Abstract
The spatial organization of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds was investigated in the present study. Wound biopsy specimens were obtained from patients diagnosed as having chronic venous leg ulcers, and bacterial aggregates in these wounds were detected and located by the use of peptide nucleic acid-based fluorescence in situ hybridization and confocal laser scanning microscopy (CLSM). We acquired CLSM images of multiple regions in multiple sections cut from five wounds containing P. aeruginosa and five wounds containing S. aureus and measured the distance of the bacterial aggregates to the wound surface. The distance of the P. aeruginosa aggregates to the wound surface was significantly greater than that of the S. aureus aggregates, suggesting that the distribution of the bacteria in the chronic wounds was nonrandom. The results are discussed in relation to our recent finding that swab culturing techniques may underestimate the presence of P. aeruginosa in chronic wounds and in relation to the hypothesis that P. aeruginosa bacteria located in the deeper regions of chronic wounds may play an important role in keeping the wounds arrested in a stage dominated by inflammatory processes.
Chronic wounds, such as diabetic foot ulcers, pressure ulcers, and venous leg ulcers, are an increasing problem worldwide. One to 2% of the population in developed countries develops chronic wounds, a condition associated with severe patient suffering, the loss of employment, a reduced quality of life, and high costs to the health care system (13). Detailed knowledge about chronic wounds is required in order to develop better wound treatment and management strategies.
A normal wound healing process involves four main phases: (i) coagulation, (ii) inflammation, (iii) cell proliferation and repair of the matrix, and (iv) epithelialization and remodeling of the scar tissue (23). However, chronic wounds are believed to be captured in the inflammatory phase, where persistent influx and elevated activity of polymorphonuclear neutrophils (PMNs) occur (1). Although PMNs play a critical role in the host defense and wound healing, they release cytolytic enzymes, free oxygen radicals, inflammatory mediators, and matrix metalloproteases, which cause local tissue damage in the host (22, 23, 26).
It is known that the microflora of chronic wounds comprises multiple species. In a bacterial profiling study, Gjødsbol et al. found that chronic venous leg ulcers harbored Staphylococcus aureus (in 93.5% of the ulcers), Enterococcus faecalis (71.7%), Pseudomonas aeruginosa (52.2%), coagulase-negative staphylococci (45.7%), Proteus species (41.3%), and anaerobic bacteria (39.1%) (12). S. aureus and P. aeruginosa are opportunistic pathogenic bacteria and are widely known to cause chronic biofilm-based infections in their hosts. S. aureus is most commonly isolated from chronic wounds (8, 12, 15, 17) and, in certain situations, can express a number of potential virulence factors and surface proteins which promote its adherence to the damaged tissue and decrease neutrophil functions and immune responses of the host (10, 11). P. aeruginosa often causes biofilm-based chronic infections and expresses virulence factors, in particular, rhamnolipid, that can eliminate the activity of PMNs (4, 16). A number of studies have demonstrated that P. aeruginosa is frequently present in chronic wounds (12, 17) and have provided evidence that the bacteria are located in aggregates enclosed in extracellular polymeric matrix material as found in biofilms (17). Furthermore, chronic wounds that harbored P. aeruginosa were larger than those that did not, and the healing process also seemed to be more severely hindered for those wounds (12, 14, 20).
Biofilms are bacterial aggregates enclosed in a self-produced extracellular polymeric matrix (6, 21, 25). In clinical environments biofilms can form on dead or living tissues, mucosal surfaces, or the surfaces of medical devices in the host. The bacteria in biofilms often display characteristics different from those of their planktonic counterparts, such as increased resistance to the activities of the host immune system and tolerance to antimicrobial treatments (7). Such characteristics are important, since biofilms are involved in many chronic bacterial infections. Recent studies have shown the presence of bacterial biofilms in chronic wounds (9, 15, 17). Although the role of biofilms in chronic wounds is not yet fully understood, it is believed that their existence may be one of the reasons for impaired wound healing (4, 16).
We previously demonstrated that there is a lack of correlation between the bacteria detected by standard culturing and those detected directly by peptide nucleic acid (PNA)-based fluorescence in situ hybridization (FISH) in chronic wound samples (17). While S. aureus was detected more frequently by swab sample cultivation than by PNA-FISH, the opposite was true for P. aeruginosa. This lack of correlation between detection by swab sample cultivation and PNA-FISH may be due to the ability of the different bacterial species to colonize different regions of chronic wounds. Swab sample cultivation identifies the microorganisms present in the surface region of the wound but may not detect microorganisms located inside the wound bed. Accordingly, in the present report, we present evidence that S. aureus primarily colonizes the region of chronic wounds which is close to the surface, whereas P. aeruginosa primarily colonizes the deeper regions of chronic wounds. The ability of P. aeruginosa to colonize the deeper regions of chronic wounds may be due to the ability of this organism to produce virulence factors which destroy PMNs (4, 16), and it may play an important role in keeping the wounds arrested in a stage dominated by inflammatory processes.
