relative percentages of wild-type (Ames) and exsK B. anthracis Ames CFU recovered from the spleens of guinea pigs 2 days after challenge with wild-type and exsK spores (103 total). Error bars indicate standard deviations. (B) Survival rates of mice challenged i.p. or i.n. with spores of wild-type or exsK Ames, plotted as the numbers of animals surviving over time postinoculation.
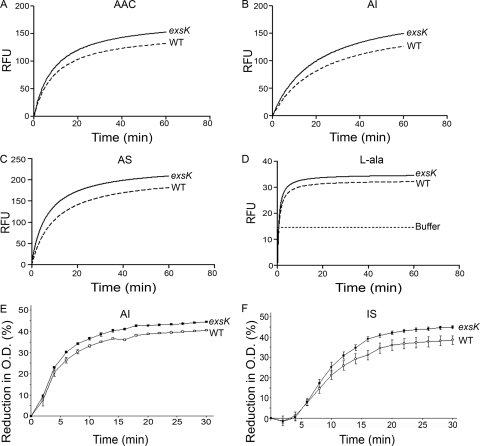
Outer-spore structures can have critical roles in germination (10, 12, 13, 17, 42). To determine whether ExsK has a role in germination, we used a fluorescent-dye uptake assay to monitor the entry of water into the spore core, which takes place during early and intermediate stages of germination (50). To stimulate germination in wild-type and exsK mutant spores in the Ames background, we used a complex medium containing alanine (AAC), a mixture of alanine and inosine (AI), a mixture of alanine and serine (AS), or l-alanine alone. When AAC, AI, or AS was used, exsK mutant spores germinated to a significantly greater extent than did wild-type spores, as indicated by a higher level of fluorescence (Fig. 7A to C). With alanine alone, germination was minimal in both exsK mutant and wild-type spores, consistent with previous findings (18). Nonetheless, the exsK mutant spores germinated slightly faster and reached a significantly higher level of fluorescence than did the wild-type parent spores (Fig. 7D). These results are not likely to be due to a fundamental inability of exsK mutant spores to fully rehydrate because, by use of phase-contrast microscopy, exsK mutant spores became fully refractile 30 min after the addition of AI, similarly to wild-type spores (data not shown). These data suggest that, directly or indirectly, ExsK modestly inhibits germination in response to several germinants.
FIG. 7.
Comparison of germination of B. anthracis wild-type and exsK mutant (Ames) spores in four germinants. (A to D) The germination of spores of the Ames wild-type (WT) and exsK spores in AAC (A), AI (B), AS (C), or l-alanine (l-ala) (D) was determined using the fluorescence microtiter assay, and regression curves for representative results are shown. P < 0.0001 for AAC and AI; P < 0.001 for l-alanine. (E and F) The germination of Sterne WT spores and exsK mutant spores in AI (E) or 1 mM inosine and 1 mM l-serine in PBS (IS) (F) was measured by the loss of OD over time. Data points represent three independent sporulations. Error bars represent standard errors of the means.
One possible explanation for an increased germination in exsK mutant spores is that ExsK impedes the flow of germinant molecules through the spore. A second possible explanation is that ExsK plays a role in the assembly or function of alanine racemase (Alr), an exosporium protein that interconverts alanine between the l- and d- forms and, as a result, can reduce the amount of active (l- form) alanine available to stimulate germination (3, 8, 14, 29, 45). To address this second possibility, we measured germination using an alanine-independent germinant (a mixture of inosine and serine) by a loss-of-OD assay (13). If ExsK controls Alr activity or deposition, then germination of exsK mutant spores should differ from germination of wild-type spores in the presence of alanine but not in the presence of inosine and serine. As expected, the results of the loss-of-OD assay were consistent with those of the fluorescent-dye uptake assay when we monitored germination with AI (Fig. 7E). When we analyzed germination in medium without alanine (using inosine and serine), we found that there was also a significant difference in germination kinetics between wild-type and exsK mutant spores, where exsK mutant spores germinated to a greater extent than did wild-type spores (Fig. 7F). Therefore, these data do not support the view that ExsK directs Alr assembly.
DISCUSSION
To elucidate the function of exosporium proteins, both the phenotype of spores lacking these proteins and the localization of the proteins within the spore compartments need to be assessed. Here we studied the previously identified exosporium protein ExsK (27, 35, 46). Strikingly, and in contrast to prior studies suggesting that exosporium (or coat) proteins have only a single location within the spore, we found that ExsK has at least two locations, i.e., on the exosporium surface and underneath the exosporium. The more interior location is likely to be the interspace, the coat, or both. The interspace and the coat are separated by a barrier, the basal layer, and it is possible that ExsK is present within this layer, as well as above and below it.
