Abstract
Point source norovirus outbreaks can be difficult to track due to high background levels of the virus in the environment and the limited strain variation in some genotyping regions. However, rapid and accurate source identification can limit the spread of a foodborne outbreak and reduce the number of cases. Harmonization of genotyping assays is critical for enabling the rapid exchange of sequence data nationally and internationally. Several regions of the genome have been proposed for this purpose, but no consensus has been reached. In the present study, two standardized genotyping protocols (region C and region D) were evaluated by nine laboratories in Canada and the United States, using a coded panel of 96 fecal specimens representing 22 different norovirus genotypes. Overall, region C typing had a success rate of 78% compared to 52% for region D; however, region D provides greater nucleotide sequence diversity for identifying new GII.4 variant strains. Significant differences in the genotyping success rate were observed among the nine participating laboratories (10% to 100%) and among the different genotypes (6% to 100%). For several genogroup II strains, reduced region D amplification correlated directly with mismatches between primer sequences and the template. Based on overall performance, we recommend the region C protocol for routine genotyping of noroviruses, while the region D protocol may be useful for identifying new GII.4 variants. Standardized genotyping protocols will enable rapid exchange of outbreak and sequence data through electronic norovirus surveillance networks.
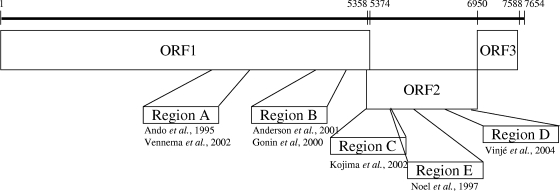
Noroviruses (NoVs) are the leading cause of viral gastroenteritis worldwide (9). In contrast to other gastroenteritis viruses, NoVs infect all age groups (3). The viruses are spread rapidly and can cause large outbreaks in a wide variety of settings, including health care facilities, restaurants, and schools (5, 6, 11, 17, 20, 23, 32). Since many countries do not have national NoV surveillance programs, the completeness and quality of reporting systems vary substantially. Outbreak surveillance systems that combine virological and epidemiological information provide an excellent mechanism for tracking differences in rates of NoV activity over time. In addition, they support identification of newly emerging strains and of common-source outbreaks so that transmission routes can be interrupted (5, 15, 18). Because NoVs cannot be routinely grown in cell culture, most clinical laboratories use either conventional or real-time reverse transcription (RT)-PCR for detection of the virus (4, 7, 13, 22, 25, 26, 29, 30). Currently, classification of NoV genotypes is based on genetic diversity within the entire capsid gene (2, 16, 33). However, most laboratories sequence a small region (200 to 344 bp) of either the polymerase (regions A and B) or major capsid (VP1; regions C, D, and E) gene (1, 2, 8, 14, 21, 27, 28) to genotype strains (Fig. 1). This practice has led to a lack of data on which region is the most useful to genotype and, consequently, a lack of standardization of genotyping methods (30).
FIG. 1.
Schematic representation of the norovirus genome and the positions of several representative regions (regions A to E) that have been used for genotyping. Numbers refer to positions in the Norwalk GI.1 virus genome (GenBank accession number M87661).
To track international NoV outbreaks, elucidate their transmission routes, and establish standardized genotyping methods in clinical virology laboratories in Canada and the United States, two genotyping protocols (region C [14] and region D [28]) that target regions within the NoV capsid gene (ORF2) were evaluated, using a coded panel of stool samples.
MATERIALS AND METHODS
Norovirus panel.
