Abstract
We report on a novel strategy for the detection of mRNA targets derived from Cryptosporidium parvum oocysts by the use of oligonucleotide-gold nanoparticles. Gold nanoparticles are functionalized with oligonucleotides which are complementary to unique sequences present on the heat shock protein 70 (HSP70) DNA/RNA target. The results indicate that the presence of HPS70 targets of increasing complexity causes the formation of oligonucleotide-gold nanoparticle networks which can be visually monitored via a simple colorimetric readout measured by a total internal reflection imaging setup. Furthermore, the induced expression of HSP70 mRNA in Cryptosporidium parvum oocysts via a simple heat shock process provides nonenzymatic amplification such that the HSP70 mRNA derived from as few as 5 × 103 purified C. parvum oocysts was successfully detected. Taken together, these results support the use of oligonucleotide-gold nanoparticles for the molecular diagnosis of cryptosporidiosis, offering new opportunities for the further development of point-of-care diagnostic assays with low-cost, robust reagents and simple colorimetric detection.
The obligate intracellular protozoan parasites of the genus Cryptosporidium cause the diarrheal illness termed cryptosporidiosis (24). Approximately 30% of adults in high-income countries and nearly all adults in resource-poor countries have serologic evidence of prior infection with this organism. However, only a small minority of people have ever been diagnosed with clinical disease. This is thought to stem from the underutilization and poor sensitivities of current diagnostic tests. For example, in the outbreak in Milwaukee, WI, in which over 400,000 people developed diarrhea associated with contamination of the water supply with Cryptosporidium parasites, fewer than 1,000 cases were confirmed on the basis of stool examination (although nearly all individuals tested developed antibodies to the organism) (3). Studies employing sensitive assays (e.g., PCR) have greatly increased the ability to recognize the burden of cryptosporidiosis. For example, Cryptosporidium DNA was found in the stools of 20% of South African children with diarrhea and three-fourths of Ugandan AIDS patients with diarrhea (18, 22).
The detection of oocysts in stool samples has served as the traditional basis for diagnosis. The organisms are usually overlooked by direct examination in the absence of special stains. Acid-fast stains (including modified the Ziehl-Neelsen and auramine O stains) or fluorescent dyes coupled to monoclonal antibodies (for the immunofluorescent-antibody test [IFAT]) are used in the standard assays used in clinical laboratories. The sensitivity of stool examination by acid-fast staining remains poor and requires an oocyst concentration of over 500,000 per ml in formed stools (23), with fewer cases being detected by acid-fast staining than by fluorescent methods (1, 12). IFAT has been reported to be up to 10 times more sensitive than acid-fast staining (5, 7, 10). Currently, the IFAT staining method is the “gold standard” for the microscopic examination of infected stool samples (2).
Microscopy-based methods are increasingly being replaced with techniques that rely on molecular recognition for the specific targeting of the pathogens of interest. The enzyme-linked immunosorbent assay (13) and PCR (16) techniques have further improved the sensitivity and specificity of detection compared to those obtained by microscopic examination. However, these newer methods have limited applicability to point-of-care use or low-resource settings due to reagent and instrumentation costs, infrastructure needs, and the need for operator training. Rapid diagnostic tests for Cryptosporidium parasites, such as lateral-flow assays (8), have not been proven to be reliable alternatives to the existing methods.
Recently, a simple colorimetric detection method that relies on the distance-dependent optical properties of oligonucleotide-functionalized gold nanoparticles has been described (6). This strategy offers a simple readout (a color change that can be visually monitored) and utilizes low-cost and robust reagents that have promising potential for point-of-care molecular diagnostics. Previous work has demonstrated that the color change associated with the target-induced formation of oligonucleotide-gold aggregates can be effectively monitored by spotting the solution on a glass slide illuminated via total internal reflection (20). The limit of detection of this approach for the detection of purified DNA from methicillin (meticillin)-resistant Staphylococcus aureus was previously shown to be <105 bacteria (20).
In the study described here, we demonstrate the detection of Cryptosporidium parvum oocysts using oligonucleotide-functionalized gold nanoparticles targeted against heat shock protein 70 (HSP70) DNA and RNA and this novel detection strategy. One advantage of targeting HSP70 is the ability to induce mRNA amplification by the use of a simple heating process that does not involve more complicated PCR-based amplification. We hypothesize that simple amplification (via the heat shock process) of targets present at very low copies (low zeptomole concentrations) in biological samples will facilitate the detection of the desired targets that are present at levels within the limit of detection of the gold aggregation assay.
