Abstract
Two chromogenic media (Chromagar VRE and chromID VRE [C-ID]) performed equally well in the direct detection of vancomycin-resistant enterococci (VRE) in stool specimens after an overnight enrichment step and a 48-h incubation period, with a sensitivity of 98.2% (56/57) for both and specificities of 96.5% (195/202) and 97.5% (197/202), respectively. However, assigning discriminatory colony color was sometimes difficult, especially on C-ID. In order to facilitate simple species identification, biochemical key reactions were implemented.
Prevalence rates of vancomycin-resistant enterococci (VRE) associated with serious clinical infections have increased worldwide over the past 15 years (3, 12). Therefore, to prevent further spread, active control measures are increasingly being implemented in hospitals. A cornerstone of those control measures is the detection of noninfected but gut-colonized patients that might serve as a source of the spread of VRE. The use of improved cultural-based methods, e.g., chromogenic media, may provide a feasible alternative for cost-effective VRE surveillance (2, 7). This study systematically compared the performance characteristics of chromID VRE (C-ID) medium (bioMérieux, Nürtingen, Germany) with those of CHROMagar VRE (CHR) medium (CHROMagar, Paris, France) with special regard to (i) selectivity, (ii) stability of colony color and growth characteristics, and (iii) the ability to recover VRE from clinical stool specimens.
In order to check the selectivity of the two media, all of the type strains (as of June 2009) of the genus Enterococcus (n = 38; vancomycin MICs ranging from 0.125 to ≥256 μg/ml) were tested. As positive controls, VRE reference strains obtained from culture collections (n = 4) and from the culture collection of the National Reference Center for Streptococci (AC; Aachen, Germany; n = 6) were used (Fig. 1). Both media efficiently suppressed the growth of pure cultures of all but one Enterococcus type strain (see Table S1 in the supplemental material). Although Enterococcus gallinarum (DSM 20623T; vancomycin MIC, 4 μg/ml) was able to grow as tiny bluish colonies on CHR medium, neither E. solitarius nor E. hirae (vancomycin MICs of 8 and ≥256 μg/ml, respectively) was able to grow on both media.
FIG. 1.
Growth of VRE reference strains (n = 10 and a negative control) in pure culture on C-ID and CHR media. Superscripts: a, lot 815923201; b, lot agx/aha; c, culture collection of the National Reference Center for Streptococci, Aachen, Germany; d, Deutsche Sammlung von Mikroorganismen; e, CHR medium, weak growth even after 48 h of incubation time; C-ID medium, efficient growth but in a rosy brown shade after 48 h of incubation; f, n.a., not applicable.
Objective color evaluation of colony appearance was performed by analyzing digital pictures taken during the 24 h of incubation at 37°C (Nikon Coolpix P5000) from colonies on the respective media with a light microscope (×10 magnification, Leitz DMRB; Leica Microsystems GmbH, Wetzlar, Germany) and a stereomicroscope (×16 magnification, Leica MZ 9.5; Leica Microsystems GmbH). Colors were assigned (center and edge of each colony separately; for details, see Table S2 in the supplemental material) and coded as Hex triplets by using WhatColor v4.71e software (http://www.hikarun.com/e).
Dilutions of VREfm (Enterococcus faecium; n = 7) and VREfs (Enterococcus faecalis; n = 2) reference strains were streaked in pure and mixed cultures onto both media and revealed the typical colony appearance described by the manufacturer (Fig. 1; see Fig. S1 in the supplemental material), except for VREfm strain AC 7075 (vancomycin MIC, 48 μg/ml), which showed a rosy brown color (Hex Code #BC8F8F).
Where the CHR medium is unable to discriminate between VREfm and VREfs, according to the manufacturer of the C-ID medium, it should be able to discriminate between VREfm and VREfs due to the production of two different colony colors after 48 h of incubation (see Fig. S1 in the supplemental material). However, after 24 h of incubation, the color shades of the respective colonies appeared to be more diverse than expected. At the edges of VREfm and VREfs colonies, seven and six color shades were identified, respectively. Two nondiscriminating colors (dark slate gray [#2F4F4F] and dim gray [#696969]; see Table S2 in the supplemental material) were observed in the centers of VREfm and VREfs colonies, thereby complicating definite VRE species identification. In general, after 24 h of incubation, it was easier to recognize VREfs colonies by their constant green color than VREfm, which exhibited different color shades; e.g., the claimed purple color of the edge/center sometimes appeared blue (dark slate blue, #483D8B) or often red (Indian red, #CD5C5C). However, the superior growth of colonies on C-ID medium compared with CHR medium after 24 h of incubation aided early detection and differentiation of VREfm and VREfs.
