Abstract
A large outbreak of hemorrhagic fever with renal syndrome (HFRS) occurred in the winter of 2006-2007 in a region southeast of Moscow in Central European Russia. Of the 422 patients with HFRS investigated in this study, 58 patients were found to be infected by Puumala virus, whereas as many as 364 were infected by Dobrava-Belgrade virus (DOBV). Early serum samples from 10 DOBV-infected patients were used for nucleic acid amplification, which was successful for 5 patients. Molecular analyses demonstrated that the causative hantavirus belongs to the DOBV-Aa genetic lineage, which is carried by the striped field mouse (Apodemus agrarius) as the natural reservoir host. Neutralization assays with convalescent-phase sera from these patients confirmed infection by DOBV-Aa; related viruses, such as the Dobrava-Slovenia virus (DOBV-Af) and the Dobrava-Sochi virus (DOBV-Ap), were neutralized at lower efficiencies. The clinical courses of the 205 patients enrolled in the study were found to be mostly mild to moderate; however, an unexpectedly high fraction (27%) of patients exhibited severe illness. One patient died from kidney failure and showed symptoms of generalized subcutaneous hemorrhage. The results provide molecular, serodiagnostic, and clinical evidence that DOBV-Aa is a common pathogen in East Europe that causes large outbreaks of HFRS.
Hemorrhagic fever with renal syndrome (HFRS) is an infectious disease caused by hantaviruses in Asia and Europe (16, 20) and most probably in Africa (13; our unpublished results). In the Americas, the autochthonous hantavirus species cause the hantavirus (cardio)pulmonary syndrome (24, 26). Hantaviruses carry trisegmented negative-strand RNA genomes and belong to the family Bunyaviridae. Different rodent species are known to be the natural hosts of the currently identified hantaviruses pathogenic for humans (16, 20). The viruses are transmitted to humans by aerosolized rodent excreta. Shrews were shown to be alternative hantavirus hosts (14, 31); however, the clinical significance of shrew-borne hantaviruses is still unclear.
The demonstration of viral nucleotide sequences in specimens derived from patients with HFRS gives a clue to the involvement of a particular virus in the infection and allows its detailed molecular phylogenetic characterization. Because of the short-term viremia typical for human hantavirus infections, viral genetic material can best be amplified from blood or plasma during the first days after the onset of clinical symptoms (16). In contrast, serotyping of neutralizing antibodies is suggested to be most significant when convalescent-phase sera from the patients are used (19).
HFRS cases in the European administrative regions of Russia are contributing to 97% of the total number of HFRS cases registered in Russia. Until recently, the majority of HFRS cases in European Russia were caused by Puumala virus (PUUV) and considerably fewer (mainly sporadic cases) were caused by Dobrava-Belgrade virus (DOBV) infections. However, during the period from 1991 to 2006, three large DOBV-associated HFRS outbreaks were registered in the central regions of European Russia (34). A detailed investigation of the 2001-2002 outbreak revealed that the striped field mouse (Apodemus agrarius) was the virus reservoir and that the A. agrarius-borne DOBV lineage (DOBV-Aa) was the causative infectious agent (15, 33).
Three DOBV genetic lineages, DOBV-Aa (6, 11, 15, 33), DOBV-Af (1, 2, 3, 7, 9, 22, 23), and DOBV-Ap (15, 33), have so far been demonstrated by molecular methods to cause HFRS in humans. The Saaremaa virus can be considered a fourth DOBV genetic lineage (21); however, it has not been detected in HFRS patients. Moreover, it has been claimed to be a separate virus species by its discoverers (25).
In the winter of 2006-2007, a large outbreak of HFRS involving 661 registered cases (according to official statistics of the Russian Ministry of Public Health) occurred in the Lipetsk region and the neighboring regions of Voronezh, Tambov, and Ryazan of Central European Russia. In the study described here, we investigated 422 of the cases to identify the causative pathogenic agent. By serodiagnostic methods, 364/422 patients were found to be infected by DOBV. For 205 of these patients, clinical data were collected and the specific clinical course of the disease was defined. Moreover, we amplified viral genetic material from the acute-phase sera of five patients and characterized the virus type as DOBV-Aa.
MATERIALS AND METHODS
Primary serodiagnostics.
About 1,500 serum samples from persons with suspected HFRS (more than 1,000 consecutive and late-convalescent-phase serum samples from seropositive patients as well as consecutive serum samples from seronegative persons) were screened by immunofluorescence assay (IFA) for the presence of hantavirus antibody, as described before (5); slides with combined antigens from Vero E6 cells infected with PUUV, DOBV, Hantaan virus (HTNV), and Seoul virus (SEOV) were used as substrates. For primary serotyping of hantavirus antibodies, all positive sera were titrated to the endpoint by IFA with slides containing monovalent antigens of all four viruses listed above. If the antibody titers to DOBV and PUUV were equal or the difference was only twofold, a late-convalescent-phase serum sample from the same patient was further serotyped by the focus-reduction neutralization test (FRNT).
Virus titration and FRNT.
For confirmation and serotyping reasons, IFA-positive serum samples obtained more than a month after disease onset were tested by FRNT. The viral stocks prepared from cell culture supernatants of infected Vero E6 cells were titrated by the focus assay with protein A-peroxidase conjugate-DAB (3,3′-diaminobenzidine)-NiCl2. For FRNT, human convalescent-phase serum samples were diluted serially in twofold steps, mixed with an equal volume of the respective virus containing 30 to 100 focus-forming units of this virus, incubated for 1 h at 37°C or overnight at 4 to 6°C, and then used to inoculate the cells. Methylcellulose (0.6%) was used to overlay the cells. After 6 to 7 days of incubation, the cells were fixed with 98% ethanol for 30 min (fresh ethanol was added after 10 min). Human convalescent-phase sera specific for the corresponding virus (DOBV, PUUV, HTNV, or SEOV) were used to detect the viral antigen as described above. A reduction in the number of foci of at least 80% was considered the criterion for virus neutralization (15).
