Abstract
As part of the Israel National Program for Prevention and Control of Tuberculosis, the molecular epidemiology of new tuberculosis cases is monitored. Prospective screening showed that about 20% of all new cases of culture-positive tuberculosis (43 of 222) in Israel in the year 2008 were caused by certain Mycobacterium tuberculosis strains of the central Asian (CAS) spoligotype lineage. The identity and similarity of these strains by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) typing form a lineage we call PETRA for polymorphic at locus ETR A. The name PETRA was given to 79 strains we have found since the year 2000, because the largest number of strains with MIRU-VNTR profiles identical other than at locus A formed three groups, including 5 of 10 strains that had deleted the ETR A region from their genomes. No PETRA strain was found to be multiple drug resistant (resistant to both isoniazid and rifampin [rifampicin]). Most patients (75% [58 of 77 patients of known origin]) infected with PETRA were of sub-Saharan African origins. The genotypes associated with the 79 PETRA lineage strains presented in this paper suggest that the PETRA lineage is a large, major contributor to new tuberculosis cases in Israel.
Mycobacterium tuberculosis is a major cause of world death and morbidity due to bacterial infection (15). It is estimated that almost a third of the world's population, 2 billion persons, has been exposed to M. tuberculosis and that M. tuberculosis causes about 9 million persons to become ill each year; of these, nearly 2 million die from tuberculosis (15). Israel is a country with a low incidence of tuberculosis, but the recent absorption of almost a fifth of the population from countries with a high incidence of tuberculosis has presented a challenge for the control, prevention, and treatment of tuberculosis. In particular, the introduction of multiple-drug-resistant (MDR) M. tuberculosis primarily from former Soviet Union countries (unpublished data) and of the non-MDR but highly infectious central Asian (CAS) spoligotype lineage MT, primarily from sub-Saharan African (SSA) countries (this study), continues to challenge the public health system in Israel.
CAS spoligotype sublineages (5, 6, 61) have been reported to be prevalent in patients from SSA countries (3, 29, 58). Extrapulmonary tuberculosis has been reported from CAS spoligotype M. tuberculosis (41). It was recently reported that the CAS spoligotype lineage contributes to the tuberculosis burden in the Middle East (3).
Genotyping on culture-positive Mycobacterium tuberculosis complex (MTC) has been performed at our National Mycobacterium Reference Laboratory since 1997 using restriction fragment length polymorphism (RFLP) typing (75, 76) as the gold standard. Later, 43-spacer spoligotyping (19) was introduced, followed by typing of 16 mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) loci (12 MIRU-VNTR [47, 66] loci, 3 exact tandem-repeat [ETR] loci [25], and the QUB11b [68] locus). For the year 2000, a retrospective analysis of all new culture-positive pulmonary MTC cases in Israel was performed using RFLP typing. From 2001 to 2005 and in 2007, MTC strains were genotyped according to clinical and epidemiological needs. Most new MTC strains of 2006 were MIRU-VNTR genotyped. Since the beginning of 2008, all new culture-positive MTC cases are being MIRU-VNTR genotyped in a prospective screen employing high-throughput capillary electrophoresis with 16 capillaries (4, 65, 67, 68).
This paper presents the results obtained from the prospective MIRU-VNTR molecular epidemiological screening of all patients with new cases of culture-positive tuberculosis in Israel in the year 2008 and in addition some results obtained from the retrospective screening of previous years in Israel. These results lead to the definition of a new M. tuberculosis MIRU-VNTR lineage (with CAS spoligotypes) which is named PETRA (for polymorphic ETR A) here. Evidence is presented that the M. tuberculosis PETRA lineage is a major contributor of tuberculosis in Israel and is of predominantly SSA origins. Some PETRA lineage strains have deleted the chromosomal region containing locus ETR A.
MATERIALS AND METHODS
Growth of MTC and drug sensitivity tests.
For the prospective screening of 2008, analysis was conducted on all new culture-positive strains isolated from material collected in 2008 and processed from 1 January 2008 through 28 February 2009. Retrospective screening of previous years, in whole or in part, was conducted on the basis of clinical and epidemiological needs. M. tuberculosis complex (MTC) strains were grown on solid Lowenstein-Jensen medium using standard methods (37). Strains with MTC biochemical and physiological characteristics were confirmed as MTC by the AccuProbe (GenProbe) test. Since 2007, a commercial strip test (Hain, Germany) (52) has been used by our laboratory for initial identification of MTC strains. We also use the commercial strip test to screen for multiple-drug-resistant status (resistance to both isoniazid and rifampin [rifampicin]). Commercial drug susceptibility test results were confirmed and expanded by the resistance ratio method (37) for isoniazid, rifampin, streptomycin, ethambutol, and pyrazinamide. Mycobacterium bovis BCG P2 was a kind gift from Hillel Bercovier, Hebrew University of Jerusalem. Mycobacterium tuberculosis strain MT14323 was a kind gift from the National Institute of Public Health and the Environment (RIVM), Bilthoven, The Netherlands. Every MIRU-VNTR typing plate analyzed by capillary electrophoresis included strain MT14323 for typing at all loci as a control.
Extraction of Mycobacterium tuberculosis DNA.
RFLP-grade DNA was extracted by the international consensus standard method (75). Initial screening for MTC strains in our lab involves commercial strip testing (Hain, Germany), which can also detect simultaneous infection with about 30 species of nontuberculous mycobacteria. It was convenient and efficient to use a fast DNA extraction method for automated high-throughput prospective MIRU-VNTR screening of all the MTC isolates from new patients. The fast DNA extraction method (used for strip testing; Hain, Germany) consisted of preparing a “crude” extract as follows. Bacteria grown on solid medium were collected with an inoculation loop and suspended in approximately 300 μl of water (molecular biology grade). The suspension was incubated for 45 min at 95°C in a water bath, transferred to an ultrasonic bath for 15 min, and then centrifuged in a microcentrifuge at full speed for 5 min. The supernatant was transferred to a new tube and stored at −20°C. This paper deals only with new MTC isolates. The few cases of mixed infection (two MIRU-VNTR alleles at one or more loci) were dropped from this study. The fast DNA extraction method (amount used, 2 μl) was also successfully employed for spoligotyping (Ocimum Biosolution Company, India).
PCR and agarose gel electrophoresis.
Quantitative agarose gel electrophoresis was performed using simplex PCR catalyzed by Qiagen HotStarTaq polymerase and accompanying reagents (catalog no. 203203; Qiagen) with the primers developed by Supply et al. (66) and Frothingham and Meeker-O'Connell (25). For example, enough mix for 2 reaction mixture volumes for 2 mM MgCl2 contained 5 μl of 10× HotStarTaq buffer (Qiagen), 4.8 μl of autoclaved H2O (catalog no. W4502; Sigma), 10 μl of 5× Q solution (Qiagen), 1 μl of 25 mM MgCl2 (Qiagen), 4 μl of 2.5 mM deoxynucleoside triphosphates, and 0.2 μl of HotStarTaq polymerase (5 U/μl) (Qiagen). To 12.5 μl of this reaction solution was added 8.5 μl of DNA containing 0.5 μl of Hain extract or 10 ng of RFLP-grade DNA. Then 2 μl each of the forward and reverse primers was added for a final volume of 25 μl. PCR was performed as follows. The lid was 99°C. PCR temperature cycling was performed as follows: (i)15 min at 95°C; (ii) 40 cycles, with 1 cycle consisting of 60 s at 94°C, 60 s at 59°C, and 90 s at 72°C; (iii) 10 min at 72°C; (iv) pause at 10°C (67). PCR product was stored at 2 to 8°C until use. Agarose gel electrophoresis was set up and performed as follows. One hundred milliliters of 2% LE agarose (SeaKem) in 1× Tris-borate-EDTA (Promega) was prepared. After the agarose was cooled to 65°C, 1 drop of ethidium bromide (0.25 μg Amresco) was added. This was poured into a 10- by 10-cm gel apparatus and allowed to solidify, and then the apparatus was loaded with samples which were electrophoresed (Bio-Rad apparatus) at 100 V for 90 min, which was immediately followed by electrophoresis at 50 V for 45 min. The PCR products from 15 different loci (or other arrangements of loci as needed) were electrophoresed along with (i) a positive control (locus MIRU-VNTR 23 with MT14323 DNA), (ii) a negative control (locus MIRU-VNTR 23 with autoclaved water [catalog no. W4502; Sigma]), and (iii) three wells (the two outer wells and middle well number 11) of base pair standards (Roche, 100-bp ladder, 100 to 1,500 bp). In our hands, only the Roche bp ladder allowed quantitative results; a bp ladder from another company was not suitable. Results were analyzed by recording the data with a Fuji/Pharmacia digital camera UV gel visualization instrument and calculating (using Total Lab v2 image analysis software) the base pairs corresponding to the migration of fluorescent ethidium bromide-labeled amplicon DNA bands relative to the migration of the standards. Translation of base pairs into copy numbers was done according to the values provided by Supply et al. (65, 66, 68). The meaning of “copy number” has been discussed by Skuce et al. (60). Another set of primers was made flanking the region encompassed by the standard primers for locus ETR A, using Primer3 software (53), that were named mla-1: forward mla-1 primer, ATA TCG ATA CAG CTA GGC ACT CCT; reverse mla-1 primer, GAA GTT CAA GAT TCC CGA TGT C. The mla-1 forward and reverse primers flanked the standard primers for amplification of the ETR A locus in order to confirm that the inability to obtain ETR A amplicons from certain strains involved chromosomal deletions.
Bioinformatics.