Panels of 96 archived fecal specimens coded and previously tested for NoV by TaqMan real-time RT-PCR (13, 26) were assembled at Health Canada, Ottawa, and at the Centers for Disease Control and Prevention (CDC), Atlanta, GA. The specimens were derived from outbreaks of acute gastroenteritis in Canada and the United States from 2001 through 2007 and were stored as whole stools at either 4°C, −20°C, or −70°C. The panel included 86 NoV-positive stool samples that had been genotyped by sequencing RT-PCR products targeting region B (1), region C (14), or region D (28), 7 NoV-negative samples, 2 rotavirus group A-positive samples, and 1 adenovirus 40/41-positive sample. The final panel included five replicates of a GI.3b sample and five replicates of a GII.4 sample as positive controls. The panel samples were frozen at −70°C and shipped on dry ice to nine participating laboratories (coded A, B, C, D, E, F, G, H, and I) in Canada and the United States. The second, smaller panel tested in laboratories C, D, E, and G was 12 RNA specimens (NoV GI.2, GI.3, GI.4, GI.6, GI.8, GI.13, GII.2, GII.3 [two samples], GII.4, GII.5, and GII.7) extracted and coded at Health Canada, Ottawa.
To reduce the number of variables in evaluating the two genotyping protocols, the participating laboratories in this study were also provided with aliquots of oligonucleotide primers (Sigma-Genosys, Oakville, Ontario, Canada) and OneStep RT-PCR kits (Qiagen, Mississauga, Ontario, Canada).
Viral RNA extraction.
All laboratories began the procedure with a 10 to 20% stool suspension clarified by centrifugation (2,000 × g for 20 min or 6,000 × g for 5 min) and used commercially available RNA extraction procedures (Table 1). Laboratory A preincubated the clarified specimens with ≥24 milli-Anson units of proteinase K (Qiagen) prior to extraction. Laboratory B used QIAshredder columns (Qiagen) prior to RNA extraction, and laboratory H pretreated the samples with Vertrel (DuPont, Mississauga, Ontario, Canada) prior to extraction. For all procedures, manufacturers' recommendations were followed.
TABLE 1.
Summary of viral RNA extraction methods used in this study
| Laboratory | Extraction buffer | Extraction procedure | Modifications |
|---|---|---|---|
| A | QIAamp viral RNA | BioRobot MDx | Pretreatment with proteinase K |
| B | MagAttract M48 | M48 robot | Pretreatment with QIAshredder columns |
| C | QIAamp viral RNA | Manual | None |
| D | QIAamp viral RNA | Manual | None |
| E | MegaZorb | Manual | None |
| F | NucliSENS easyMAG | Manual | None |
| G | MagMAX | KingFisher | None |
| H | QIAamp viral RNA | Manual | Pretreatment with Vertrel |
| I | NucliSENS easyMAG | NucliSENS easyMAG | None |
Norovirus TaqMan real-time RT-PCR detection.
TaqMan real-time RT-PCR assays for GI and GII NoVs were conducted in each laboratory according to published procedures (see Table S1 through S3 in the supplemental material) (12, 13, 22, 26). Laboratory A used a GI and GII multiplex format with QuantiTect probe RT-PCR kits (Qiagen). Approximately 2 × 104 copies of MS2 bacteriophage RNA (Roche, Mississauga, Ontario, Canada) was coextracted with each sample as an internal control to detect potential inhibitors. Laboratory B also used QuantiTect Probe RT-PCR kits (Qiagen) in monoplex reactions for GII. Laboratories C and D used Brilliant II QRT-PCR core reagent kits and One-Step RT-PCR kits (Stratagene, La Jolla, CA) in monoplex reactions. Laboratory E performed cDNA synthesis using random priming with SuperScript II kits (Invitrogen, Burlington, Ontario, Canada) prior to GI and GII multiplex real-time PCR detection with TaqMan universal DNA master mix kits (Applied Biosystems, Austin, TX). Laboratories F and G tested samples in a monoplex format using AgPath-ID RT-PCR kits (Ambion, Austin, TX). Laboratory H used the monoplex format with LightCycler RNA amplification kit (Roche, Nutley, NJ) reagents. Laboratory I performed GI and GII multiplex reactions by using Superscript III Platinum One-Step qRT-PCR kits (Invitrogen).
Conventional region C and region D RT-PCR assays for genotyping of norovirus.