MATERIALS AND METHODS
Detection strategy.
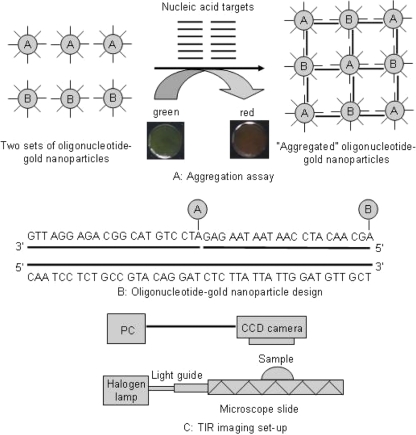
Figure 1A shows the overall detection strategy used in the study. That strategy relies on the formation of oligonucleotide-gold networks upon hybridization to target DNA or RNA. In this scheme, two sets of gold nanoparticles are functionalized with oligonucleotides that are complementary to adjacent sequences present on the nucleic acid target (Fig. 1B). If the target sequence is added to a solution containing the oligonucleotide-functionalized gold nanoparticles, complementary binding occurs, forming a network of oligonucleotide-gold nanoparticles. The target-induced aggregation results in a colorimetric or spectral shift due to the change in surface plasmon resonance as a result of the close proximity of the gold nanoparticles. The color change (from green to red) can then be visualized when the samples are spotted on a waveguide coupled to a total internal reflection setup (Fig. 1C).
FIG. 1.
Schematic diagram of the oligonucleotide-gold nanoparticle aggregation assay and detection strategy. (A) Aggregation of the nanoparticle probes in the presence of HSP70 target DNA or RNA to form oligonucleotide-gold nanoparticle networks and the green to red color shift associated with this transition; (B) the sequence of the oligonucleotide-gold nanoparticle probes designed for detection of the HSP70 targets from C. parvum; (C) schematic diagram of the total internal reflection imaging setup used for detection of assay results.
Preparation of oligonucleotide-functionalized gold nanoparticles.
Colloidal gold nanoparticles (particle diameter, 50 nm; 4.5 × 1010 particles per ml) were obtained from Ted Pella, Inc. (Redding, CA). Two sets of modified oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and had the sequences shown in Table 1. The oligonucleotide probes were functionalized with thiol groups (carbon 6 S-S), hexaethylene glycol (18-atom), and poly(A) (15-nucleotide) spacers, followed by a 21-nucleotide sequence complementary to adjacent sequences of the HSP70 nucleic acid targets. To functionalize the gold nanoparticles, the modified oligonucleotides (50 μl, 100 μM) were reduced with an ∼20-fold molar excess of Tris(2-carboxyethyl)phosphine hydrochloride (Pierce Chemicals, Rockford, IL) for 30 min and added to 1.0 ml of 50-nm-diameter colloidal gold nanoparticles. The oligonucleotide-gold conjugates were slowly aged by increasing the concentration of phosphate-buffered saline (PBS) for 24 h until a 1× PBS concentration was reached. Excess oligonucleotides were removed by centrifugation (two times) of the conjugates at 12,000 × g for 30 min. The oligonucleotide-gold probes were resuspended in 1× PBS to a final concentration of 100 pM. The concentration of oligonucleotide-gold probes was determined from UV-visible spectroscopy measurements (Cary 50 spectrometer; Varian, Inc., Palo Alto, CA) at a wavelength of 520 nm. These absorbance values were then related to the nanoparticle probe concentration by using Beer's law (A = ɛbc, where A is absorbance, ɛ at 520 nm is 1.6 × 1010 M−1 cm−1, b is the path length of the sample, and c is the concentration of compound in solution) and the molar absorptivity for 50-nm gold nanoparticles (20). The surface loading of oligonucleotides per 50-nm gold nanoparticle was not directly measured in this study but has previously been established to be approximately 50 pmol/cm2 by the use of similar conjugation protocols (11). The oligonucleotide-coated gold probes were stable at room temperature for at least 6 months.
TABLE 1.