Tools provided by the National Center for Biotechnology Information (NCBI) were used to obtain and examine the sequences of strains H37Rv Rv1917c and RD Rv1917c with chromosomal deletions and insertion of IS6110. These tools were BLAST2 sequences, nucleotide, and entrez gene. In addition, the PubMed literature search tool was employed. All NCBI tools were utilized at the central website, http://www.ncbi.nlm.nih.gov. Primers flanking the standard ETR A locus region were constructed with the software program Primer3 (53) found at the website http://frodo.wi.mit.edu. Note that the address does not include www. The MIRU-VNTR and spoligotype profiles of the PETRA lineage were identified as being a sublineage of the CAS spoligotype lineage by comparison with data in the MIRU-VNTRplus database (5): http://www.miru-vntrplus.org/MIRU/index.faces. In addition, further CAS MIRU-VNTR types and associated spoligotypes were identified in the literature (3, 6, 7, 58, 61, 62).
High-throughput capillary electrophoresis using the AB3100 Prism with 16 capillaries.
Capillary electrophoresis with 16 capillaries allowed the construction of a high-throughput system based on multiplex PCR (4, 65, 67, 68) and automatic electrophoresis and recording of results. Analysis of results was facilitated by GeneMapper software version 4 (Applied Biosystems), which after initial determination of offsets by comparison with agarose gel results and results published in the literature (4, 65) and subsequent assignation of corresponding bins allowed speedy assignation of alleles (copy numbers) to peaks. The primers were constructed and labeled by the method of Supply et al. (68) with the following exceptions. MIRU 4 was prepared by the method of Supply et al. (67). ETR B was prepared by the method of Frothingham and Meeker-O'Connell (25) with the forward primer labeled with VIC. ETR C was prepared by the method of Frothingham and Meeker-O'Connell (25) with the forward primer labeled with 6-carboxyfluorescein (FAM). QUB11b was prepared by the method of Supply et al. (68) but with the reverse primer labeled with PET (red color; Applied Biosystems). In addition, the MIRU 23 primers (68) were also made with forward primer labeled with PET. Five colors were used: 6-FAM (blue) (represented by f in primer designations), VIC (green; Applied Biosystems) (represented by v in primer designations), NED (yellow; Applied Biosystems) (represented by n in primer designations), PET (red) (represented by p in primer designations) and LIZ (orange, for the standard bp ladder LIZ1200 purchased from Applied Biosystems). An identical MgCl2 concentration for all primer sets was sought for multiplex PCR in order to maximize the high-throughput aspects of MIRU-VNTR screening. For our primer sets, the optimal concentration was 3 mM MgCl2 using Qiagen HotStarTaq polymerase and kit components of polymerase buffer, MgCl2 and Q buffer. These primer sets (panels) were as follows: (i) 4f-26v-40n (68), (ii) Cf-16v-31n, (iii) 2f-23v-39n (68), (iv) 20f-24v-27n (68), (v) 10f-An, (vi) Bv-QUB11bp, and (vii) 23p. Primer set 7 was of course for a simplex PCR, but 3 mM MgCl2 was also used for this PCR. The water used was autoclaved water (catalog no. W4502; Sigma). The deoxynucleoside triphosphates were used at a final concentration of 0.2 mM each. The final reaction mixture volume was 20 μl. PCR was performed as previously described (67). DNA for PCR was usually extracted by the fast DNA extraction method, although RFLP-grade DNA (2 ng in 2 μl) (75) and DNA extracted by heat (45 min at 95°C) (47) could also be employed. The advantage of using the fast-method DNA extracts was to connect directly with the routine throughput of the diagnostic laboratory, which had switched to Hain diagnostic tests for identification of Mycobacterium species. Undiluted crude fast DNA extract (2 μl) was used for primer sets 2, 4, and 5. For the remaining primer sets, crude fast DNA extract diluted 1:50 in 1× Tris-EDTA (Sigma) was employed. Mixes, reaction mixture volumes, and electrophoresis conditions were adapted from those used by Supply et al. (68) and from the Institute Pasteur Manual kindly provided by Philip Supply (65). Capillary electrophoresis runs were for 2,100 s using POP4 (Applied Biosystems) and 36-cm-long capillaries. Currently, for capillary electrophoresis, we use an upgraded machine, Applied Biosystems 3130xl. The 96-well microplate setup for our routine consists of 14 strains analyzed in wells A1 to A6, wells B1 to B6, to wells F7 to F12. Wells 1 to 6 and wells 7 to 12 contain the DNA for a given strain and multiplex panels 1 to 6, respectively. Wells G7 to G12 contain DNA from fast DNA extracted from strain MT14323. Wells H7 to H9 contain negative-control water instead of DNA, with multiplex panel 3. Wells H10 to H12 contain positive-control fast DNA extracted from strain MT14323 with simplex panel 7. After PCR (PCR product could be stored at 2 to 8°C for at least 2 weeks), 1 μl of PCR product was added to 10 μl of mix for a total volume of 11 μl. The mix was composed of 8.5 volumes of formamide (HI-DI Applied Biosystems), 0.5 volume of LIZ1200 size standard (Applied Biosystems), and 0.5 volume of MIRU 23p MT14323 (1 part of MIRU 23p MT14323 diluted with 3 parts of autoclaved water [catalog no. W4502; Sigma]) (MIRU 23p MT14323 from one well of wells H10 to H12) as an internal standard. Thus, each plate for electrophoresis has an MT14323 control for all loci (wells G7 to G12), and an internal MIRU 23p MT14323 control (red peak) in each well, in addition to the LIZ1200 size standard in each well. This is a high-throughput operation, using multidispenser pipettes (for example, to dispense the 10 μl of mix into each one of the 96 wells of the electrophoresis plate) or multichannel pipettes whenever possible (for example, to transfer 1 μl of PCR product from each well of the PCR plate to the corresponding well of the electrophoresis plate).
MIRU-VNTR results were stored in an Excel database (Microsoft) and in the Bionumerics version 3.5 database (Applied Maths BVBA, Belgium) by associating the database information with the genotypic profiles stored using the characters module. Clustering MIRU-VNTR results was performed by using the categorical coefficient and the unweighted-pair group method using average linkages (UPGMA) algorithm to generate a dendrogram, or analysis was done by generating a minimum spanning tree (MST) using the following default options: coefficient, categorical; priority, highest number of single-locus variants (SLVs); creation of complexes, maximum neighbor distance two changes and minimum size 2 types.
IS6110 RFLP typing.
Restriction fragment length polymorphism typing was performed using standard international conventions (75) as detailed in the RIVM manual (76) kindly provided by Dick van Soolingen and Kristin Kremer of the RIVM, Bilthoven, The Netherlands. It is important to note that most of the electrophoresis is performed overnight at 20 V. Electrophoresis of PvuII-digested M. tuberculosis DNA is performed until a standard running marker band has migrated to 7.0 cm (±0.4 cm). The DNA fragments in the 0.8% agarose (SeaKem LE) gel are then vacuum blotted onto a nitrocellulose membrane (Hybond N+) and fixed to the membrane using a UV cross-linker instrument. The membrane is hybridized first with probe for insertion sequence IS6110 and then, according to the protocol, hybridized with probe for the internal standards that were placed in every well. In addition to the internal standards, we run three MT14323 M. tuberculosis standards (two outside lanes and the middle lane) on every gel. The IS6110-containing band and internal standard bands were visualized using the GE Healthcare kit for enhanced chemiluminescence (ECL kit). The autoradiograms were scanned using international conventions (75, 76), and the bands were stored in the Bionumerics database, version 3.5. RFLP profiles were stored in the Bionumerics database, version 3.5, associated with the corresponding strain information. Dendrograms were produced using the Dice coefficient with UPGMA clustering (results not shown).
Spoligotyping.
Spacer oligonucleotide typing (spoligotyping) for 43 spacers of the direct repeat region (9, 56, 61) was performed using a kit according to the manufacturer's instructions (Isogen/Omicium). The Taq polymerase (catalog no. M1861; Promega) was kept in storage buffer A. Results were visualized using octal or character representation of the 43 spacers (19). Spoligotyping results were stored in the Bionumerics database, version 3.5, using the characters module, and clustered using the categorical coefficient and the UPGMA algorithm, or analyzed with the MST option using the default options described above.
RESULTS
PETRA lineage consensus MIRU-VNTR typing profile.
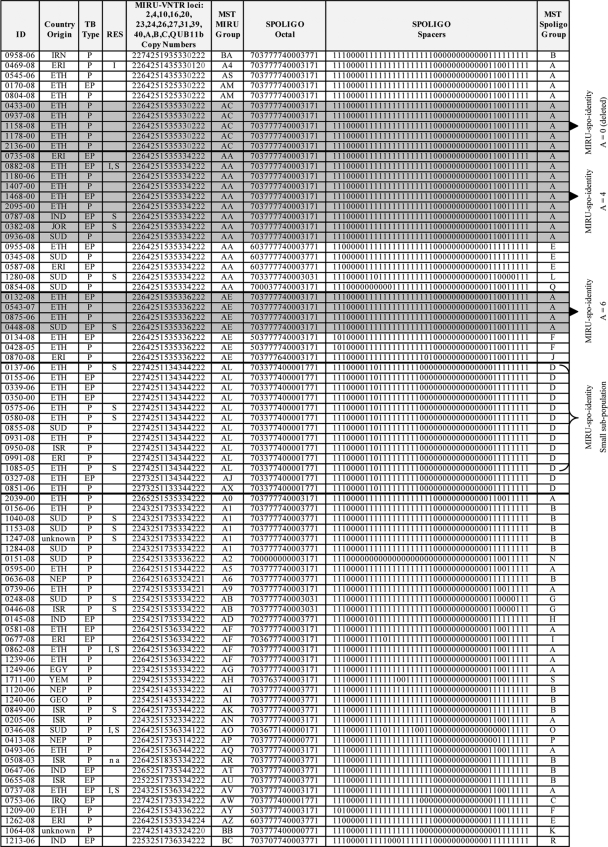
A total of 79 cases were found to have the M. tuberculosis PETRA lineage profile described below (Table 1). The consensus MIRU-VNTR profiles of 16 loci were as follows (shown in the form locus:copy number): 02:2; 04:2; 10:variable; 16:variable; 20:2; 23:5; 24:1; 26:variable; 27:3 (rarely 1 or 2); 31:variable; 39:3; 40:variable; A:4, 6, or no amplicon; B:2 (rarely 1); C:2; and QUB11b:variable. Most profiles come from the years 2000, 2006, and 2008 because those were the years that systematic complete screening (retrospective for 2000 [RFLP], prospective for year 2008 [MIRU-VNTR]) or partial screening (retrospective for year 2006 [MIRU-VNTR]) was undertaken. The MIRU-VNTR profiles are strikingly similar to the profiles exhibited by the 10 PETRA strains that lack the ETR A locus (Table 2 and Fig. 1). The invariant allele for locus 24, copy number 1, is characteristic of “modern” tuberculosis, and is reportedly typical for the CAS strains (64).
