Abstract
Adherent-invasive Escherichia coli (AIEC) pathovar strains, which are associated with Crohn's disease, share many genetic and phenotypic features with extraintestinal pathogenic E. coli (ExPEC) strains, but little is known about the level of genetic similarity between the two pathovars. We aimed to determine the frequency of strains with the “AIEC phenotype” among a collection of ExPEC strains and to further search for a common phylogenetic origin for the intestinal and extraintestinal AIEC strains. The adhesion, invasion, and intramacrophage replication capabilities (AIEC phenotype) of 63 ExPEC strains were determined. Correlations between virulence genotype and AIEC phenotype and between intestinal/extraintestinal origin, serotype, and phylogroup were evaluated for the 63 ExPEC and 23 intestinal AIEC strains. Phylogenetic relationships between extraintestinal and intestinal AIEC strains were determined using multilocus sequence typing (MLST) and pulsed-field gel electrophoresis. Only four (6.35%) ExPEC strains, belonging to the O6:H1, O83:H1, and O25:H4 serotypes, were classified as having an AIEC phenotype. These strains were found to be genetically related to some intestinal AIEC strains of the same serotypes as revealed by MLST. No particular virulence gene sets correlated with the intestinal/extraintestinal origin of the strains or with the AIEC phenotype, whereas the gene sets did correlate with the serogroup. We identified two intestinal AIEC strains and one extraintestinal AIEC strain belonging to the O25:H4 serotype that also belonged to the emerging and virulent clonal group ST131. In conclusion, the ExPEC and AIEC pathovars share similar virulence gene sets, and certain strains are phylogenetically related. However, the majority of ExPEC strains did not behave like AIEC strains, thus confirming that the AIEC pathovar possesses virulence-specific features that, to date, are detectable only phenotypically.
Members of the Enterobacteriaceae family, especially Escherichia coli, have been repeatedly suggested to play a role in the origin and/or perpetuation of Crohn's disease (CD). In part, this suggestion was based on the higher abundance of this bacterium in CD patients than in control subjects (4, 10, 20, 23, 28, 29, 32, 41, 48, 51). Although considerable effort has been devoted to the search for intestinal pathogenic E. coli strains associated with CD, to date none of the six previously described pathovars (27) has been implicated in this condition. Darfeuille-Michaud et al. (18) observed that E. coli strains with adhesion and invasion properties colonized the ileal mucosae of CD patients more frequently than those of control subjects. Darfeuille-Michaud et al. further characterized these strains and proposed a new potential E. coli pathovar associated with CD, which was designated adherent-invasive E. coli (AIEC) (10). The implication of AIEC in CD is becoming increasingly relevant because several independent studies from different countries have reported a higher prevalence of invasive E. coli in CD patients (4, 17, 33, 34, 47).
The main characteristics of AIEC are (i) the ability to adhere to and invade intestinal epithelial cells, (ii) the ability to survive and replicate expansively within macrophages without triggering host cell death and inducing the release of tumor necrosis factor alpha (21), and (iii) the lack of known invasive determinants (17). Recently, Glasser and Darfeuille-Michaud (22) proposed a model explaining the mechanism of pathogenesis for AIEC strains. The AIEC strains isolated to date are clonally diverse and belong to distinct serotypes. Moreover, despite the fact that they fall primarily into the B2 phylogroup, AIEC strains belonging to the A, B1, and D phylogroups have also been isolated (4, 33-35, 47). Although no specific virulence factors have been described for this pathovar, AIEC strains carry many virulence-associated genes characteristic of extraintestinal pathogenic E. coli (ExPEC) strains, which suggests that the AIEC pathovar could be closely related to the ExPEC pathovar (4, 17, 34).
The aim of this work was to determine the frequency of strains with the “AIEC phenotype” among E. coli strains that cause extraintestinal infections, including uropathogenic E. coli (UPEC), septicemic E. coli, and neonatal meningitis E. coli strains. To achieve this objective, we determined the ability of a collection of ExPEC strains to adhere to and invade intestinal epithelial cells, as well as their capacity to survive and replicate within macrophages. In parallel, we compared the distributions of virulence-associated genes among ExPEC and AIEC strains. Furthermore, we searched for a common phylogenetic origin of the ExPEC strains that had an AIEC phenotype (referred to in this study as extraintestinal AIEC) and a collection of AIEC strains isolated mainly from the intestinal mucosae of CD patients (intestinal AIEC).
MATERIALS AND METHODS
Bacterial strains.
The present study involved a collection of 86 E. coli strains, some of which were the same as those reported in previously published studies (5, 7, 8, 17, 34, 39, 40, 42) (Table 1). Sixty-three (73.3%) were obtained from human extraintestinal infections (28 from urinary tract infections [UTIs], 21 from sepsis, 12 from meningitis, 1 from intra-abdominal pus, and 1 from a wound infection), and 23 were obtained from the intestinal mucosae of patients with CD (16 strains) or ulcerative colitis (1 strain) and the intestinal mucosae of control subjects (without inflammatory bowel disease [non-IBD]) (6 strains). Control subjects were asymptomatic and did not present inflammation and/or evidence of polyps during colonoscopy. Among CD patients, 39% had Crohn's colitis, 35% had Crohn's ileitis, and 26% had ileal/colonic disease. Further information about the sources of intestinal AIEC strains can be obtained from reference 34). The prototype AIEC strain LF82 was included in this group of 23 intestinal AIEC strains.
TABLE 1.
Collection of ExPEC and intestinal AIEC strains used in this study
| Strain | Origina | Infectionb | Country | AIEC phenotypec | I_ ADHd |
I_INVe |
I_REPLf |
Serotypeg | Phylogroup | ExPEC-likeh | β-Lactamase | Virulence gene carriage | Referencei | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |||||||||||