RT-PCR, cloning, and sequencing.
For RNA extraction, 0.1 ml of patient whole blood was added to 0.9 ml of Trizol reagent (Invitrogen, Karlsruhe, Germany), and the mixture was stored at −80°C. Total RNA was then extracted by a standard protocol (4). RNA was reverse transcribed by use of a SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR; Invitrogen), according to the manufacturer's instructions. Hantavirus RNA was detected by a nested RT-PCR (5 Prime GmbH, Hamburg, Germany) with two S-segment-specific primer pairs, as described before (30). A nested RT-PCR with DOBV M-segment-specific primers (primer DOB-M11, 5′-CTCCGCAAGAAATAGCAGT-3′; primer M2029R, 5′-CCATGIGCITTITCIKTCCA-3′; primer M907F, 5′-GTTGCAACTTATTCAATTG-3′; primer M1990R, 5′-TCIGMTGCISTIGCIGCCCA-3′) was also performed to amplify the M-segment fragment. Cloning and sequencing of the amplified products were performed as described previously (12). At least three recombinant plasmids were sequenced in both directions to determine the consensus sequences.
Sequence and phylogenetic analyses.
The sequences obtained were aligned and further analyzed by the maximum-likelihood phylogenetic method with the TREE-PUZZLE software package (27) and the neighbor-joining phylogenetic method with the PAUP* software package (32), as described previously (15).
Nucleotide sequence accession numbers.
The sequences of the following viruses obtained in the present study have been deposited in the GenBank database under the indicated accession numbers: DOBV/Lip2/hu, GQ205393 and GQ205399; DOBV/Lip4/hu, GQ205394; DOBV/Lip5/hu, GQ205395; DOBV/Lip6/hu, GQ205396; DOBV/Lip7/hu, GQ205397 and GQ205400; and DOBV/Lip17/Aa, GQ205398.
For the comparisons, the sequence data for the following virus isolates were obtained from the GenBank sequence database (the GenBank accession numbers are given in parentheses): DOBV/SK/Aa (AY961615 and AY961616), DOBV/East Slovakia/862Aa/97 (AJ269550, AY168578), DOBV/Esl/29Aa/01 (AY533118), DOBV/Esl/81Aa/01 (AY533120), DOBV/Kurkino/44Aa/98 (AJ131672), DOBV/Kurkino/53Aa/98 (AJ131673), Saaremaa/160V (AJ009773, AJ009774), DOBV/Lipetsk/Aa (EU188452, EU188453), DOBV/Omsk172/Aa (EU562989), DOBV/Omsk189/Aa (EU562990), DOBV/Omsk180/Aa (EU562991), DOBV/GER/293/Aa (GQ205401), DOBV/GER239/Aa (GQ205405), DOBV/GER118/Aa (GQ205407), DOBV/GER1064/Aa (GQ205404), DOBV/H169 (AY533117), DOBV/Saaremaa/90Aa/97 (AJ009775), DOBV/Slovenia (L41916, L33685), DOBV/East Slovakia/400Af/98 (AY168576, AY168577), DOBV/Ano-Poroia/9Af/99 (AJ410615, AJ410616), DOBV/Ano-Poroia/13Af/99 (AJ410619), DOBV/Sochi/Ap (EU188449, EU188450), HTNV strain 76-118 (M14626, M14627), HTNV strain Z10 (AB027108, AB027076), HTNV strain SC-1 (AY675353, AY675349), SEOV strain 80-39 (AY273791, NC_005237), SEOV strain L99 (AF288299, AF035833), SEOV strain Z37 (AF187082, AF187081), Sangassou virus strain SA14 (DQ268650), and Thailand virus strain Thai749 (L08756).
RESULTS
Outbreak characteristics.
The 2006-2007 HFRS outbreak occurred in four regions of Central European Russia (the Lipetsk, Voronezh, Ryazan, and Tambov regions) and involved 661 officially reported cases. These four regions are located southeast of the city of Moscow, and the whole outbreak area had a north-south dimension of about 630 km and a west-east dimension of about 385 km. Serological screening of 422 patients with HFRS (investigated in this study) revealed 364 and 58 infections caused by DOBV and PUUV, respectively. For 205 DOBV-infected patients, we were able to collect more detailed information about the patients' age, gender, and clinical course (Table 1). Specimens from 10 patients with serologically confirmed DOBV infection from two districts in the south of the Lipetsk region and an adjacent district of the Voronezh region were taken for further molecular analysis. The residences of the 10 patients in these three districts were located about 70 to 110 km south or southeast of the city of Lipetsk.
TABLE 1.
Age, gender, and clinical course of DOBV-infected patients with HFRS from the 2006-2007 outbreak monitoreda
| Characteristic | % of persons |
|---|---|
| Gender | |
| Males | 62.4 |
| Females | 37.6 |
| Age (yr) | |
| ≤14 | 6.9 |
| 15-19 | 8.3 |
| 20-29 | 12.1 |
| 30-39 | 22.5 |
| 40-49 | 20.4 |
| 50-59 | 14.9 |
| ≥60 | 14.9 |
| Severity of clinical courseb | |
| Mild | 34.6 |
| Moderate | 38.6 |
| Severe | 26.8 |
Data are for 205 patients.