Samples positive by TaqMan real-time RT-PCR were amplified by both a region C and a region D conventional RT-PCR assay, using a OneStep RT-PCR kit (Qiagen), which was provided by the coordinating laboratories. For region C, 5 μl of extracted RNA was amplified with 0.4 μM of each oligonucleotide primer, G1SKF and G1SKR for GI, or G2SKF and G2SKR for GII (14), in a final reaction volume of 25 μl. After 30 min of RT at 42°C, followed by heat activation of Taq polymerase for 15 min at 95°C, PCR consisting of 40 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s followed by a final extension for 5 min at 72°C was performed. For region D, 5 μl of extracted RNA was amplified with 0.32 μM of each oligonucleotide primer, CapA, CapB1, and CapB2 for GI, or CapC, CapD1, and CapD3 for GII (28). RNA was reverse transcribed at 42°C for 60 min, followed by heating at 95°C for 15 min. Thermocycling conditions consisted of 40 cycles at 94°C for 60 s, 40°C for 60 s, and 72°C for 60 s followed by a final extension at 72°C for 10 min.
DNA sequencing.
All RT-PCR products of the expected size (region C, 330 bp [GI] or 344 bp [GII]; region D, 177 bp [GI] or 253 bp [GII]) were purified and bidirectionally sequenced, using a BigDye Terminator kit (Applied Biosystems) (laboratories A, B, E, F, G and I) on an ABI Prism 3100, 3130, or 3130XL genetic analyzer (Applied Biosystems). RT-PCR products from laboratories C and D were sequenced at the Genome Quebec Innovation Center, and RT-PCR products from laboratory H were purified using a QIAquick gel extraction kit and sequenced using a Beckman DTCS Quick Start kit and CEQ 8000 genetic analyzer (Beckman Coulter, Inc., Fullerton, CA).
Data analysis.
All results from each participating laboratory were submitted to the coordinating laboratories at Health Canada and the CDC. Results were included in the data analysis when they fulfilled the following criteria: (i) each region C- or region D-positive RT-PCR product was confirmed by sequencing and (ii) the sequence was not identical to that from another panel sample (this condition was waived for known replicates). In order to focus the analysis on the genotyping assays, only samples that tested positive by TaqMan real-time RT-PCR in at least 4 of 5 (GI; 18 samples) or 7 of 9 (GII; 49 samples) laboratories were included. Region C and region D sequences were genotyped by comparison with NoV reference sequences (33), using Pearson correlation similarity settings. BioNumerics (Applied Maths, Austin, TX) was used to construct neighbor-joining trees with bootstrap values derived from 2,000 replicates.
A permutation test was used to compare region C typing with region D typing, keeping the number of samples and number of positives fixed within each lab. The ability of a specific laboratory to detect a virus using region C typing or region D typing was assessed by using a sign test (10) on the pairwise differences of the indicator of detection among the labs. These pairwise comparisons were used to group labs that were not significantly different at a significance level of 0.01. This value was chosen to reduce the chance of false positives.
RESULTS
Quality control.
To assess the accuracy and reproducibility of the results submitted from each participating laboratory, we first examined the results for the NoV-negative (n = 10) and -positive (GI, n = 5; GII, n = 5) control samples that had been randomly dispersed in the stool panel. Seven of the 10 negative-control samples tested negative for NoV by TaqMan real-time RT-PCR in all laboratories. The remaining three samples tested positive for NoV GII (threshold cycle [CT] values ranged from 33.0 to 40.9) in one (H), two (C and E), or three (B, E, and I) laboratories. One of these six test results was confirmed, but the region C sequence obtained was identical to that of an adjacent panel sample. Thus, none of the false positives met the criteria for inclusion in the data analysis. Five (E, F, G, H, and I) of the nine laboratories detected the GI.3b-positive control samples by real-time RT-PCR (CT range, 27.7 to 38.0). Therefore, only GI data from these partner laboratories was included for further analysis. All partner laboratories detected the GII.4-positive control samples by real-time RT-PCR (CT range, 18.7 to 34.5); therefore, all GII genotyping data were included in the analysis.