TABLE 1.
Probabilities of the appearance of MIRU-VNTR alleles (copy numbers) in the 79 PETRA lineage strains
| MIRU-VNTR locus | Probability of copy number (allele)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NPPb | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 02 | 1.000 | |||||||||
| 04 (ETR D) | 1.000 | |||||||||
| 10 | 0.013 | 0.089 | 0.101 | 0.570 | 0.215 | 0.000 | 0.013 | |||
| 16 | 0.013 | 0.127 | 0.835 | 0.025 | ||||||
| 20 | 1.000 | |||||||||
| 23 | 1.000 | |||||||||
| 24 | 1.000 | |||||||||
| 26 | 0.165 | 0.000 | 0.013 | 0.063 | 0.570 | 0.013 | 0.152 | 0.013 | 0.013 | |
| 27 | 0.013 | 0.025 | 0.962 | |||||||
| 31 (ETR E) | 0.013 | 0.013 | 0.165 | 0.709 | 0.101 | |||||
| 39 | 1.000 | |||||||||
| 40 | 0.013 | 0.025 | 0.747 | 0.215 | ||||||
| ETR A | 0.127 | 0.000 | 0.000 | 0.000 | 0.759 | 0.000 | 0.114 | |||
| ETR B | 0.025 | 0.975 | ||||||||
| ETR C | 1.000 | |||||||||
| QUB11b | 0.025 | 0.025 | 0.937 | 0.000 | 0.013 | |||||
The probability (number of strains/number of total strains) of the appearance of MIRU-VNTR copy number (allele) in 79 PETRA lineage strains is shown. For each MIRU-VNTR locus, the probabilities add up to 1 total for the different copy numbers or alleles. The major alleles are indicated by boldface type, and the invariant alleles are shown in italic boldface type.
NPP, no PCR product.
TABLE 2.
Evidence for deletion of ETR A in certain PETRA lineage strainsa
| Status | Strain | PCR product of ETR A by AGEb |
ETR A copy no. by CEc | PCR product of mla-1 by AGE |
PCR product of ETR B by AGE |
Copy no. by CE |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp | Copy no. | Proto-bp | bp | Proto-bp | bp | Copy no. | Proto-bp | ETR B | QUB11b | |||
| mla PETRA | 0433-00 | NB | NB | NB | 234 | 2 | 235 | 2 | 2 | |||
| 1178-00 | NB | NB | NB | 231 | 2 | 235 | 2 | 2 | ||||
| 2136-00 | NB | NB | NB | 232 | 2 | 235 | 2 | 2 | ||||
| 0545-06 | NB | NB | NB | 233 | 2 | 235 | 2 | 2 | ||||
| 0958-06 | NB | NB | NB | 232 | 2 | 235 | 2 | 2 | ||||
| 0170-08 | NB | NB | NB | 227 | 2 | 235 | 2 | 2 | ||||
| 0469-08 | NB | NB | NB | 176 | 1 | 178 | 1 | 0 | ||||
| 0804-08 | NB | NB | NB | 232 | 2 | 235 | 2 | 2 | ||||
| 0937-08 | NB | NB | NB | 227 | 2 | 235 | 2 | 2 | ||||
| 1158-08 | NB | NB | NB | 233 | 2 | 235 | 2 | 2 | ||||
| PETRA | 1468-00 | 497 | 4 | 495 | 4 | 748 | 753 | 230 | 2 | 235 | 2 | 2 |
| 0428-05 | 637 | 6 | 645 | 6 | 887 | 903 | 230 | 2 | 235 | 2 | 2 | |
| 0339-06 | 500 | 4 | 495 | 4 | 755 | 753 | 232 | 2 | 235 | 2 | 2 | |
| 0382-08 | 491 | 4 | 495 | 4 | 748 | 753 | 229 | 2 | 235 | 2 | 2 | |
| Beijing (PETRA-like) | 0674-06 | 494 | 4 | 495 | 4 | 753 | 753 | 231 | 2 | 235 | 2 | 2 |
| 0653-08 | 488 | 4 | 495 | 4 | 750 | 753 | 229 | 2 | 235 | 2 | 6 | |
| Beijing (classic) | 0419-04 | 494 | 4 | 495 | 4 | 746 | 753 | 228 | 2 | 235 | 2 | 6 |
| 0944-06 | 497 | 4 | 495 | 4 | 746 | 753 | 236 | 2 | 235 | 2 | 6 | |
| Control | BCG P2 | 573 | 5 | 570 | 5 | 840 | 828 | 410 | 5 | 406 | 5 | 3 |
| MT14323 | 421 | 3 | 420 | 3 | 672 | 678 | 232 | 2 | 235 | 2 | 2 | |
The chromosomal region containing ETR A in the putative PPE protein-coding RD Rv1917c gene is deleted in some PETRA lineage strains. PETRA lineage strains lacking locus ETR A (missing locus A [mla] [no amplicons made on PCR with the standard primer pair]) also lack an enlarged region amplified with primers (mla-1) flanking the standard ETR A region primers. Note that the ETR A repeat size for a copy is 75 bp. Locus QUB11b is also found in M. tuberculosis RD Rv1917c, upstream of the ETR A locus. The flanking mla-1 primers enlarge the ETR A region by 258 bp (170 bp upstream and 88 bp downstream according to H37Rv sequence data).
The PCR product of ETR A by agarose gel electrophoresis (AGE) is shown. Abbreviations: bp, base pairs; proto-bp, prototype (expected) base pairs; copy no., MIRU-VNTR copy number; NB, no band.
The PCR product of ETR A by capillary electrophoresis (CE) is shown. NB, no band.
FIG. 1.
Data for the 79 PETRA lineage strains of this study. The leftmost column (ID) contains the strain identification codes (a number followed by the year of isolation). The next column shows the country of origin of the patient with tuberculosis as follows: IRN, Iran; ERI, Eritrea; ETH, Ethiopia; IND, India; JOR, Jordan; SUD, Sudan; NEP, Nepal; ISR, Israel; EGY, Egypt; YEM, Yemen; GEO, Georgia; IRQ, Iraq. The type of tuberculosis specimen (TB type) is shown as follows: P, pulmonary; EP, extrapulmonary. In the RES column, resistance to first line anti-tuberculosis drugs (RES) is shown as follows: I, isoniazid; S, streptomycin; n a, information not available. The MIRU-VNTR loci column shows the MIRU-VNTR copy number profile for each strain, by locus in the following order: 2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39, 40, A, B, C, and QUB11b, which are clustered in Fig. 2. Note that 0 indicates an absence of the amplicon (terminology which facilitates data manipulation by Excel and Bionumerics). The column labeled MST MIRU Group shows the MIRU-VNTR minimum spanning tree (MST) group as visualized in Fig. 4. The SPOLIGO Octal column shows the octal equivalent of the 43-spacer spoligotype. The column labeled SPOLIGO Spacers shows the character representation of the 43-spacer spoligotype profiles for each strain, which are clustered in Fig. 3, as follows: 1 for hybridization and 0 for no hybridization. The MST Spoligo Group column shows the spoligotype MST group as visualized in Fig. 5. The strains with gray shaded backgrounds have identical spoligotypes and MIRU-VNTR types that vary only at locus ETR A. To the right of the figure, the MIRU-VNTR subclusters that also share spoligotype identity (MIRU-spo-identity) are shown. ETR A amplicons of various copy numbers are shown (for example, amplicons of copy number 4 are represented by A = 4). Since it is difficult in places to distinguish among zero, Q, and capital O, we note for the sake of clarity that the country of origin for strain 0753-06 is Iraq (IRQ), the MST MIRU group for strain 2039-00 is A0, the MST MIRU group for strain 0346-08 is AO, the MST MIRU group for strain 0493-06 is AQ, the MST spoligo group for strain 0854-08 is Q, and the MST spoligo group for strain 0346-08 is O.
Drug sensitivity test results.
No PETRA lineage member was multiple drug resistant (resistant to at least both isoniazid and rifampin), nor were any PETRA lineage members resistant to rifampin or ethambutol (Fig. 1). One strain was resistant to isoniazid alone, 14 strains were resistant to streptomycin alone, and 4 strains were resistant to both isoniazid and streptomycin (Fig. 1). In summary, 19 of the 78 strains (24%) for which information was available, were resistant to one or more of the first-line antituberculosis drugs isoniazid and/or streptomycin.
ETR A deletion in the PETRA lineage.