Nucleic acid sequences of the probes used to functionalize gold nanoparticles for the aggregation assay and primers used to obtain target DNA and RNA
| Probe or primer | Sequencec |
|---|---|
| Cryptosporidium probe A | 5′-thiol-A15-PEG-ATCCTGTACGGCAGAGGATTG-3′a |
| Cryptosporidium probe B | 5′-thiol-A15-PEG-AGCAACATCCAATAATAAGAG-3′ |
| Cryptosporidium 42-ntb DNA | 5′-CAATCCTCTGCCGTACAGGATCTCTTATTATTGGATGTTGCT-3′ |
| Cryptosporidium 42-nt RNA | 5′-CAAUCCUCUGCCGUACAGGAUCUCUUAUUAUUGGAUGUUGCU-3′ |
| Cryptosporidium primers (DNA target) | 5′-TCCTCTGCCGTACAGGATCTCTTA-3′ (1217 forward) |
| 5′-TGCTGCTCTTACCAGTACTCTTATCA-3′ (1537 reverse) | |
| Entamoeba primers (control target) | 5′-GAGAGCAGCAAAGAACCAAATTGC (175 forward) |
| 5′-TCTGCAGCAATCTTATCAGCA-3′ (1709 reverse) |
PEG, polyethylene glycol.
nt, nucleotide.
The numbers in parentheses correspond to the nucleotide number in the full sequence (HSP-0) a which the primer is located.
Preparation of DNA and RNA targets with increasing complexity.
The oligonucleotide-gold probes designed for the detection of the Cryptosporidium HSP70 targets were evaluated by testing the assay with various targets of increasing complexity, including short 42-nucleotide single-stranded DNA (ssDNA) and single-stranded RNA (ssRNA) synthetic targets, double-stranded DNA (dsDNA) targets obtained by reverse transcription-PCR (RT-PCR) amplification, RNA targets obtained by total RNA purification, and mRNA targets purified from C. parvum oocysts. The 42-nucleotide ssDNA and ssRNA targets were obtained from Integrated DNA Technologies and have the sequences shown in Table 1. The synthetic targets are present in the HSP70 sequence starting at position 1214. The Cryptosporidium DNA target (312 bp) corresponding to HSP70 (GenBank accession no. U69698) was obtained by RT-PCR with the Superscript one-step system (Invitrogen) and a 0.5-μg template of total RNA purified from 5 × 106 C. parvum parasites. The conditions for the RT-PCR were 50°C for 15 min; 94°C for 2 min; 40 cycles at 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s; and 1 cycle at 72°C for 10 min. The Cryptosporidium RNA target was obtained by total RNA purification of 5 × 106 C. parvum parasites. The DNA and RNA targets from Entamoeba dispar were used as control targets. The Entamoeba DNA target (1,535 bp) corresponding to HSP70 (GenBank accession no. XM_001735280.1) was obtained by RT-PCR from total RNA purified from E. dispar parasites. The conditions for the RT-PCR were 50°C for 15 min; 94°C for 2 min; 35 cycles at 94°C for 15 s, 55°C for 30 s, 72°C for 2 min; and 1 cycle at 72°C for 10 min. The primers used in the RT-PCR for the detection of C. parvum and E. dispar parasites are shown in Table 1.
Lastly, total mRNA targets from C. parvum oocysts were purified by the standard protocol provided with the Dynabeads mRNA direct kit (Invitrogen). A 10-μl aliquot of various concentrations of oocysts (0 to 1.0 × 106 oocysts) was initially heat shocked at 42°C for 20 min. The lysis buffer from the kit was then added, and the heat-shocked oocysts were then lysed by freeze-thaw (five cycles of placement in 95% ethanol-dry ice and then placement in a 65°C water bath at 1-min intervals). After lysis, the released mRNA was then purified by using oligo(dT) magnetic particles, on the basis of the standard protocol provided with the kit. The mRNA bound on the magnetic beads was released by heating the solution (10 μl) at 70°C for 2 min, resuspended in ice-cold 10 mM Tris-HCl buffer, and immediately placed on ice prior for use in the aggregation assay.
Aggregation assays with oligonucleotide-functionalized gold nanoparticles.