Comparison of genotyping results.
The rate of GI and GII real-time RT-PCR detection in each laboratory ranged from 73% (36/49 GII, laboratory D) to 100% (49/49 GII, laboratories A, B, and I) (Table 2). Genotyping rates ranged from 44% (laboratory H) to 100% (laboratories F and I) for region C and from 10% (laboratory C) to 94% (laboratory F) for region D (Table 2). Significant differences were noted among testing laboratories for each of the standardized genotyping protocols (P < 0.01) (Table 2).
TABLE 2.
Norovirus detection and genotyping results from nine laboratories for a coded panel of 96 stool specimens
| Laboratorya | No. of samples positive for indicated genotype by real-time RT-PCR: |
No. (%) of samples positive for indicated genotype by region C typingb: |
No. (%) of samples positive for indicated genotype by region D typingb: |
|||
|---|---|---|---|---|---|---|
| GI | GII | GI | GII | GI | GII | |
| A | 0 | 49 | 46 (94%) d | 44 (90%) j | ||
| B | 0 | 49 | 45 (92%) d | 33 (67%) kl | ||
| C | 0 | 48 | 31 (65%) cf | 5 (10%) mn | ||
| D | 0 | 36 | 28 (78%) def | 21 (58%) lm | ||
| E | 18 | 47 | 13 (72%) bc | 26 (55%) f | 9 (50%) hi | 7 (15%) n |
| F | 17 | 46 | 17 (100%) b | 44 (96%) d | 16 (94%) g | 35 (76%) k |
| G | 18 | 40 | 13 (72%) bc | 35 (88%) de | 15 (83%) gh | 20 (50%) kl |
| H | 18 | 45 | 8 (44%) c | 24 (53%) f | 7 (39%) i | 14 (31%) n |
| I | 18 | 49 | 18 (100%) b | 43 (88%) d | 16 (89%) gh | 33 (67%) kl |
Laboratories A through D were excluded from the GI analysis because they did not detect the positive controls.
Percentages in parentheses are based on the number of real-time RT-PCR positives from each individual laboratory. Values followed by the same lowercase letter or letters are not significantly different (P < 0.01).
A small, nucleic acid panel (n = 12) was also tested in laboratories C, D, E, and G. The results indicate that the overall conclusions from this study are sound, with the region C assay typing 100% of GI samples and 78% of GII samples and the region D assay typing 94% of GI samples and 56% of GII samples (data not shown). Interestingly, laboratories C and E had much better typing results with RNA than with stool samples, indicating that extraction and detection methods contribute significantly to the observed lab-to-lab variability (data not shown).
To compare the differences in genotyping rates between the region C and D assays, we assembled and stratified the data from all laboratories by genotype (Table 3). All genotypes were amplified and sequenced by at least one lab. Genotyping rates ranged from 59% (GII.10) to 100% (GI.4) for region C and from 6% (GII.14) to 100% (GI.2 and GI.3) for region D (Table 3). For strains GI.1, GII.2, GII.3, GII.4, GII.5, GII.7, GII.10, and GII.14, region C amplification was more efficient than region D amplification (P < 0.05) (Table 3).
TABLE 3.