According to the criteria presented previously (15, 17), which consider the presence and the extent of the typical clinical symptoms (fever, headache, vision disturbance, abdominal pain, hemorrhagic signs, oliguria) and the range of values of the most important laboratory markers (maximum blood urea and creatinine levels, leukocyte count).
Characterization of viral nucleotide sequences.
By using blood from 10 patients (patients Lip1 to Lip10) sampled as early as 1 to 9 days after the onset of disease, a nested RT-PCR (based on S-segment sequences) was performed. Specific nucleotide sequences were amplified from 5/10 samples. The laboratory codes of these patients/blood specimens/nucleotide sequences were Lip2/hu, Lip4/hu, Lip5/hu, Lip6/hu, and Lip7/hu.
Table 2 shows the nucleotide and amino acid identity values for the 559-bp sequence of the viral S segment investigated. The identities between the five sequences of human origin ranged from 89.6 to 100% at the nucleotide level and 98.3 to 100% at the amino acid level. The sequence identity of the group of Lip2/hu to Lip7/hu sequences with the sequence of cell culture virus isolate DOBV-Lipetsk/Aa and an amplified sequence of a DOBV isolate from Kurkino (which is located about 250 km from the region investigated here) was 90.1 to 91.7% at the nucleotide level and 97.8 to 99.4% at the amino acid level. Slightly lower levels of identity were found with cell culture isolate DOBV-SK/Aa (which originated from a place in Slovakia that is about 1,400 km from the Lipetsk region). Even less similar (83.8 to 87.2% at the nucleotide level, 96.2 to 97.8% at the amino acid level) were sequences of the virus lineage DOBV-Af (from A. flavicollis), the Saaremaa virus (which is known to resemble DOBV-Af in its S-segment sequence [8, 10]), and the virus lineage DOBV-Ap (from A. ponticus).
TABLE 2.
Comparisons of identities of virus sequences from the patients
| Gene region and strain | % Identity with straina: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| S segment (559 bp) | |||||||||||
| 1. Lip2/hu | 89.9 | 99.6 | 90.3 | 90.3 | 90.5 | 90.5 | 88.5 | 86.7 | 86.9 | 85.1 | |
| 2. Lip4/hu | 98.3 | 89.6 | 98.9 | 98.9 | 90.6 | 90.6 | 89.9 | 85.5 | 85.1 | 83.8 | |
| 3. Lip5/hu | 100 | 98.3 | 89.9 | 89.9 | 90.1 | 90.1 | 88.5 | 86.7 | 87.2 | 85.1 | |
| 4. Lip6/hu | 99.4 | 98.9 | 99.4 | 100 | 91.7 | 91.7 | 90.6 | 85.8 | 85.3 | 84.2 | |
| 5. Lip7/hu | 99.4 | 98.9 | 99.4 | 100 | 91.7 | 91.7 | 90.6 | 85.8 | 85.3 | 84.2 | |
| 6. Lipetsk/Aa | 98.9 | 98.3 | 98.9 | 99.4 | 99.4 | 99.2 | 90.8 | 87.1 | 86.7 | 86.7 | |
| 7. Kurk44/Aa | 98.3 | 97.8 | 98.3 | 98.9 | 98.9 | 99.4 | 90.5 | 87.1 | 87.1 | 86.4 | |
| 8. SK/Aa | 98.9 | 98.3 | 98.9 | 99.4 | 99.4 | 98.9 | 98.3 | 84.7 | 84.7 | 85.6 | |
| 9. Slo/Af | 97.8 | 96.2 | 97.8 | 97.3 | 97.3 | 96.7 | 96.2 | 97.8 | 87.8 | 90.5 | |
| 10. Saa160/Aa | 96.7 | 96.2 | 96.7 | 96.2 | 96.2 | 95.6 | 95.1 | 96.7 | 96.7 | 85.3 | |
| 11. Sochi/Ap | 96.2 | 96.2 | 96.2 | 96.7 | 96.7 | 96.2 | 95.6 | 97.3 | 97.3 | 95.6 | |
| M segment (1,044 bp) | |||||||||||
| 1. Lip2/hu | 99.1 | 93.2 | 86.3 | 83.4 | 85.9 | 80.3 | |||||
| 5. Lip7/hu | 99.4 | 93.4 | 86.5 | 83.1 | 85.9 | 80.3 | |||||
| 6. Lipetsk/Aa | 99.1 | 98.5 | 86.3 | 82.8 | 85.3 | 80.3 | |||||
| 8. SK/Aa | 97.4 | 96.8 | 97.6 | 82.2 | 86.0 | 79.4 | |||||
| 9. Slo/Af | 94.8 | 94.2 | 94.8 | 95.1 | 80.8 | 80.1 | |||||
| 10. Saa160/Aa | 96.8 | 96.2 | 96.5 | 97.1 | 96.2 | 78.3 | |||||
| 11. Sochi/Ap | 90.4 | 90.2 | 90.7 | 91.0 | 93.3 | 90.7 | |||||
For each gene region, the data above the diagonal spaces are nucleotide sequence identities and the data below the diagonal spaces are amino acid sequence identities.
It was also possible to amplify and analyze viral M-segment sequences from two patients (patients Lip2 and Lip7). The levels of identity between Lip2/hu and Lip7/hu for the 1,044-bp sequence analyzed were 99.1% and 99.4% at the nucleotide and amino acid levels, respectively (Table 2). Cell culture isolate DOBV-Lipetsk/Aa was found to be most similar. DOBV-SK/Aa, DOBV-Af, Saaremaa virus, and DOBV-Ap exhibited lower levels of nucleotide and amino acid identity to Lip2/hu and Lip7/hu (80.1 to 86.5%, respectively, at the nucleotide level and 90.2 to 97.4%, respectively, at the amino acid level).