In general, VRE grew better on C-ID than on CHR agar plates. For example, on the CHR medium, the borders of tiny colonies were diffuse and lacked obvious edges because of the contrast with the “milky” background of the medium (Fig. 1). Therefore, VRE colonies grown on the CHR medium were more difficult to detect for subculturing than were VRE colonies grown on the C-ID medium.
For routine evaluation, stool specimens (n = 259, Table 1) obtained from patients at risk of being colonized with VRE, i.e., from oncology and intensive care units and the postsurgical gastroenterology ward, were used. Preliminary experiments in which stool specimens were plated directly or after prior dilution in isotonic saline failed to obtain acceptable results due to the overwhelming growth of contaminating bacteria and yeasts, resulting in low sensitivity (data not shown). Therefore, a stool specimen enrichment culture step, done as described recently (2, 6), was applied before parallel inoculation of both chromogenic media for comparative testing. These media were evaluated for the growth of suspected VRE colonies after 24 and 48 h of incubation by following the instructions of the manufacturers (C-ID medium, bluish green or violet colonies; CHR medium, rose/mauve colonies). Gram staining of suspected VRE colonies and a 5-min pyrrolidonyl arylamidase spot paper test (PYRase) were performed (5) for confirmation of enterococci. Further identification to the species level was done by testing susceptibility to 50 μg furazolidone and 10 μg mupirocin with Rosco Neo-Sensitabs on Mueller-Hinton agar (Mast Diagnostika) and a rapid d-xylose fermentation test with Rosco Diatabs (Rosco Diagnostica, Taastrup, Denmark) (8, 14) by following the instructions given by the manufacturer (Diatabs User's Guide, 2009). The reaction pattern obtained allowed the confirmation of E. faecium or E. faecalis or identification of intrinsically low-level VRE like E. casseliflavus and E. gallinarum (Identification of Enterococci application sheet; Rosco) (see Table S3 in the supplemental material). For confirmation, resistance to the antimicrobials vancomycin and teicoplanin (Etest; AB Biodisk, Solna, Sweden) was tested by applying CLSI breakpoints. The presence of vancomycin resistance genes were determined by multiplex PCR (4, 10).
TABLE 1.
Results of stool sample screening for VRE (n = 259) revealed by parallel inoculation of the two chromogenic media from the same vancomycin enrichment broth after 48 h of incubation
| Performance characteristic | No. of samples |
|
|---|---|---|
| C-ID | CHR | |
| Growth of VREa | 56 | 56 |
| Missed VREfmb | 1 | 1 |
| Sterile | 79 | 130 |
| Growth but not VRE | 123 | 72 |
| Yeasts | 55 (dull green) | 39 (white) |
| Gram-negative rods | 2 | 1 |
| Gram-positive coccic | 17 | 13 |
| Gram-positive rods | 22 (various colors) | 14 (various colors) |
| Weak growth, not differentiated | 26 | 4 |
| Molds | 1 | 1 |
| Calculated % sensitivity/specificity | 98.2/97.5 | 98.2/96.5 |
True positives.
False negatives.
Comprising isolates that could be mistaken as VRE due to a medium purple colony color on C-ID (E. faecalis, n = 1; Pediococcus dextrinicus, n = 4) and a rose-to-mauve colony color on CHR (E. faecalis, n = 2; E. faecium, n = 1; Enterococcus sp., n = 2; P.dextrinicus, n = 2) and therefore judged as false positives.
Parallel screening of 259 stool samples on both chromogenic media revealed 54 VREfm and 2 E. gallinarum isolates (Table 1), representing a colonization rate with VRE of 21%. The genetic relatedness of the VREfm isolates was determined by using multiple-locus variable-number tandem repeat analysis (MLVA) as described previously (11). Six different MLVA types (MTs) were determined among the 49 VREfm isolates analyzed, including MT-1 (n = 2), MT-7 (n = 36), MT-11 (n = 1), MT-12 (n = 2), MT-159 (n = 7), and MT-332 (n = 1) (Table 2). Based on different repeat combinations for VNTR-7, -8, and -10, all of the MTs, except MT-332, were found to belong to clonal complex 17 (CC17) (Table 2) (11, 13). CC17, which is based on multilocus sequence typing, is associated with nosocomial outbreaks and infections and is clearly distinct from complexes composed of human-community- and animal-derived isolates. There was no correlation between the six different MTs and the respective color shades exhibited on the CHR and C-ID media.
TABLE 2.