We adapted the method of detection of genomic DNA from methicillin-resistant Staphylococcus aureus bacterial samples (20) with slight modifications. One modification involved the elimination of Mg2+ from the hybridization mixture since Mg2+ enhances the hydrolysis of mRNA in the presence of divalent ions (15). Our results suggest an improvement in the limit of detection of 2 to 3 orders of magnitude in the absence of Mg2+ divalent ions from the hybridization buffer (data not shown). For the 42-nucleotide ssDNA and ssRNA targets: 3 μl of gold nanoparticles functionalized with probe A (50 pM) and 3 μl of gold nanoparticles functionalized with probe B (50 pM) were mixed with 4 μl of hybridization mixture (a stock solution of 16% dextran sulfate, 20% formamide, and 3.75 mM MgCl2 for DNA and the same solution ingredients but no MgCl2 for RNA). Five microliters of the DNA or RNA targets at various dilutions was then added to the solution containing gold nanoparticles functionalized with probes A and B and the hybridization mixture. The solution was initially heated above the melting temperature (70°C) for 2 min in a thermocycler (Bio-Rad), incubated in a 37°C water bath for 20 min, and then brought down to room temperature and allowed to react for 1 to 2 h. For the HSP70 dsDNA target (from the RT-PCR) and RNA target (from total RNA purification), 0.6 μl of gold nanoparticles functionalized with probe A (25 pM) and 0.6 μl of gold nanoparticles functionalized with probe B (25 pM) were mixed with 0.8 μl of the hybridization mixture (no MgCl2 for RNA). One microliter of the DNA or RNA target at various dilutions was added to the solution. The solution was heated above the melting temperature (70°C for 15 min for RNA, 95°C for 2 min for DNA), incubated in a 37°C water bath for 20 min, and then brought down to room temperature and allowed to react for 1 to 2 h. For the HSP70 mRNA targets derived from C. parvum oocysts, 3 μl of gold nanoparticles functionalized with probe A (10 pM) and 3 μl of gold nanoparticles functionalized with probe B (10 pM) were mixed with 4 μl of the RNA hybridization buffer. Five microliters of the purified mRNA from C. parvum was then added to the solution containing gold nanoparticles functionalized with probes A and B and the hybridization mixture. The solution was initially heated above the melting temperature (70°C) for 15 min in a thermocycler (Bio-Rad), incubated in a 37°C water bath for 20 min, and then brought down to room temperature and allowed to react for 1 to 2 h.
Imaging of oligonucleotide-gold networks.
To confirm the presence or absence of oligonucleotide-gold networks, a 1-μl aliquot from the aggregation assay mixture (containing the targets and oligonucleotide-functionalized gold nanoparticles) was spotted on aldehyde-coated glass slides (Cel Associates). The slides were then coupled to a light guide illuminated with a halogen light source (Edmund Optics) to generate evanescent waves and allow total internal reflection. The total internal reflection images were then obtained with a charge-coupled-device color camera (MRC5; Zeiss) mounted on a standard stereoscope (SZ61; Olympus). By using ImageJ image analysis software (17), the color images were separated into red, green, and blue color channels. Triplicate regions of interest were randomly selected within each assay spot, and the average intensity of the red channel was measured in each region of interest. For statistical analysis, data were exported to the Microsoft Excel program with Analyze-It software (Analyze-It Software Ltd., Leeds, United Kingdom). A one-way analysis of variance (ANOVA) test was performed to compare the mean red spectral intensity for experimental samples containing target nucleic acid from the negative controls. ANOVA P values of less than 0.05 were accepted as statistically significant. Image contrast and saturation were adjusted for visualization and printing purposes only and were applied universally to all image panels in each of Fig. 2 to 5.
FIG. 2.
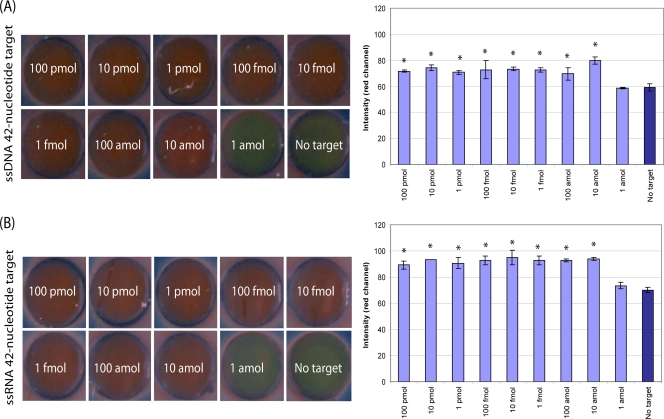
Total internal reflection images of synthetic DNA (A) and RNA (B) targets at various dilutions subjected to the aggregation assay and corresponding bar graphs of the measurable signal intensity in the red channel determined by the use of ImageJ software. Each spot is approximately 1.5 mm in diameter and has been resized to fill the full field of view. Image contrast and saturation were adjusted equally for all image panels for visualization and printing purposes only. *, a statistically significant difference in the mean signal intensity compared to that for the negative control with no target DNA or RNA, as determined by a one-way ANOVA (P < 0.05).