MATERIALS AND METHODS
Tissue sample collection and preparation.
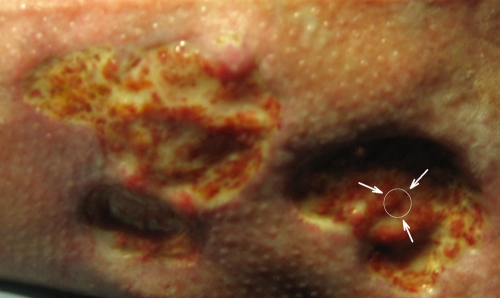
Nine patients diagnosed with chronic venous leg ulcers were included in the study. As described below, four patients had S. aureus-containing wounds, four patients had P. aeruginosa-containing wounds, and one patient had a wound that contained both S. aureus and P. aeruginosa. Material (4-mm punch biopsy specimens) from chronic venous leg ulcers (Fig. 1) was obtained with the acceptance of the patients and in accordance with biomedical project protocols H-B-2008-023 and KA-20051011, which were approved by the Danish Scientific Ethical Board. Wound biopsy material was collected by a surgical team before cleansing and surgical preparation of the wound (2). The material was immediately transferred to phosphate-buffered saline with 4% paraformaldehyde and stored at 5°C before further preparation for microscopic investigation. The biopsy material for microscopic investigation was embedded in paraffin, cut into 4-μm sagittal sections, and mounted on glass slides. Prior to microscopic investigation, the paraffin was removed from the tissue sections by immersing the glass slides twice in xylene (total, 10 min), twice in 99.9% ethanol (total, 6 min), twice in 96% ethanol (total, 6 min), and three times in distilled sterile water (total, 9 min).
FIG. 1.
Sampling region on a chronic venous leg ulcer. Biopsy specimens were taken from a central region within the wounds. The arrows point to a representative sampling region.
PNA-FISH and conventional tissue staining.
The deparaffinized tissue sections were analyzed by means of conventional hematoxylin and eosin (H&E) staining and FISH with PNA probes. The PNA probe in hybridization solution (AdvanDx, Inc., Woburn, MA) was added dropwise to each tissue section, which was then covered with a coverslip and hybridized in a PNA-FISH workstation (AdvanDx, Inc.), which was covered with a lid, at 55°C for 90 min. Three separate PNA probe solutions were used: (i) a Texas Red (TxR)-labeled P. aeruginosa-specific probe, (ii) a fluorescein isothiocyanate (FITC)-labeled S. aureus-specific probe, and (iii) a mixture of the TxR-labeled P. aeruginosa-specific probe and the FITC-labeled S. aureus-specific probe. The slides with tissue sections were washed in a wash solution (AdvanDx, Inc.) at 55°C for 30 min, air dried, mounted with Vectashield mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories), and covered with a coverslip. The tissue sections were examined as described below.
Image acquisition and analysis.
Microscopic observations of the tissue sections were performed with an epifluorescence microscope (Olympus, Hamburg, Germany) or a TCS-SP5 confocal laser scanning microscope (Leica Microsystems, Mannheim, Germany) equipped with an argon laser and a helium-neon laser for excitation of the fluorophores. Multichannel simulated fluorescence projection images were generated by using the IMARIS software package (Bitplane AG, Zurich, Switzerland) and were further processed for display by using PhotoShop software (Adobe). Subtraction of the background from the images was performed with the IMARIS software package to remove the host tissue autofluorescence. The images were converted to eight-bit gray-scale images by using ImageJ (version 1.41o) software (http://rsb.info.nih.gov/ij/index.html), and the moment calculator tool (http://rsb.info.nih.gov/ij/plugins/moments.html) of the same software was used to locate the center of mass of the bacterial population displayed on the images.
Statistical evaluation.
To evaluate whether the data obtained from the distance measurement of P. aeruginosa and S. aureus aggregates to the wound surface were statistically significant, an unpaired t test was performed. P values of ≤0.05 were considered significant. The statistical program Stat-View (SAS Institute Inc., Cary, NC) was used to calculate P values.