The first 10 PETRA strains listed in Table 2, missing locus A (mla) PETRA, failed to amplify an amplicon (primers mla-1) for a larger flanking ETR A-containing region (larger by 258 bp). This highly suggests that there is a chromosomal deletion of the sequence containing the ETR A locus and of nearby flanking regions. In contrast, the control strains all amplified the ETR A region with standard primers, as well as the larger ETR A-containing region flanked by the mla-1 primers. Table 2 lists these control strains in the following order: (i) four PETRA family members of ETR A copy number 4 or 6, (ii) two Beijing family members with PETRA-like MIRU-VNTR profiles, (iii) two Beijing family members with classic Beijing MIRU-VNTR profiles, (iv) BCG P2, and (v) MT14323. The QUB11b region, which is located in gene Rv1917c upstream of the ETR A-containing region, was with one exception (strain 0469-08) amplified by the mla PETRA strains (Table 2). This suggests that deletion of the ETR A-containing region involved only partial deletion of the M. tuberculosis RD Rv1917c gene for at least 9 of the 10 mla PETRA strains. Amplification of the QUB11b region for strain 0469-08 failed in all independent experiments, and the reason for the possible QUB11b deletion needs to be determined. Indeed, only sequencing of the regions spanning the ETR A deletions will confirm and explain these deletions (for example, see references 27 and 43). No data were gathered concerning the region farther downstream of the ETR A locus-containing region. It can be concluded that there is probably a chromosomal deletion of the ETR A region in the Rv1917c gene of the 10 PETRA strains listed first in Table 2 and Fig. 1. Further investigation, precise determination of the regions flanking the deletions, is needed to determine whether or not other genes are codeleted with the ETR A deletions. Locus ETR B was amplified for all strains as a positive control, and the expected ETR B PCR product was produced by all strains.
Bioinformatics search for examples of deleted ETR A regions.
Two examples (unpublished) relevant to our work were submitted by other investigators to GenBank; these were examples of the Rv1917c gene disrupted by insertion and deletion of the mobile element IS6110 (submitted 20 January 2005 to the National Institute for Medical Research, United Kingdom, by C. Menendez). Isolate 9375 can be accessed with the identifier GI:71056981, and isolate 8088 can be accessed with the identifier GI:71056983. Both isolates were part of a clinical study of M. tuberculosis strains in the South Asian community of the United Kingdom. Using “BLAST2 sequences” to probe the Menendez et al. GenBank information for the IS6110 indel Rv1917c sequences, it can be observed that isolate 9375 had deleted the QUB11b and ETR A-containing regions from the gene. However, it retained at least some of the region above QUB11b, below ETR A, and between QUB11b and ETR A. Isolate 8088 had deleted the QUB11b and ETR A-containing regions from the gene, and it also retained at least some of the regions between QUB11b and ETR A and below ETR A. However, in contrast to isolate 9375, isolate 8088 had deleted all of the Rv1917c sequences found above the QUB11b region. This information combined with our results, although lacking detailed sequences of the RD Rv1917c regions of mla PETRA isolates, suggests that Rv1917c is a target for IS6110 insertion/deletion, and that IS6110 insertion/deletion can cause the elimination of the ETR A locus (including the flanking standard primers). The IS6110 mediated ETR-A deletions reported in GenBank appear to be different from the deletions observed in our PETRA ETR A-deleted isolates, since our isolates (with one exception) retained the QUB11b region. It is also interesting to note that the two isolates were obtained from the South Asian community and therefore may have been of CAS spoligotype lineage. Neither transmissibility nor pathogenicity was abrogated in M. tuberculosis PETRA lineage strains with ETR A-deleted regions or in the two M. tuberculosis strains with reported ETR A deletions in GenBank. It can be inferred from these results that IS6110 insertion/deletion is a possible, and even probable, cause of the PETRA lineage chromosomal deletions of the RD Rv1917c ETR A-containing region. However, definitive characterization will require sequencing of the postdeletion chromosomal region.
Genotypic and demographic analyses of M. tuberculosis PETRA lineage strains.
Figure 1 summarizes data for all 79 PETRA strains including strain identification (in the form strain number-year of isolation), country of origin of the patient, type of tuberculosis specimen, resistance to first-line antituberculosis drugs, MIRU-VNTR profiles for 16 loci (also shown in Fig. 2 and 3), MIRU-VNTR minimum spanning tree group (MST shown in Fig. 4), 43-spacer octal spoligotypes (also shown in Fig. 2 and 3), 43-spacer character spoligotypes (0 for no hybridization, 1 for hybridization), and spoligotype minimum spanning tree group (MST shown in Fig. 5). Figure 1 is arranged in sections for convenient reference and can be summarized as a whole or by section. The top four sections are comprised of four of the largest PETRA MIRU-VNTR identity clusters. Subclusters that also share spoligotype identity are indicated to the right of the figure. The countries of origin of 75% (58 of 77 for whom information was available) of the patients infected with PETRA lineage strains were in sub-Saharan Africa. The patients were from the following countries in SSA: Ethiopia (39 patients [51%]), Sudan (12 patients [16%]), and Eritrea (7 patients [9%]). The remaining patients were from India (4 patients [5%]), Israel (6 patients [7.7%]), Nepal (3 patients [4%]) (total of 13 patients [16.7%] from India, Israel, and Nepal) and then from Egypt, Georgia, Iran, Iraq, Jordan, and Yemen, each with 1 case (1.3% each) for a total of 6 patients (7.8%) from the last six countries.
FIG. 2.
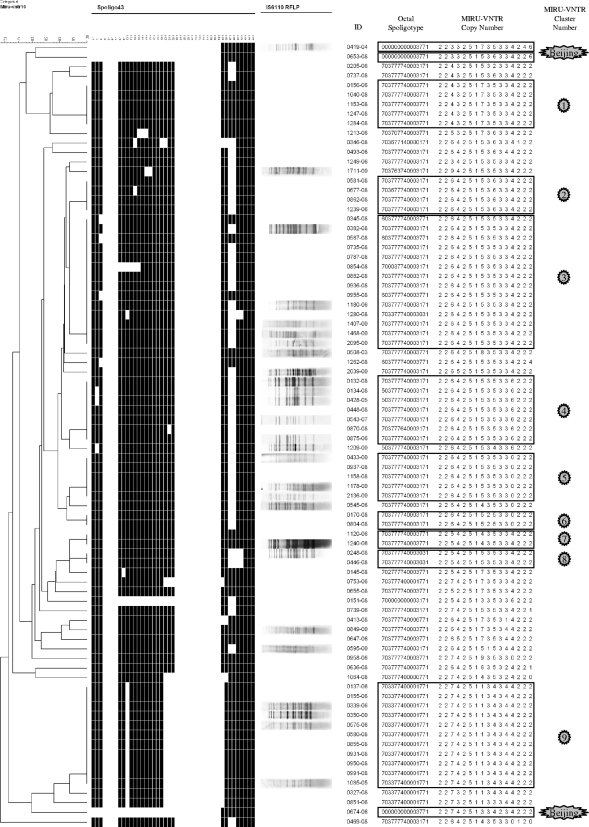
Dendrogram of the 79 PETRA lineage strains clustered on their MIRU-VNTR profiles, using Bionumerics software with the categorical coefficient and the UPGMA algorithm. Three Beijing strains were included for comparison. The strain designations are shown in the ID (identification) column. Octal spoligotype and MIRU-VNTR copy number are also shown in Fig. 1.
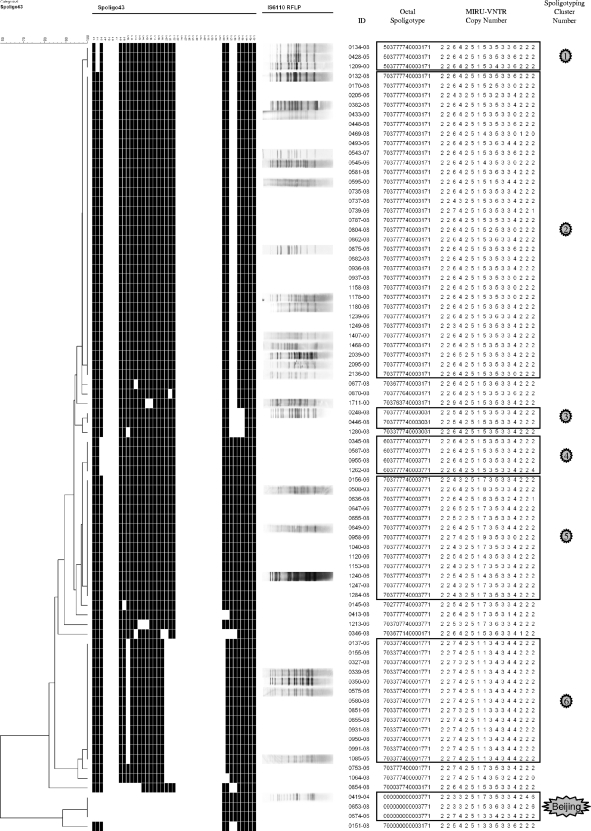
FIG. 3.
Dendrogram of the 79 PETRA lineage strains clustered on their 43 spacer spoligotype profiles, using Bionumerics software with the categorical coefficient and the UPGMA algorithm. Three Beijing strains were included for comparison. Strain designations are shown in the ID (identification) column. Octal spoligotype and MIRU-VNTR copy number are also shown in Fig. 1.
FIG. 4.
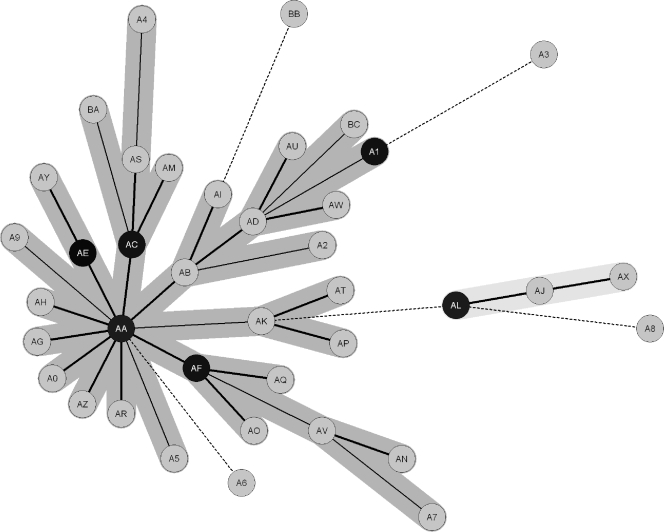
Minimum spanning tree calculated on MIRU-VNTR data for the 79 PETRA lineage strains and 3 Beijing family strains for comparison. The MST was constructed using Bionumerics software with the following default settings: (i) priority rule, highest number of SLVs; (ii) coefficient, categorical; and (iii) for creation of complexes, maximum neighbor distance of two changes and minimum size of two types. The length of the branches reflects the distance between the types, and the thickness, dotting, and graying of the branch lines also reflect distance between the nodes (BioNumerics version 3.5 user manual; Applied Maths BVBA, Sint-Martens-Latem, Belgium).