| 15802 | EI | Intra-abdominal pus | Canada | − | 2.2 | 2.7 | 0.009 | 0.005 | O25:H4 | B2 | − | CTX-M-15 | iucD, fimH | 42 | ||
| EC-1 | EI | Wound infection | Spain | − | 1.2 | 0.4 | 0.026 | 0.009 | O6:H1 | B2 | + | − | papC, papGIII, sfa- focDE, hlyA, cnf1, fimH | 40 | ||
| SM22 | EI | Meningitis | United States | − | 0.2 | 0.3 | 0.005 | 0.006 | O6:H1 | B2 | − | − | iucD, fimH | 5 | ||
| H1166 | EI | Meningitis | France | − | 1.8 | 0.8 | 0.060 | 0.057 | O6:H1 | B2 | + | − | papC, papGII, sfa-focDE, iucD, hlyA, fimH | 5 | ||
| SM18 | EI | Meningitis | United States | − | 0.1 | 0.1 | 0.000 | 0.000 | O7:H− | D | + | − | papC, papGI-GII, neuC, iucD, fimH | 5 | ||
| SM21 | EI | Meningitis | United States | − | 0.8 | 1.3 | 0.007 | 0.003 | O16:H6 | B2 | + | − | papC, papGII, neuC, iucD, fimH, fimAvMT78 | 5 | ||
| SM43 | EI | Meningitis | France | − | 0.1 | 0.1 | 0.013 | 0.018 | O6:H1 | B2 | + | − | sfa-focDE, iucD, hlyA, cnf1, fimH | 5 | ||
| SM57 | EI | Meningitis | United States | − | 0.6 | 0.2 | 0.013 | 0.004 | O83:H7 | B2 | + | − | sfa-focDE, neuC, iucD, ibeA, fimH, fimAvMT78 | 5 | ||
| SM63 | EI | Meningitis | United States | − | 0.5 | 0.1 | 0.024 | 0.016 | O1:H7 | B2 | + | − | papC, papGI, neuC, iucD, fimH, fimAvMT78 | 5 | ||
| SM69 | EI | Meningitis | France | − | 1.8 | 0.4 | 0.018 | 0.011 | O18:H7 | B2 | + | − | papC, sfa-focDE, neuC, iucD, hlyA, cnf1, ibeA, fimH, fimAvMT78 | 5 | ||
| SM72 | EI | Meningitis | France | − | 0.2 | 0.0 | 0.011 | 0.005 | O45:H7 | B2 | + | − | papC, papGII, sfa-focDE, neuC, iucD, fimH, fimAvMT78 | 5 | ||
| SM148 | EI | Meningitis | France | − | 0.1 | 0.1 | 0.009 | 0.012 | O18:H7 | B2 | + | − | sfa-focDE, neuC, iucD, ibeA, fimH, fimAvMT78 | 5 | ||
| SM168 | EI | Meningitis | France | − | 0.8 | 0.3 | 0.013 | 0.011 | O18:H7 | B2 | + | − | sfa-focDE, neuC, iucD, hlyA, cnf1, ibeA, fimH, fimAvMT78 | 5 | ||
| SM177 | EI | Meningitis | France | − | 0.0 | 0.0 | 0.000 | 0.000 | O1:H− | D | + | − | papC, papGI-GII, neuC, iucD, hlyA | 5 | ||
| H1088 | EI | Sepsis | Spain | − | 1.8 | 2.0 | 0.043 | 0.053 | O25:H4 | B2 | − | − | iucD, fimH | This study | ||
| H109 | EI | Sepsis | Spain | − | 0.0 | 0.0 | 0.002 | 0.003 | O6:H10 | A | − | − | iucD | This study | ||
| H685 | EI | Sepsis | Spain | − | 0.5 | 0.1 | 0.021 | 0.002 | O25:H4 | B2 | − | − | ibeA, fimH | This study | ||
| H6166 | EI | Sepsis | France | − | 0.7 | 0.2 | 0.023 | 0.025 | O45:H7 | B2 | + | − | papC, papGII, sfa-focDE, neuC, iucD, hlyA, cnf1, fimH, fimAvMT78 | 5 | ||
| H6290 | EI | Sepsis | France | − | 0.5 | 0.1 | 0.018 | 0.004 | O45:H7 | B2 | + | − | papC, neuC, iucD, fimH, fimAvMT78 | 5 | ||
| FV7561 | EI | Sepsis | Spain | − | 1.3 | 0.5 | 0.041 | 0.030 | O25:H4 | B2 | + | CTX-M-15 | afa-draBC, iucD, fimH | 42 | ||
| H169 | EI | Sepsis | Spain | − | 1.5 | 0.0 | 0.029 | 0.009 | O18:H7 | B2 | + | − | sfa-focDE, neuC, iucD, ibeA, fimH, fimAvMT78 | 39 | ||
| PP16 | EI | Sepsis | Spain | + | 1.2 | 1.0 | 0.104 | 0.081 | 1,045.9 | 541.2 | O83:H1 | B2 | + | − | ibeA, fimH, fimAvMT78 | This study |
| H102A | EI | Sepsis | Spain | − | 0.8 | 0.3 | 0.031 | 0.013 | O83:H1 | B2 | + | − | ibeA, fimH, fimAvMT78 | This study | ||
| H126 | EI | Sepsis | Spain | − | 0.3 | 0.3 | 0.027 | 0.011 | O83:H31 | B2 | + | − | iucD, ibeA, fimH | This study | ||
| H106A | EI | Sepsis | Spain | − | 0.2 | 0.2 | 0.001 | 0.001 | O6:H31 | B2 | + | − | papC, papGIII, sfa-focDE, iucD, hlyA, cnf1, fimH | This study | ||
| PP215 | EI | Sepsis | Spain | + | 0.8 | 0.6 | 0.453 | 0.350 | 1,425.4 | 229.4 | O6:H1 | B2 | + | − | papC, papGII, sfa-focDE, iucD, hlyA, cnf1, fimH | This study |
| H219B | EI | Sepsis | Spain | − | 0.2 | 0.1 | 0.006 | 0.009 | O25:H4 | B2 | + | − | iucD, ibeA, fimH, fimAvMT78 | This study | ||
| H676A | EI | Sepsis | Spain | − | 0.1 | 0.1 | 0.003 | 0.004 | O25:H4 | B2 | + | − | papC, papGIII, iucD, hlyA, ibeA, fimH, fimAvMT78 | This study | ||
| H68a | EI | Sepsis | Spain | − | 0.1 | 0.1 | 0.005 | 0.000 | O6:H7 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, ibeA, fimH | 8 | ||
| H778 | EI | Sepsis | Spain | − | 1.7 | 1.8 | 0.033 | 0.039 | O25:H4 | B2 | + | − | afa-draBC, iucD, ibeA, fimH | This study | ||
| H810A | EI | Sepsis | Spain | − | 0.3 | 0.1 | 0.016 | 0.013 | O25:H4 | B2 | + | − | iucD, ibeA, fimH | This study | ||
| H858 | EI | Sepsis | Spain | − | 3.6 | 2.0 | 0.083 | 0.018 | O25:H4 | B2 | + | − | afa-draBC, ibeA, fimH | This study | ||
| PP209 | EI | Sepsis | Spain | − | 0.6 | 0.6 | 0.029 | 0.002 | O6:H1 | B2 | + | − | papC, papGII-GIII, sfa-focDE, iucD, hlyA, cnf1, fimH | This study | ||
| PP42 | EI | Sepsis | Spain | − | 0.0 | 0.1 | 0.019 | 0.009 | O25:H4 | B2 | − | − | ibeA, fimH | This study | ||
| PP89 | EI | Sepsis | Spain | − | 0.0 | 0.0 | 0.030 | 0.014 | O6:H10 | A | − | − | iucD | This study | ||
| EC-2 | EI | UTI | Spain | − | 3.0 | 0.1 | 0.038 | 0.032 | ONT:HNT | B1 | − | − | fimH | 40 | ||
| VC1 | EI | UTI | France | − | 1.8 | 1.9 | 0.035 | 0.035 | O25:H4 | B2 | − | − | iucD, fimH | This study | ||
| HDE3 | EI | UTI | France | − | 0.1 | 0.0 | 0.004 | 0.001 | O25:H4 | B2 | − | CTX-M-15 | iucD, fimH | 42 | ||
| 17102 | EI | UTI | Canada | − | 5.2 | 0.9 | 0.078 | 0.043 | O25:H4 | B2 | − | CTX-M-15 | iucD, fimH | 42 | ||
| OL52A | EI | UTI | Spain | − | 0.0 | 0.0 | 0.004 | 0.005 | O101:H− | A | − | − | iucD | 7 | ||
| OL80A | EI | UTI | Spain | − | 0.2 | 0.2 | 0.008 | 0.004 | O51:H49 | B2 | − | − | fimH, fimAvMT78 | 7 | ||
| OB59A | EI | UTI | Spain | − | 0.2 | 0.1 | 0.011 | 0.006 | O83:H1 | B2 | − | − | ibeA, fimH | 7 | ||
| COR227 | EI | UTI | Spain | − | 0.6 | 0.8 | 0.037 | 0.010 | O6:H25 | B1 | − | − | fimH | 7 | ||
| FV7563 | EI | UTI | Spain | + | 6.9 | 5.9 | 0.129 | 0.072 | 470.0 | 264.0 | O25:H4 | B2 | + | CTX-M-15 | afa-draBC, iucD, fimH | 42 |
| FV7569 | EI | UTI | Spain | − | 6.2 | 2.5 | 0.083 | 0.054 | O25:H4 | B2 | + | CTX-M-15 | afa-draBC, iucD, fimH | 42 | ||
| FV7588 | EI | UTI | Spain | − | 2.7 | 1.0 | 0.060 | 0.028 | O25:H4 | B2 | + | CTX-M-15 | afa-draBC, iucD, fimH | 42 | ||
| OL96A | EI | UTI | Spain | + | 5.2 | 5.0 | 0.388 | 0.159 | 457.5 | 259.3 | O6:H1 | B2 | + | − | papC, papGII, sfa-focDE, iucD, hlyA, cnf1, fimH | 7 |
| OL61A | EI | UTI | Spain | − | 0.5 | 0.2 | 0.026 | 0.014 | O75:H7 | B2 | + | − | sfa-focDE, neuC, hlyA, cnf1, ibeA, fimH | 7 | ||
| OL37A | EI | UTI | Spain | − | 0.1 | 0.1 | 0.000 | 0.000 | O1:H1 | D | + | − | papC, neuC, iucD, fimH, fimAvMT78 | 7 | ||