Our conclusion that ExsK is present in at least two locations within the spore stems from flow cytometry and immunofluorescence data. However, the interior location of ExsK is not reflected in the pattern of fluorescence in cells bearing ExsK-GFP. This inconsistency is unlikely due to an inability of a GFP fusion protein at this location to be detected, as we have previously detected fluorescence from GFP fusions located beneath the exosporium (13, 28). In fact, we know that ExsK-GFP was deposited beneath the exosporium because we detected ExsK-GFP on the surfaces of sonicated, exosporium-deficient exsK-gfp spores by flow cytometry and IFM with anti-ExsK serum (data not shown). The deposition of ExsK-GFP beneath the exosporium may result in cleavage or some other type of inactivation of the GFP domain. Although we did not detect a cleavage product in mature spores by Western blot analysis (data not shown), it is possible that the cleavage product was not detected because it was cross-linked to a high-molecular-mass species that did not migrate into the gel.
A second important finding from our work is a previously undiscovered role for BclA in the formation of protein complexes within the exosporium. BclA is required for wild-type levels of the 25-kDa ExsK complex and for the assembly of complexes migrating at 37, 50, and 250 kDa. Therefore, BclA is an important factor in ExsK complex formation.
Our data suggest a revised model for the maturation of the B. anthracis exosporium surface (Fig. 8). In this view, both BclA and ExsK begin deposition to the exosporium surface relatively late in sporulation, at a time when most spores have assembled a complete basal layer shell (12, 13). Both proteins first deposit at the mother-cell-proximal spore pole and then assemble around the rest of the spore surface, at least partially under the control of ExsFA/BxpB (13). BclA partially occludes the surface exposure of ExsK, consistent with the likelihood that these two molecules are in close proximity on the spore surface. exsK mutant spores possess BclA-dependent hairlike projections (“the nap”) (based on transmission electron microscopy analysis [data not shown]), and therefore, ExsK is not required for BclA assembly. BclA, however, is required for the appearance of high-molecular-mass forms of ExsK. It is reasonable to speculate that proximity between ExsK and BclA on the spore surface is required for this effect of BclA. The ability of BclA to affect ExsK cross-linking does not depend on entirely normal BclA deposition, as exsFA (bxpB) mutant spores have essentially normal cross-linked ExsK. BclA may serve as a scaffold or jig that facilitates proper placement of ExsK on the exosporium surface and thereby allows an as-yet-unknown protein to create cross-links.
FIG. 8.
Model of B. anthracis spore maturation. BclA and ExsK are deposited to the spore late in sporulation, with BclA deposited prior to ExsK. Both proteins assemble first to the mother-cell-proximal pole and then around the spore surface in an ExsFA/BxpB-dependent manner. Due to the partial occlusion of ExsK by BclA, it is likely that these two proteins are adjacent to each other on the spore surface. Once deposited to the spore surface, ExsK assembles into high-molecular-mass complexes in a BclA-dependent manner. Because ExsK assembles to the exosporium and underlying spore compartments after the formation of a complete basal layer, ExsK may be synthesized by ribosomes in the mother cell as well as by ribosomes trapped within the interspace. In mature spores, ExsK remains asymmetrically distributed and is more concentrated at the mother-cell-proximal pole (depicted as dark-red triangles at the pole and light-red triangles around the spore). Gray ovals, BclA; gray-striped arrows, assembly of BclA around the spore surface.
ExsK assembles below the exosporium as well as on its surface. Since much, if not most, ExsK is synthesized after the basal layer is a closed shell, it is most likely that the interior-located ExsK is synthesized by ribosomes trapped within the interspace (12).
This model differs from previous models in two important ways. First, we argue that the features of the exosporium surface depend on interactions between at least two surface proteins, BclA and ExsK. In particular, BclA controls both the surface accessibility and the complex formation of ExsK. Therefore, no individual protein defines spore surface properties. Second, we propose that an exosporium protein is present in two compartments within the spore. A key prediction of our model is that BclA and ExsK interact, directly or indirectly, in the process of exosporium formation and in the mature spore. Future experiments will address this potential interaction between ExsK and BclA.
We found that ExsK has a modest but significant negative role in germination. Our data do not support the possibility that the exosporium protein Alr has a significant role in the effect of ExsK on germination and, therefore, argue for an as-yet-unidentified role for ExsK in germination. We do not know the mechanistic basis for the effect of ExsK on germination. A simple possibility is that ExsK provides a kinetic barrier to the entry of germinant into the spore. Alternatively, ExsK may direct the assembly of molecules with a negative role in germination, such as inosine-uridine nucleoside hydrolase (IunH) (25).