FIG. 5.
Minimum spanning tree calculated on spoligotype data for the 79 PETRA strains and 3 Beijing strains for comparison. The MST was constructed using Bionumerics software with the following default settings: (i) priority rule, highest number of SLVs; (ii) coefficient, categorical; and (iii) for creation of complexes, maximum neighbor distance of two changes, minimum size of two types. The length of the branches reflects the distance between the types, and the thickness, dotting, and graying of the branch lines also reflect the distance between the nodes (BioNumerics version 3.5 user manual; Applied Maths BVBA, Sint-Martens-Latem, Belgium).
Of the 79 PETRA lineage strains shown in Figure 1, 23 (29%) were isolated from various extrapulmonary specimens. Nine of the 10 strains lacking ETR A (90%) were isolated from specimens from patients with pulmonary tuberculosis, and 1 (10%) was from an extrapulmonary specimen (Fig. 1, top section); therefore, the chromosomal deletions of ETR A did not attenuate the affected strains. Possible confounding factors that prevent clear interpretation of these data would be the patient's genetic and health backgrounds.
The countries of origin of the patients infected with PETRA lineage strains from the year 2008 (Fig. 1) were predominantly SSA, accounting for 33 of the 41 patients (80%) of known origins. The patients were from the following countries in SSA: Ethiopia (14 patients [34.1%]), Sudan (12 patients [29.3%]), and Eritrea (7 patients [17.1%]). The remaining countries were India (2 patients [4.9%]), Israel (3 patients [7.3%]), Nepal (2 patients [4.9%]), and Jordan (1 patient [2.4%]). The tuberculosis cases from patients from Sudan are recent cases, appearing only in the year 2008.
The top section of Fig. 1 shows the 10 PETRA lineage strains lacking locus ETR A (missing locus a [mla]). Nine of these strains (the exception being strain 0958-06, from a patient originating from Iran) have the same spoligotype and are of SSA origin. Five of these strains (strains 0433-00, 0937-08, 1158-08, 1178-00, and 2136-00 [indicated by gray shaded background]) have identical spoligotypes and MIRU-VNTR profiles, and all were isolated from patients originating from Ethiopia.
Continuing down the list of strains in Fig. 1, the next section shows 14 PETRA strains with MIRU-VNTR profiles identical to the profiles of the 5 identical mla PETRA strains, except that locus ETR A yields amplicons of copy number 4. Nine of these strains with amplicons of copy number 4 (A = 4) (strains 0735-08, 0882-08, 1180-06, 1407-00, 1468-00, 2095-00, 0787-08, 0382-08, and 0936-08) also have spoligotypes identical to those of the five identical mla PETRA strains and are indicated by gray shaded background. Seven of these nine strains are from patients of SSA origins (five from Ethiopia, one from Eritrea, and one from Sudan), and two have non-SSA origins, one from India and one from Jordan.
The next lower section of the Fig. 1 shows PETRA lineage strains with MIRU-VNTR profiles identical to the profiles of the five identical ETR A-deleted PETRA strains, except that locus ETR A yields amplicons of copy number 6. Four of these A = 6 strains (strains 0132-08, 0543-07, 0875-06, and 0448-08 [indicated by gray shaded background]) also have spoligotypes identical to those of the five identical ETR A-deleted PETRA strains and nine identical A = 4 PETRA strains. These four strains are of SSA origin (three strains from Ethiopia and one from Sudan).
In summary, the 18 strains indicated by gray shaded background in Fig. 1 all share MIRU-VNTR typing profiles that vary only at locus ETR A, and all share identical spoligotypes. Sixteen of the 18 strains (89%) are from patients of SSA origins, 13 from Ethiopia, 2 from Sudan, and 1 from Eritrea. It is worth noting for those new to MIRU-VNTR copy number notation that copy number 0 is in standard usage reserved for amplicons with no repeats (68), but the assigned usage of copy number 0 for the absence of an amplicon as in this paper greatly facilitates the manipulation of MIRU-VNTR data by Excel and Bionumerics algorithms.
Continuing down Fig. 1, the next section presents 13 PETRA lineage strains with identical spoligotypes (MST spoligo group D [Fig. 5]), 11 of which have identical MIRU-VNTR types (MST MIRU group AL), and 2 (strains 0327-08 and 0851-06) of which have very similar MIRU-VNTR typing profiles to the majority (MST MIRU groups AJ and AX, respectively [Fig. 4]). These 13 strains constitute a separate “small” subpopulation in both MIRU-VNTR type and spoligotype MSTs (Fig. 4, MST MIRU-VNTR type identity groups AL, AJ, and AX, and Fig. 5, MST spoligotype identity group D, plus two other identity groups of one profile each). The spoligotyping profiles in the MST identity group D are identical to the profile published for a large cluster observed in the SSA country Malawi (the MIRU-VNTR profiles were not published) (29). In Israel, 12 of the 13 PETRA lineage strains with the D spoligotype came from patients of SSA origins: 10 from Ethiopia, 1 from Eritrea, and 1 from Sudan, and 1 strain was from a patient of non-SSA origin, Israel.
The last section of Fig. 1 shows the remaining strains listed in alphanumeric order of their MIRU-VNTR MST groups (displayed in Fig. 4). Strain 0151-08 (patient origin is Sudan) has a spoligotype that lacks more than the usual amount of missing spacers and thus approaches the Beijing spoligotype. This is visualized in Fig. 5 by the MST spoligotype identity circle N (the MST group of strain 0151-08) being the closest neighbor to MST identity circle M (the Beijing family spoligotype MST group). On the other hand, the MIRU-VNTR identity circle (A2 for strain 0151-08) shown in Fig. 4 presents strain 0151-08 as a part of the major PETRA lineage subpopulation.
Three Beijing family members (strains 0419-04, 0674-06, and 0653-08 [Table 2]) were included for comparison in the clustering shown in Fig. 2 and 3 and the MSTs shown in Fig. 4 and 5.
All three Beijing strains were found in the same classical Beijing spoligotype MST identity group M shown in Fig. 5. Beijing strain 0419-04 is a classic Beijing strain (39, 40) belonging to our RFLP typing cluster 14 (Table 2 and Fig. 2), which is identical to the previously reported RFLP typing F cluster (50). It is labeled “A3” in Fig. 4, separate from the main large MIRU-VNTR MST population in Fig. 4. Beijing strain 0674-06 is a PETRA-like Beijing in its MIRU-VNTR profile (Table 2 and Fig. 2). It is labeled “A8,” separate from the “small” MIRU-VNTR MST subpopulation in Fig. 4. Beijing strain 0653-08 is a PETRA-like Beijing strain in its MIRU-VNTR profile (Table 2 and Fig. 2). It is labeled “A7” and importantly is included in the “large” PETRA lineage MIRU-VNTR MST subpopulation in Fig. 4, although at the perimeter of the tree.
Figure 2 shows the Bionumerics version 3.5-generated dendrogram (categorical coefficient, UPGMA) of the 79 PETRA family MIRU-VNTR profiles, along with their associated spoligotypes and partial RFLP typing profiles. Profiles from three Beijing family strains (strains 0419-04, 0653-08, and 0674-06) were included for comparison. By comparison with the MIRU-VNTRplus database (5) and published MIRU-VNTR profiles and spoligotyping profiles (3, 6, 7, 61, 62), we determined that our PETRA spoligotypes are identical to those found in the CAS spoligotype lineage and that our PETRA consensus MIRU-VNTR types (Table 1 and Fig. 1) therefore constitute a lineage of CAS MIRU-VNTR types. Our 16-locus MIRU-VNTR panel yielded nine MIRU-VNTR clusters (identical MIRU-VNTR profiles), three of which clustered together different spoligotypes (clusters 2, 3, and 4 of Fig. 2). Therefore, the 16-locus panel is less discriminatory than RFLP for molecular typing of PETRA lineage strains, although it still may have value for identifying epidemiological links (8, 74). A 24-locus MIRU-VNTR panel has recently been reported to separate each of many different CAS spoligotypes (6). However, the fact that the 16-locus MIRU-VNTR panel gave nine clusters containing 52 strains, while clustering on spoligotypes (categorical coefficient, UPGMA, Fig. 3) yielded six clusters containing 67 strains shows that the 16-locus MIRU-VNTR typing is more discriminatory than the 43-spacer spoligotyping for clustering PETRA lineage strains. Thirty-two of the 43 PETRA lineage strains from 2008 (74%) were found in eight of the nine MIRU-VNTR clusters (Fig. 2). Each of the 32 clustered PETRA 2008 strains was found in cluster with at least one other PETRA strain from 2008 (Fig. 2). This is highly suggestive of recent transmission, but not necessarily transmission in Israel. Twenty-six of the 32 clustered PETRA 2008 strains (81%) were from patients of SSA origins (Fig. 1 and Fig. 2). As already noted, in 2008, there were 33 SSA origin PETRA strains; therefore, 79% (26 of 33) of these SSA origin PETRA strains were clustered by MIRU-VNTR typing (Fig. 1 and 2).