Phylogenetic analyses.
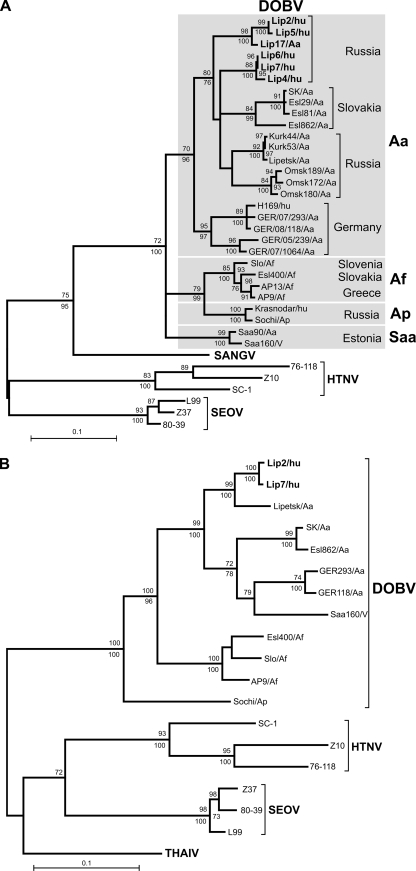
Evolutionary trees based on both partial S- and M-segment sequences were constructed by the maximum-likelihood and neighbor-joining phylogenetic methods. Analysis of the S-segment sequences (Fig. 1A) showed that all patient-associated sequences belonged to the DOBV-Aa lineage. Within that lineage, they formed two separate clusters: Lip2/hu and Lip5/hu on the one hand and Lip4/hu, Lip6/hu, and Lip7/hu on the other. Interestingly, both clusters were markedly distant from the Lipetsk/Aa virus, isolated during the HFRS outbreak in the Lipetsk region in 2001-2002. On the other hand, the Lip2/hu and Lip5/hu sequences shared a common ancestor with the Lip17/Aa sequence, obtained from an A. agrarius mouse captured in the outbreak region. Altogether, two main clades could be recognized within the DOBV-Aa lineage; one was formed by the sequences from Russia and Slovakia and the second was formed by the sequences from Germany (Fig. 1A). Analysis of the M segment (Fig. 1B) confirmed the association of the novel patient-associated sequences with the DOBV-Aa lineage.
FIG. 1.
Maximum-likelihood phylogenetic trees of DOBV showing the phylogenetic placement of patient-associated sequences from the Lipetsk 2006-2007 outbreak (in boldface) on the basis of the partial S-segment nucleotide sequence (559 bp, positions 377 to 935) (A) and the partial M-segment nucleotide sequence (1,044 bp, positions 925 to 1,968) (B). In the analysis of the S segment, for which more data were available than for theMsegment, the different DOBV lineages are marked by gray boxes. The scale bar indicates an evolutionary distance of 0.1 substitutions per position in the sequence. The trees were computed with the TREE-PUZZLE package by using the Tamura-Nei evolutionary model. The values at the tree branches are the PUZZLE support values. The values below the branches are the bootstrap values of the corresponding neighborjoining tree (Tamura-Nei evolutionary model) calculated with PAUP* software from 1,000 bootstrap replicates. No differences in the tree topology were found between the maximum-likelihood and the neighbor-joining trees in the case of the M-segment tree. In the case of the S-segment trees, only minor differences that did not change the conclusions were found (trifurcations in the DOBV-Aa clade were further resolved, SK/Aa clustered with Esl29/Aa, H168/hu clustered with GER/08/118/Aa, and the DOBV-Saa clade formed an outgroup to the DOBV-Aa group in the neighborjoining tree; but this topology was not statistically supported). SANGV, Sangassou virus; THAIV, Thailand virus.
Serotyping of convalescent-phase sera.
As mentioned above, the DOBV-Aa sequences from five patients were amplified and characterized. Convalescent-phase sera were obtained from these patients, and neutralizing antibodies were typed by FRNT under biosafety level 3 conditions. The assay determined the neutralization of three viruses prevalent in European Russia: PUUV, the Lipetsk virus isolate (DOBV-Aa), and the Sochi isolate (DOBV-Ap). In addition, three reference hantaviruses—HTNV, SEOV, and the Belgrade virus isolate (DOBV-Af)—were included in the analysis. The results of this study clearly show that DOBV-Aa was the most efficiently neutralized by the patients' sera (Table 3). Compared with the next most closely related virus lineages, DOBV-Ap from A. ponticus and DOBV-Af from A. flavicollis, the difference in neutralizing antibody titers was at least fourfold. Less related viruses, such as PUUV, HTNV, and SEOV, were neutralized even less. The results of serotyping agree with the molecular findings that DOBV-Aa caused HFRS in the patients.
TABLE 3.