| OL65A | EI | UTI | Spain | − | 1.4 | 1.2 | 0.003 | 0.004 | O6:H1 | B2 | + | − | sfa-focDE, iucD, hlyA, cnf1, fimH | 7 | ||
| OB112A | EI | UTI | Spain | − | 0.5 | 0.3 | 0.005 | 0.000 | O2:H− | B2 | + | − | papC, papGII-GIII, sfa-focDE, iucD, hlyA, cnf1, fimH | 7 | ||
| OB29A | EI | UTI | Spain | − | 0.2 | 0.2 | 0.029 | 0.023 | O6:H1 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, fimH | 7 | ||
| OB4A | EI | UTI | Spain | − | 0.8 | 0.2 | 0.017 | 0.009 | O6:H1 | B2 | + | − | sfa-focDE, hlyA, cnf1 | 7 | ||
| OL100A | EI | UTI | Spain | − | 0.2 | 0.1 | 0.000 | 0.000 | O18:H− | B2 | + | − | papC, papGII, sfa-focDE, iucD, hlyA, fimH | 7 | ||
| OB64A | EI | UTI | Spain | − | 0.3 | 0.3 | 0.002 | 0.003 | O75:H5 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, ibeA, fimH | 7 | ||
| OL16A | EI | UTI | Spain | − | 0.4 | 0.4 | 0.000 | 0.000 | xO6:H− | B2 | + | − | papC, papGII, sfa-focDE, neuC, iucD, hlyA, cnf1, fimH | 7 | ||
| OB103A | EI | UTI | Spain | − | 1.2 | 0.8 | 0.016 | 0.008 | O6:H1 | B2 | + | − | sfa-focDE, hlyA, cnf1, fimH | 7 | ||
| OL85A | EI | UTI | Spain | − | 0.4 | 0.2 | 0.024 | 0.015 | O2:H1 | B2 | + | − | papC, papGIII, sfa-focDE, iucD, hlyA, cnf1, fimH | 7 | ||
| OB23A | EI | UTI | Spain | − | 1.1 | 0.1 | 0.072 | 0.018 | O6:H1 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, fimH | 7 | ||
| OB123A | EI | UTI | Spain | − | 3.7 | 0.5 | 0.039 | 0.008 | O6:H1 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, fimH | 7 | ||
| OB79A | EI | UTI | Spain | − | 0.5 | 0.2 | 0.014 | 0.008 | O83:H1 | B2 | + | − | ibeA, fimH, fimAvMT78 | 7 | ||
| OL64A | EI | UTI | Spain | − | 0.0 | 0.0 | 0.000 | 0.000 | O6:H31 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, fimH | 7 | ||
| OL118A | EI | UTI | Spain | − | 0.2 | 0.1 | 0.003 | 0.004 | O6:H31 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, fimH | 7 | ||
| AIEC25 | I | C-CD (colon) | Spain | + | 2.8 | 1.3 | 0.482 | 0.129 | 775.93 | 128.3 | O6:H31 | B2 | + | − | papC, papGIII, hlyA, cnf1, fimH | 34 |
| AIEC21 | I | I-CD (colon) | Spain | + | 17.0 | 7.8 | 0.109 | 0.013 | 1,297.1 | 625.2 | O6:H1 | B2 | + | − | papC, papGII, iucD, hlyA, cnf1, fimH | 34 |
| AIEC12 | I | IC-CD (colon) | Spain | + | 22.3 | 3.9 | 0.142 | 0.017 | 93.697 | 55.93 | O26:H− | B2 | + | − | papC, afa-draBC, iucD, fimH | 34 |
| AIEC20 | I | IC-CD (ileum) | Spain | + | 14.2 | 6.2 | 0.125 | 0.098 | 343.89 | 244.6 | O11:H18 | D | + | − | papC, afa-draBC, iucD, fimH | 34 |
| AIEC17 | I | I-CD (ileum+colon) | Spain | + | 21.6 | 17.5 | 0.266 | 0.055 | 1,053 | 75 | ONT:HNT | D | + | − | neuC, ibeA, fimH | 34 |
| AIEC05 | I | CD (ileum+colon) | Spain | + | 9.4 | 2.2 | 0.202 | 0.042 | 704.91 | 714 | O1:H− | B2 | + | − | papC, papGII, neuC, fimH | 34 |
| AIEC02 | I | CD (colon) | Spain | + | 0.9 | 1.0 | 0.802 | 0.035 | 2,187.8 | 4.794 | O8:H21 | B2 | + | − | papC, neuC, fimH | 34 |
| AIEC01 | I | I-CD (ileum) | Spain | + | 15.9 | 9.3 | 0.284 | 0.106 | 1,566.7 | 1,060 | O6:H1 | B2 | + | − | sfa-focDE, iucD, fimH | 34 |
| AIEC09 | I | IC-CD (colon) | Spain | + | 5.4 | 4.0 | 0.216 | 0.010 | 2,562.3 | 240.6 | ONT:H− | B2 | + | − | papC, sfa-focDE, iucD, hlyA, cnf1, fimH, fimAvMT78 | 34 |
| AIEC24 | I | IC-CD (colon) | Spain | + | 2.0 | 1.4 | 0.309 | 0.138 | 1,625.6 | 115.6 | ONT:H− | A | + | − | iucD, fimH, fimAvMT78 | 34 |
| AIEC23 | I | C-CD (ileum) | Spain | + | 9.7 | 0.7 | 0.568 | 0.148 | 2,362.1 | 250.2 | O5:HNT | A | − | − | fimH | 34 |
| AIEC11 | I | I-CD (ileum) | Spain | + | 4.4 | 3.4 | 0.508 | 0.081 | 847.95 | 512.8 | O22:H1 | B2 | + | − | afa-draBC, iucD, fimH | 34 |
| AIEC15-1 | I | I-CD (ileum) | Spain | + | 10.0 | 1.4 | 0.305 | 0.159 | 659.75 | 437 | O22:H1 | B2 | − | − | fimH | 34 |
| AIEC14-1 | I | I-CD (ileum+colon) | Spain | + | 9.8 | 5.2 | 0.238 | 0.011 | 800.69 | 252.4 | O22:H1 | B2 | − | − | fimH | 34 |
| AIEC16-2 | I | I-CD (ileum) | Spain | + | 9.7 | 3.6 | 1.400 | 0.424 | 921.05 | 489.7 | O22:H1 | B2 | − | − | fimH | 34 |
| AIEC13 | I | UC (colon) | Spain | + | 7.9 | 4.3 | 1.400 | 0.000 | 225.91 | 181.6 | O25:H4 | B2 | + | − | papC, papGIII, iucD, hlyA, cnf1, ibeA, fimH | 34 |
| AIEC19 | I | Non-IBD (colon) | Spain | + | 2.4 | 0.7 | 0.111 | 0.016 | 1,568.1 | 1,726 | ONT:H− | A | + | − | iucD, fimH, fimAvMT78 | 34 |
| AIEC07 | I | Non-IBD (ileum) | Spain | + | 20.0 | 13.4 | 0.565 | 0.392 | 1,692.6 | 296.8 | O22:H7 | B1 | + | − | papC, iucD, fimH | 34 |
| AIEC04 | I | Non-IBD (ileum) | Spain | + | 21.6 | 8.9 | 0.320 | 0.016 | 584.69 | 418.5 | O6:HNT | B2 | + | − | papC, sfa-focDE, iucD, hlyA, cnf1, fimH, fimAvMT78 | 34 |
| AIEC10 | I | Non-IBD (ileum) | Spain | + | 5.9 | 1.0 | 0.226 | 0.192 | 1,413.7 | 51.37 | O159:H34 | A | − | − | fimH | 34 |
| AIEC06 | I | Non-IBD (colon) | Spain | + | 10.2 | 3.4 | 0.177 | 0.019 | 1,717.7 | 307.9 | O6:H5 | B2 | + | − | papC, papGIII, sfa-focDE, hlyA, cnf1, fimH | 34 |
| AIEC08 | I | Non-IBD (colon) | Spain | + | 1.1 | 0.2 | 0.172 | 0.066 | 104.75 | 49.71 | O25:H4 | B2 | + | − | papC, papGIII, iucD, ibeA, fimH | 34 |
| LF82 | I | I-CD (ileum) | France | + | 25.7 | 15.7 | 2.261 | 1.349 | 776.88 | 304.8 | O83:H1 | B2 | + | − | ibeA, fimH, fimAvMT78 | 17 |
EI, extraintestinal; I, intestinal.
I-CD, Crohn's ileitis; IC-CD, ileocolonic disease; C-CD, Crohn's colitis; UC, ulcerative colitis; non-IBD: controls without inflammatory bowel disease. For those strains of intestinal origin, specific zones from which the strains have been isolated along the intestinal tract are indicated in parentheses.
Those strains with an I_INV of >0.1 and an I_REPL of >100% were classified as AIEC strains in the present study.
Adhesion ability, calculated as the mean number of bacteria per I407 cell after 3 h of incubation. Results are from triplicate determinations.
Invasion ability, calculated as the percentage of inoculum surviving after 1 h of gentamicin treatment: I_INV (%) = (intracellular bacteria/4 × 106 bacteria inoculated) × 100. Results are from triplicate determinations.
Intramacrophage replication ability, calculated as the percentage of intracellular bacteria at 24 h postinfection relative to that after 1 h of gentamicin treatment: I_REPL (%) = (CFU ml−1 at 24 h/CFU ml−1 at 1 h) × 100. Results are from triplicate determinations.
H−, nonmotile strain.
Strains with ≥2 virulence-associated genes regardless of the presence of fimH.
References indicate the origin of the strain isolation.