Distribution of MTs and clonality results of 49 VREfm isolates determined by MLVA
| MT | MLVA profileb (no. of repeats): |
No. of isolates | CC17-specific MTsa | |||||
|---|---|---|---|---|---|---|---|---|
| VNTR-1 | VNTR-2 | VNTR-7 | VNTR-8 | VNTR-9 | VNTR-10 | |||
| 1 | 5 | 7 | 3 | 3 | 2 | 3 | 2 | + |
| 7 | 5 | 7 | 3 | 3 | 2 | 2 | 36 | + |
| 11 | 6 | 7 | 3 | 3 | 2 | 3 | 1 | + |
| 12 | 5 | 7 | 3 | 3 | 1 | 3 | 2 | + |
| 159 | 5 | 7 | 3 | 3 | 1 | 2 | 7 | + |
| 332 | 6 | 4 | 3 | 1 | 2 | 1 | 1 | − |
+, MTs with the following repeat profiles for VNTR-7, -8, and -10: 3-3-3, 3-2-3, 3-3-2, 4-3-3, 3-4-3, 4-2-3, and 3-3-1 (13).
VNTR, variable-number tandem repeat locus.
Since each chromogenic medium failed to detect one VRE strain, which was successfully detected by the other, the sensitivity of both media was 98.2% (56/57). Although the specificities of both media were high, 96.5% (195/202) for CHR medium and 97.5% (197/202) for C-ID medium, there is a risk of false-positive VRE identification when solely based on Gram staining and colony pigmentation on chromogenic medium (Table 1). Most strikingly, growth of Pediococcus species (vancomycin MIC, ≥256 μg/ml) on both media was visually indistinguishable from that of VRE (medium purple colonies on C-ID medium, rose-to-mauve colonies on CHR medium). PYRase testing and the two diagnostic agar diffusion susceptibility tests (furazolidone and mupirocin) were extremely helpful in immediately identifying enterococci and thereby ruling out Pediococcus. All Pediococcus spp. are PYRase negative and intrinsically resistant to vancomycin. Pediococcus species are regularly cultured from human stool specimens, and infections have been seen in patients with chronic underlying diseases, as well as those with a history of abdominal surgery (1, 9). Therefore, Pediococcus spp. have to be considered opportunistic pathogens and it can be assumed that they are selected under vancomycin therapy, similar to VRE.
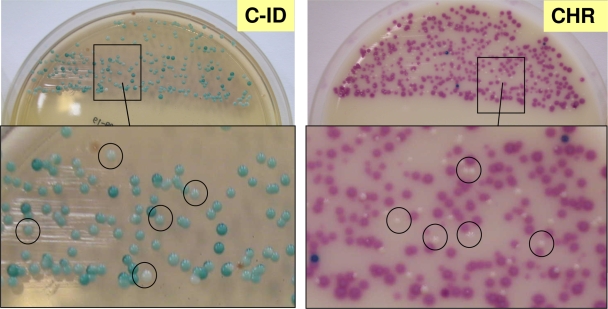
The rapid xylose fermentation test enabled identification to the species level in the case of two E. gallinarum isolates (see Table S3 in the supplemental material) that grew on both media with misleading colony colors. Furthermore, yeasts grew very well on both media, producing white colonies on CHR medium and slightly dull green colonies on C-ID medium, a colony color that could be confused with that of VREfs (Fig. 2). However, yeasts were easily differentiated from VRE by subjecting suspected colonies to Gram staining or microscopic inspection.
FIG. 2.
Mixed cultures of VREfs and yeasts on C-ID and CHR media after 48 h of incubation. (CHR) VREfs, mauve; yeasts, white (examples circled). (C-ID) VREfs, green; yeasts, dull green (examples circled).
The vanA genotype was identified in all of the VREfm isolates, with a single exception. The latter one appeared to exhibit the VanB phenotype (teicoplanin MIC of 1 μg/ml), and PCR confirmed the vanB genotype. Vancomycin-susceptible (MIC, ≤4 μg/ml) E. faecium or E. faecalis was rarely isolated from chromogenic media (CHR medium, n = 3; C-ID medium, n = 1) inoculated with enriched stool specimens. This might be due to overgrowth of other bacteria and consecutive exhaustion of selective substrates.
Our study has clearly shown that the C-ID and CHR media do not fulfill all of the expectations regarding rapid VRE detection, especially when stool specimens are plated directly or after predilution. Overwhelming growth of different bacteria and yeasts on both media hampers medium evaluation when stool samples are plated directly, as does the innate color of stool samples (see Fig. S2 in the supplemental material). Thus, in our experience, stool specimens are inappropriate for direct plating onto the C-ID and CHR media. Our study confirmed that overnight precultivation of stool specimens to enable selective enrichment is essential before specimen plating onto these chromogenic media (2, 6).