Characterization of patient sera by IFA and FRNT
| Serum sample no.a | Day after disease onset | IFA (reciprocal antibody titer): |
FRNT (reciprocal endpoint titer of neutralizing antibody) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PUUV | DOBV-Aa Lipetsk | SEOV | HTNV | PUUV | DOBV-Ap Sochi | DOBV-Aa Lipetsk | DOBV-Af Bel-1 | SEOV | HTNV | ||
| 5342 Lip2 | 2 | <16 | 64 | 16 | 32 | ||||||
| 8 | <16 | 4,096 | 1,024 | 2,048 | |||||||
| 36 | 256 | 8,192 | 4,096 | 2,048 | <40 | 1,280 | 10,240 | 640 | 40 | 320 | |
| 5357 Lip4 | 1 | <16 | 256 | 64 | 128 | ||||||
| 5 | <16 | 1,024 | 512 | 256 | |||||||
| 98 | 256 | 4,096 | 1,024 | 2,048 | <40 | 640 | 5,120 | 640 | 80 | 40 | |
| 5309 Lip5 | 9 | <16 | 4,096 | 1,024 | 2,048 | ||||||
| 72 | 1,024 | 4,096 | 2,048 | 4,096 | <40 | 1,280 | 5,120 | 320 | 80 | 320 | |
| 5310 Lip6 | 6 | <16 | 4,096 | 1,024 | 1,024 | ||||||
| 91 | 64 | 2,048 | 1,024 | 512 | <40 | 320 | 2,560 | 320 | 40 | 40 | |
| 5618 Lip7 | 7 | <16 | 2,048 | 512 | 512 | ||||||
| 104 | 1,024 | 4,096 | 2,048 | 4,096 | <40 | 320 | 2,560 | 320 | 160 | 320 | |
| 5268 | 6 | 128 | 8,192 | 1,024 | 2,048 | ||||||
| 142 | 2,048 | 4,096 | 1,024 | 1,024 | <40 | 1,280 | 10,240 | 2,560 | 40 | 80 | |
| 5284 | 5 | 256 | 4,096 | 2,048 | 1,024 | ||||||
| 48 | 2,048 | 4,096 | 1,024 | 512 | <40 | 1,280 | 2,560 | 640 | 40 | 40 | |
| 5305 | 7 | 1,024 | 4,096 | 1,024 | 512 | ||||||
| 32 | 1,024 | 4,096 | 2,048 | 4,096 | <40 | 1,280 | 5,120 | 1,280 | 160 | 320 | |
| 5564 | 4 | 2,048 | 8,192 | 2,048 | 2,048 | ||||||
| 85 | 1,024 | 4,096 | 512 | 512 | <40 | 640 | 2,560 | 5,120 | 40 | 160 | |
| 5576 | 9 | 256 | 1,024 | 512 | 256 | ||||||
| 97 | 512 | 4,096 | 2,048 | 1,024 | <40 | 2,560 | 5,120 | 1,280 | 160 | 80 | |
| 5597 | 3 | <16 | 64 | 32 | 32 | ||||||
| 7 | 64 | 4,096 | 1,024 | 1,024 | |||||||
| 68 | 4,096 | 8,192 | 2,048 | 4,096 | <40 | 2,560 | 10,240 | 2,560 | 320 | 320 | |
Samples with “LipN” following the serum sample number are those for which the hantavirus nucleotide sequences were obtained in this study. An additional six serum samples were randomly chosen.
Moreover, an additional six randomly chosen serum samples were included in the serotyping FRNT (Table 3). In agreement with the results described above, four serum samples showed at least fourfold higher neutralizing titers against the Lipetsk virus isolate than against the other hantaviruses. In two cases, the difference was only twofold. Surprisingly, in one of these two cases, the highest titer was found against prototypical virus DOBV-Af.
Clinical characteristics and case fatality.
The most prominent clinical data for 205 DOBV-infected patients are summarized in Table 4. According to the standard criteria used in the Russian Federation (15, 17), the clinical severity of the illness of the patients with HFRS was classified as mild, moderate, and severe in 34.6%, 38.6%, and 26.8% of the patients, respectively (Table 1). The most meaningful clinical symptoms and biochemical findings for the five patients whose diagnoses were achieved by molecular methods are presented for each patient in Table 5.
TABLE 4.
Clinical outcome of HFRS for 205 DOBV-infected patients
| Selected clinical criterion | % of patients | Avg duration (days) |
|---|---|---|
| Fever | 100 | 6.8 ± 3.2 |
| Abdominal pain | 23.4 | 5 ± 3.3 |
| Vision disturbance | 2.4 | 4.2 ± 3.1 |
| Vomiting | 1.9 | 2.7 ± 1.4 |
| Nausea | 24.4 | 3.5 ± 1.5 |
| Diarrhea | 5.4 | 2.6 ± 1.1 |
| Hyperemia of the face | 22 | 5.6 ± 2.4 |
| Hemorrhagic sclerae | 14.6 | 7.9 ± 4.1 |
| Hypertension | 28.8 | 3.5 ± 1.8 |
| Liver enlargement | 3.4 | 4.4 ± 1.6 |
| Oliguria < 500 ml | 53.7 | 3.5 ± 1.9 |
| Anuria < 200 ml | 0.5 | 2.0 ± 0.0 |
| Serum creatinine elevation | 90.1 | NDa |
ND, not determined.
TABLE 5.
Clinical characteristics of the five molecularly diagnosed patients
| Characteristic | Result for patient no.: |
||||
|---|---|---|---|---|---|
| Lip2 5342a | Lip4 5357a | Lip5 5309b | Lip6 5310b | Lip7 5618c | |
| Age (yr)/sexd | 65/M | 69/F | 57/F | 7/F | 37/M |
| Clinical course | |||||
| Clinical severity | Mild | Moderate | Moderate | Mild | Moderate |
| Acute onset | + | + | + | + | + |
| Avg duration (days) ofe: | |||||
| Fever | 4 | 6 | 6 | 3 | 6 |
| Headache | 2 | 11 | 15 | 4 | 9 |
| Vision disturbance | − | − | − | − | − |
| Muscle and joint pain | − | − | 11 | 2 | 4 |
| Low back pain | 4 | − | 11 | − | 9 |
| Vomiting | − | 2 | − | − | − |
| Oliguria < 500 ml | − | 2 | 2 | − | 2 |
| Hypotonia | − | − | 2 | − | − |
| Hypertonia | − | − | 2 | − | − |
| Polyuria | − | − | 3 | 2 | − |
| Blood analysisf | |||||
| Maximum blood urea (2.8-7.2 mmol/liter) | 5.3 | 11.2 | 10.8 | 8.5 | 11.4 |
| Maximum blood creatinine (42-97 μmol/liter) | 104.2 | 164.5 | 271.8 | 115 | 180.0 |
| Maximum white blood cell count (3.5 × 109-10.1 × 109/liter) | 5.4 | 13.3 | 7.6 | 6.2 | 13.4 |
| Minimum white blood cell count (3.5 × 109-10.1 × 109/liter) | 4.6 | 4.3 | 4.0 | 4.2 | 6.5 |
| Minimum platelet count (140 × 109-360 × 109/liter) | NDg | ND | 160.0 | 230.0 | ND |
Lipetsk region, Dobrinskyi district.