Adhesion and invasion assays with Intestine-407 epithelial cells.
The Intestine-407 epithelial cell line was used for the adhesion and invasion assays (ATCC CCL-6). Cell culture, adhesion, and invasion assays were performed as described previously (10). Briefly, 24-well plates containing 4 × 105 cells/well that had been incubated for 20 h were infected at a multiplicity of infection of 10. Duplicate plates, one for the adhesion assay and one for the invasion assay, were incubated for 3 h at 37°C with 5% CO2. For the bacterial adhesion assays, the cell monolayers were washed five times with phosphate-buffered saline and then lysed with 1% Triton X-100. Adherent bacteria were quantified by plating them on nutrient agar. Plating was performed over a maximum period of 30 min in order to avoid bacterial lysis by Triton X-100. Adherence ability (I_ADH) was determined by calculating the mean number of bacteria per cell. For the bacterial invasion assays, the monolayers were washed twice with phosphate-buffered saline after 3 h of infection, and fresh cell culture medium containing 100 μg ml−1 of gentamicin was added and left for 1 h to kill extracellular bacteria. After cell lysis with 1% Triton X-100, the number of intracellular bacteria was determined by plating. Invasive ability was expressed as the percentage of the initial inoculum that became intracellular: I_INV (%) = (intracellular bacteria/4 × 106 bacteria inoculated) × 100.
Survival and replication in J774 macrophages.
The macrophage-like J774A.1 cell line (ATCC TIB-67) was used as a model in E. coli survival and replication assays. Cell culture was performed as described previously (21). E. coli isolates with known adherence and invasion properties were checked for their ability to survive and replicate inside macrophages as previously described (17). Macrophages were seeded at 2 × 105 cells per well in two 24-well plates and incubated for 20 h. After incubation, the medium was replaced with fresh medium and bacteria were seeded at a multiplicity of infection of 10. To promote internalization of bacteria by the macrophages, the samples were centrifuged at 900 rpm for 10 min and incubated for an additional 10 min at 37°C with 5% CO2. Nonphagocytosed bacteria were killed with gentamicin (20 μg ml−1). Intracellular bacteria were quantified in the same manner as described for the invasion assays after 1 and 24 h of infection. The results are expressed as the mean percentages of bacteria recovered at 1 and 24 h postinfection: I_REPL (%) = (CFU ml−1 at 24 h/CFU ml−1 at 1 h) × 100. Those strains with an I_INV of >0.1 and an I_REPL of >100% were classified as AIEC strains in the present study.
Phylotyping and virulence genotyping by PCR.
Determination of the major E. coli phylogenetic group (A, B1, B2, or D) was performed as described by Clermont et al. (16).
The presence of virulence genes was analyzed as described elsewhere (34). Primers specific for 10 genes and operons that encode extraintestinal virulence factors characteristic of ExPEC were used. These genes included those for adhesins (pyelonephritis-associated pili [papC], S and F1C fimbriae [sfa-focDE], Dr-binding adhesins [afa-draBC], and type 1 fimbriae [fimH and fimAvMT78, the strain MT78 avian pathogenic variant of fimA]), two toxins (hlyA and cnf1), and one aerobactin (iucD). The analyzed genes also included two protectin/invasion-encoding genes corresponding to the K1 kps variant (neuC) and the invasion of brain endothelium gene (ibeA). The papC-positive strains were tested for the papGI, papGII, and papGIII alleles. The E. coli strains were also screened for specific genes found in diarrheagenic E. coli pathovars (stx1, stx2, eae, bfpA, ipaH, pCDV432, eltA, and est).
Additional virulence genes (cdtB, cytolethal distending toxin; bmaE, M fimbriae; gafD, G fimbriae; sat, secreted autotransporter toxin; cvaC, microcin [colicin] V; traT, serum resistance associated; malX, pathogenicity island marker; usp, uropathogenic-specific protein; focG, F1C fimbriae; sfaS, S fimbriae; iroN, salmochelin receptor; kpsMII, group 2 capsule; and kpsMIII, group 3 capsule) were investigated for those strains included in Fig. 2 and 3. The amplification procedures have been documented elsewhere (see reference 37 and references therein).
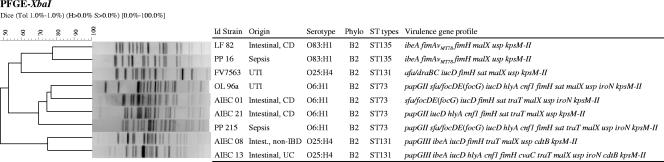
FIG. 2.
Consensus UPGMA dendrogram generated from the Dice coefficients of XbaI PFGE profiles of the four extraintestinal AIEC strains detected in this study (OL96a, PP215, PP16, and FV7563) and of the five intestinal AIEC strains with similar serotypes. Serotype, phylogroup, ST, and virulence-associated genes are specified. UC, ulcerative colitis; non-IBD, controls without IBD.
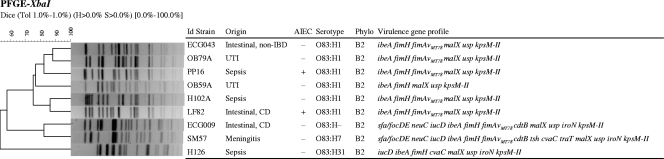
FIG. 3.
Consensus UPGMA dendrogram generated from the Dice coefficients of XbaI PFGE profiles of six O83 ExPEC strains and three O83 intestinal E. coli strains. AIEC phenotype, serotype, phylogroup, and virulence-associated genes are specified. Non-IBD: controls without IBD. The ECG-043 and ECG-009 strains were used only in this section; their characteristics are described elsewhere (34).
Serotyping.
Determination of O and H antigens was carried out using the method previously described by Guinée et al. (24).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described elsewhere (15). Agarose-embedded DNA was digested with 0.2 U/μl XbaI (Roche) according to the manufacturer's instructions. The XbaI-digested genomic DNA was analyzed on a 1% agarose gel in 0.5× Tris-boric acid-EDTA buffer at 14°C using CHEF MAPPER (Bio-Rad). The gel was run for 21.30 h at 6 V/cm, with initial and final switch times of 2.16 s and 54.17 s, respectively. The gel was stained with ethidium bromide (1 μg/ml), observed using a Gel Doc 2000 system (Bio-Rad), and analyzed with the BioNumerics fingerprinting software (Applied Maths, St-Martens-Latem, Belgium). Cluster analysis of the Dice similarity indices based on the unweighted pair group method using arithmetic averages (UPGMA) was performed to generate a dendrogram describing the relationships among the PFGE profiles.
MLST.
Multilocus sequence typing (MLST) was carried out as previously described (53). Gene amplification and sequencing of the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were performed using the primers and protocol specified at the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli). The sequences were reviewed by visual inspection with the BioEdit sequence alignment editor (version 7.0.9; Ibis Biosciences). The ClustalW2 program was used to align the sequences. The allelic profiles of the seven gene sequences, the sequence types (STs), and the sequence complexes (Clpx) (defined as STs that are linked by distances of one or two allelic differences) were obtained via the electronic database at the E. coli MLST website.
Statistical analyses.
Fisher's exact test (for small contingency tables) or Pearson's χ2 test (for frequencies of higher than five within cells) was used to measure the significance of frequency values using SPSS 15.0 software.
Correspondence analysis was used to determine if a particular distribution of virulence-associated genes correlated with the serogroup, phylogroup, AIEC phenotype, ExPEC-like genotype (more than two virulence genes in addition to fimH), origin of the strains (extraintestinal/intestinal), and/or disease caused (intra-abdominal pus, wound infection, sepsis, meningitis, UTI, and IBD). The input variables were the presence/absence of virulence genes (papC, sfa-focDE, afa-draBC, hlyA, cnf1, iucD, neuC, ibeA, fimH, and fimAvMT78), and all 86 E. coli strains were included in the analysis. Correspondence analysis was performed with the CANOCO program (version 4.5 for Windows) using biplot scaling (52). To corroborate the significance of the dispersion of the samples in the plot according to the serogroup, phylogroup, AIEC phenotype, ExPEC-like genotype, and origin of the strains, an analysis-of-variance test was applied using Tukey's post hoc test for multiple comparisons of those variables comprising more than two subgroups of samples. For quantitative variables, such as adhesion (I_ADH), invasion (I_INV), and intramacrophage replication (I_REPL) indices, the Pearson correlation coefficient was used.
RESULTS
Presence of AIEC-like strains among ExPEC strains.
