Abstract
Over the past few years, a number of new nucleic acid extraction methods and extraction platforms using chemistry combined with magnetic or silica particles have been developed, in combination with instruments to facilitate the extraction procedure. The objective of the present study was to investigate the suitability of these automated methods for the isolation of Toxoplasma gondii DNA from amniotic fluid (AF). Therefore, three automated procedures were compared to two commercialized manual extraction methods. The MagNA Pure Compact (Roche), BioRobot EZ1 (Qiagen), and easyMAG (bioMérieux) automated procedures were compared to two manual DNA extraction kits, the QIAamp DNA minikit (Qiagen) and the High Pure PCR template preparation kit (Roche). Evaluation was carried out with two specific Toxoplasma PCRs (targeting the 529-bp repeat element), inhibitor search PCRs, and human beta-globin PCRs. The samples each consisted of 4 ml of AF with or without a calibrated Toxoplasma gondii RH strain suspension (0, 1, 2.5, 5, and 25 tachyzoites/ml). All PCR assays were laboratory-developed real-time PCR assays, using either TaqMan or fluorescent resonance energy transfer probes. A total of 1,178 PCRs were performed, including 978 Toxoplasma PCRs. The automated and manual methods were similar in sensitivity for DNA extraction from T. gondii at the highest concentration (25 Toxoplasma gondii cells/ml). However, our results showed that the DNA extraction procedures led to variable efficacy in isolating low concentrations of tachyzoites in AF samples (<5 Toxoplasma gondii cells/ml), a difference that might have repercussions since low parasite concentrations in AF exist and can lead to congenital toxoplasmosis.
Molecular methods play an important role in the microbiological diagnosis of infectious diseases due to their high sensitivity and specificity (1, 5). Among these, PCR is now recognized as an essential diagnostic tool for congenital toxoplasmosis (1, 8, 11, 19, 20) as well as for toxoplasmosis in immunocompromised individuals (9, 16) and ocular toxoplasmosis (22, 26). Multicenter comparative evaluations have shown sensitivity differences in the PCR detection of Toxoplasma gondii in amniotic fluid (AF), especially when parasite loads were low (<10 T. gondii cells/ml [T/ml]) (2, 14). Since it has been reported that about half of AF samples infected by T. gondii contain fewer than 10 T/ml (8), it is important to use procedures that allow for the detection of such parasite concentrations.
Among the different steps that participate in ensuring a reliable molecular detection method, DNA extraction is crucial (3, 12, 24, 27). Indeed, prior to amplification, biological samples (tissue biopsy samples, body fluid samples, tissue scrapings, etc.) must be prepared not only to extract and concentrate the DNA but also to eliminate proteins, lipids, polysaccharides, and other potential inhibitors of the DNA polymerase. DNA extraction consists of nucleic acid isolation, purification, and concentration in an eluted product, and many commercial systems have now become available, replacing in-house methods in many laboratories (25).
One of the general aims of the French National Reference Centre for Toxoplasmosis (Centre National de Référence de la Toxoplasmose) is to determine the best molecular detection strategies for Toxoplasma and to recommend them to diagnosis laboratories. In this sense, a specific objective is to better define the importance of DNA extraction in the whole PCR process. This prompted us to compare the performance of widely used commercial DNA extraction kits for T. gondii detection using a strict experimental protocol. We carried out a prospective and multicenter study to evaluate five commercial DNA extraction procedures, two manual and three automated kits, for the isolation of T. gondii DNA in AF. The yield of DNA extraction was assessed by subsequent DNA amplification, combining two Toxoplasma-specific real-time PCR assays using either TaqMan or fluorescence resonance energy transfer (FRET) probe detection, as well as one human beta-globin PCR and one inhibitor search PCR. To detect fine differences among the extraction methods, we decided to work with low concentrations of the parasite, down to 5 tachyzoites/ml (2, 14).
MATERIALS AND METHODS
The study was conducted between April and June 2007 in four different hospital laboratories with recognized proficiency in the molecular detection of T. gondii.
Mimic sample preparation.