Lipetsk region, Usmanskyi district.
Voronezh region, Vernehavskyi district (directly on the border with the Lipetsk region).
M, male; F, female.
−, the adverse event did not occur.
The range of normal values is given in parentheses.
ND, not done.
One fatal case was registered during the 2006-2007 HFRS outbreak, which led us calculate a case fatality index of 1/364, or 0.3%. The deceased patient was a 34-year-old man who had lived in a village in the Lipetsk region. Two days after the onset of disease he had a high fever, weakness, and vomiting and he was admitted to the hospital in serious condition. He developed hypotension and acute renal failure with oliguria and later developed generalized subcutaneous hemorrhage and hematuria. Laboratory findings included leukocytosis, thrombocytopenia, and a high serum creatinine concentration of 480 μmol/liter. DOBV-specific antibody titers, as determined by IFA, ranged from 1:256 on day 4 to 1:1,024 on day 10, whereas the anti-PUUV titers remained constant (1:16). Despite the provision of intensive care, including hemodialysis and artificial pulmonary ventilation, the patient died.
DISCUSSION
The central area of European Russia is known as a zone of endemicity where DOBV outbreaks sporadically occur (15, 18, 33). The recent outbreak of 2006-2007, which had about 600 registered cases of HFRS, has for the first time allowed the characterization of the outbreak in a comprehensive study that included both molecular and serological diagnostic methods and the clinical characterization of patients. The data let us conclude that at least a majority of the cases in this latest HFRS outbreak in the Lipetsk region of Russia (European Russia, southeast of Moscow) were caused by the transmission of DOBV-Aa, a virus carried by A. agrarius as the natural host.
The serological screening of 422 patients with HFRS from the outbreak gave indications of DOBV infection in 364 cases and PUUV infection in 58 cases. The number of PUUV infections detected in this geographical area also appears to be higher than the number detected in other years (E. A. Tkachenko, unpublished data). One could speculate that the climate conditions and food resources in the preceding year were particularly favorable for the growth of rodent populations. This could have led to the overpopulation not only of the DOBV-Aa host, A. agrarius, but also of other rodent species, including the PUUV host, Myodes glareolus, and to the more frequent virus spillover to humans. The awareness of the large hantavirus outbreak, however, might have led to a better compilation of HFRS cases, including those caused by PUUV infections, which usually escape detection.
Serum samples from 10 of the DOBV-infected patients were obtained very early after the onset of disease (days 1 to 9) and were used for nucleic acid amplification. Virus RNA from five patients was amplified and investigated. The first DOBV-Aa sequence to have been characterized originated from a patient in northern Germany (11), but this is the first molecular analysis of DOBV-Aa from a group of patients. As found in the sequence comparisons (Table 2) and molecular phylogenetic analyses (Fig. 1A), the S-segment sequences formed two different clusters. A previously determined sequence of the Lipetsk/Aa isolate (15) even formed a third cluster with the Kurkino strains. This indicates a high degree of sequence variability in a small geographical area of about 75 km in diameter, suggesting the long-term survival and the evolution of DOBV-Aa in this region.
The serum samples from the five patients whose diagnosis was made by molecular analysis were used for the typing of neutralizing antibodies to confirm whether FRNT is also able to determine the causative agent in the case of such closely related viruses. DOBV-Aa was best neutralized by the sera of all five patients, followed by neutralization of the related Sochi virus (DOBV-Ap), which is also prevalent in Russia, and the Belgrade virus (DOBV-Af). Among an additional six randomly selected serum samples, five serum samples were also best neutralized by DOBV-Aa. It should be mentioned that in a previous study with sera from patients in Central Europe, we found less pronounced neutralization differences between DOBV-Aa and DOBV-Af (12). The better performance of FRNT in this study could probably be explained by the fact that the local virus (the Lipetsk/Aa virus isolate for patients from the Lipetsk region) was used to type the sera, while in the previous study, the more distant SK/Aa isolate from Slovakia was employed to type the sera of German patients.
Our results confirm our previous observations (12, 15, 28, 29, 30) that DOBV-Aa infections mainly cause mild or moderate clinical courses of HFRS. However, as in the previous DOBV-Aa outbreak in Central European Russia (in the winter of 2001-2002 [15]), we again found an unexpectedly high number of severe cases of HFRS (27% in both outbreaks). In the 2001-2002 outbreak, the case fatality rate was determined to be about 0.9% (15). In the current study, the case fatality rate was found to be 0.3%. In contrast, infections by the other DOBV lineages, DOBV-Ap and DOBV-Af, mostly cause moderate or severe clinical courses of HFRS and the cases fatality rates are about 6% (15) and 12% (3, 12), respectively. It would be worthwhile to investigate whether the different clinical severities of infections by DOBV-Aa, DOBV-Af, and DOBV-Ap that have so far been observed in Germany and European Russia (DOBV-Aa infections), the Balkan region of Europe (DOBV-Af infections), and southern European Russia (DOBV-Ap infections) are caused by the various genetic susceptibilities of the human resident populations and/or subtle but functionally important genetic differences between the DOBV members. In the latter case, it would be interesting to determine which dissimilarities in the genetic makeup of the viruses are responsible for their different virulences for humans.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (grant KR1293/2-4) and the Slovak scientific grant agency VEGA (grant 2/0189/09).