FIG. 5.
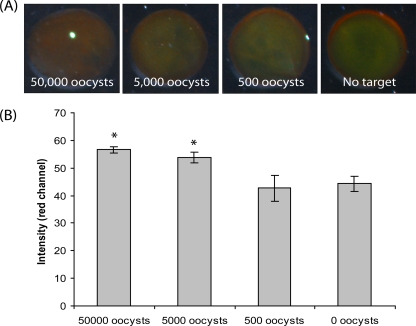
Total internal reflection images of mRNA harvested from C. parvum oocysts (heat shocked) subjected to the aggregation assay (A) and the corresponding signal intensity in the red channel measured by use of ImageJ software (B). *, a statistically significant difference in the mean signal intensity over the that for negative control (0 oocysts, no target) by a one-way ANOVA (P < 0.01).
RESULTS
Using the oligonucleotide-tagged gold nanoparticle detection scheme illustrated in Fig. 1, we explored the analytical performance of the C. parvum aggregation assay with DNA and RNA targets of increasing complexity. Figure 2 provides representative total internal reflection images of slides spotted with assay aggregates identifying the 42-nucleotide ssDNA and ssRNA synthetic targets, which were present at various amounts ranging from 0 to 100 pmol. For both the DNA and the RNA target sequences, the formation of the oligonucleotide-gold nanoparticle networks was confirmed via a color change from green (the absence of the target) to red (the presence of sufficient amounts of target) with a detection limit of ∼10 amol, or 6 million copies. Increases in the target amount from 10 amol to 100 pmol were not associated with a greater signal intensity under the assay conditions used.
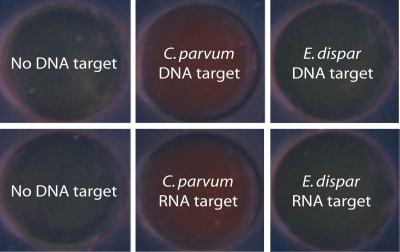
dsDNA targets obtained by RT-PCR amplification and ssRNA targets obtained by the purification of total RNA were also subjected to the aggregation assay. The formation of a red color in the presence of HSP70 DNA and RNA targets from C. parvum oocysts again confirmed the gold nanoparticle aggregation and the successful detection of target nucleotides, while in the absence of DNA or RNA targets, a green color associated with unaggregated oligonucleotide-gold nanoparticles was observed (Fig. 3). Moreover, negative control HSP70 DNA and HSP70 RNA targets from E. dispar also exhibited a persistent green color due to the absence of regions complementary to the oligonucleotide-gold nanoparticles.
FIG. 3.
Total internal reflection images of dsDNA targets (HSP70 from RT-PCR) and ssRNA targets (from total RNA purification) from C. parvum oocysts subjected to the aggregation assay. The specificity of the oligonucleotide-gold probes was tested with E. dispar DNA and RNA targets. The amount of DNA and RNA targets used in each case was adjusted to ∼10 fmol.
To further assess the specificity of the oligonucleotide-gold nanoparticles, a BLAST analysis of the complementary regions (42 nucleotides) of the HSP70 target was performed. The sequence was found to be conserved in other species of Cryptosporidium parasites, including C. hominis, that commonly infect humans and that also cause cryptosporidiosis (data not shown). No cross-reactivity (E value, <5e−07) was observed for other common intestinal parasites, suggesting that the current probe design will likely detect other infectious Cryptosporidium species with a high degree of specificity.