T. gondii tachyzoites (RH strain) were collected from infected mouse ascites samples, purified by passaging the ascites fluid through 3.0-μm-pore-size polycarbonate membrane filters (Nuclepore-Costar, France), washed in phosphate-buffered saline (pH 7.4), and numerated using a Malassez cell (six independent cell counts were performed). One AF sample, collected from a Toxoplasma-seronegative patient treated for hydramnios who provided written consent, was used to prepare all the mimic samples analyzed in this study. AF samples (4 ml) were centrifuged at 1,300 × g for 10 min, and each one was resuspended as a 190-μl aliquot. A calibrated suspension of parasites (10 μl) was then added to each of these aliquots to obtain concentrations of 1, 2.5, 5, and 25 T/ml. In practice, a stock solution of 10,000 tachyzoites/ml was prepared and diluted by five- and tenfold in phosphate-buffered saline to obtain suspensions of 2,000 and 1,000 tachyzoites/ml, respectively. The suspension of 2,000 tachyzoites/ml was diluted by fivefold to obtain a suspension of 400 tachyzoites/ml. Each parasite suspension was checked using Malassez cell counting. After centrifugation of 400 ml of total AF (1,300 × g for 10 min), resuspension in 19 ml, and distribution into 100 aliquots of 190 μl, 10 μl of each parasite suspension was added to the AF aliquot (190 μl) as follows. (i) A total of 10 μl of the 400-tachyzoites/ml suspension was added to 30 aliquots to obtain the 200-μl samples of concentrated AF with 1 T/ml. (ii) A total of 10 μl of the 1,000-tachyzoites/ml suspension was added to 15 aliquots to obtain AF samples with 2.5 T/ml. (iii) A total of 10 μl of the 2,000-tachyzoites/ml suspension was added to 15 aliquots to obtain AF samples with 5 T/ml. (iv) A total of 10 μl of the 10,000-tachyzoites/ml suspension was added to 10 aliquots to obtain AF samples with 25 T/ml. During the distribution of tachyzoite suspensions into the aliquots, the suspensions were mixed regularly. Then, 10 μl of phosphate-buffered saline was added to 30 aliquots to obtain AF samples with no parasites (negative controls). These samples, composed of 200 μl of concentrated AF (corresponding to 4 ml of total fluid) with or without Toxoplasma tachyzoites, were dispatched at 4°C in less than 24 h to three different laboratories and stored at 4°C before DNA extraction.
DNA extraction.
All DNA extractions were performed on the same day in three participating laboratories. The following three automated DNA extraction methods were tested in one laboratory: (i) the MagNA Pure Compact system using nucleic acid isolation kit I (Roche); (ii) the BioRobot EZ1 system using the EZ1 DNA tissue kit (Qiagen); and (iii) the NucliSens easyMAG system using the NucliSens magnetic kit (bioMérieux). Two manual extraction kits, the QIAamp DNA minikit (Qiagen) using the biological fluid protocol and the High Pure PCR template preparation kit (Roche), were used in two other laboratories (Table 1). For each sample, DNA was extracted according to the manufacturers' recommendations and was eluted in 100 μl of elution buffer, except for the NucliSens protocol, in which DNA was eluted in 110 μl. Twenty samples were extracted by each extraction method, comprising six control samples with no Toxoplasma sp. and 14 samples at 1 to 25 T/ml (Table 2). DNA extracts (n = 100) were immediately stored at −20°C and simultaneously sent at 4°C to the three laboratories involved in their molecular analysis. DNA extracts were stored at 4°C, and all the PCRs were performed within the next 15 days.
TABLE 1.
DNA extraction method characteristics
| Method (company) | Kit (company) | Protocol | Maximum no. of specimens/run | Elution buffer used |
|---|---|---|---|---|
| MagNA Pure Compact (Roche) | Nucleic acid isolation kit I (Roche) | Total nucleic acid plasma, 100-400 μla | 8 | NGc |
| BioRobot EZ1 (Qiagen) | EZ1 DNA tissue kit (Qiagen) | EZ1 DNA tissuea | 6 | Tris-EDTA |
| NucliSens easyMAG (bioMérieux) | NucliSens magnetic kit (bioMérieux) | Boom nucleic acid extractionb | 24 | 3 mM borate, pH 8.5 |
| Manual extraction | QIAamp DNA minikit (Qiagen) | Biological fluid | Sterile distilled water | |
| High Pure PCR template prepn kit (Roche) | Sterile distilled water |
A pretreatment using proteinase K is not included and must be done before starting extraction.
The lysis step is included in the protocol.
NG, not given.
TABLE 2.