We thank Christina Grübel for help with the graphics.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Antoniadis, A., A. Stylianakis, A. Papa, S. Alexiou-Daniel, A. Lampropoulos, S. T. Nichol, C. J. Peters, and C. F. Spiropoulou. 1996. Direct genetic detection of Dobrava virus in Greek and Albanian patients with hemorrhagic fever with renal syndrome. J. Infect. Dis. 174:407-410. [DOI] [PubMed] [Google Scholar]
- 2.Avsic-Zupanc, T., S. Y. Xiao, R. Stojanovic, A. Gligic, G. van der Groen, and J. W. LeDuc. 1992. Characterization of Dobrava virus: a hantavirus from Slovenia, Yugoslavia. J. Med. Virol. 38:132-137. [DOI] [PubMed] [Google Scholar]
- 3.Avsic-Zupanc, T., M. Petrovec, P. Furlan, R. Kaps, F. Elgh, and A. Lundkvist. 1999. Hemorrhagic fever with renal syndrome in the Dolenjska region of Slovenia—a 10-year survey. Clin. Infect. Dis. 28:860-865. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Dzagurova, T., E. Tkachenko, R. Slonova, L. Ivanov, E. Ivanidze, S. Markeshin, A. Dekonenko, B. Niklasson, and A. Lundkvist. 1995. Antigenic relationships of hantavirus strains analysed by monoclonal antibodies. Arch. Virol. 140:1763-1773. [DOI] [PubMed] [Google Scholar]
- 6.Dzagurova, T., E. Tkachenko, V. Bashkirtsev, N. Okulova, N. Apekina, A. Bernshtein, N. Korotina, A. Smirnov, Y. Yunicheva, I. Hodyakova, G. Slusareva, D. Trankvilevskii, and O. Medvdkina. 2008. Isolation and typing of hantavirus strains caused HFRS in European Russia, p. 142-150. In Medical virology. Proceedings of the Chumakov Institute of Poliomyelitis and Viral Encephalitides, vol. XXV. Chumakov Institute of Poliomyelitis and Viral Encephalitides, Moscow, Russia. (In Russian.)
- 7.Gligic, A., N. Dimkovic, S. Y. Xiao, G. J. Buckle, D. Jovanovic, D. Velimirovic, R. Stojanovic, M. Obradovic, G. Diglisic, J. Micic, D. M. Asher, J. W. LeDuc, R. Yanagihara, and D. C. Gajdusek. 1992. Belgrade virus: a new hantavirus causing severe hemorrhagic fever with renal syndrome in Yugoslavia. J. Infect. Dis. 166:113-120. [DOI] [PubMed] [Google Scholar]
- 8.Henttonen, H., P. Buchy, Y. Suputtamongkol, S. Jittapalapong, V. Herbreteau, J. Laakkonen, Y. Chaval, M. Galan, G. Dobigny, N. Charbonnel, J. Michaux, J. F. Cosson, S. Morand, and J. P. Hugot. 2008. Recent discoveries of new hantaviruses widen their range and question their origins. Ann. N. Y. Acad. Sci. 1149:84-89. [DOI] [PubMed] [Google Scholar]
- 9.Jakab, F., J. Sebok, E. Ferenczi, G. Horváth, and G. Szucs. 2007. First detection of Dobrava hantavirus from a patient with severe haemorrhagic fever with renal syndrome by SYBR green-based real time RT-PCR. Scand. J. Infect. Dis. 39:902-906. [DOI] [PubMed] [Google Scholar]
- 10.Klempa, B., H. A. Schmidt, R. Ulrich, S. Kaluz, M. Labuda, H. Meisel, B. Hjelle, and D. H. Kruger. 2003. Genetic interaction between distinct Dobrava hantavirus subtypes in Apodemus agrarius and A. flavicollis in nature. J. Virol. 77:804-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klempa, B., M. Schutt, B. Auste, M. Labuda, R. Ulrich, H. Meisel, and D. H. Kruger. 2004. First molecular identification of human Dobrava virus infection in central Europe. J. Clin. Microbiol. 42:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klempa, B., M. Stanko, M. Labuda, R. Ulrich, H. Meisel, and D. H. Kruger. 2005. Central European Dobrava hantavirus isolate from a striped field mouse (Apodemus agrarius). J. Clin. Microbiol. 43:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klempa, B., E. Fichet-Calvet, E. Lecompte, B. Auste, V. Aniskin, H. Meisel, C. Denys, L. Koivogui, J. ter Meulen, and D. H. Kruger. 2006. Hantavirus in African wood mouse, Guinea. Emerg. Infect. Dis. 12:838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klempa, B., E. Fichet-Calvet, E. Lecompte, B. Auste, V. Aniskin, H. Meisel, P. Barrière, L. Koivogui, J. ter Meulen, and D. H. Kruger. 2007. Novel hantavirus sequences in shrew, Guinea. Emerg. Infect. Dis. 13:520-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klempa, B., E. A. Tkachenko, T. K. Dzagurova, Y. V. Yunicheva, V. G. Morozov, N. M. Okulova, G. P. Slyusareva, A. Smirnov, and D. H. Kruger. 2008. Hemorrhagic fever with renal syndrome caused by 2 lineages of Dobrava hantavirus, Russia. Emerg. Infect. Dis. 14:617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruger, D. H., R. Ulrich, and A. Lundkvist. 2001. Hantavirus infections and their prevention. Microbes Infect. 3:1129-1144. [DOI] [PubMed] [Google Scholar]