RESULTS
Initially, we identified P. aeruginosa and S. aureus in biopsy material from chronic wounds by the use of PNA-FISH with species-specific probes. On the basis of those identities, we selected five wounds containing P. aeruginosa and five wounds containing S. aureus (one of the wounds selected contained both P. aeruginosa and S. aureus). In order to study the spatial distribution of P. aeruginosa and S. aureus in these wounds, we cut five sections sagittally at 50-μm intervals from each wound biopsy specimen and performed PNA-FISH of these sections with P. aeruginosa- and S. aureus-specific probes. We subsequently acquired images at three different regions on each section by confocal laser scanning microscopy (CLSM). The bacteria were predominantly present as large aggregates. To get a measure of the distance of the bacteria to the wound surface, we located the center of mass of the bacterial aggregates identified on each image by using the moment calculator tool of ImageJ software and measured its distance to the wound surface. This analysis showed that the S. aureus aggregates were located close to the wound surface, whereas the P. aeruginosa aggregates were located deeper in the wound bed (P < 0.0001) (Table 1). The centers of mass of the S. aureus aggregates were primarily located at a distance of 20 to 30 μm to the wound surface, whereas the centers of mass of the P. aeruginosa aggregates were primarily located at a distance of 50 to 60 μm to the wound surface (Fig. 2). Figure 3 shows representative CLSM images of the locations of P. aeruginosa and S. aureus in the chronic wounds. The range of the distribution of P. aeruginosa and S. aureus was limited, so colocalization of the two bacterial species was rare. In order to visualize host cells and bacteria in the wound biopsy specimens, we performed combined PNA-FISH and DAPI staining as well as H&E staining of the biopsy specimens from wounds containing P. aeruginosa or S. aureus. As shown in Fig. 4, the analyzed sections from wounds with P. aeruginosa had a higher number of PMNs than the sections from wounds with S. aureus, suggesting that wounds infected with P. aeruginosa may have a higher degree of inflammation than wounds infected with S. aureus.
TABLE 1.
Average distance of bacterial aggregates to the surface of wound samples
| Wound biopsy specimen | Bacterial species detected by PNA-FISH | Avg distance to wound surface (μm)b |
|---|---|---|
| LGA02 | S. aureus | 28.3 (6.6) |
| BIJ04 | S. aureus | 8.8 (1.7) |
| HAH08 | S. aureus | 28.1 (5.0) |
| M2a | S. aureus | 26.1 (5.1) |
| Pt17 | S. aureus | 23.7 (3.7) |
| M3a | P. aeruginosa | 57.5 (9.4) |
| Pt11 | P. aeruginosa | 50.0 (13.4) |
| Pt20 | P. aeruginosa | 53.5 (9.9) |
| Pt23B | P. aeruginosa | 68.7 (11.2) |
| Pt31 | P. aeruginosa | 46.1 (6.0) |
Specimens M2 and M3 are biopsy specimens obtained from the same wound. Both S. aureus and P. aeruginosa were detected in these biopsy specimens. In the case of specimen M2, the distance of S. aureus to the wound surface was analyzed, whereas in the case of specimen M3, the distance of P. aeruginosa to the wound surface was analyzed.
The center of mass of the bacterial aggregates on each image was located, and its distance to the wound surface was measured. The average distances of the center of mass to the wound surface were obtained from 15 images acquired for each wound sample. The values in parentheses are standard deviations.
FIG. 2.
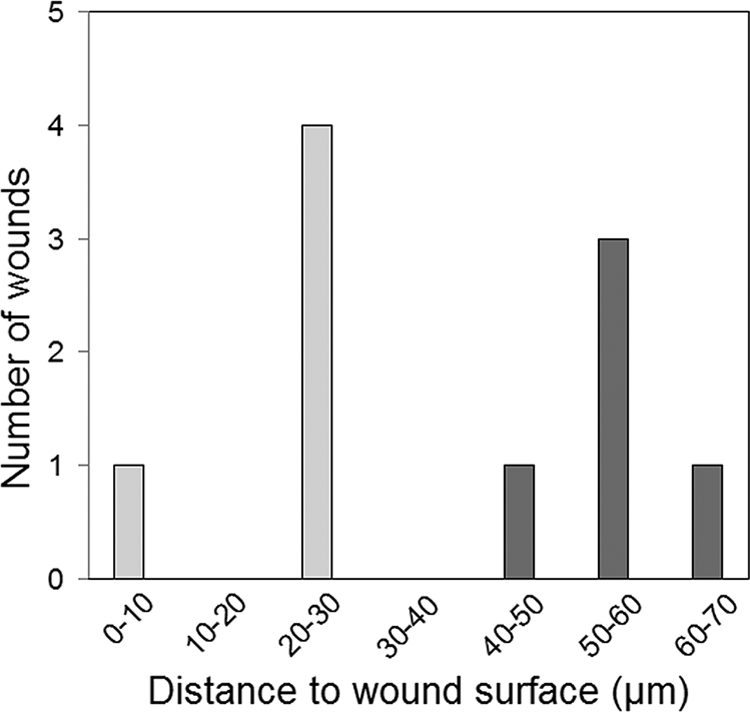
Distribution of the distances from the wound surface to the center of mass of S. aureus aggregates (light gray shading) or P. aeruginosa aggregates (dark gray shading). The distances are average values obtained from the analysis of 15 images for each wound sample.