Cumulative results from nine participating laboratories for comparisons of region C and region D genotyping and correlation with primer-template mismatchesa
| Genotype | No. of samples positive and typed by real-time RT-PCR | No. (%) of samples positive by region C typingb | No. (%) of samples positive by region D typingb | No. of region C mismatches | No. of region D mismatches |
|---|---|---|---|---|---|
| GI.1 | 23 | 18 (78%)c | 15 (65%) | 0 | 0 |
| GI.2 | 5 | 4 (80%) | 5 (100%) | 0 | 1 |
| GI.3 | 5 | 3 (60%) | 5 (100%) | 2 | 1 |
| GI.3b | 24 | 16 (67%) | 15 (63%) | 2 | 0 |
| GI.4 | 10 | 10 (100%) | 5 (50%) | 2 | 0 |
| GI.6 | 5 | 4 (80%) | 3 (60%) | 1 | 1 |
| GI.7 | 5 | 4 (80%) | 4 (80%) | 3 | 0 |
| GI.13 | 10 | 6 (60%) | 8 (80%) | 3 | N/A |
| GI.14 | 5 | 4 (80%) | 3 (60%) | 3 | N/A |
| GII.1 | 9 | 7 (78%) | 7 (78%) | 0 | 1 |
| GII.2 | 25 | 22 (88%)c | 8 (32%) | 0 | 2 |
| GII.3 | 74 | 71 (96%)c | 54 (73%) | 0 | 1 |
| GII.4 | 141 | 117 (83%)c | 86 (61%) | 0 | 2 |
| GII.5 | 18 | 15 (83%)c | 9 (50%) | 0 | 2 |
| GII.6 | 42 | 29 (69%) | 27 (64%) | 0 | 1 |
| GII.7 | 18 | 15 (83%)c | 3 (17%) | 1 | 3 |
| GII.10 | 17 | 10 (59%)c | 2 (12%) | 2 | 4 |
| GII.12 | 8 | 5 (63%) | 2 (25%) | 0 | 2 |
| GII.14 | 18 | 14 (78%)c | 1 (6%) | 1 | 4 |
| GII.16 | 9 | 9 (100%) | 6 (67%) | 0 | 2 |
| GII.17 | 8 | 6 (75%) | 3 (38%) | 0 | 4 |
| GII.18 | 7 | 2 (29%) | 4 (57%) | 1 | 2 |
Sequences from twenty-two different norovirus genotypes were used for reference. Reference strains: GI.1 (GenBank accession number M87661), GI.2 (L07418), GI.3 (U04469), GI.3b (AF414405), GI.4 (AB042808), GI.6 (AF093797), GI.7 (AB058535 and AJ277609), GI.13 (AB112132), GI.14 (AB112100), GII.1 (U07611), GII.2 (X81879), GII.3 (U02030), GII.4 (X76716), GII.5 (AF397156 and AJ277607), GII.6 (AB039776 and AJ277620), GII.7 (AF414409 and AJ277608), GII.10 (AF504671), GII.12 (AB045603 and AJ277618), GII.14 (AY130761), GII.16 (AY502010), GII.17 (AY502009), GII.18 (AY823305). N/A, sequence information not available for the reference strain.
Percentages in parentheses are based on the number of real-time RT-PCR positives for each individual genotype.
There was a statistically significant (P < 0.05) difference between the efficiency of the region C and region D amplification protocols for these genotypes.
To examine whether a lack of primer-template homology could explain differences in detection rates between region C and region D primers, we determined the number of mismatches between the oligonucleotide primers and GI and GII reference strains. Region C primers, which detected 83% of GII.7 strains, had only one mismatch with the GII.7 reference strain, whereas region D primers, which detected 17% of the GII.7 strains, had three mismatches (Table 3). The correlation between primer mismatches and detection rate was maintained for strains GII.2, GII.3, GII.4, GII.5, GII.10, and GII.14 (Table 3). Interestingly, region C primers detected 87% and region D primers detected 74% of the GI.1 strains (P < 0.05), while there were no mismatches with the GI.I reference strain in either primer set (Table 3).
Comparative resolution of region C and region D strain differentiation.
We compared the phylogenetic analyses of the panel samples to those of standard genotypes (33). A few discrepancies between the genotypes assigned by region C and region D protocols were noted. Most (15/18, 83%) GI viruses grouped identically. One sample was assigned to the GI.3 genotype using region D and to the GI.14 genotype using region C. Two other samples were assigned to the GI.8 genotype using region D and to the GI.13 genotype using region C. The GII samples were assigned to the same cluster by both protocols (48/49, 98%), with the exception of a GII.18 strain that was untypeable by region D.