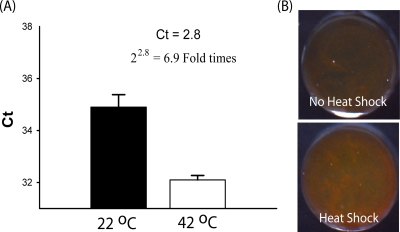
The use of the heat shock process to induce the expression of HSP70 mRNA targets in viable purified C. parvum oocysts was next examined. By RT-PCR, a 6.9-fold induction of HSP70 mRNA was demonstrated by the use of heat shock treatment for 30 min at 42°C (Fig. 4A). This biologically amplified HSP70 mRNA was also detected by the gold aggregation assay, as evidenced by the formation of a more intense red color when mRNA targets from heat-shocked C. parvum oocysts were used than when untreated oocysts were used (Fig. 4B). The ability of the oligonucleotide-gold probes to target purified mRNA harvested from various dilutions of purified C. parvum oocysts was explored by using the heat shock treatment. As shown in Fig. 5, nanoparticle aggregation was demonstrated by the formation of a red hue (Fig. 5A), with as few as 5,000 copies of purified C. parvum oocysts successfully being detected (Fig. 5B) (P < 0.01).
FIG. 4.
The induction of HSP70 mRNA using the heat shock process was confirmed by real-time PCR assay (A) and the gold aggregation assay (B). The amount of oocysts subjected to heat shock for the aggregation assay was ∼50,000. Ct, threshold cycle.
DISCUSSION
In this study, we demonstrated a method for the detection of HSP70 mRNA targets from C. parvum oocysts using an oligonucleotide-gold aggregation assay with a simple colorimetric readout and imaging modality. The oligonucleotide-gold nanoparticle probes (21 nucleotides each) were complementary to two adjacent target regions (∼42 nucleotides) on the HSP70 mRNA from C. parvum (2,495 nucleotides) which are not expected to cross-react with other organisms. The detection method with oligonucleotide-gold nanoparticles was initially tested with short synthetic ssDNA and ssRNA targets to determine the limit of detection of the assay, which was approximately 10 amol (Fig. 2). This value is lower by 2 to 3 orders of magnitude compared to that of fluorescence-based methods.
Interestingly, the presence of additional amounts of the target, at concentrations above 10 amol, did not result in a measurable increase in the intensity of scattering in the red channel of the aggregation assay (Fig. 2). Additional studies are necessary to fully characterize the dynamic range of the assay and its sensitivity to variations in assay conditions. A number of factors can influence the optical properties of the gold nanoparticle networks, including the target-to-probe ratio and the minimum number of gold nanoparticle probes needed in order to obtain a measurable signal. Under the assay conditions reported here, the increased red scattering in the total internal reflection images provides an indication of whether the amount of target DNA and RNA exceeds a threshold. This is consistent with previously published results, which also suggest that the use of spectroscopy as an alternative to total internal reflection imaging may prove useful for quantitating the target and increasing the sensitivity and dynamic range of the assay (20). Future studies are needed to explore the use of spectroscopy and to optimize the target-to-probe and probe-to-probe ratios in order to characterize and extend the dynamic range and sensitivity of nanoparticle-based assays for the detection of C. parvum.
In addition to synthetic DNA and RNA targets, the detection strategy with oligonucleotide-gold nanoparticles was able to detect longer target sequences in amplified dsDNA and purified RNA derived from C. parvum oocysts (Fig. 3). Furthermore, when they were tested against DNA/RNA targets from other pathogenic microorganisms, such as E. dispar, the oligonucleotide-gold nanoparticle probes designed for the detection of Cryptosporidium species did not exhibit a color shift, indicating no gold nanoparticle networks formed in the presence of nonspecific DNA and RNA. BLAST analysis further confirmed that the probes are highly specific to Cryptosporidium species and do not show signs of cross-reactivity with other intestinal parasites.