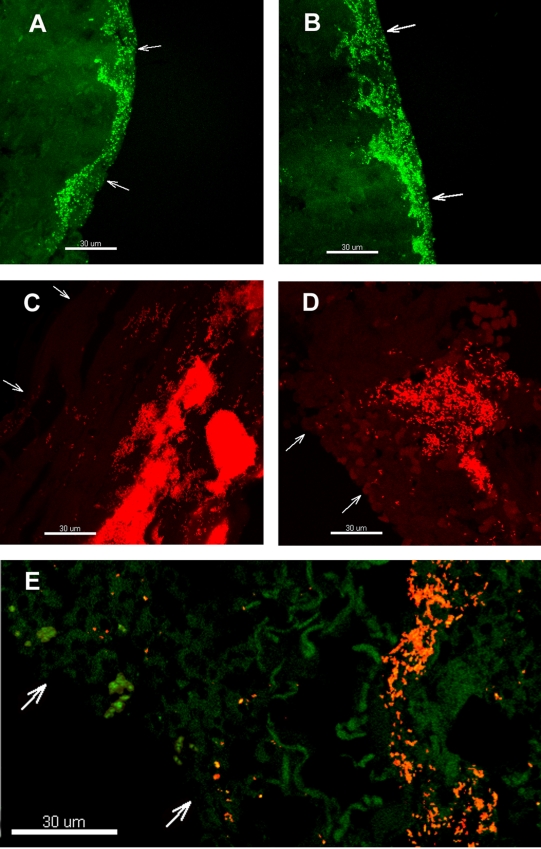
FIG. 3.
Representative CLSM images of S. aureus (A and B), P. aeruginosa (C and D), and both organisms (E) in chronic wounds. The bacteria were detected by PNA-FISH with an FITC-labeled S. aureus-specific probe (green) or a TxR-labeled P. aeruginosa-specific probe (red), or a mixture of the two probes. Arrows point to the wound surfaces. Bars, 30 μm.
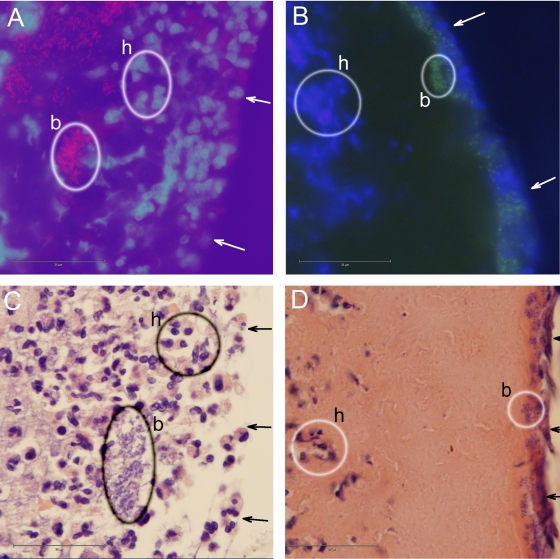
FIG. 4.
Epifluorescence micrographs (A and B) and bright-field micrographs (C and D) showing red PNA-FISH-stained P. aeruginosa cells (A), green PNA-FISH-stained S. aureus cells (B), blue DAPI-stained host cells (A and B), H&E-stained host cells and P. aeruginosa cells (C), and H&E-stained host cells and S. aureus cells (D). Some of the bacteria and host cells are encircled and labeled b and h, respectively. Arrows point to the wound surfaces. Bars, 35 μm.