Phylogenetic analyses of 17 GII.4 region D sequences demonstrated that 10 strains belong to the Farmington Hills/2002 variant, 2 strains to the Hunter/2004 variant, 2 strains to the Laurens/2006a variant, and 3 strains to the GII.4 Minerva/2006b variant (Fig. 2B), with nucleotide sequence identities ranging from 87 to 95%. This sequence variation between GII.4 strains is higher than for region C sequences, where nucleotide identities ranged from 96 to 98%. The region C and region D sequences of one GII.4 panel sample (046) fell in different clusters (Fig. 2A).
FIG. 2.
Neighbor-joining analysis of the sequences obtained from GII.4-positive panel samples with CDC reference sequences for (A) region C and (B) region D analysis of GII.4 noroviruses. Bootstrap scores for branches are shown as percentages of 2,000 replicates. Sequences used as standards were GII.4 Camberwell/<1996 (GenBank accession number AF145896), GII.4 Grimsby/1996 (AJ004864), GII.4 Farmington Hills/2002 (AY502023), GII.4 Hunter/2004 (DQ078794), GII.4 Laurens/2006a (not in database), and GII.4 Minerva/2006b (not in database).
DISCUSSION
We compared two different NoV genotyping protocols (region C and region D) and evaluated their performances in nine laboratories in Canada and the United States on a coded stool panel. Overall, 78% of the GI samples were genotyped by the region C assay compared to 52% by the region D assay.
In a previous study, the region D protocol, which targets a more-variable region of the VP1 gene than region C, demonstrated an excellent correlation with genotyping of GI and GII NoVs without needing to resort to complete VP1 sequencing (28). The initial publications of the region C and region D protocols reported genotyping rates of 100% and 95%, respectively (14, 28). Our data may reflect the increased diversity of the contemporary GII strains included in our stool panel. A previous comparison of different NoV RT-PCR assays gave a similar range of sensitivities (52 to 73%) for assays targeting a partial region of the polymerase gene (30). For several panel strains, a direct correlation between the number of nucleotide mismatches of the region D primers and the reference sequences for each genotype and the failure to genotype strains was found. However, for strains that were not amplified and did not have mismatches between primers and reference sequences, G/C content and secondary structure may have affected amplification efficiency. Alternatively, a simple substitution in the primer target region of the field strain compared to the prototype strain may explain this. Data from our study will be used to refine the region D primers, which were developed in 2003, to improve the efficiency of this protocol in genotyping strains as the ability to differentiate GII.4 variant strains becomes greater.
The lack of a standardized nomenclature for NoV genotyping using different regions of the genome led several participating laboratories to assign identical sequences to different genotypes (data not shown). Based on the amino acid sequence of the complete VP1 gene, NoVs can be classified into five genogroups and at least 32 genotypes (8 in GI.1-8, 19 in GII, 2 in GIII, 2 in GIV, and 1 in GV) (19, 31, 33). However, since several genotypes (e.g., GI.6) form distinct clusters at the nucleotide level, further standardization based on partial nucleotide sequences awaits international agreement.
An important limitation of this study was the variation in viral RNA extraction methods used by the different participating laboratories. In addition to the differences in extraction methods, not all laboratories employed the same GI and GII real-time RT-PCR protocols, which may explain in part the differences in assay sensitivity as well as why several laboratories were unable to detect the GI.3b-positive control strain. To reduce variation in RNA extraction methods, we sent out a small panel of nucleic acids to four laboratories and found that the variability between region C and region D genotyping was significantly reduced, supporting the overall conclusions of this study.
Several electronic surveillance systems for NoV outbreaks have recently been initiated, including CaliciNet in the United States, ViroNet in Canada, and a global norovirus surveillance network called NoroNet (24). Standardized genotyping protocols for NoV are fundamental for accurate genotyping as well as for rapid exchange of NoV sequences for early identification of newly emerging strains and international norovirus outbreaks.