Number of planned DNA extractions, Toxoplasma-specific PCR, inhibitor detections, and human beta-globin PCRs, according to the parasite concentration for each extraction method
| AF concn (T/ml) | No. of extractions done per method | No. of Toxoplasma PCRs |
No. of inhibitor detections |
No. of beta-globin quantifications |
||||
|---|---|---|---|---|---|---|---|---|
| Per extraction and per assay | Per assay | Per total no. of PCRs | Per extraction PCR | Per total no. of PCRs | Per extraction | Per total no. of PCRs | ||
| 0 | 6 | 1 | 6 | 12 | 1 | 6 | 1 | 6 |
| 1 | 6 | 8 | 48 | 96 | 1 | 6 | 1 | 6 |
| 2.5 | 3 | 8 | 24 | 48 | 1 | 3 | 1 | 3 |
| 5 | 3 | 6 | 18 | 36 | 1 | 3 | 1 | 3 |
| 25 | 2 | 4 | 8 | 16 | 1 | 2 | 1 | 2 |
| Total | 20 | 104 | 208 | 20 | 20 | |||
DNA amplification.
A total of 1,240 PCRs were planned, with 1,040 Toxoplasma-specific PCRs, 100 inhibitor amplification detection PCRs, and 100 human beta-globin PCRs. All in all, because a DNA volume less than 100 μl was collected from some extractions, only 1,178 PCRs were carried out. Of these, 978 were Toxoplasma PCRs, including 448 reactions with DNA extracted from 1-T/ml AF samples.
Toxoplasma-specific PCR.
Toxoplasma DNA amplification targeted the 529-bp genomic repeat element described by Homan et al. (13) and was performed with two different real-time PCR assays, using TaqMan probe detection on an ABI Prism 7000 instrument (Applied Biosystems) in one laboratory and FRET probes on a LightCycler I system (Roche) in the second laboratory. The first PCR assay was performed in a 25-μl reaction volume, including 5 μl of DNA, 0.8 μM of each primer (forward primer, 5′ CCTCTCCGACTCTCGTC 3′, and reverse primer, 5′ TCCTCCAGCCGTCTTGGA 3′), 0.2 μM of TaqMan probe (6-carboxyfluorescein-5′ CACGCCACCCCCTCA 3′-minor groove binder) and 1× TaqMan universal master mix with uracil N-glycosylase (Applied Biosystems). The second PCR assay was performed as previously described (18).
The number of amplification reactions varied according to the parasite concentration and was higher for lower concentrations (a maximum of eight reactions were carried out in each PCR assay at the 1-T/ml concentration) (Table 2). A total of 208 PCRs were planned for each DNA extraction method (Table 2).
Inhibitor detection.
PCR inhibition was checked in each DNA extract by amplifying a noncompetitive internal control, composed of an exogenous DNA inserted in a plasmid (pCR2.1 vector; Invitrogen), in the presence of 5 μl of extract. Therefore, 1 to 10 copies of plasmid, 0.4 μM of primers, and 0.05 μM of TaqMan probe targeting the internal control were added to one TaqMan Toxoplasma PCR. This small quantity of exogenous DNA is assumed to avoid the misamplification of 0.2 Toxoplasma per reaction. Internal control amplification (expressed as threshold cycle values) in extracts from AF samples was compared to that in extracts from negative control samples (distilled sterile water). A delay over three PCR cycles (1 log) was considered to be significant inhibition.
Human beta-globin quantification.
A quantitative beta-globin PCR assay was performed with 2 μl of DNA extracts using LightCycler control kit DNA (Roche) on a LightCycler I instrument (Roche). One reaction was performed for each DNA extract, according to the manufacturer's instructions. Results were expressed as the number of human genomes per microliter.
Statistical analysis.
The frequencies of positive Toxoplasma PCRs obtained with the different extraction methods were compared using the chi-square test. In cases of small sample sizes, Fisher's exact test was used. The mean human beta-globin DNA concentrations found by PCR were compared among the different extraction methods using the Kruskal-Wallis and Mann-Whitney tests. A probability of 0.05 or less was considered to be significant. The reproducibility of the DNA extraction methods was assessed by analyzing the coefficient of variation of human beta-globin DNA concentrations in the extracts.
RESULTS
Comparative assessment of DNA extraction methods using Toxoplasma PCR assays.