The genetic and phenotypic characteristics of the 63 ExPEC and 23 intestinal AIEC strains used in this study are listed in Table 1. Strains belonging to serogroups O1 (n = 3), O2 (n = 2), O6 (n = 21), O7 (n = 1), O16 (n = 1), O18 (n = 5), O25 (n = 16), O45 (n = 3), O51 (n = 1), O75 (n = 2), O83 (n = 6), O101 (n = 1), and ONT (n = 1), which were obtained from extraintestinal infections, were selected to be compared with a collection of intestinal AIEC strains belonging to serogroups O1 (n = 1), O5 (n = 1), O6 (n = 5), O8 (n = 1), O11 (n = 1), O22 (n = 5), O25 (n = 2), O26 (n = 1), O83 (n = 1), O159 (n = 1), and ONT (n = 4).
After determining the capacity of ExPEC strains to adhere to and invade intestinal epithelial cells and their ability to survive and replicate within macrophages, we classified four strains (6.35%) as AIEC strains (Table 2). These strains are referred to as “extraintestinal AIEC” in this study. Two of these strains were isolated from patients suffering from sepsis, and the other two stains were isolated from UTIs. The extraintestinal AIEC strains belonged to the O6:H1 (two strains), O25:H4, and O83:H1 serotypes. These serotypes comprised 21.7%, 8.7%, and 4.3% of intestinal AIEC strains, respectively. Thus, the majority of the ExPEC strains that were tested did not exhibit the phenotypic features that characterize the AIEC pathovar.
TABLE 2.
Frequency of ExPEC strains with AIEC phenotype
| Extraintestinal infection | Total no. of strains | AIEC frequency |
|
|---|---|---|---|
| No. | % | ||
| Intra-abdominal pus | 1 | 0 | 0 |
| Meningitis | 12 | 0 | 0 |
| Sepsis | 21 | 2 | 9.5 |
| UTI | 28 | 2 | 7.1 |
| Wound infection | 1 | 0 | 0 |
| Total | 63 | 4 | 6.35 |
Distribution of virulence genes in AIEC and ExPEC strains.
The distribution of virulence-associated genes in ExPEC strains was similar to that obtained for AIEC strains isolated from the human intestinal mucosae, with the exception of the sfa-focDE operons, which were more prevalent among ExPEC strains (P = 0.013) (Table 3). The distribution of phylogroups was also similar, with B2 being the most abundant phylogroup (85.7% and 69.6% of ExPEC and AIEC strains, respectively). Regarding the AIEC strains, all of the strains studied in the present work harbored the fimH gene. The papC and iucD genes were also prevalent, being present in more than 50% of the AIEC strains. The papGII and papGIII alleles were the most frequent alleles found among ExPEC and AIEC strains.
TABLE 3.
Frequency of virulence-associated genes by phenotype (AIEC or non-AIEC) and origin (extraintestinal or intestinal)a
| Virulence gene | No. (%) of strains |
P | No. (%) of strains |
P | ||
|---|---|---|---|---|---|---|
| Non-AIEC (n = 59) | AIEC (n = 27) | Extraintestinal (n = 63) | Intestinal (n = 23) | |||
| papC | 25 (42.4) | 14 (51.9) | NS | 27 (42.9) | 12 (52.2) | NS |
| papGIb | 1 (4.0) | 0 | NS | 1 (3.7) | 0 | NS |
| papGII | 6 (24) | 3 (21.4) | NS | 7 (25.9) | 2 (16.7) | NS |
| papGIII | 11 (44) | 4 (28.6) | NS | 11 (40.7) | 4 (33.3) | NS |
| papGI-II | 2 (8.0) | 0 | NS | 2 (7.4) | 0 | NS |
| papGII-III | 2 (8.0) | 0 | NS | 2 (7.4) | 0 | NS |
| sfa-focDE | 27 (45.8) | 6 (22.2) | 0.031 | 29 (46.0) | 4 (17.4) | 0.013 |
| afa-draBC | 5 (8.5) | 4 (14.8) | NS | 6 (9.5) | 3 (13.0) | NS |
| fimH | 54 (91.5) | 27 (100.0) | NS | 58 (92.1) | 23 (100.0) | NS |
| fimAvMT78 | 16 (27.1) | 6 (22.2) | NS | 17 (27.0) | 5 (21.7) | NS |
| neuC | 15 (25.4) | 3 (11.1) | NS | 15 (23.8) | 3 (13.0) | NS |
| iucD | 39 (66.1) | 15 (55.6) | NS | 42 (66.7) | 12 (52.2) | NS |
| ibeA | 19 (32.2) | 5 (18.5) | NS | 20 (31.7) | 4 (17.4) | NS |
| hlyA | 25 (42.4) | 8 (29.6) | NS | 27 (42.9) | 6 (26.1) | NS |
| cnf1 | 21 (35.6) | 8 (29.6) | NS | 23 (36.5) | 6 (26.1) | NS |
AIEC includes intestinal and extraintestinal AIEC strains; non-AIEC includes only ExPEC strains. Extraintestinal includes AIEC and non-AIEC ExPEC strains; intestinal includes only intestinal AIEC strains. NS, not significant.
For papG alleles, percentages are calculated with respect to papC-positive strains.
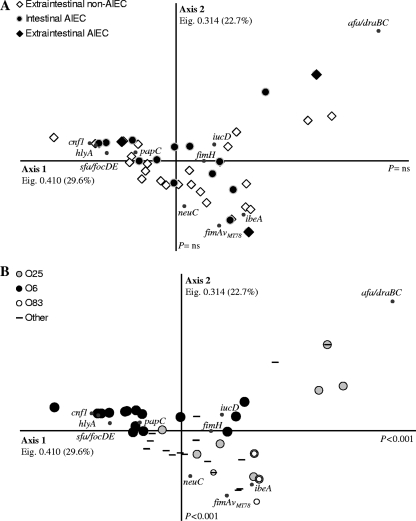
Correspondence analysis for the presence of virulence genes in the strains corroborated these observations (Fig. 1). Neither the intestinal/extraintestinal origin nor the AIEC phenotype was able to explain the segregation of the strains, thus indicating that AIEC and ExPEC pathovars had similar genotypes (Fig. 1A). Moreover, no correlation with adhesion, invasion, or intramacrophage replication indices was detected. Similarly, no segregation was observed between strains that caused different diseases or between strains with distinct phylogenetic origins. A more representative collection of strains from all phylogroups and from all types of extraintestinal infections would be necessary to corroborate this observation. The virulence gene profiles of the strains were associated primarily with the serogroup, as shown in Fig. 1B. The majority of O6 strains appeared to be segregated from the O83 and O25 serogroups by axis 1 (P < 0.001), whereas axis 2 separated the O83 strains from the majority of O6 and O25 strains (P < 0.001). These results indicate that O6 and O83 strains clearly clustered separately in the correspondence analysis by their virulence gene profile, whereas O25 strains showed a higher variability of virulence gene sets. Two main clusters of O25 strains appeared in the correspondence analysis plot. The one situated in the upper right side of the plot grouped afa-draBC-positive O25 strains, whereas the O25 strains clustering closer to O6 and O83 strains were afa-draBC negative. In particular, those virulence genes that had a better correlation with the O6 serogroup were hlyA, with a prevalence of 80.8% within the serogroup; cnf1 and sfa-focDE, each with a prevalence of 76.9%; and papC, with a prevalence of 65.4%. Among the O83 strains, 100% were positive for ibeA and 71.4% were positive for fimAvMT78. Finally, iucD and ibeA were present in 83.3% and 50% of the O25 strains, respectively. Moreover, six out of the nine (66.7%) strains that were positive for afa-draBC belonged to the O25 serogroup (P = 0.003).
FIG. 1.
Correspondence analysis of the distribution of 10 virulence-associated genes (papC, sfa-focDE, afa-draBC, fimH, fimAvMT78, neuC, iucD, ibeA, cnf1, and hlyA) in 63 ExPEC strains and 23 intestinal AIEC strains. Eigenvalues (Eig.) and percentages of variance are provided for each axis. (A) Extraintestinal/intestinal origin of the strains and AIEC phenotype. (B) The serogroup was the sole factor that explained the segregation of the strains (only the most frequent serogroups in our collection [O6, O25, and O83] are specified). Axis 1 explains the segregation of O6 strains from the strains belonging to the O83 and O25 serogroups (P < 0.001), whereas axis 2 segregated O83 strains from the O6 and O25 serogroups (P < 0.001).
Although they showed distinct phenotypes (in terms of adhesion, invasion, and intracellular replication abilities), the AIEC and ExPEC strains shared similar serotypes, phylogenetic origins, and virulence-associated gene distributions.
Clonality and phylogenetic relationships among O6:H1, O25:H4, and O83:H1 extraintestinal and intestinal AIEC strains.