The aggregation assay also successfully demonstrated an ability to detect purified HSP70 mRNA from purified C. parvum oocysts in the absence of enzymatic amplification. Previous work has demonstrated the detection of HSP70 mRNA from Cryptosporidium parasites for various downstream applications, including RT-PCR (19) and nucleic acid sequence-based amplification (4) isothermal amplification assays. HSP70 mRNA was chosen as the target for various reasons. First, the production of HSP70 mRNA can be induced by simple heating, therefore providing an inherent biological amplification of the desired target. Previous studies indicate a 1,000- to 10,000-fold increase in the amount of HSP70 mRNA recovered from Drosophila species under heat stress (14). Second, HSP70 mRNA is a good indicator of oocyst viability; only viable oocysts will produced HSP70 mRNA by the use of heat shock (9, 19). Third, previous studies suggest that the half-life of HSP70 mRNA (∼50 min) extracted from human cell lines (HeLa and 283 cells) is increased at least 10-fold upon heat shock, suggesting the increased stability of the targets (21). Lastly, other HSP targets can easily be identified from other related parasites (Giardia, Entamoeba, Cyclospora, and Isospora species), allowing the potential for the multiplexed detection of these agents by use of the heat shock process for target amplification and the aggregation assay for detection. In summary, the strategy of using the heat shock process for amplification and the aggregation assay for detection showed the ability to detect HSP70 mRNA purified from as few as 5,000 oocysts (Fig. 5).
Conclusion.
Having demonstrated in the present study the functionality of the gold nanoparticle aggregation assays for the detection of purified C. parvum oocysts, further studies to define the assay performance and the limit of detection in clinically relevant stool samples are now warranted. In those studies, we will explore various methods of concentrating the oocysts from stool specimens. The most common preconcentration method involves the use of size-selective membranes, followed by the ethyl acetate sedimentation technique. The use of magnetic separation is also another interesting approach for the purification of oocysts by the use of immunomagnetic techniques. In both cases, the rate of recovery of the target specimens is not well established since it is not critical for currently used downstream applications (PCR based) due to amplification of the targets. The gold nanoparticle strategy proposed here will require high recovery rates to maximize the analytical sensitivity of the assay. If it is needed, an alternative assay approach could integrate additional amplification techniques, such as isothermal PCR, prior to the formation of DNA-linked nanoparticle networks and the detection of amplified products by the simple colorimetric-based aggregation assays.
Acknowledgments
We acknowledge Robert Kester and Tomasz Tkaczyk for the initial setup of the total internal reflection imaging device.
This work was funded through NIH-Western Regional Center for Excellence (grant U54 AI057156).
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Alles, A. J., M. A. Waldron, L. S. Sierra, and A. R. Mattia. 1995. Prospective comparison of direct immunofluorescence and conventional staining methods for detection of Giardia and Cryptosporidium spp. in human fecal specimens. J. Clin. Microbiol. 33:1632-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chappell, C. L., and P. C. Okhuysen. 2002. Cryptosporidiosis. Curr. Opin. Infect. Dis. 15:523-527. [DOI] [PubMed] [Google Scholar]
- 3.Cicirello, H. G., K. S. Kehl, D. G. Addiss, M. J. Chusid, R. I. Glass, J. P. Davis, and P. L. Havens. 1997. Cryptosporidiosis in children during a massive waterborne outbreak in Milwaukee, Wisconsin: clinical, laboratory and epidemiologic findings. Epidemiol. Infect. 119:53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connelly, J. T., S. R. Nugen, W. Borejsza-Wysocki, R. A. Durst, R. A. Montagna, and A. J. Baeumner. 3 March 2008. Human pathogenic Cryptosporidium species bioanalytical detection method with single oocyst detection capability. Anal. Bioanal Chem. [Epub ahead of print.] [DOI] [PubMed]
- 5.Dagan, R., D. Fraser, J. El-On, I. Kassis, R. Deckelbaum, and S. Turner. 1995. Evaluation of an enzyme immunoassay for the detection of Cryptosporidium spp. in stool specimens from infants and young children in field studies. Am. J. Trop. Med. Hyg. 52:134-138. [DOI] [PubMed] [Google Scholar]