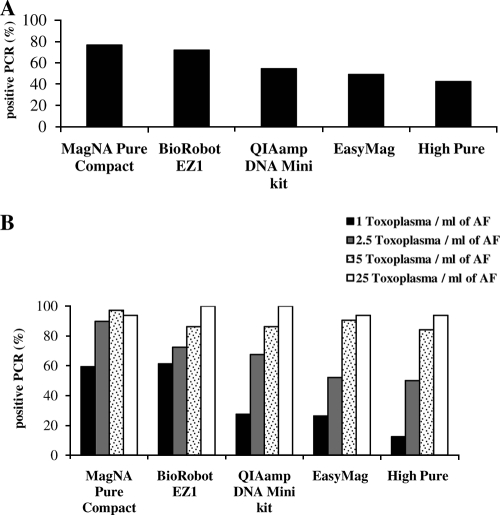
Since we worked with low concentrations of the parasite and a proportion of the PCRs are negative when the sensitivity limit of the method is reached (6), the results were expressed as a score representing the number of positive reactions divided by the total number of reactions performed during the study for a given extraction method (Fig. 1; Table 3).
FIG. 1.
Overall percentage of positive Toxoplasma PCRs for each extraction method, combining TaqMan and FRET PCR in positive AF samples (A) or according to Toxoplasma concentration per ml of AF (B).
TABLE 3.
Performance of the five DNA extraction methods
| Extraction method | PCR results |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Toxoplasma RT-PCR (tachyzoites/ml of AF)a |
% of positive inhibitor detection PCR results | Beta-globin quantification (mean value of equivalent human genome/μl of extract [±SD]) |
|||||||||
| % of positive Toxoplasma TaqMan PCR results for indicated parasite concn |
% of positive Toxoplasma FRET PCR results for indicated parasite concn |
Quantification | Coefficient of variation | ||||||||
| 25 | 5 | 2.5 | 1 | 25 | 5 | 2.5 | 1 | ||||
| MagNA Pure Compact | 87.5 | 94.4 | 83.3 | 60.4 | 100 | 100 | 95.8 | 58.3 | 3.2 | 1,127.5 (±357.1) | 30.8 |
| BioRobot EZ1 | 100 | 88.9 | 69.6 | 79.2 | 100 | 83.3 | 75 | 42.2 | 0 | 857.7 (±241.2) | 27.4 |
| QIAamp DNA minikit | 100 | 77.8 | 63.6 | 31.1 | 100 | 94.4 | 70.8 | 23.9 | 0 | 1,506 (±279.5) | 18 |
| NucliSens easyMAG | 100 | 88.9 | 50 | 25 | 87.5 | 92.3 | 54.2 | 27.7 | 0 | 335.4 (±88.7) | 25.8 |
| High Pure PCR template prepn kit | 100 | 83.3 | 45.8 | 13.3 | 87.5 | 85.7 | 56.3 | 10.7 | 13.7 | 1,320.4 (±433.0) | 32 |
RT-PCR, real-time PCR.
The 30 control samples without Toxoplasma (six samples extracted by each of the five extraction methods) were found to be PCR negative with both Toxoplasma-specific assays.
The study of the 70 spiked samples showed a significant difference in the PCR scores among the five extraction methods, whether using TaqMan PCR (P < 10−8) or FRET PCR (P < 10−4) or combining both sets of data (P < 10−12) (Fig. 1A). When we analyzed the results as a function of the Toxoplasma concentrations, a significant difference was found in samples containing 1 T/ml and 2.5 T/ml of AF (P values of <10−13 and <10−3, respectively) (Fig. 1B). At these low concentrations, the MagNA Pure Compact and BioRobot EZ1 systems provided the best Toxoplasma PCR results. At the concentration of 1 T/ml, these two automated methods yielded 42.2% to 79.2% positive reactions (Table 3). The other extraction methods provided less than 32% positive reactions. At the 2.5 T/ml concentration, the differences in PCR scores among the five extraction methods were found to be significant but were even more so with FRET PCR (P < 0.02) than when using TaqMan PCR (P = 0.05). At this concentration, the MagNA Pure Compact, the BioRobot EZ1, and the manual QIAamp DNA minikit yielded 63.6% to 95.8% positive reactions (Table 3). The two other extraction methods gave less than 57% positive results (Table 3). From 5 T/ml upwards, the five extraction procedures gave equivalent PCR results for whichever assay was used (no statistically significant difference). All the methods yielded more than 77.8% positive reactions (Table 3). There were no significant differences between the results obtained with either Toxoplasma-specific PCR assays, which were actually very similar when considering them together or for each concentration (Table 3).
Detection of PCR inhibitors.