The MST calculated on 16-locus MIRU-VNTR typing results, using Bionumerics version 3.5 default parameters, is shown in Fig. 4. The PETRA lineage strains were found in one large subpopulation and one small subpopulation as already described concerning the results shown in Fig. 1. The 10 PETRA strains with deleted ETR A were not separated into a subpopulation different from the “wild-type” PETRA strains (capable of producing ETR A amplicons), but they did form their own “branch” of the larger subpopulation (Fig. 1 and 4, MST identity circles AC, AM, AS, A4, and BA). These results support the suitability of the 16-locus MIRU-VNTR panel for screening for PETRA lineage membership. One Beijing family member with a PETRA-like MIRU-VNTR profile (strain 0653-08, identity circle A7) was also included in the PETRA population. This is not necessarily an artifact, and its implications are considered below in the Discussion. On the other hand, this indicates that the 16-locus MIRU-VNTR panel will in rare cases give unclear results or even false-positive results with respect to PETRA versus Beijing assignation. It is not certain that even a 24-locus MIRU-VNTR screen (68, 80) could avoid this difficulty (31). Therefore, use of the 16-locus MIRU-VNTR panel (the original 12 standard loci, ETR loci A, B, and C, and QUB11b) necessitates complementary screening by 43-spacer spoligotyping for definitive identification as a PETRA lineage (this paper) or Beijing family (10, 39, 40, 77) strain.
The MST calculated on 43-spacer spoligotyping results using Bionumerics version 3.5 default parameters, is shown in Fig. 5. The spoligotype MST divided the PETRA strains into one large subpopulation and one small subpopulation as already described concerning the results shown in Fig. 1. This division can also be visualized in the Fig. 3 dendrogram of spoligotype clusters. These two populations were essentially equivalent to the two major subpopulations of the MST based on MIRU-VNTR profiles. As expected, the Beijing strains formed another separate subpopulation (Fig. 5, MST spoligotype identity circle M; also see Fig. 3). PETRA 43-spacer spoligotypes were clearly shown to be of CAS lineage (3, 6, 7, 58, 61, 62), as visualized in Fig. 3 and Fig. 5. Since MIRU-VNTR resolves MST subpopulations similar to the spoligotype-resolved MST subpopulations, it can be an important aid in determining a spoligotype lineage, such as the CAS lineage.
DISCUSSION
Discovery of PETRA.
Prospective screening of every new culture-positive Mycobacterium tuberculosis complex case in Israel in the year 2008 was a direct benefit of automatic, high-throughput, 16-capillary electrophoresis of multiplex panels of 16 MIRU-VNTR loci (4, 47, 65, 67, 68). In addition, retrospective screening had been undertaken (partially by agarose gel electrophoresis) of isolates submitted in previous years, primarily from the years 2000 (in which a complete retrospective RFLP typing screen of new pulmonary Mycobacterium tuberculosis was performed) and 2006 (a year of partial systematic retrospective MIRU-VNTR screening). This screening and especially the high-throughput MIRU-VNTR screening enabled discovery and characterization of a major contributor to new tuberculosis cases in Israel, responsible for about 20% (43 of 222) of new culture-positive cases in 2008. The contributor was found to be a new consensus MIRU-VNTR lineage (Table 1 and Fig. 1) of the central Asian (CAS) spoligotype lineage (5, 6, 61, 62) which was termed “PETRA” for “polymorphic ETR A.”
CAS spoligotypes: considerations and origins with a focus on the PETRA lineage.
Recently, other groups of investigators have reported that the CAS spoligotype lineage contributes to the tuberculosis burdens of various nations in the Middle East (attributed to population influx from Africa [3]), Africa (29, 58), and Asia (7, 33, 63). In Israel as reported in this paper, 77 (out of a total of 79) of the PETRA lineage strains were isolated from patients for whom information regarding country origin was available. Out of the 77 patients, 58 (75%) originated from SSA. Interestingly, other investigators reporting on possible venues for CAS genotypes did not observe significant numbers of CAS genotypes in their sample populations from Turkey (2, 21), Ethiopia (14), and East Asia (RD Rv1519 deletion confers immune subverting phenotype [49]) (77).
PETRA strains were isolated from both pulmonary and extrapulmonary samples. Extrapulmonary tuberculosis has been reported from CAS spoligotype MT (41). The role, if any, for PETRA lineage ETR A in extrapulmonary tuberculosis requires further work for elucidation.
Since the majority of our PETRA strains, 75%, originate from SSA countries, we anticipate that the distribution of PETRA is global, although we are the first to describe this lineage. Indeed, 25% of the PETRA strains have non-SSA origins (Fig. 1). A similar situation was recently described in which a Mycobacterium tuberculosis RD lineage was a major cause of tuberculosis in Rio de Janeiro, Brazil (43), and was subsequently found to be globally disseminated (27) as a sublineage of the Latin American-Mediterranean (LAM) Mycobacterium tuberculosis spoligotype lineage. MIRU-VNTR combined with spoligotyping was recently used to define a new Mycobacterium tuberculosis lineage responsible for over 40% of smear-positive pulmonary tuberculosis cases in Cameroon during the time period of the study (51).
Usually, screening for Beijing family members on the recommended loci (40) allows differentiation of Beijing and PETRA lineage members. However, certain strains give inconclusive results and require spoligotyping to resolve the issue. This is reasonable, as clustering on MIRU-VNTR loci (24 loci) has shown that Beijing and CAS lineages are closely related (4, 68). It has been suggested that the CAS lineage is the ancestor to the Beijing lineage (61). One of our PETRA strains (strain 0151-08) has a spoligotype that closely resembles that of Beijing members (Fig. 1, 2, 3, and 5); this spoligotype has been assigned to the CAS lineage (5).
Putative mechanism for deletion of ETR A in PETRA strains.
IS6110 has been inferred to insert into preferential nonrandom sites (23) and to mediate certain deletion polymorphisms in Mycobacterium tuberculosis (22, 24, 57). IS6110-mediated deletion provides a likely mechanism for the deletions of ETR A in M. tuberculosis RD Rv1917c in the IS6110 indel sequences submitted by Menendez et al. to GenBank, GI:71056981 and GI:71056983. The Menendez et al. submissions were sequences recovered from virulent clinical isolates and thus complement our findings that PETRA isolates with deleted ETR A remain virulent. However, the deletions of ETR A in PETRA lineage members must be characterized in sequence-specific detail before any firm conclusions can be reached concerning the mechanisms responsible for the deletions. IS6110-mediated deletion of the RvD2 region in clinical isolates did not attenuate virulence, and by inference, deletion of RvD2 was not responsible for attenuation of H37Ra versus the virulent H37Rv (42, 81). Therefore, attenuation of virulence very much depends on which sequences are deleted. For example, deletion (not necessarily involving IS6110) of RD1 from M. bovis BCG (13) is the primary cause of attenuation of BCG (34). The effect of IS6110 insertion is further complicated by the ability of IS6110 to provide a promoter sequence for the transcription of genes (9). Small differences in genotypes from strains isolated from the same patient (46, 57) may have IS6110-mediated associations (57).
From our limited RFLP typing data, it can be seen that PETRA lineage strains have many copies of IS6110 (profiles with many bands [Fig. 2 and 3]). However, it is interesting that despite the many different RFLP typing profiles with large numbers of bands (from strains with large numbers of chromosomal copies of IS6110), the PETRA MIRU-VNTR lineage is the first lineage and so far the only lineage to be shown to include a significant number of strains that lack ETR A.
Effect of the ETR A locus on the putative PPE34 protein.
The ETR A region is found in the gene Rv1917c which is a coding sequence for the putative transmembrane PPE34 protein (PPE stands for Pro-Pro-Glu) (17, 55, 79). Thus, our discovery of PETRA strains that delete the ETR A region suggests the intriguing, but currently speculative, possibility that the PETRA lineage is subject to evolutionary pressure on its putative PPE34 protein.
As reported in this work, PETRA is the first MIRU-VNTR lineage, a sublineage of the CAS spoligotype lineage, shown to be clearly predisposed toward chromosomal deletions of the Rv1917c ETR A locus (Table 2 and Fig. 1, 2, and 3). Unlike the PPE gene-associated deletion events that resulted in attenuated nonpathogenic M. bovis BCG (12, 13, 32), the deletion of putative PPE34-coding, Rv1917c ETR A-containing chromosomal regions reported herein left the virulence of these PETRA lineage members intact, although possibly modified. A total of 10 ETR A-deleted PETRA strains were observed, 5 during the prospective screen of 2008, 2 from 2006, and 3 from the year 2000. About 12% of the PETRA lineage members in 2008 (5 of 43) were missing locus ETR A (mla in Table 2 and Fig. 1). Loss of ETR A has immediate implications for standardized MIRU-VNTR epidemiological screening, for which ETR A is one of the recommended screening loci (68).
Previous works reported that the Rv1917c gene is transcribed (mRNA was detected) in M. tuberculosis (55, 79). However, to date, there is no evidence that the putative M. tuberculosis PPE34 protein is transmembrane and surface exposed, although indirect evidence with a recombinant PPE34 in M. smegmatis suggests this behavior (55). Bioinformatics prediction of PPE protein structure also suggests that the putative PPE34 when found in M. tuberculosis will be transmembrane and surface bound. Many of the MPTR PPE proteins have a predicted beta-barrel outer membrane protein structure (reviewed in reference 26). An obvious but important question is whether there is a difference in PPE34 expression, location, antigenic potential, or function between PETRA strains with ETR A deletions compared to the PETRA strains that possess ETR A. Zvi et al. (82) have reviewed antigenic and functional M. tuberculosis PPE proteins that could serve as vaccine candidates. It would be desirable to know whether putative PPE34 peptides are suitable candidates for vaccines.
The fact that in some PETRA lineage strains, the ETR A locus and flanking primers are deleted with the PETRA-associated deletion of part of the chromosomal region encompassed by M. tuberculosis RD Rv1917c results in the failure to detect the ETR A locus in epidemiologic MIRU-VNTR PCR screening. Deletion of the target region has been reported to cause the failure of a commercial assay to detect Mycobacterium tuberculosis DNA in sputum samples (28) and to cause negative results when attempting to detect the mtp40 sequence (plcA) by PCR (78). As a positive contribution, deletion profiling has been used to type Mycobacterium tuberculosis complex species (16, 35, 38) and also has shown the close relationship between the Beijing and CAS lineages (7).