MLST is a DNA sequencing-based method that has become a popular tool for characterizing pathogenic microorganisms, including E. coli (53). Using MLST, the genetic relatedness of isolates can be compared, and closely related organisms can be grouped together in clonal complexes. We compared the four extraintestinal AIEC strains with five intestinal AIEC strains of identical serotypes using MLST in order to check for a possible phylogenetic relationship among them. Interestingly, the strains segregated into three distinct STs according to their serotype, irrespective of their intestinal/extraintestinal origin. In particular, the CD-associated strains AIEC01 and AIEC21, the UPEC strain OL96a, and the sepsis-associated strain PP215 all belonged to the O6:H1 serotype and the B2 phylogroup, and they carried the same combination of alleles across the seven sequenced loci corresponding to ST73 of the ST73 Clpx. Additionally, AIEC strain LF82, isolated from a CD patient, and the septicemic strain PP16 belonged to phylogroup B2, ST135 (no Clpx association). Finally, two intestinal O25:H4 AIEC strains isolated from a patient with ulcerative colitis (AIEC13) and a non-IBD control (AIEC08) and the UPEC FV7563 strain (O25:H4 CTX-M-15 positive) all belonged to phylogroup B2 and displayed ST131 (no Clpx association).
As shown in Fig. 2, all of the intestinal and extraintestinal AIEC strains belonging to the O6:H1 (ST73), O83:H1 (ST135), and O25:H4 (ST131) serotypes (and ST types) harbored the pathogenicity-associated island marker malX and the uropathogenic-specific protein gene usp, and they all possessed a group II polysaccharide capsule gene (kpsMII). In contrast, the secreted autotransporter toxin (sat) gene was detected in the four AIEC strains with the O6:H1 serotype (ST73) and also in one O25:H4 (ST131) extraintestinal AIEC strain. The serum resistance-associated gene (traT) was identified in three AIEC strains belonging to the O6:H1 serotype (ST73) and in two intestinal AIEC strains with the O25:H4 serotype (ST131).
We compared the XbaI PFGE macrorestriction profiles of the intestinal and extraintestinal AIEC strains sharing the same ST and phylogroup. PFGE is a highly discriminatory method and is useful for detecting small DNA differences that rapidly accumulate in the bacterial genome. We used this tool to better differentiate the compared strains by identifying clusters with different similarity values. As expected, most strains of the same serotype, phylogenetic group, and ST grouped together in the dendrogram (Fig. 2). Thus, the macrorestriction analysis demonstrated that the four strains of serotype O6:H1 B2 ST73 clustered together with 69.8% similarity. In particular, OL96a, AIEC21, and AIEC01 grouped with 74.6% similarity. The two O83:H1 B2 ST135 strains (intestinal LF82 and ExPEC PP16) exhibited a similarity value of 77.8%. Finally, the two intestinal O25:H4 B2 ST131 strains (AIEC08 and AIEC13) grouped together in the dendrogram (75% similarity), while the UPEC FV7563 isolate (CT-X-M15 afa-draBC) appeared to be very different, exhibiting only 48.1% similarity.
Further PFGE analysis introducing additional intestinal and extraintestinal O83 strains demonstrated that a diversity of pulsotypes existed among this serogroup, which segregated according to their flagellar H type and virulence genotype. Thus, the six strains of serotype O83:H1 (including intestinal and extraintestinal AIEC strains and intestinal and extraintestinal non-AIEC strains) grouped together with 75.2% similarity (Fig. 3). Two clusters with similarities of >85% displayed a close genetic relationship; in particular, the AIEC strain LF82 clustered together with the sepsis-associated strain H102A with 86.5% similarity, and the ECG043 intestinal strain clustered with the UTI strain OB79A (88.2% similarity).
Therefore, although the majority of ExPEC strains did not exhibit an AIEC phenotype, a minority of strains that did have this phenotype were genetically related to some intestinal AIEC strains, as revealed by MLST and, for certain strains, by PFGE.
DISCUSSION
Despite the fact that the AIEC pathovar has been repeatedly associated with CD (4, 17, 33, 34, 47), some uncertainty exists regarding (i) the genetic relationship between AIEC strains and other pathogenic and nonpathogenic E. coli strains, (ii) its particular identity as pathovar, and (iii) the putative involvement of AIEC strains in extraintestinal diseases in addition to their suspected role in IBD. For that reason, the aim of this work was to determine the AIEC phenotypes of a collection of ExPEC strains and further search for a common phylogenetic origin for the intestinal and extraintestinal AIEC strains.
Given the genetic similarity between the AIEC and ExPEC strains with regard to their virulence gene profiles and phylogenetic origins (mainly B2 and D phylogroups [30, 45]), we suspected that a high proportion of ExPEC strains could also be classified as AIEC strains, but only 4 out of 63 (6.35%) ExPEC strains from our collection were found to share the phenotypic characteristics that describe the AIEC pathovar. These results suggest that the AIEC pathovar comprise a particular group of E. coli strains that are closely related to the ExPEC pathovar but are distinguishable by phenotypic traits (4), which give to the pathovar a particular identity. Unfortunately, no specific genes involved in the adhesion, invasion, or intramacrophage replication abilities of AIEC strains have been discovered to date. Although some genes and regulatory processes have been implicated in the pathogenesis of the prototypic AIEC strain LF82 (2, 3, 9-12, 43, 44), most of these genes are present in the nonpathogenic E. coli strain K-12, thus indicating that differences in gene expression or small sequence variations of these genes might contribute to the AIEC phenotype.
A high diversity of serotypes and virulence gene profiles exists among ExPEC strains, which complicates their classification into pathotypes. Although correspondence analysis segregated the strains by their serogroup, AIEC and non-AIEC strains of intestinal and extraintestinal origins were present in all clusters, thus indicating that a variety of seropathotypes can also be found among AIEC strains. In particular, those virulence genes that best correlated with O6 strains were papC, sfa-focDE, cnf1, and hlyA, whereas fimAvMT78 and ibeA correlated with O83 strains and afa-draBC, iucD, and ibeA correlated with O25 strains. Nevertheless, some genes (malX, usp, and kpsMII) were constantly found in all O6, O25, and O83 AIEC strains, of both intestinal and extraintestinal origins. These genes have been already described for AIEC strain LF82 (50).
Several studies providing a complete description of the virulence-associated genes of a variety of AIEC strains have been published to date, and all coincide in showing that the AIEC pathovar shows homology to human ExPEC strains (4, 9, 13, 17, 33, 34, 50). The virulence genes fimH, fimAvMT78, lpfA, papC, papGII, afa-draBC, sfa-focDE, ColV plasmid, iucD, iss, kpsMII, neuC, ibeA, malX, usp, chuA, hlyA, and cnf1 and UPEC pathogenicity islands IV536, VI536, ICFT073, and IICFT073, characteristic of ExPEC strains, have been detected at distinct frequencies in AIEC strains. In addition, virulence genes of other pathogenic Enterobacteriaceae, such as Salmonella (ratA), Yersinia (pMT1, fyuA, and irp1 and -2), and Vibrio (hcp), have been detected in LF82 and other AIEC strains (4). The presence and prevalence of papC, afa-draBC, and fimH within the AIEC collection used in this study agree with previous descriptions of AIEC or intramucosal E. coli strains isolated by other researchers (4, 17, 33). In contrast, whereas some AIEC strains in this study carried sfa-focDE, cnf1, and hlyA, these virulence genes have not been detected in other collections of invasive E. coli strains (33). In particular, the virulence factors hlyD/cnf1 (pathogenicity island IIJ96) have been reported to be present in the genomes of 40% of E. coli strains isolated from colorectal cancer, whereas they were absent in eight strains isolated from CD patients (13). In contrast, we detected six cnf1- and hlyA-positive AIEC strains, five of which were isolated from colon specimens and one from the ileum of a healthy individual. The heterogeneity of gene profiles found in different studies can be explained by the great genetic diversity among AIEC strains, by the fact that patients came from different geographical regions, or because the E. coli collections used are not representative enough of the real E. coli diversity present in CD patients. Nevertheless, such genes have been also detected in nonpathogenic E. coli strains and are thought to actually be contributing to fitness or colonization (25).
A portion of AIEC strains, including the prototype AIEC LF82, showed virulence genes (fimAvMT78, neuC, ibeA, or cdt) that are frequent among avian pathogenic E. coli (APEC) strains belonging to the subcluster B2-1 defined by Moulin-Schouleur et al. (38). Interestingly, these B2-1 APEC strains were reported to be genetically and phenotypically close to certain human ExPEC strains as revealed by MLST, serotyping, and genotyping. The authors suggested that little or no host specificity exists among these groups of human and avian E. coli strains, and thus APEC might constitute a zoonotic risk. Because previous reports have already addressed the similarity between the two pathovars (4, 9, 13), in conjunction with the fact that AIEC-like strains have been detected in granulomatous Boxer dogs and cow mastitis, determining the distribution of AIEC strains in different healthy and diseased animals is a research area that could contribute to the understanding of AIEC epidemiology, host specificity, and possible routes of transmission.