In conclusion, this study is an important step toward the standardization of NoV genotyping methods for clinical virology laboratories. Further efforts should also include viral RNA extraction methods. Based on overall performance, the region C protocol is recommended for routine genotyping of noroviruses, while the region D protocol has a higher resolution to better distinguish GII.4 variant strains.
Supplementary Material
Acknowledgments
The authors thank Reuben Chen, Carole Danis, AliReza Eshaghi, Emily Hopkins, Anu Shukla, and Pierre Ward for expert technical assistance. We thank Sabah Bidawid, Theodore Kuschak, Lai-King Ng, Neil Simonsen, and Niki Vegh for expert advice. We also thank Franco Pagotto and Anne Reid for critical evaluations of the manuscript and helpful comments. This project was financially supported by the Canadian Public Health Laboratory Network, Health Canada, and the Centers for Disease Control and Prevention.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the funding agency or the Centers for Disease Control and Prevention (CDC). This article did receive clearance through the appropriate channels at the CDC prior to submission.
Footnotes
Published ahead of print on 21 October, 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Anderson, A. D., A. G. Heryford, J. P. Sarisky, C. Higgins, S. S. Monroe, R. S. Beard, C. M. Newport, J. L. Cashdollar, G. S. Fout, D. E. Robbins, S. A. Seys, K. J. Musgrave, C. Medus, J. Vinjé, J. S. Bresee, H. M. Mainzer, and R. I. Glass. 2003. A waterborne outbreak of Norwalk-like virus among snowmobilers—Wyoming, 2001. J. Infect. Dis. 187:303-306. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like” viruses. J. Infect. Dis. 181[Suppl. 2]:S336-S348. [DOI] [PubMed] [Google Scholar]
- 3.Atmar, R. L., and M. K. Estes. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35:275-290. [DOI] [PubMed] [Google Scholar]
- 4.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2007. Norovirus activity—United States, 2006-2007. MMWR Morb. Mortal. Wkly. Rep. 56:842-846. [PubMed] [Google Scholar]
- 6.David, S. T., L. McIntyre, L. MacDougall, D. Kelly, S. Liem, K. Schallie, A. McNabb, A. Houde, P. Mueller, P. Ward, Y. L. Trottier, and J. Brassard. 2007. An outbreak of norovirus caused by consumption of oysters from geographically dispersed harvest sites, British Columbia, Canada, 2004. Foodborne Pathog. Dis. 4:349-358. [DOI] [PubMed] [Google Scholar]
- 7.Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79-87. [DOI] [PubMed] [Google Scholar]
- 8.Gonin, P., M. Couillard, and M. A. d'Halewyn. 2000. Genetic diversity and molecular epidemiology of Norwalk-like viruses. J. Infect. Dis. 182:691-697. [DOI] [PubMed] [Google Scholar]
- 9.Green, K. Y. 2007. Caliciviridae: the Noroviruses, p. 949-979. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Hollander, M., and D. A. Wolfe. 1973. Non-parametric statistical methods. John Wiley, New York, NY.