DISCUSSION
Although the microflora of chronic wounds is polymicrobial and heterogeneous, S. aureus and P. aeruginosa are among the bacteria that are most frequently isolated from these wounds (8, 12, 15). In the present study, we characterized the distribution of P. aeruginosa and S. aureus in nine chronic wounds: four wounds with S. aureus, four wounds with P. aeruginosa, and one wound with both S. aureus and P. aeruginosa. Analysis of the images obtained by PNA-FISH and CLSM indicated that P. aeruginosa was located significantly deeper in the wound bed than S. aureus.
We previously investigated samples from 22 chronic venous leg ulcers for the presence of bacteria by standard culturing and PNA-FISH (17). By swab sample cultivation, we found that 12 of the wounds were colonized with S. aureus, whereas 5 of them were colonized with P. aeruginosa. Conversely, by using PNA-FISH, P. aeruginosa was detected in nine of the wounds, while S. aureus was detected in only two of them. Our present finding that P. aeruginosa is located in the deeper regions of the wound bed offers an explanation for the different results obtained by swab sample cultivation and PNA-FISH. Because the swab sample culture technique detects bacteria in the upper region of the wounds, bacteria that primarily colonize the deeper regions may not be detected.
For a good healing response, the bacterial load of chronic wounds needs to be optimally managed. Topical antimicrobials can in some cases effectively control superficial bacterial burdens if the infection is localized but may not be appropriate for highly infected wounds. Systemic antibiotics may be effective in some cases of severe infection with tissue invasion (23). The use of a nanocrystalline silver dressing was shown to decrease the superficial bacterial burden, as assessed by surface swab investigation, but had no effect on the bacterial burden of the deep wound compartment, as measured by tissue biopsy (24). Thus, it is of great importance to define the spatial organization of the bacterial species within a chronic wound for the most effective management of the infection. A relevant picture of the spatial organization of the bacteria in a chronic wound might be obtained by using molecular methods, such as denaturing gradient gel electrophoresis (2, 8) and FISH (16), in combination with traditional culturing of swab as well as biopsy samples.
The biofilm mode of growth provides bacteria characteristics different from those of their planktonic counterparts, such as protection against the activities of the host immune system and increased tolerance to antimicrobial treatments (7). P. aeruginosa bacteria in biofilms express quorum-sensing-controlled virulence factors that can kill or eliminate the activity of host immune cells. It has been shown that rhamnolipid, a leukocidal toxin produced by P. aeruginosa, causes rapid necrosis of PMNs in vitro (16). Bjarnsholt and colleagues proposed that rhamnolipid offers a protective shield against the activities of host immune cells and demonstrated that aggregates of P. aeruginosa in chronic wounds were surrounded by host cells, possibly PMNs, but were not penetrated (5, 17), similar to what was observed in in vitro biofilms of P. aeruginosa overlaid with freshly isolated PMNs (4). The bacteria in chronic wounds are expected to compete with each other for the available nutrients. The ability of P. aeruginosa to migrate via type IV pili and flagellum-mediated motility in biofilms (3, 18, 19) and to produce virulence factors that can eliminate the activity of host defense systems (4, 16) may explain the presence of these bacteria in the deeper regions of chronic wounds. The destruction of PMNs by virulence factors produced by P. aeruginosa bacteria located in the deeper regions of chronic wounds may be one of the factors that causes a persistent influx of PMNs and that keeps the wound in an inflammatory stage. However, more research is required before specific bacterial species in specific modes of growth can be identified as the causative agents in chronic wounds.
Acknowledgments
We thank Jette Pedersen from the Bartholin Institute, University of Copenhagen, and Anne Jørgensen from the Department of Pathology, University of Copenhagen, for help with preparing and microscopy of the tissue sections. AdvanDx, Inc., is gratefully acknowledged for providing the PNA probes.
M.G. received financial support for the present study from the Danish Strategic Research Council. T.B. received financial support from the Carlsberg Foundation and the Lundbeck Foundation).