The role of PPE proteins in antigenic variation of Mycobacterium tuberculosis has been strongly suspected at least since the publication of the whole-genome sequence of M. tuberculosis H37Rv (17). Whole-genome comparisons of fully sequenced strain H37Rv with fully sequenced strain H37Ra (81) and fully sequenced strain CDC 1551 (24) have shown that the PPE gene family significantly contributes to the observed polymorphism. In three other whole-genome comparison studies, technical issues related to methodology prevented informative results on PPE gene family members (36, 54, 71). The 68-member glycine-rich PPE (Pro-Pro-Glu motif) family was one of the new families revealed in the first published analysis of the complete genome of Mycobacterium tuberculosis H37Rv (17). The ETR A locus (25) is found in the translated open reading frame of the H37Rv Rv1917c gene coding for M. tuberculosis Rv1917c mRNA (55, 79) and for the putative PPE34 protein (17, 26, 55). Rv1917c has been assigned to the major polymorphic tandem repeat (MPTR) subfamily (17, 26). It has been suggested (26) that the PPE MPTR subfamily originated from a duplication from ESAT-6 (esx) gene cluster region 5. The ESAT-6 system is well-known for being deleted from M. bovis BCG in the RD1 deletion, which is largely responsible for the attenuation of BCG (12, 32). In addition, this provides the basis for a commercial test employing selected RD1 peptides for diagnosis of active tuberculosis (30). Mycobacterial “core” genes (45) and the Rv1917c gene (26) have been searched for in nontuberculous mycobacteria. Some apparent homology to Rv1917c was found in Mycobacterium asiaticum, Mycobacterium gordonae, Mycobacterium marinum, and Mycobacterium ulcerans, but no detailed comparisons were made to the M. tuberculosis Rv1917c gene (26).
Various PPE proteins have been shown to be immunogenic (for example, PPE41 [reviewed in reference 1], PPE44 [11], PPE55 [59], and PPE68 [20]), cell surface associated (for example, recombinant PPE34 in Mycobacterium smegmatis [55]), and secreted (for example, PPE41 [1]). Clusters of PE and PPE genes of Mycobacterium tuberculosis have been reported to be organized in operons (73). As already mentioned, PE (for example, PE-PGRS33 [70]) and PPE (for example, putative PPE34 [55]) proteins may be surface expressed and potentially contribute a large amount of the antigenic variation that affects immunity (17, 82). A recent paper reporting genome-wide transcriptional profiling of chronic tuberculosis (69) did not focus on the PE/PPE protein families. However, an earlier paper summarizing whole-genome Mycobacterium tuberculosis PE/PPE expression patterns under various environmental conditions (79), reported that expression of the gene for PPE34 was repressed on day 14 of stationary phase.
Previous reports on the structure, location, antigenic potential, and function of PPE proteins and the proclivity of their DNA coding regions to be associated with chromosomal deletions suggest that it is likely that the Rv1917c putative PPE34 protein will be transmembrane, surface exposed, antigenic, and functional. If further investigation shows this to be the case, then the chromosomal deletions of ETR A in RD Rv1917c could indicate that M. tuberculosis PETRA is under evolutionary pressure at PPE34. Such pressure could occur through normal immune surveillance or host genetic variability in innate immunity such as that recently documented for Toll-like receptors (48) or perhaps variability in other receptors, such as dectin-1 (72), or even other types of genetic variability (18). The possible interaction between host genotype and M. tuberculosis genotype has been noted by the authors of a recent article (44).
Conclusions.
Culture-positive tuberculosis in Israel includes two challenging groups of strains. These are (i) the multiple drug resistance-associated Beijing family strains (unpublished data), primarily from former Soviet Union countries, and (ii) the PETRA MIRU-VNTR lineage (this paper): non-MDR central Asian (CAS) spoligotype lineage M. tuberculosis, primarily from sub-Saharan African countries, causing about 20% of new cases of tuberculosis in Israel in 2008.
The genotypes associated with the 79 PETRA lineage strains presented in this paper strengthen the conviction that there is a large, ongoing spread (but not necessarily transmitted, in whole or in part, in Israel) of tuberculosis caused by the PETRA lineage.
Acknowledgments
This work was performed under the auspices of the Israel National Program for the Prevention and Control of Tuberculosis run by the Ministry of Health. We thank Daniel Chemtob, Head of the Department of Tuberculosis and AIDS, for his support.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Abdallah, A. M., T. Verboom, F. Hannes, M. Safi, M. Strong, D. Eisenberg, R. J. Musters, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, J. Luirink, and W. Bitter. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62:667-679. [DOI] [PubMed] [Google Scholar]
- 2.Aktas, E., T. Zozio, F. B. Comert, C. Kulah, O. Aydin, N. Rastogi, and C. Sola. 2007. A first insight into the genetic diversity and population structure of Mycobacterium tuberculosis in Zonguldak, Turkey. Clin. Microbiol. Infect. 14(1):55-59. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hajoj, S. A. M., T. Zozio, F. Al-Rabiah, V. Mohammad, M. Al-Nasser, C. Sola, and N. Rastogi. 2007. First insight into the population structure of Mycobacterium tuberculosis in Saudi Arabia. J. Clin. Microbiol. 45:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783-789. [DOI] [PubMed] [Google Scholar]
- 5.Allix-Béguec, C., D. Harmesen, T. Weniger, P. Supply, and S. Niemann. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allix-Béguec, C., M. Fauville-Dufaux, and P. Supply. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banu, S., S. V. Gordon, S. Palmer, R. Islam, S. Ahmed, K. M. Alam, S. T. Cole, and R. Brosch. 2004. Genotypic analysis of Mycobacterium tuberculosis in Bangladesh and prevalence of the Beijing strain. J. Clin. Microbiol. 42:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barlow, E. L., D. M. Gascoyne-Binzi, S. H. Gillespie, A. Dickens, S. Qamer, and P. M. Hawkey. 2001. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:2453-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-51. [DOI] [PubMed] [Google Scholar]
- 11.Bonanni, D., L. Rindi, N. Lari, and C. Garzelli. 2005. Immunogenicity of mycobacterial PPE44 (Rv2770c) in Mycobacterium bovis BCG-infected mice. J. Med. Microbiol. 54:443-448. [DOI] [PubMed] [Google Scholar]
- 12.Brodin, P., L. Majlessi, L. Marsollier, M. I. De Jonge, D. Bottai, C. Demangel, J. Hinds, O. Neyrolles, P. D. Butcher, C. Leclerc, S. T. Cole, and R. Brosch. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. D. Santos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 104:5596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruchfeld, J., G. Aderaye, I. B. Palme, B. Bjorvatn, S. Ghebremichael, S. Hoffner, and L. Lindquist. 2002. Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates from Ethiopian pulmonary tuberculosis patients with and without human immunodeficiency virus infection. J. Clin. Microbiol. 40:1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2007. World TB Day—March 24, 2007. MMWR Morb. Mortal. Wkly. Rep. 56:245.17380106 [Google Scholar]
- 16.Cole, S. T. 2002. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology 148:2919-2928. [DOI] [PubMed] [Google Scholar]
- 17.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 18.Cooke, G. S., S. J. Campbell, S. Bennett, C. Lienhardt, K. P. W. J. McAdam, G. Sirugo, O. Sow, P. Gustafson, F. Mwangulu, P. van Helden, P. Fine, E. G. Hoal, and A. V. S. Hill. 2008. Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am. J. Respir. Crit. Care Med. 178:203-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillbaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 20.Demangel, C., P. Brodin, P. J. Cockle, R. Brosch, L. Majlessi, C. Leclerc, and S. T. Cole. 2004. Cell envelope protein PPE68 contributes to Mycobacterium tuberculosis RD1 immunogenicity independently of a 10-kilodalton culture filtrate protein and ESAT-6. Infect. Immun. 72:2170-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durmaz, R., T. Zozio, S. Gunal, C. Allix, M. Fauville-Dufaux, and N. Rastogi. 2007. Population-based molecular epidemiological study of tuberculosis in Malatya, Turkey. J. Clin. Microbiol. 45:4027-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang, Z., D. T. Kenna, C. Doig, D. N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 2001. Molecular evidence for independent occurrence of IS6110 insertions at the same sites of the genome of Mycobacterium tuberculosis in different clinical isolates. J. Bacteriol. 183:5279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Bernter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 26.Gey van Pittius, N. C., S. L. Sampson, H. Lee, Y. Kim, D. van Helden, and R. M. Warren. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson, A. L., R. C. Huard, N. C. Gey van Pittius, L. C. O. Lazzarini, F. Driscoll, N. Kurepina, T. Zozio, C. Sola, S. M. Spindola, A. L. Kritski, D. Fitzgerald, K. Kremer, H. Mardassi, P. Chitale, J. Brinkworth, D. G. de Viedma, B. Gicuel, J. W. Pape, D. van Soolingen, B. N. Kreiswirth, R. M. Warren, P. D. van Helden, N. Rastogi, P. N. Suffys, J. Lapa e Silva, and J. L. Ho. 2008. Application of sensitive and specific molecular methods to uncover global dissemination of the major RDRio sublineage of the Latin American-Mediterranean Mycobacterium tuberculosis spoligotype family. J. Clin. Microbiol. 46:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilpin, C. M., D. J. Dawson, G. O'Kane, J. G. Armstrong, and C. Coulter. 2002. Failure of commercial ligase chain reaction to detect Mycobacterium tuberculosis DNA in sputum samples from a patient with smear-positive pulmonary tuberculosis due to a deletion of the target region. J. Clin. Microbiol. 40:2305-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glynn, J. R., A. C. Crampin, H. Traore, S. Chaguluka, D. T. Mwafulirwa, S. Alghamdi, B. M. M. Ngwira, M. D. Yates, F. D. Drobniewski, and P. E. M. Fine. 2008. Determinants of cluster size in large, population-based molecular epidemiology study of tuberculosis, northern Malawi. Emerg. Infect. Dis. 14:1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goletti, D., D. Vincenti, S. Carrara, O. Butera, F. Bizzoni, G. Bernardini, M. Amicosante, and E. Girardi. 2005. Selected RD1 peptides for active tuberculosis diagnosis: comparison of a gamma interferon whole-blood enzyme-linked immunosorbent assay and an enzyme-linked immunospot assay. Clin. Diagn. Lab. Immunol. 12:1311-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanekom, M., G. D. van der Spuy, N. C. Gey van Pittius, C. R. E. McEvoy, K. G. P. Hoek, S. L. Ndabambi, A. M. Jordaan, T. C. Victor, P. D. van Helden, and R. M. Warren. 2008. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J. Clin. Microbiol. 46:3338-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harboe, M., T. Oettinger, H. G. Wiker, I. Rosenkrands, and P. Andersen. 1996. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect. Immun. 64:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasan, T., M. Tanveer, A. Kanji, Q. Hasan, S. Ghebremichael, and R. Hasan. 2006. Spoligotyping of Mycobacterium tuberculosis isolates from Pakistan reveals predominance of Central Asian strain 1 and Beijing isolates. J. Clin. Microbiol. 44:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 100:12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huard, R. C., L. C. de Oliveira Lazzarini, W. R. Butler, D. van Soolingen, and J. L. Ho. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology—a guide for the level III laboratory. Centers for Disease Control, Public Health Service, U.S. Department of Health and Human Services, Atlanta, GA.