- 17.Leshchinskaia, E. V., E. A. Tkachenko, E. V. Ryltseva, V. A. Petrov, S. M. Ianovskii, T. A. Gasanova, V. F. Bistrovskii, T. V. Mogila, G. S. Kovalskii, V. Z. Priven, R. Muhametov, A. N. Bondarenko, O. F. Brizgacheva, and T. I. Astahova. 1990. Characteristics of endemic foci of hemorrhagic fever with renal syndrome in various regions of the USSR. Vopr. Virusol. 35:42-45. (In Russian.) [PubMed] [Google Scholar]
- 18.Lundkvist, A., N. Apekina, Y. Myasnikov, O. Vapalahti, A. Vaheri, and A. Plyusnin. 1997. Dobrava hantavirus outbreak in Russia. Lancet 350:781-782. [DOI] [PubMed] [Google Scholar]
- 19.Lundkvist, A., M. Hukic, J. Horling, M. Gilljam, S. Nichol, and B. Niklasson. 1997. Puumala and Dobrava viruses causes hemorrhagic fever with renal syndrome in Bosnia-Hercegovina: evidence of highly cross-neutralizing antibody responses in early patient sera. J. Med. Virol. 53:51-59. [PubMed] [Google Scholar]
- 20.Maes, P., J. Clement, I. Gavrilovskaya, and M. van Ranst. 2004. Hantaviruses: immunology, treatment, and prevention. Viral Immunol. 17:481-497. [DOI] [PubMed] [Google Scholar]
- 21.Maes, P., B. Klempa, J. Clement., J. Matthijnssens, D. C. Gajdusek, D. H. Krüger, and M. van Ranst. 2009. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect. Genet. Evol. 9:813-820. [DOI] [PubMed] [Google Scholar]
- 22.Papa, A., and A. Antoniadis. 2001. Hantavirus infections in Greece—an update. Eur. J. Epidemiol. 17:189-194. [DOI] [PubMed] [Google Scholar]
- 23.Papa, A., K. Nemirov, H. Henttonen, J. Niemimaa, A. Antoniadis, A. Vaheri, A. Plyusnin, and O. Vapalahti. 2001. Isolation of Dobrava virus from Apodemus flavicollis in Greece. J. Clin. Microbiol. 39:2291-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters, C. J., and A. S. Khan. 2002. Hantavirus pulmonary syndrome: the new American hemorrhagic fever. Clin. Infect. Dis. 34:1224-1231. [DOI] [PubMed] [Google Scholar]
- 25.Plyusnin, A., A. Vaheri, and A. Lundkvist. 2006. Saaremaa hantavirus should not be confused with its dangerous relative, Dobrava virus. J. Clin. Microbiol. 44:1608-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 28.Schutt, M., P. Gerke, H. Meisel, R. Ulrich, and D. H. Kruger. 2001. Clinical characterization of Dobrava hantavirus infections in Germany. Clin. Nephrol. 55:371-374. [PubMed] [Google Scholar]
- 29.Schutt, M., H. Meisel, D. H. Krüger, R. Ulrich, K. Dalhoff, and C. Dodt. 2004. Life-threatening Dobrava hantavirus infection with unusually extended pulmonary involvement. Clin. Nephrol. 62:54-57. [DOI] [PubMed] [Google Scholar]
- 30.Sibold, C., R. Ulrich, M. Labuda, A. Lundkvist, H. Martens, M. Schutt, P. Gerke, K. Leitmeyer, H. Meisel, and D. H. Kruger. 2001. Dobrava hantavirus causes hemorrhagic fever with renal syndrome in Central Europe and is carried by two different Apodemus mice species. J. Med. Virol. 63:258-267. [PubMed] [Google Scholar]
- 31.Song, J. W., L. J. Baek, C. S. Schmaljohn, and R. Yanagihara. 2007. Thottapalayam virus, a prototype shrew-borne hantavirus. Emerg. Infect. Dis. 13:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swofford, D. L. 2002. PAUP* (phylogenetic analysis using parsimony) (* and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 33.Tkachenko, E., A. Bernshteyn, T. Dzagurova, V. Bashkirtsev, N. Sedova, A. Malkin, E. Gorbachkova, A. Balakirev, S. Drozdov, I. Sicora, I. Shukina, S. Savelyev, I. Bogatova, D. Trankvilevskiy, M. Chubirko, R. Nurgaleeva, G. Minin, I. Zagidullin, A. Ivanova, V. Zhukov, V. Morozov, S. Khaibulina, and S. Morzunov. 2005. Comparative analysis of epidemic HFRS outbreaks caused by Puumala and Dobrava viruses. Epidemiol. Vaccine Prophylaxis 4:28-34. (In Russian.) [Google Scholar]
- 34.Tkachenko, E., T. Dzagurova, V. Bashkirtsev, A. Bernshtein, N. Apekina, N. Sedova, N. Okulova, N. Korotina, P. Nabatnikov, A. Malkin, V. Morozov, Y. Yunicheva, B. Klempa, and D. H. Kruger. 2007. Epidemiology of HFRS in European Russia, p.17. Abstr. VII Int. Conf. HFRS, HPS, and hantaviruses.