In summary, our findings show that exosporium assembly involves a significantly greater degree of complexity than previously understood. First, we found that exosporium proteins can be present at an additional location, below the exosporium surface, suggesting that proteins acting at the surface can have additional roles. Second, we characterized a novel spore surface protein and, importantly, showed how its maturation depends on the well-characterized surface protein BclA. Finally, our data argue for a more complex role for the exosporium in germination, which includes the assembly of Alr as well as other negative regulatory functions. Given the pivotal role of cell surfaces in interactions with the environment, these results provide a deeper understanding of spore functions.
Acknowledgments
This work was supported by grant AI050260 from the National Institutes of Health (K.L.K.); by the Medical Biological Defense Research Program, U.S. Army Medical Research and Materiel Command, project 1.1A0010-07.RDB (USAMRIID) (S.L.W.); and by an in-house laboratory innovative research award from the Department of the Army under project 92489 (J.B.).
Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals (30b). The facilities where this research was conducted are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Baillie, L., S. Hibbs, P. Tsai, G. L. Cao, and G. M. Rosen. 2005. Role of superoxide in the germination of Bacillus anthracis endospores. FEMS Microbiol. Lett. 245:33-38. [DOI] [PubMed] [Google Scholar]
- 2.Ball, D. A., R. Taylor, S. J. Todd, C. Redmond, E. Couture-Tosi, P. Sylvestre, A. Moir, and P. A. Bullough. 2008. Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol. Microbiol. 68:947-958. [DOI] [PubMed] [Google Scholar]
- 3.Barlass, P. J., C. W. Houston, M. O. Clements, and A. Moir. 2002. Germination of Bacillus cereus spores in response to L-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 148:2089-2095. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, T., S. Little, A. G. Stover, and A. Driks. 1999. Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 181:7043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozue, J., C. K. Cote, K. L. Moody, and S. L. Welkos. 2007. Fully virulent Bacillus anthracis does not require the immunodominant protein BclA for pathogenesis. Infect. Immun. 75:508-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozue, J., K. L. Moody, C. K. Cote, B. G. Stiles, A. M. Friedlander, S. L. Welkos, and M. L. Hale. 2007. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect. Immun. 75:4498-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmbhatt, T. N., B. K. Janes, E. S. Stibitz, S. C. Darnell, P. Sanz, S. B. Rasmussen, and A. D. O'Brien. 2007. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect. Immun. 75:5233-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokova, O. N., S. A. McPherson, C. T. Steichen, and C. L. Turnbough, Jr. 2009. The spore-specific alanine racemase of Bacillus anthracis and its role in suppressing germination during spore development. J. Bacteriol. 191:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote, C. K., J. Bozue, K. L. Moody, T. L. DiMezzo, C. E. Chapman, and S. L. Welkos. 2008. Analysis of a novel spore antigen in Bacillus anthracis that contributes to spore opsonization. Microbiology 154:619-632. [DOI] [PubMed] [Google Scholar]
- 10.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driks, A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 10:251-254. [DOI] [PubMed] [Google Scholar]
- 12.Giorno, R., J. Bozue, C. Cote, T. Wenzel, K. S. Moody, M. Mallozzi, M. Ryan, R. Wang, R. Zielke, J. R. Maddock, A. Friedlander, S. Welkos, and A. Driks. 2007. Morphogenesis of the Bacillus anthracis spore. J. Bacteriol. 189:691-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giorno, R., M. Mallozzi, J. Bozue, K. S. Moody, A. Slack, D. Qiu, R. Wang, A. Friedlander, S. Welkos, and A. Driks. 2009. Localization and assembly of proteins comprising the outer structures of the Bacillus anthracis spore. Microbiology 155:1133-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould, G. W. 1966. Stimulation of l-alanine-induced germination of Bacillus cereus spores by d-cycloserine and O-carbamyl-d-serine. J. Bacteriol. 92:1261-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, England.