Reaction inhibitors were detected only in samples extracted with the manual High Pure PCR template preparation kit (13/18 samples) (Table 3) and the automated nucleic acid isolation kit I on the MagNA Pure Compact (4/20 samples) (Table 3). A high degree of inhibition (a threshold cycle delay over six PCR cycles, i.e., 2 logs) was found more often in samples extracted using the High Pure PCR template preparation kit (12 samples) than in those extracted using nucleic acid isolation kit I (1 sample).
Comparative assessment of DNA extraction methods using a human beta-globin PCR assay.
The human beta-globin gene was detected in all DNA extracts obtained using the five procedures. It was quantified to check the integrity of the human DNA and the extraction quality (22, 25). A significant difference was observed in the mean beta-globin DNA concentrations among the methods (P < 10−13) (Table 3). The manual and automated kits commercialized by Roche and the manual QIAamp DNA minikit yielded statistically equivalent results that were better than those yielded by the two other kits (P < 0.05) (Table 3). The QIAamp DNA minikit showed the best reproducibility, and the results obtained with the High Pure PCR template preparation kit were the least reproducible.
DISCUSSION
Molecular methods are now commonly applied to the diagnosis of toxoplasmosis, particularly for the early diagnosis of congenital toxoplasmosis, for which a molecular test with high sensitivity is required (8, 11, 14, 19, 20). Since about half of AF samples infected by T. gondii have been described as containing less than 10 parasites/ml (8), it is important to use DNA extraction procedures that allow for the detection of low parasite concentrations. Toxoplasma PCR assays need to be evaluated, as exemplified by a recent multicenter study for molecular detection of T. gondii in AF samples that showed differences in sensitivity among the assays, particularly with samples presenting low parasite concentrations (14). The lowest concentration (5 parasites/ml of lyophilized AF) was not correctly identified in 39.5% of the data sets, demonstrating the need for improvement in the sensitivity of T. gondii molecular detection methods.
Given that DNA extraction is the first step in the PCR detection of a pathogen and is likely to be restrictive, it is probable that the choice of DNA extraction method will affect performances of PCR assays differently. Only one previous study (10) has compared the influences of two DNA extraction methods (QIAamp DNA minikit and MagNA Pure Compact) on the sensitivities of various PCR assays. No significant difference between extraction methods was found using T. gondii tachyzoites (1 to 104 parasites per sample), but differences were observed using 2-ml spiked blood samples (10 to 104 parasites per sample). With these samples, the conventional PCR detection limit was lower when DNA was extracted with the QIAamp DNA minikit (10 versus 100 parasites per sample) than with the MagNA Pure Compact system (10). In agreement with our results, Edvinsson et al. observed differences between the two extraction methods, with biological samples (AF or blood) containing low tachyzoite concentrations, i.e., ≤5 T/ml (10).
Here we show that the use of different commercial DNA extraction techniques can lead to variable efficacy in the detection of low tachyzoite concentrations in AF samples (<5 T/ml). Two automated methods, MagNA Pure Compact using nucleic acid isolation kit I and BioRobot EZ1 using the EZ1 DNA tissue kit, produced the best results, with a mean result of 60% positive reactions at the lowest concentration (1 T/ml). At 2.5 T/ml, these two automated procedures still yielded the best results, followed by the QIAamp manual method (mean results, 89.5%, 72.3%, and 67.2% positive reactions, respectively) (Fig. 1B). From 5 T/ml upwards, the five extraction methods studied here led to equivalent PCR results. In addition, the use of two different high-performance PCR assays (with high analytical sensitivities) carried out at two independent laboratories with the French National Reference Centre for Toxoplasmosis (Centre National de Référence de la Toxoplasmose) allowed us to minimize possible bias due to eventual incompatibility between the DNA extraction substrate and the master mix used for PCR.
Differences in performances among the five extraction procedures were found not only in their ability to extract parasite DNA but also when we evaluated their effectiveness in extracting human DNA. In spite of using the same AF for all samples, there was a significant difference in the amount of human DNA amplified. Two automated procedures—NucliSens easyMAG and BioRobot EZ1—exhibited the lowest yields in extracting human DNA, which could be related to the lysis step and/or the quantity of silica particles. The ability to detect human DNA as well as Toxoplasma DNA in AF samples requires an effective cell wall lysis step, for which easyMAG's protocol is the only one that has a lysis step based on a buffer containing chaotropic salt without pretreatment using proteinase K. Since silica particles are known to have a limited capacity to bind DNA (4), an insufficient quantity in these two automated methods (silica particles must be added manually in easyMAG's protocol) could lead to their binding capacity being exceeded. Manual methods and the MagNA Pure Compact system have already been reported to show no significant differences in the extraction of human DNA (10).