- 6.Elghanian, R., J. J. Storhoff, R. C. Mucic, R. L. Letsinger, and C. A. Mirkin. 1997. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277:1078-1081. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, L. S., and R. Y. Shimizu. 1997. Evaluation of nine immunoassay kits (enzyme immunoassay and direct fluorescence) for detection of Giardia lamblia and Cryptosporidium parvum in human fecal specimens. J. Clin. Microbiol. 35:1526-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia, L. S., R. Y. Shimizu, S. Novak, M. Carroll, and F. Chan. 2003. Commercial assay for detection of Giardia lamblia and Cryptosporidium parvum antigens in human fecal specimens by rapid solid-phase qualitative immunochromatography. J. Clin. Microbiol. 41:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallier-Soulier, S., and E. Guillot. 2003. An immunomagnetic separation-reverse transcription polymerase chain reaction (IMS-RT-PCR) test for sensitive and rapid detection of viable waterborne Cryptosporidium parvum. Environ. Microbiol. 5:592-598. [DOI] [PubMed] [Google Scholar]
- 10.Ignatius, R., M. Eisenblatter, T. Regnath, U. Mansmann, U. Futh, H. Hahn, and J. Wagner. 1997. Efficacy of different methods for detection of low Cryptosporidium parvum oocyst numbers or antigen concentrations in stool specimens. Eur. J. Clin. Microbiol. Infect. Dis. 16:732-736. [DOI] [PubMed] [Google Scholar]
- 11.Javier, D. J., N. Nitin, M. Levy, A. Ellington, and R. Richards-Kortum. 2008. Aptamer-targeted gold nanoparticles as molecular-specific contrast agents for reflectance imaging. Bioconjug. Chem. 19:1309-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston, S. P., M. M. Ballard, M. J. Beach, L. Causer, and P. P. Wilkins. 2003. Evaluation of three commercial assays for detection of Giardia and Cryptosporidium organisms in fecal specimens. J. Clin. Microbiol. 41:623-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katanik, M. T., S. K. Schneider, J. E. Rosenblatt, G. S. Hall, and G. W. Procop. 2001. Evaluation of ColorPAC Giardia/Cryptosporidium rapid assay and ProSpecT Giardia/Cryptosporidium microplate assay for detection of Giardia and Cryptosporidium in fecal specimens. J. Clin. Microbiol. 39:4523-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindquist, S., and R. Petersen. 1990. Selective translation and degradation of heat-shock messenger RNAs in Drosophila. Enzyme 44:147-166. [DOI] [PubMed] [Google Scholar]
- 15.Liu, W. F., A. Zhang, Y. Cheng, H. M. Zhou, and Y. B. Yan. 2007. Effect of magnesium ions on the thermal stability of human poly(A)-specific ribonuclease. FEBS Lett. 581:1047-1052. [DOI] [PubMed] [Google Scholar]
- 16.Parr, J. B., J. E. Sevilleja, A. Samie, C. Alcantara, S. E. Stroup, A. Kohli, R. Fayer, A. A. Lima, E. R. Houpt, and R. L. Guerrant. 2007. Detection and quantification of Cryptosporidium in HCT-8 cells and human fecal specimens using real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 76:938-942. [PMC free article] [PubMed] [Google Scholar]
- 17.Rasband, W. 2006, posting date. ImageJ, image processing and analysis in Java. U.S. National Institutes of Health, Bethesda, MD.
- 18.Samie, A., P. O. Bessong, C. L. Obi, J. E. Sevilleja, S. Stroup, E. Houpt, and R. L. Guerrant. 2006. Cryptosporidium species: preliminary descriptions of the prevalence and genotype distribution among school children and hospital patients in the Venda region, Limpopo Province, South Africa. Exp. Parasitol. 114:314-322. [DOI] [PubMed] [Google Scholar]
- 19.Stinear, T., A. Matusan, K. Hines, and M. Sandery. 1996. Detection of a single viable Cryptosporidium parvum oocyst in environmental water concentrates by reverse transcription-PCR. Appl. Environ. Microbiol. 62:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storhoff, J. J., A. D. Lucas, V. Garimella, Y. P. Bao, and U. R. Muller. 2004. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 22:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theodorakis, N. G., and R. I. Morimoto. 1987. Posttranscriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol. Cell. Biol. 7:4357-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumwine, J. K., A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, S. M. Rich, G. Widmer, X. Feng, and S. Tzipori. 2003. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am. J. Trop. Med. Hyg. 68:710-715. [PubMed] [Google Scholar]
- 23.Weber, R., R. T. Bryan, H. S. Bishop, S. P. Wahlquist, J. J. Sullivan, and D. D. Juranek. 1991. Threshold of detection of Cryptosporidium oocysts in human stool specimens: evidence for low sensitivity of current diagnostic methods. J. Clin. Microbiol. 29:1323-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White, A. C., Jr. 2005. Cryptosporidiosis (Cryptosporidium hominis, Cryptosporidium parvum, other species), p. 3215-3228. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 6th ed. Elsevier Churchill Livingstone, Philadelphia, PA.