- 38.Kong, Y., M. D. Cave, L. Zhang, B. Foxman, C. F. Marrs, J. H. Bates, and Z. H. Yang. 2006. Population-based study of deletions in five different genomic regions of Mycobacterium tuberculosis and possible clinical relevance of the deletions. J. Clin. Microbiol. 44:3940-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kremer, K., B. K. Y. Au, P. C. W. Yip, R. Skuce, P. Supply, K. M. Kam, and D. van Soolingen. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lari, N., L. Rindi, R. Cristofani, N. Rastogi, E. Tortoli, and C. Garzelli. 2009. Association of Mycobacterium tuberculosis complex isolates of BOVIS and Central Asian (CAS) genotypic lineages with extrapulmonary disease. Clin. Microbiol. Infect. 15:538-543. [DOI] [PubMed] [Google Scholar]
- 42.Lari, N., L. Rindi, and C. Garzelli. 2001. Identification of one insertion site of IS6110 in Mycobacterium tuberculosis H37Ra and analysis of the RvD2 deletion in M. tuberculosis clinical isolates. J. Med. Microbiol. 50:805-811. [DOI] [PubMed] [Google Scholar]
- 43.Lazzarini, L. C. O., R. C. Huard, N. L. Boechat, H. M. Gomes, M. C. Oelemann, N. Kurepina, E. Shashkina, F. C. Q. Mello, A. L. Gibson, M. J. Virginio, A. G. Marsico, W. R. Butler, B. N. Kreiswirth, P. N. Suffys, J. R. Lapa e Silva, and J. L. Ho. 2007. Discovery of a novel Mycobacterium tuberculosis lineage that is a major cause of tuberculosis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 45:3891-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma, X., Y. Liu, B. B. Gowen, E. A. Graviss, A. G. Clark, and J. M. Musser. 2007. Full-exon resequencing reveals Toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One 2:e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 46.Martin, A., M. Herranz, M. J. R. Serrano, E. Bouza, and D. G. de Viedma. 2007. Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol. 7:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazars, E., S. Lesjean, A.-L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Misch, E. A., and T. R. Hawn. 2008. Toll-like receptor polymorphisms and susceptibility to human disease. Clin. Sci. 114:347-360. [DOI] [PubMed] [Google Scholar]
- 49.Newton, S. M., R. J. Smith, K. A. Wilkinson, M. P. Nicol, N. J. Garton, K. J. Staples, G. R. Stewart, J. R. Wain, A. R. Martineau, S. Fandrich, T. Smallie, B. Foxwell, A. Al-Obaidi, J. Shafi, K. Rajakumar, B. Kampmann, P. W. Andrew, L. Ziegler-Heitbrock, M. R. Barer, and R. J. Wilkinson. 2006. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc. Natl. Acad. Sci. USA 103:15594-15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niemann, S., S. Rusch-Gerdes, and E. Richter. 1997. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J. Clin. Microbiol. 35:3015-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, J. Thonnon, V. Vincent, and M. C. Gutierrez. 2004. Molecular characteristics of strains of the Cameroon family, the major group of Mycobacterium tuberculosis in a country with a high prevalence of tuberculosis. J. Clin. Microbiol. 42:5029-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richter, E., M. Weizenegger, S. Rusch-Gerdes, and S. Niemann. 2003. Evaluation of genotype MTBC assay for differentiation of clinical Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 41:2672-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 54.Salamon, H., M. Kato-Maeda, P. M. Small, J. Drenkow, and T. R. Gingeras. 2000. Detection of deleted genomic DNA using a semiautomated computational analysis of GeneChip data. Genome Res. 10:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampson, S. L., P. Lukey, R. M. Warren, P. D. van Helden, M. Richardson, and M. J. Everett. 2001. Expression, characterization and subcellular localization of the Mycobacterium tuberculosis PPE gene Rv1917c. Tuberculosis 81:305-317. [DOI] [PubMed] [Google Scholar]
- 56.Sampson, S. L., R. M. Warren, M. Richardoson, T. C. Victor, A. M. Jordaan, G. D. van der Spuy, and P. D. Helden. 2003. IS6110-mediated deletion polymorphism in the direct repeat region of clinical isolates of Mycobacterium tuberculosis. J. Bacteriol. 185:2856-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sampson, S. L., M. Richardson, P. D. van Helden, and R. M. Warren. 2004. IS6110-mediated deletion polymorphism in isogenic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 42:895-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharaf-Eldin, G. S., N. S. Saeed, M. E. Hamid, A. M. Jordaan, G. D. Van der Spuy, R. M. Warren, P. D. Van Helden, and T. C. Victor. 2002. Molecular analysis of clinical isolates of Mycobacterium tuberculosis collected from patients with persistent disease in the Khartoum region of Sudan. J. Infect. 44:244-251. [DOI] [PubMed] [Google Scholar]
- 59.Singh, K. K., Y. Dong, S. A. Patibandla, D. N. McMurray, V. K. Arora, and S. Laal. 2005. Immunogenicity of the Mycobacterium tuberculosis PPE55 (Rv3347c) protein during incipient and clinical tuberculosis. Infect. Immun. 73:5004-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 61.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 62.Streicher, E. M., T. C. Victor, G. van der Spuy, C. Sola, N. Rastogi, P. D. van Helden, and R. M. Warren. 2007. Spoligotype signatures in the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 45:237-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun, Y. J., R. Bellamy, A. S. G. Lee, S. T. Ng, S. Ravindran, S. Y. Wong, C. Locht, P. Supply, and N. I. Paton. 2004. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J. Clin. Microbiol. 42:1986-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun, Y. J., A. S. Lee, S. T. Ng, S. Ravindran, K. Kremer, R. Bellamy, S. Y. Wong, D. van Soolingen, P. Supply, and N. I. Paton. 2004. Characterization of ancestral Mycobacterium tuberculosis by multiple genetic markers and proposal of genotyping strategy. J. Clin. Microbiol. 42:5058-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Supply, P. 2005. Multilocus variable number tandem repeat genotyping of Mycobacterium tuberculosis. Technical guide. Institut de Biologie/Institut Pasteur de Lille, Lille, France.
- 66.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 67.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Talaat, A. M., S. K. Ward, C.-W. Wu, E. Rondon, C. Tavano, J. P. Bannantine, R. Lyons, and S. A. Johnston. 2007. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 189:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talarico, S., M. D. Cave, C. F. Marrs, B. Foxman, L. Zhang, and Z. Yang. 2005. Variation of the Mycobacterium tuberculosis PE_PGRS33 gene among clinical isolates. J. Clin. Microbiol. 43:4954-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. L. G. de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsoni, S. V., and G. D. Brown. 2008. Beta-glucose and dectin-1. Ann. N. Y. Acad. Sci. 1143:45-60. [DOI] [PubMed] [Google Scholar]
- 73.Tundup, S., Y. Akhter, D. Thiagarajan, and S. E. Hasnain. 2006. Clusters of PE and PPE genes of Mycobacterium tuberculosis are organized I operons: evidence that PE Rv2431c is co-transcribed with PPE Rv2430c and their gene products interact with each other. FEBS Lett. 20:1285-1293. [DOI] [PubMed] [Google Scholar]
- 74.van Deutekom, H., P. Supply, P. E. de Haas, E. Willery, S. P. Hoijng, C. Locht, R. A. Coutinho, and D. van Soolingen. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 43:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Soolingen, D., and K. Kremer. 1999. Manual for restriction fragment length polymorphism (RFLP) typing. RIVM, Bilthoven, The Netherlands.
- 77.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vera-Cabrera, L., M. A. Hernandez-Vera, O. Welsh, W. M. Johnson, and J. Castro-Garza. 2001. Phospholipase region of Mycobacterium tuberculosis is a preferential locus for IS6110 transposition. J. Clin. Microbiol. 39:3499-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voskuil, M. I., D. Schnappinger, R. Rutherford, Y. Liu, and G. K. Schoolnik. 2004. Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis 84:256-262. [DOI] [PubMed] [Google Scholar]
- 80.Wada, T., S. Maeda, A. Hase, and K. Kobayashi. 2007. Evaluation of variable numbers of tandem repeat as molecular epidemiological markers of Mycobacterium tuberculosis in Japan. J. Med. Microbiol. 56:1052-1057. [DOI] [PubMed] [Google Scholar]
- 81.Zheng, H., L. Lu, B. Wang, S. Pu, X. Zhang, G. Zhu, W. Shi, L. Zhang, H. Wang, S. Wang, G. Zhao, and Y. Zhang. 2008. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PloS One 3:e2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zvi, A., N. Ariel, J. Fulkerson, J. C. Sadoff, and A. Shafferman. 2008. Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med. Genomics 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]