- 11.Jones, E. L., A. Kramer, M. Gaither, and C. P. Gerba. 2007. Role of fomite contamination during an outbreak of norovirus on houseboats. Int. J. Environ. Health Res. 17:123-131. [DOI] [PubMed] [Google Scholar]
- 12.Jothikumar, N., J. A. Lowther, K. Henshilwood, D. N. Lees, V. R. Hill, and J. Vinjé. 2005. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 71:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 15.Koopmans, M., H. Vennema, H. Heersma, E. van Strien, Y. van Duynhoven, D. Brown, M. Reacher, and B. Lopman. 2003. Early identification of common-source foodborne virus outbreaks in Europe. Emerg. Infect. Dis. 9:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koopmans, M., J. Vinjé, E. Duizer, M. de Wit, and Y. van Duijnhoven. 2001. Molecular epidemiology of human enteric caliciviruses in The Netherlands. Novartis Found. Symp. 238:197-214. [DOI] [PubMed] [Google Scholar]
- 17.Korsager, B., S. Hede, H. Boggild, B. E. Bottiger, and K. Molbak. 2005. Two outbreaks of norovirus infections associated with the consumption of imported frozen raspberries, Denmark, May-June 2005. Euro. Surveill. 10:E050623.1. [DOI] [PubMed] [Google Scholar]
- 18.Kroneman, A., H. Vennema, J. Harris, G. Reuter, C. H. von Bonsdorff, K. O. Hedlund, K. Vainio, V. Jackson, P. Pothier, J. Koch, E. Schreier, B. E. Bottiger, and M. Koopmans. 2006. Increase in norovirus activity reported in Europe. Euro. Surveill. 11:E061214.1. [DOI] [PubMed] [Google Scholar]
- 19.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 13:1071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maunula, L., I. T. Miettinen, and C. H. von Bonsdorff. 2005. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 11:1716-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noel, J. S., T. Ando, J. P. Leite, K. Y. Green, K. E. Dingle, M. K. Estes, Y. Seto, S. S. Monroe, and R. I. Glass. 1997. Correlation of patient immune responses with genetically characterized small round-structured viruses involved in outbreaks of nonbacterial acute gastroenteritis in the United States, 1990 to 1995. J. Med. Virol. 53:372-383. [DOI] [PubMed] [Google Scholar]
- 22.Pang, X. L., J. K. Preiksaitis, and B. Lee. 2005. Multiplex real time RT-PCR for the detection and quantitation of norovirus genogroups I and II in patients with acute gastroenteritis. J. Clin. Virol. 33:168-171. [DOI] [PubMed] [Google Scholar]
- 23.Schmid, D., E. Gschiel, M. Mann, S. Huhulescu, W. Ruppitsch, G. Bohm, J. Pichler, I. Lederer, G. Hoger, S. Heuberger, and F. Allerberger. 2007. Outbreak of acute gastroenteritis in an Austrian boarding school, September 2006. Euro. Surveill. 12:224. [PubMed] [Google Scholar]
- 24.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinjé, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII. 4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 25.Straub, T. M., K. Honer zu Bentrup, P. Orosz-Coghlan, A. Dohnalkova, B. K. Mayer, R. A. Bartholomew, C. O. Valdez, C. J. Bruckner-Lea, C. P. Gerba, M. Abbaszadegan, and C. A. Nickerson. 2007. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 13:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trujillo, A. A., K. A. McCaustland, D. P. Zheng, L. A. Hadley, G. Vaughn, S. M. Adams, T. Ando, R. I. Glass, and S. S. Monroe. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vennema, H., E. de Bruin, and M. Koopmans. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233-235. [DOI] [PubMed] [Google Scholar]
- 28.Vinjé, J., R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116:109-117. [DOI] [PubMed] [Google Scholar]
- 29.Vinjé, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 30.Vinjé, J., H. Vennema, L. Maunula, C. H. von Bonsdorff, M. Hoehne, E. Schreier, A. Richards, J. Green, D. Brown, S. S. Beard, S. S. Monroe, E. de Bruin, L. Svensson, and M. P. Koopmans. 2003. International collaborative study to compare reverse transcriptase PCR assays for detection and genotyping of noroviruses. J. Clin. Microbiol. 41:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Q. H., M. G. Han, S. Cheetham, M. Souza, J. A. Funk, and L. J. Saif. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yee, E. L., H. Palacio, R. L. Atmar, U. Shah, C. Kilborn, M. Faul, T. E. Gavagan, R. D. Feigin, J. Versalovic, F. H. Neill, A. L. Panlilio, M. Miller, J. Spahr, and R. I. Glass. 2007. Widespread outbreak of norovirus gastroenteritis among evacuees of Hurricane Katrina residing in a large “megashelter” in Houston, Texas: lessons learned for prevention. Clin. Infect. Dis. 44:1032-1039. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.