Concerning specificity, when pure and mixed cultures were plated, both media performed very well by suppressing the growth of all of the type strains of the genus Enterococcus and supporting only the growth of VRE reference strains. The proposed key reactions are helpful for the reliable characterization of suspicious isolates and are highly recommended in order to avoid false-positive results. Confirmation of the vancomycin resistance of suspected VRE colonies is also advisable.
The evaluation of the colony color of CHR medium revealed that the color shades of colonies during the different growth phases were stable, which is in contrast to the C-ID medium, a medium that should allow differentiation of VREfs from VREfm by color after 48 h of incubation. With C-ID medium, differentiation of the two VRE species in a mixed culture can be complicated by the appearance of numerous comparable color shades, especially in the center of the colony (see Table S2 in the supplemental material). Furthermore, the development of the purple color of VREfm was uneven after 24 h of incubation. Only incubation for up to 48 h generated the expected pair of distinguishable colors.
In conclusion, the incorporation of an enrichment culture step, along with confirmation of antibiotic resistance, and the use of the proposed key reactions to confirm species identity are highly recommended for the accurate identification of VRE from stool specimens. Unfortunately, however, implementation of these additional steps, especially the enrichment step, does slow down the identification of VRE. Nevertheless, the use of one of the two chromogenic media tested represents a cost-effective screening approach for VRE in stool samples, thereby enabling subsequent implementation of barrier precautions that should ultimately lead to a reduction in life-threatening infections.
Supplementary Material
Acknowledgments
We thank Kirstin Beckers, Eline Maicot, and Sabine von Oy for their excellent technical assistance.
Footnotes
Published ahead of print on 7 October 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Barton, L. L., E. D. Rider, and R. W. Coen. 2001. Bacteremic infection with Pediococcus: vancomycin-resistant opportunist. Pediatrics 107:775-776. [DOI] [PubMed] [Google Scholar]
- 2.Delmas, J., F. Robin, C. Schweitzer, O. Lesens, and R. Bonnet. 2007. Evaluation of a new chromogenic medium, ChromID VRE, for detection of vancomycin-resistant enterococci in stool samples and rectal swabs. J. Clin. Microbiol. 45:2731-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshpande, L. M., T. R. Fritsche, G. J. Moet, D. J. Biedenbach, and R. N. Jones. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 58:163-170. [DOI] [PubMed] [Google Scholar]
- 4.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufhold, A., R. Lütticken, and U. Schwien. 1989. Few-minutes tests for the identification of group A streptococci and enterococci with chromogenic substrates. Zentralbl. Bakteriol. 272:191-195. [DOI] [PubMed] [Google Scholar]
- 6.Kuch, A., K. Stefaniuk, T. Ozorowski, and W. Hryniewicz. 2009. New selective and differential chromogenic agar medium chromID VRE, for screening vancomycin-resistant Enterococcus species. J. Microbiol. Methods 77:124-126. [DOI] [PubMed] [Google Scholar]
- 7.Ledeboer, N. A., K. Das, M. Eveland, C. Roger-Dalbert, S. Mailler, S. Chatellier, and W. M. Dunne. 2007. Evaluation of a novel chromogenic agar medium for isolation and differentiation of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis isolates. J. Clin. Microbiol. 45:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manero, A., and A. R. Blanch. 1999. Identification of Enterococcus spp. with a biochemical key. Appl. Environ. Microbiol. 65:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mastro, T. D., J. S. Spika, P. Lozano, J. Appel, and R. R. Facklam. 1990. Vancomycin-resistant Pediococcus acidilactici: nine cases of bacteremia. J. Infect. Dis. 161:956-960. [DOI] [PubMed] [Google Scholar]
- 10.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, and F. R. Cockerill III. 1997. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 35:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Top, J., L. M. Schouls, M. Bonten, and R. Willems. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Top, J., R. Willems, and M. Bonten. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297-308. [DOI] [PubMed] [Google Scholar]
- 13.Top, J., R. Willems, S. van der Velden, M. Asbroek, and M. Bonten. 2008. Emergence of clonal complex 17 Enterococcus faecium in The Netherlands. J. Clin. Microbiol. 46:214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyrrell, G. J., L. Turnbull, L. M. Teixeira, J. Lefebvre, G. Carvalho Mda, R. R. Facklam, and M. Lovgren. 2002. Enterococcus gilvus sp. nov. and Enterococcus pallens sp. nov. isolated from human clinical specimens. J. Clin. Microbiol. 40:1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.