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Agren, M. S., W. H. Eaglstein, M. W. J. Ferguson, K. G. Harding, K. Moore, U. K. Saarialo-Kere, and G. S. Schultz. 2000. Causes and effects of the chronic inflammation in venous leg ulcers. Acta Dermatol. Venerol. Suppl. 210:3-17. [PubMed] [Google Scholar]
- 2.Andersen, A., K. E. Hill, P. Stephens, D. W. Thomas, B. Jorgensen, and K. A. Krogfelt. 2007. Bacterial profiling study using skin grafting, standard culture and molecular bacteriological methods. J. Wound Care 16:171-175. [DOI] [PubMed] [Google Scholar]
- 3.Barken, K. B., S. J. Pamp, L. Yang, M. Gjermansen, J. J. Bertrand, M. Klausen, M. Givskov, C. B. Whitchurch, J. N. Engel, and T. Tolker-Nielsen. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331-2343. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt, T., P. Ø. Jensen, M. Burmølle, M. Hentzer, J. A. Haagensen, H. P. Hougen, H. Calum, K. G. Madsen, C. Moser, S. Molin, N. Høiby, and M. Givskov. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373-383. [DOI] [PubMed] [Google Scholar]
- 5.Bjarnsholt, T., K. Kirketerp-Møller, P. O. Jensen, K. G. Madsen, R. Phipps, K. Krogfelt, N. Hoiby, and M. Givskov. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2-10. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 8.Davies, C. E., K. E. Hill, M. J. Wilson, P. Stephens, C. M. Hill, K. G. Harding, and D. W. Thomas. 2004. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J. Clin. Microbiol. 42:3549-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, S. C., C. Ricotti, A. Cazzaniga, E. Welsh, W. H. Eaglstein, and P. M. Mertz. 2008. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 16:23-29. [DOI] [PubMed] [Google Scholar]
- 10.Fedtke, I., F. Gotz, and A. Peschel. 2004. Bacterial evasion of innate host defences: the Staphylococcus aureus lesson. Int. J. Med. Microbiol. 294:189-194.. [DOI] [PubMed] [Google Scholar]
- 11.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948-958. [DOI] [PubMed] [Google Scholar]
- 12.Gjødsbol, K., J. J. Christiensen, T. Karlsmark, B. Jørgensen, B. M. Klein, and K. A. Krogfelt. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int. Wound J. 3:225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottrup, F. 2004. A specialized wound healing center concept: importance of a multidisciplinary department structure and surgical treatment facilities in the treatment of chronic wounds. Am. J. Surg. 187:38S-43S. [DOI] [PubMed] [Google Scholar]
- 14.Halbert, A. R., M. C. Stacey, J. B. Rohr, and A. Jopp-McKay. 1992. The effect of bacterial colonization on venous leg ulcer healing. Australas. J. Dermatol. 33:75-80. [DOI] [PubMed] [Google Scholar]
- 15.James, G. A., E. Swogger, R. Wolcott, E. Pulcini, P. Secor, J. Sestrich, J. W. Costerton, and P. S. Stewart. 2008. Biofilms in chronic wounds. Wound Repair Regen. 16:37-44. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, P. Ø., T. Bjarnsholt, R. Phipps, T. B. Rasmussen, H. Calum, L. Christoffersen, C. Moser, P. Williams, T. Pressler, M. Givskov, and N. Høiby. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum sensing controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329-1338. [DOI] [PubMed] [Google Scholar]
- 17.Kirketerp-Møller, K., P. O. Jensen, M. Fazli, K. G. Madsen, J. Pedersen, C. Moser, T. Tolker-Nielsen, N. Hoiby, M. Givskov, and T. Bjarnsholt. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 19.Klausen, M., A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of microbial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 20.Madsen, S. M., H. Westh, L. Danielsen, and V. T. Rosdahl. 1996. Bacterial colonization and healing of venous leg ulcers. APMIS 104:895-899. [DOI] [PubMed] [Google Scholar]
- 21.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagase, H., and J. F. Woessner, Jr. 1999. Matrix metalloproteinases. J. Biol. Chem. 274:21491-21494. [DOI] [PubMed] [Google Scholar]
- 23.Schultz, G. S., R. G. Sibbald, V. Falanga, E. A. Ayello, C. Dowsett, K. Harding, M. Romanelli, M. C. Stacey, L. Teot, and W. Vanscheidt. 2003. Wound bed preparation: a systematic approach to wound management. Wound Repair Regen. 11:S1-28. [DOI] [PubMed] [Google Scholar]
- 24.Sibbad, R. G., A. C. Browne, P. Coutts, and D. Queen. 2001. Screening evaluation of an ionized nanocrystalline silver dressing in chronic wound care. Ostomy Wound Manage. 47:38-43. [PubMed] [Google Scholar]
- 25.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 26.Yager, D. R., and B. C. Nwomeh. 1999. The proteolytic environment of chronic wounds. Wound Repair Regen. 7:433-441. [DOI] [PubMed] [Google Scholar]