Reaction inhibitions are another crucial factor influencing the performances of a PCR diagnostic assay (3), which are known to be highly dependent upon the DNA extraction procedure used for a given biological sample but which are also linked to the PCR protocol chosen to detect the inhibition factors. Here we detected inhibitors only in samples extracted using the two extraction procedures commercialized by Roche, particularly with the High Pure PCR template preparation kit. This did not significantly reduce the performance of the MagNA Pure Compact method, probably because of the analysis of the samples of AF, a biological fluid known to present low levels of inhibitors. Other authors reported the presence of inhibitors using the MagNA Pure Compact system (17, 21), which may explain the reduction of PCR sensitivity reported when Toxoplasma DNA was extracted from blood using the MagNA Pure Compact system rather than the QIAamp DNA minikit (10). We suggest that poor adaptation between extraction procedures and master mixes from different manufacturers could lead to different PCR results, potentially including inhibition of DNA polymerase. These phenomena remain a minor hindrance when weakly inhibiting samples (i.e., AF, cerebrospinal fluid, or aqueous humor samples) are used and as long as they are not contaminated with blood. Finally, we found no evidence of carryover contamination in nucleic acid extracts by either automated or manual methods.
Our findings are corroborated by several previous studies that compared manual and commercial nucleic acid extraction methods in microbiology (7, 10, 17, 21, 23, 25, 27). Manual extraction methods were found to be comparable to (7, 10, 21) or less sensitive than (17, 23, 27) automated methods. Moreover, the results varied according to the samples tested (pure microbial suspensions or biological samples) (10). Despite the use of magnetic particles in all the automated procedures evaluated herein, their performances were found to be unequal. In particular, the NucliSens easyMAG method performed less well than the others, giving lower genome copy numbers and lower percentages of positive real-time PCR results, as previously reported for cytomegalovirus (23) and RNA/DNA respiratory virus analysis (7). Another study compared the MagNA Pure Compact system, the NucliSens miniMAG extraction instrument (bioMérieux), the NucliSens easyMAG method, and the BioRobot EZ1 system for the isolation of polyomavirus BK virus and the human beta-actin gene from urine specimens (25). The rate of viral detection was 100% using the four systems, but the miniMAG yielded the largest amounts of viral nucleic acids from the urine specimen spiked with viral DNA. The rates of human gene detection were 100% (NucliSens miniMAG and BioRobot EZ1), 96% (MagNA Pure Compact), and 94.7% (NucliSens easyMAG), but the amount of extracted human DNA was not evaluated. In another study, the miniMAG system was found to give excellent results (100% sensitivity) for the isolation of severe acute respiratory syndrome coronavirus RNA in stool samples (17).
As PCR is now widely used for the biological diagnosis of infectious diseases, the use of optimal extraction and amplification methods is becoming increasingly important. We report that the automated methods studied here, with the exception of the easyMAG system, yield significantly more Toxoplasma DNA from AF samples than the manual methods. We were concerned about the reaction inhibition observed with the Roche systems, which may increase the difficulty with detecting parasite DNA at very low concentrations. However, the reaction inhibition observed with the Roche system, for which we cannot rule out a misadaptation between the extraction process and the amplification step, may increase the difficulty with identifying parasite DNA at very low concentrations. Moreover, the following two major points should be kept in mind. (i) The physical properties of the microorganism targeted should be taken into account, especially when the lysis protocol does not include proteinase K pretreatment and might provide low yields, as observed with the easyMAG method. Here, Toxoplasma, like the other members of the Apicomplexa phylum, is known to have a number of prominent structural elements that make it more resistant to the detergent lysis step (15). (ii) The high specificity of real-time PCR chemistries may render the optimization of the molecular tool more complex and more specific, particularly the combination of the “right” extraction plus the “right” amplification. Our use of two independent PCR assays, producing similar results, allowed us to minimize a possible bias due to incompatibilities in this combination. Ideally, an evaluation of several combined extraction and PCR methods, each optimized in combination with the other, should be undertaken for each microorganism and sample type being examined.