Noticeably, some strains belonging to the same phylogenetic group, having identical serotypes and virulence gene profiles (for example, the five O83:H1 B2 ST135 strains harboring the fimH, fimAvMT78, ibeA, malX, usp, and kpsMII genes), and having a close genetic relationship as determined by PFGE (Fig. 3) displayed different adhesion, invasion, and intramacrophage replication abilities and thus different AIEC phenotypes. Similarly, in a previous study we observed that isolates from a given subject had identical PFGE profiles but differed from their AIEC phenotype (34). This observation led us to postulate that the AIEC phenotype is achieved by differences in gene expression, the existence of single-nucleotide polymorphisms, or the loss or gain of DNA by horizontal gene transfer. We agree with the hypothesis that AIEC strains are members of the ExPEC pathovar, which usually reside the intestine (46) but have evolved taken advantage of the particular “IBD microenvironment” (49). However, we would remark that the genetic determinants implicated in the AIEC phenotype are at least not detectable by PFGE, MLST, or virulence genotyping of known virulence factors. Baumgart et al. (4) and Bronowski et al. (13) have performed genome subtraction in order to search for unknown AIEC-specific genes. However, these studies were designed to compare strains that are in fact very different from each other (they used as “drivers” nonpathogenic E. coli and UPEC strains), thus obtaining a large number of subtracted genes in addition to those related to the AIEC phenotype. Given the high genetic variability among E. coli strains, a more targeted discrimination, searching for differences between genetically close strains that differ only in their AIEC phenotype, would probably reduce the number of differences, and only those genes most involved in producing the AIEC phenotype would appear in the subtraction library.
It should be emphasized that the four extraintestinal AIEC strains detected in our collection, which belonged to the O6:H1, O25:H4, and O83:H1 serotypes, were found to belong to the same clonal groups as some intestinal AIEC strains with the same serotypes, as revealed by MLST. These results suggest that some intestinal AIEC could cause extraintestinal infections or vice versa. Interestingly, one of the most representative clones from our AIEC collection, O6:H1-ST73, is a frequent cause of UTIs and septicemia. The possible implication of intestinal pathogenic E. coli in extraintestinal infections has been suggested (31). A recent study reports that 6.9% of the strains from a collection of 225 ExPEC strains exhibited a diffuse-adhering phenotype, which is characteristic of the intestinal pathogenic pathovar diffuse-adhering E. coli (1). Moreover, the authors also detected several virulence genes, principally from enteroaggregative E. coli, in some ExPEC strains, thus indicating that certain ExPEC strains may carry virulence properties of diarrheagenic E. coli. All of these observations suggest that certain AIEC strains might be involved in extraintestinal infections.
ExPEC strains may cause a wide range of extraintestinal infections, especially in immunocompromised patients and in persons exposed to high infective doses or persons who have coinfections (45). Whether ExPEC strains should be considered true pathogens or merely opportunistic commensal bacteria remains controversial (19, 36, 45). The same question is also posed for AIEC strains. The presence of AIEC strains in control subjects suggests that additional host and/or environmental factors are needed for these bacteria to cause an infection even though these strains should have the virulence features needed to cause disease. Among the host factors, CD-specific genetic susceptibility loci, such as NOD2/CARD15 (implicated in peptidoglycan recognition, tolerance to bacteria, and bacterial clearance) and the autophagy-related genes ATG16L1 and IRGM, could be involved in the management of AIEC infections (22).
We identified two intestinal AIEC strains and one extraintestinal AIEC strain, all of which were of the O25:H4 serotype, that belonged to the emerging and virulent clonal group ST131 (6, 14, 26, 31, 42). Currently, this clonal pathotype is the most prevalent among human ExPEC strains. Nicolas-Chanoine et al. (42) recently reported the intercontinental emergence of an ExPEC O25:H4-ST131 clone that produces the extended-spectrum β-lactamase CTX-M-15. In the present study, we found that only one (FV7563) of the seven assayed ExPEC O25:H4-ST131 strains producing CTX-M-15 had an AIEC phenotype. This extraintestinal AIEC strain showed a different virulence profile and a very different macrorestriction profile compared to the two intestinal AIEC strains (AIEC08 and AIEC13) of the same clonal group. This is the first time that strains belonging to clone ST131 have been shown to harbor the papC, papGIII, ibeA, cnf1, and hlyA genes (6, 26, 42).
To conclude, the present study demonstrated that the ExPEC and AIEC pathovars share similar virulence gene sets and that certain strains are phylogenetically related. However, the majority of ExPEC strains did not behave like AIEC strains, thus confirming that the AIEC pathovar is related to the ExPEC pathovar but possesses virulence-specific features. These observations suggest that the AIEC phenotype might be encoded by unknown virulence factors or might be the result of differential expression of key genes. Further investigation of the genes implicated in the AIEC phenotype is necessary.
Acknowledgments
This work was partially supported by the Spanish Ministry of Education and Science (SAF2006-00414 and AGL-2008-02129), the Spanish Ministry of Health and Consumer Affairs (REIPI RD06/0008-1018 and FIS PI060059), the Autonomous Government of Galicia (Xunta de Galicia, PGIDIT065TAL26101PR and 07MRU036261PR), and the European Union (Programme Alban E05D055472BR and PEN project FOOD-CT-2006-36256). A. Mora acknowledges the Ramón y Cajal program from the Ministerio de Educación y Ciencia de España.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Abe, C. M., F. A. Salvador, I. N. Falsetti, M. A. M. Vieira, J. Blanco, J. E. Blanco, M. Blanco, A. M. O. Machado, W. P. Elias, R. T. Hernandes, and T. A. T. Gomes. 2008. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 52:397-406. [DOI] [PubMed] [Google Scholar]
- 2.Barnich, N., J. Boudeau, L. Claret, and A. Darfeuille-Michaud. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781-794. [DOI] [PubMed] [Google Scholar]
- 3.Barnich, N., M.-A. Bringer, L. Claret, and A. Darfeuille-Michaud. 2004. Involvement of lipoprotein NlpI in the virulence of adherent invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect. Immun. 72:2484-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgart, M., B. Dogan, M. Rishniw, G. Weitzman, B. Bosworth, R. Yantiss, R. H. Orsi, M. Wiedmann, P. McDonough, S. G. Kim, D. Berg, Y. Schukken, E. Scherl, and K. W. Simpson. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1:403-418. [DOI] [PubMed] [Google Scholar]
- 5.Bidet, P., F. Mahjoub-Messai, J. Blanco, J. Blanco, M. Dehem, Y. Aujard, E. Bingen, and S. Bonacorsi. 2007. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J. Infect. Dis. 196:297-303. [DOI] [PubMed] [Google Scholar]
- 6.Blanco, M., M. P. Alonso, M.-H. Nicolas-Chanoine, G. Dahbi, A. Mora, J. E. Blanco, C. Lopez, P. Cortes, M. Llagostera, V. Leflon-Guibout, B. Puentes, R. Mamani, A. Herrera, M. A. Coira, F. Garcia-Garrote, J. M. Pita, and J. Blanco. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63:1135-1141. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, M., J. Blanco, M. Alonso, and J. Blanco. 1996. Virulence factors and O groups of Escherichia coli isolates from patients with acute pyelonephritis, cystitis and asymptomatic bacteriuria. Eur. J. Epidemiol. 12:191-198. [DOI] [PubMed] [Google Scholar]
- 8.Blanco, M., J. Blanco, M. Alonso, and J. Blanco. 1994. Virulence factors and O groups of Escherichia coli strains isolated from cultures of blood specimens from urosepsis and non-urosepsis patients. Microbiologia 10:249-256. [PubMed] [Google Scholar]
- 9.Boudeau, J., N. Barnich, and A. Darfeuille-Michaud. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272-1284. [DOI] [PubMed] [Google Scholar]
- 10.Boudeau, J., A.-L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bringer, M.-A., N. Barnich, A.-L. Glasser, O. Bardot, and A. Darfeuille-Michaud. 2005. HtrA stress protein is involved in intramacrophagic replication of adherent and invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect. Immun. 73:712-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bringer, M.-A., N. Rolhion, A.-L. Glasser, and A. Darfeuille-Michaud. 2007. The oxidoreductase DsbA plays a key role in the ability of the Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 to resist macrophage killing. J. Bacteriol. 189:4860-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bronowski, C., S. L. Smith, K. Yokota, J. E. Corkill, H. M. Martin, B. J. Campbell, J. M. Rhodes, C. A. Hart, and C. Winstanley. 2008. A subset of mucosa-associated Escherichia coli isolates from patients with colon cancer, but not Crohn's disease, share pathogenicity islands with urinary pathogenic E. coli. Microbiology 154:571-583. [DOI] [PubMed] [Google Scholar]
- 14.Cagnacci, S., L. Gualco, E. Debbia, G. Schito, and A. Marchese. 2008. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J. Clin. Microbiol. 46:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC. 2004. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). PulseNet PFGE manual, p. 5.1-5.3, 1-13. CDC, Atlanta, GA.