- 16.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a σE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555-588. [DOI] [PubMed] [Google Scholar]
- 18.Ireland, J. A., and P. C. Hanna. 2002. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis ΔSterne endospores: gerS mediates responses to aromatic ring structures. J. Bacteriol. 184:1296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487-502. [DOI] [PubMed] [Google Scholar]
- 20.Klobutcher, L. A., K. Ragkousi, and P. Setlow. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 103:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, K., K. Hashiguchi, K. Yokozeki, and S. Yamanaka. 1998. Molecular cloning of the transglutaminase gene from Bacillus subtilis and its expression in Escherichia coli. Biosci. Biotechnol. Biochem. 62:1109-1114. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, K., Y. Kumazawa, K. Miwa, and S. Yamanaka. 1996. ɛ-(γ-Glutamyl) lysine cross-links of spore coat proteins and transglutaminase activity in Bacillus subtilis. FEMS Microbiol. Lett. 144:157-160. [Google Scholar]
- 23.Kroos, L., and Y. T. Yu. 2000. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3:553-560. [DOI] [PubMed] [Google Scholar]
- 24.La Duc, M. T., M. Satomi, and K. Venkateswaran. 2004. Bacillus odysseyi sp. nov., a round-spore-forming bacillus isolated from the Mars Odyssey spacecraft. Int. J. Syst. Evol. Microbiol. 54:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Liang, L., X. He, G. Liu, and H. Tan. 2008. The role of a purine-specific nucleoside hydrolase in spore germination of Bacillus thuringiensis. Microbiology 154:1333-1340. [DOI] [PubMed] [Google Scholar]
- 26.Little, S. F., and G. B. Knudson. 1986. Comparative efficacy of Bacillus anthracis live spore vaccine and protective antigen vaccine against anthrax in the guinea pig. Infect. Immun. 52:509-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, H., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates III, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallozzi, M., J. Bozue, R. Giorno, K. S. Moody, A. Slack, C. Cote, D. Qiu, R. Wang, P. McKenney, E. M. Lai, J. R. Maddock, A. Friedlander, S. Welkos, P. Eichenberger, and A. Driks. 2008. Characterization of a Bacillus anthracis spore coat-surface protein that influences coat-surface morphology. FEMS Microbiol. Lett. 289:110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKevitt, M. T., K. M. Bryant, S. M. Shakir, J. L. Larabee, S. R. Blanke, J. Lovchik, C. R. Lyons, and J. D. Ballard. 2007. Effects of endogenous d-alanine synthesis and autoinhibition of Bacillus anthracis germination on in vitro and in vivo infections. Infect. Immun. 75:5726-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monroe, A., and P. Setlow. 2006. Localization of the transglutaminase cross-linking sites in the Bacillus subtilis spore coat protein GerQ. J. Bacteriol. 188:7609-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Moody, K. L., A. Driks, G. L. Rother, C. K. Cote, E. E. Brueggmann, H. Hines, A. M. Friedlander, and J. Bozue. Assembly and localization of a Bacillus anthracis spore coat protein that undergoes processing. Microbiology, in press. [DOI] [PubMed]
- 30b.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 31.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohye, D. F., and W. G. Murrell. 1973. Exosporium and spore coat formation in Bacillus cereus T. J. Bacteriol. 115:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliva, C. R., M. K. Swiecki, C. E. Griguer, M. W. Lisanby, D. C. Bullard, C. L. Turnbough, Jr., and J. F. Kearney. 2008. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc. Natl. Acad. Sci. USA 105:1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piggot, P., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC.
- 35.Redmond, C., L. W. Baillie, S. Hibbs, A. J. Moir, and A. Moir. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355-363. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 38.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 39.Shatalin, K. Y., and A. A. Neyfakh. 2005. Efficient gene inactivation in Bacillus anthracis. FEMS Microbiol. Lett. 245:315-319. [DOI] [PubMed] [Google Scholar]
- 40.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steichen, C. T., J. F. Kearney, and C. L. Turnbough, Jr. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J. Bacteriol. 187:5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steichen, C. T., J. F. Kearney, and C. L. Turnbough, Jr. 2007. Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol. Microbiol. 64:359-367. [DOI] [PubMed] [Google Scholar]
- 43.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 44.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2005. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J. Bacteriol. 187:5122-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Titball, R. W., and R. J. Manchee. 1987. Factors affecting the germination of spores of Bacillus anthracis. J. Appl. Bacteriol. 62:269-273. [DOI] [PubMed] [Google Scholar]
- 46.Todd, S. J., A. J. Moir, M. J. Johnson, and A. Moir. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 185:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vary, P. S. 1994. Prime time for Bacillus megaterium. Microbiology 140:1001-1013. [DOI] [PubMed] [Google Scholar]
- 48.Weaver, J., T. J. Kang, K. W. Raines, G. L. Cao, S. Hibbs, P. Tsai, L. Baillie, G. M. Rosen, and A. S. Cross. 2007. Protective role of Bacillus anthracis exosporium in macrophage-mediated killing by nitric oxide. Infect. Immun. 75:3894-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiner, M. A., T. D. Read, and P. C. Hanna. 2003. Identification and characterization of the gerH operon of Bacillus anthracis endospores: a differential role for purine nucleosides in germination. J. Bacteriol. 185:1462-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welkos, S. L., C. K. Cote, K. M. Rea, and P. H. Gibbs. 2004. A microtiter fluorometric assay to detect the germination of Bacillus anthracis spores and the germination inhibitory effects of antibodies. J. Microbiol. Methods 56:253-265. [DOI] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Zilhão, R., R. Isticato, L. O. Martins, L. Steil, U. Volker, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2005. Assembly and function of a spore coat-associated transglutaminase of Bacillus subtilis. J. Bacteriol. 187:7753-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]