Acknowledgments
We are grateful for Emilie Fréalle (Lille Hospital) for her steady interest and fruitful discussions and Filomena Naji and Michèle Wauquier (Lille Hospital), Aline Boulon, Catherine Ducloyer, and Christelle Rousseau (Hôpital Cochin), Yvette Solé (Institut de Puériculture et de Périnatalogie), and Sylvie Matern and Rachel Huber (Hôpitaux Universitaires de Strasbourg) for their substantial contributions involving the molecular biology techniques. We thank R. Pierce for his attentive critical reading of the manuscript and Linda Northrup for her editing of the manuscript.
This study was initiated and funded by the Molecular Biology pole of the French National Reference Centre for Toxoplasmosis (Centre National de Référence de la Toxoplasmose).
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Bastien, P. 2002. Molecular diagnosis of toxoplasmosis. Trans. R. Soc. Trop. Med. Hyg. 96:S205-S215. [DOI] [PubMed] [Google Scholar]
- 2.Bastien, P., E. Jumas-Bilak, E. Varlet-Marie, P. Marty, and the ANOFEL Toxoplasma-PCR Quality Control Group. 2007. Three years of multi-laboratory external quality control for the molecular detection of Toxoplasma gondii in amniotic fluid in France. Clin. Microbiol. Infect. 13:430-433. [DOI] [PubMed] [Google Scholar]
- 3.Bastien, P., G. W. Procop, and U. Reischl. 2008. Quantitative real-time PCR is not more sensitive than “conventional” PCR. J. Clin. Microbiol. 46:1897-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretagne, S., and J. M. Costa. 2006. Towards a nucleic acid-based diagnosis in clinical parasitology and mycology. Clin. Chim. Acta 363:221-228. [DOI] [PubMed] [Google Scholar]
- 6.Chabbert, E., L. Lachaud, L. Crobu, and P. Bastien. 2004. Comparison of two widely used PCR primer systems for detection of Toxoplasma in amniotic fluid, blood, and tissues. J. Clin. Microbiol. 42:1719-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, K. H., W. C. Yam, C. M. Pang, K. M. Chan, S. Y. Lam, K. F. Lo, L. L. Poon, and M. J. Peiris. 2008. Comparison of the NucliSens easyMAG and Qiagen BioRobot 9604 nucleic acid extraction systems for diagnosis of RNA and DNA respiratory viruses on nasopharyngeal aspirates. J. Clin. Microbiol. 46:2195-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa, J. M., P. Ernault, E. Gautier, and S. Bretagne. 2001. Prenatal diagnosis of congenital toxoplasmosis by duplex real-time PCR using fluorescence resonance energy transfer hybridization probes. Prenat. Diagn. 21:85-88. [DOI] [PubMed] [Google Scholar]
- 9.Dupouy-Camet, J., S. L. de Souza, C. Maslo, A. Paugam, A. G. Saimot, R. Benarous, C. Tourte-Schaefer, and F. Derouin. 1993. Detection of Toxoplasma gondii in venous blood from AIDS patients by polymerase chain reaction. J. Clin. Microbiol. 31:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edvinsson, B., S. Jalal, C. E. Nord, B. S. Pedersen, B. Evengard, and the ECSMID Study Group on Toxoplasmosis. 2004. DNA extraction and PCR assays for detection of Toxoplasma gondii. APMIS 112:342-348. [DOI] [PubMed] [Google Scholar]
- 11.Filisetti, D., M. Gorcii, E. Pernot-Marino, O. Villard, and E. Candolfi. 2003. Diagnosis of congenital toxoplasmosis: comparison of targets for detection of Toxoplasma gondii by PCR. J. Clin. Microbiol. 41:4826-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldschmidt, P., S. Degorge, C. Saint-Jean, H. Yera, F. Zekhnini, L. Batellier, L. Laroche, and C. Chaumeil. 2008. Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. Br. J. Ophthalmol. 92:112-115. [DOI] [PubMed] [Google Scholar]
- 13.Homan, W. L., M. Vercammen, J. De Braekeleer, and H. Verschueren. 2000. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int. J. Parasitol. 30:69-75. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser, K., A. M. Van Loon, H. Pelloux, J. Ferrandiz, S. Picot, P. Wallace, and F. Peyron. 2007. Multicenter proficiency study for detection of Toxoplasma gondii in amniotic fluid by nucleic acid amplification methods. Clin. Chim. Acta 375:99-103. [DOI] [PubMed] [Google Scholar]