- 16.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darfeuille-Michaud, A., J. Boudeau, P. Bulois, C. Neut, A.-L. Glasser, N. Barnich, M.-A. Bringer, A. Swidsinski, L. Beaugerie, and J.-F. Colombel. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127:412-421. [DOI] [PubMed] [Google Scholar]
- 18.Darfeuille-Michaud, A., C. Neut, N. Barnich, E. Lederman, P. Di Martino, P. Desreumaux, L. Gambiez, B. Joly, A. Cortot, and J. Colombel. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405-1413. [DOI] [PubMed] [Google Scholar]
- 19.Dobrindt, U. 2005. (Patho-)genomics of Escherichia coli. Int. J. Med. Microbiol. 295:357-371. [DOI] [PubMed] [Google Scholar]
- 20.Fujita, H., Y. Eishi, I. Ishige, K. Saitoh, T. Takizawa, T. Arima, and M. Koike. 2002. Quantitative analysis of bacterial DNA from Mycobacteria spp., Bacteroides vulgatus, and Escherichia coli in tissue samples from patients with inflammatory bowel diseases. J. Gastroenterol. 37:509-516. [DOI] [PubMed] [Google Scholar]
- 21.Glasser, A.-L., J. Boudeau, N. Barnich, M.-H. Perruchot, J.-F. Colombel, and A. Darfeuille-Michaud. 2001. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69:5529-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glasser, A.-L., and A. Darfeuille-Michaud. 2008. Abnormalities in the handling of intracellular bacteria in Crohn's disease: a link between infectious etiology and host genetic susceptibility. Arch. Immunol. Ther. Exp. 56:237-244. [DOI] [PubMed] [Google Scholar]
- 23.Gophna, U., K. Sommerfeld, S. Gophna, W. F. Doolittle, and S. J. O. Veldhuyzen van Zanten. 2006. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guinée, P. A., C. M. Agterberg, and W. H. Jansen. 1972. Escherichia coli O antigen typing by means of a mechanized microtechnique. Appl. Microbiol. 24:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hejnova, J., U. Dobrindt, R. Nemcova, C. Rusniok, A. Bomba, L. Frangeul, J. Hacker, P. Glaser, P. Sebo, and C. Buchrieser. 2005. Characterization of the flexible genome complement of the commensal Escherichia coli strain A0 34/86 (O83:K24:H31). Microbiology 151:385-398. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. R., M. Menard, B. Johnston, M. A. Kuskowski, K. Nichol, and G. G. Zhanel. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 28.Keighley, M. R., Y. Arabi, F. Dimock, D. W. Burdon, R. N. Allan, and J. Alexander-Williams. 1978. Influence of inflammatory bowel disease on intestinal microflora. Gut 19:1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleessen, B., A. Kroesen, H. Buhr, and M. Blaut. 2002. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 37:1034-1041. [DOI] [PubMed] [Google Scholar]
- 30.Kotlowski, R., C. N. Bernstein, S. Sepehri, and D. O. Krause. 2007. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manges, A., H. Tabor, P. Tellis, C. Vincent, and P. Tellier. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 14:1575-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangin, I., R. Bonnet, P. Seksik, L. Rigottier-Gois, M. Sutren, Y. Bouhnik, C. Neut, M. D. Collins, J.-F. Colombel, P. Marteau, and J. Dore. 2004. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol. Ecol. 50:25-36. [DOI] [PubMed] [Google Scholar]
- 33.Martin, H. M., B. J. Campbell, C. A. Hart, C. Mpofu, M. Nayar, R. Singh, H. Englyst, H. F. Williams, and J. M. Rhodes. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127:80-93. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Medina, M., X. Aldeguer, M. Lopez-Siles, F. González-Huix, C. López-Oliu, G. Dahbi, J. E. Blanco, J. Blanco, L. J. Garcia-Gil, and A. Darfeuille-Michaud. 2009. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm. Bowel Dis. 15:872-882. [DOI] [PubMed] [Google Scholar]
- 35.Masseret, E., J. Boudeau, J. F. Colombel, C. Neut, P. Desreumaux, B. Joly, A. Cortot, and A. Darfeuille-Michaud. 2001. Genetically related Escherichia coli strains associated with Crohn's disease. Gut 48:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mokady, D., U. Gophna, and E. Ron. 2005. Virulence factors of septicemic Escherichia coli strains. Int. J. Med. Microbiol. 295:455-462. [DOI] [PubMed] [Google Scholar]
- 37.Mora, A., C. Lopez, G. Dahbi, M. Blanco, J. Blanco, M. P. Alonso, A. Herrera, R. Mamani, S. Bonacorsi, M. Moulin-Schouleur, and J. Blanco. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiology. 9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moulin-Schouleur, M., M. Reperant, S. Laurent, A. Bree, S. Mignon-Grasteau, P. Germon, D. Rasschaert, and C. Schouler. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 45:3366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moulin-Schouleur, M., C. Schouler, P. Tailliez, M.-R. Kao, A. Bree, P. Germon, E. Oswald, J. Mainil, M. Blanco, and J. Blanco. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44:3484-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naves, P., G. del Prado, L. Huelves, M. Gracia, V. Ruiz, J. Blanco, G. Dahbi, M. Blanco, M. del Carmen Ponte, and F. Soriano. 2008. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 45:86-91. [DOI] [PubMed] [Google Scholar]
- 41.Neut, C., J. F. Colombel, F. Guillemot, A. Cortot, P. Gower, P. Quandalle, M. Ribet, C. Romond, and J. C. Paris. 1989. Impaired bacterial flora in human excluded colon. Gut 30:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicolas-Chanoine, M.-H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Canica, Y.-J. Park, J.-P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273-281. [DOI] [PubMed] [Google Scholar]
- 43.Rolhion, N., N. Barnich, L. Claret, and A. Darfeuille-Michaud. 2005. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 187:2286-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rolhion, N., F. A. Carvalho, and A. Darfeuille-Michaud. 2007. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Mol. Microbiol. 63:1684-1700. [DOI] [PubMed] [Google Scholar]
- 45.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 46.Russo, T. A., A. Stapleton, S. Wenderoth, T. M. Hooton, and W. E. Stamm. 1995. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J. Infect. Dis. 172:440-445. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki, M., S. V. Sitaraman, B. A. Babbin, P. Gerner-Smidt, E. M. Ribot, N. Garrett, J. A. Alpern, A. Akyildiz, A. L. Theiss, A. Nusrat, and J.-M. A. Klapproth. 2007. Invasive Escherichia coli are a feature of Crohn's disease. Lab. Investig. 87:1042-1054. [DOI] [PubMed] [Google Scholar]
- 48.Seksik, P., L. Rigottier-Gois, G. Gramet, M. Sutren, P. Pochart, P. Marteau, R. Jian, and J. Dore. 2003. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sepehri, S., R. Kotlowski, C. N. Bernstein, and D. O. Krause. 2009. Phylogenetic analysis of inflammatory bowel disease associated Escherichia coli and the fimH virulence determinant. Inflamm. Bowl Dis. doi: 10.1002/ibd.20966. [DOI] [PubMed]
- 50.Simpson, K. W., B. Dogan, M. Rishniw, R. E. Goldstein, S. Klaessig, P. L. McDonough, A. J. German, R. M. Yates, D. G. Russell, S. E. Johnson, D. E. Berg, J. Harel, G. Bruant, S. P. McDonough, and Y. H. Schukken. 2006. Adherent and invasive Escherichia coli is associated with granulomatous colitis in Boxer dogs. Infect. Immun. 74:4778-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swidsinski, A., J. Weber, V. Loening-Baucke, L. P. Hale, and H. Lochs. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43:3380-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.ter Braak, C. J. F., and P. Smilauer. 1998. CANOCO reference manual and user's guide to Canoco for Windows: software for canonical community ordination (version 4). Plant Research International, Wageningen, The Netherlands.
- 53.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. Wieler, H. Karch, P. Reeves, M. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]