- 15.Mann, T., E. Gaskins, and C. Beckers. 2002. Proteolytic processing of TgIMC1 during maturation of the membrane skeleton of Toxoplasma gondii. J. Biol. Chem. 277:41240-41246. [DOI] [PubMed] [Google Scholar]
- 16.Martino, R., S. Bretagne, H. Einsele, J. Maertens, A. J. Ullmann, R. Parody, U. Schumacher, C. Pautas, K. Theunissen, C. Schindel, C. Muñoz, N. Margall, C. Cordonnier, and the Infectious Disease Working Party of the European Group for Blood and Marrow Transplantation. 2005. Early detection of Toxoplasma infection by molecular monitoring of Toxoplasma gondii in peripheral blood samples after allogeneic stem cell transplantation. Clin. Infect. Dis. 40:67-78. [DOI] [PubMed] [Google Scholar]
- 17.Petrich, A., J. Mahony, S. Chong, G. Broukhanski, F. Gharabaghi, G. Johnson, L. Louie, K. Luinstra, B. Willey, P. Akhaven, L. Chui, F. Jamieson, M. Louie, T. Mazzulli, R. Tellier, M. Smieja, W. Cai, M. Chernesky, S. E. Richardson, and the Ontario Laboratory Working Group for the Rapid Diagnosis of Emerging Infections. 2006. Multicenter comparison of nucleic acid extraction methods for detection of severe acute respiratory syndrome coronavirus RNA in stool specimens. J. Clin. Microbiol. 44:2681-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reischl, U., S. Bretagne, D. Krüger, P. Ernault, and J. M. Costa. 2003. Comparison of two DNA targets for the diagnosis of Toxoplasmosis by real-time PCR using fluorescence resonance energy transfer hybridization probes. BMC Infect. Dis. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert-Gangneux, F., M. F. Gavinet, T. Ancelle, J. Raymond, C. Tourte-Schaefer, and J. Dupouy-Camet. 1999. Value of prenatal diagnosis and early postnatal diagnosis of congenital toxoplasmosis: retrospective study of 110 cases. J. Clin. Microbiol. 37:2893-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romand, S., M. Wallon, J. Franck, P. Thulliez, F. Peyron, and H. Dumon. 2001. Prenatal diagnosis using polymerase chain reaction on amniotic fluid for congenital toxoplasmosis. Obstet. Gynecol. 97:296-300. [DOI] [PubMed] [Google Scholar]
- 21.Schuurman, T., R. de Boer, R. Patty, M. Kooistra-Smid, and A. van Zwet. 2007. Comparative evaluation of in-house manual, and commercial semi-automated and automated DNA extraction platforms in the sample preparation of human stool specimens for a Salmonella enterica 5′-nuclease assay. J. Microbiol. Methods 71:238-245. [DOI] [PubMed] [Google Scholar]
- 22.Simon, A., P. Labalette, I. Ordinaire, E. Fréalle, E. Dei-Cas, D. Camus, and L. Delhaes. 2004. Use of fluorescence resonance energy transfer hybridization probes to evaluate quantitative real-time PCR for diagnosis of ocular toxoplasmosis. J. Clin. Microbiol. 42:3681-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soetens, O., C. Vauloup-Fellous, I. Foulon, P. Dubreuil, B. De Saeger, L. Grangeot-Keros, and A. Naessens. 2008. Evaluation of different cytomegalovirus (CMV) DNA PCR protocols for analysis of dried blood spots from consecutive cases of neonates with congenital CMV infections. J. Clin. Microbiol. 46:943-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens, S. J., S. A. Verkuijlen, A. J. Brule, and J. M. Middeldorp. 2002. Comparison of quantitative competitive PCR with LightCycler-based PCR for measuring Epstein-Barr virus DNA load in clinical specimens. J. Clin. Microbiol. 40:3986-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang, Y. W., S. E. Sefers, H. Li, D. J. Kohn, and G. W. Procop. 2005. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 43:4830-4833. (Erratum, 43:5833.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villard, O., D. Filisetti, F. Roch-Deries, J. Garweg, J. Flament, and E. Candolfi. 2003. Comparison of enzyme-linked immunosorbent assay, immunoblotting, and PCR for diagnosis of toxoplasmic chorioretinitis. J. Clin. Microbiol. 41:3537-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson, D., B. Yen-Lieberman, U. Reischl, I. Warshawsky, and G. W. Procop. 2004. Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens. J. Clin. Microbiol. 42:5913-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]