Abstract
The patient-to-patient transmission of highly prevalent Pseudomonas aeruginosa clones which are associated with enhanced disease progression has led to strict segregation policies for cystic fibrosis (CF) patients in many countries. However, little is known about the population structure of P. aeruginosa among CF patients. The aim of the present cross-sectional study was to determine the prevalence and genetic relatedness of P. aeruginosa isolates from CF patients who visited two major CF centers in The Netherlands in 2007 and 2008. These patients represented 45% of the Dutch CF population. P. aeruginosa carriage in the respiratory tract was determined by standard microbiological culture techniques, and all phenotypically different isolates in the first specimens recovered in 2007 and 2008 were genotyped by multilocus sequence typing. A total of 313 (57%) of 551 patients whose samples were cultured carried P. aeruginosa. Two sequence types (STs), ST406 and ST497, were found in 15% and 5% of the patients, respectively, and 60% of the patients harbored a strain that was also found in at least two other patients. The risk ratios for carrying ST406 and ST497 were 17.8 (95% confidence interval [CI], 7.2 to 43.6) for those aged between 15 and 24 years and 6 (95% CI, 1.4 to 26.1) for those aged >25 years. ST406 and ST497 were not genetically linked to previously described epidemic clones, which were also not found in this CF population. The population structure of P. aeruginosa in Dutch CF patients is characterized by the presence of two prevalent STs that are associated with certain age groups and that are not genetically linked to previously described epidemic clones.
Pseudomonas aeruginosa is a ubiquitous, versatile bacterium that can infect humans as well as plants and animals. The species is infamous for causing nosocomial infections in immunocompromised patients and patients in intensive care units and is a major cause of morbidity and mortality in patients with cystic fibrosis (CF) (26).
The widely held belief that CF patients acquire P. aeruginosa strains mainly from their inanimate environment, with most patients being colonized by unique strains, has been challenged by reports indicating that P. aeruginosa clones may frequently be transmitted between CF patients (3, 6, 18, 19, 23, 24). Some of these clones, such as the Liverpool epidemic strain and the Melbourne epidemic strain, have been associated with enhanced disease progression and higher rates of mortality, respectively (1, 13). In The Netherlands, the patient-to-patient transmission of P. aeruginosa was documented during a summer camp (4). These findings have led to strict segregation policies for CF patients in many countries, including The Netherlands. However, despite these studies, there is little information on the population structure of P. aeruginosa within populations of CF patients. We therefore investigated the prevalence and genetic relatedness of P. aeruginosa isolates compared to those of the international known genotypes in an unbiased cohort representing 45% of the CF population in The Netherlands in 2007 and 2008.
MATERIALS AND METHODS
Patients and design.
All CF patients who visited the Wilhemina Children's Hospital/University Medical Centre Utrecht (UMCU) in Utrecht, The Netherlands, in 2007 or the Haga Teaching Hospital (Haga) in The Hague, The Netherlands, from August 2007 to June 2008 were included in this cross-sectional study.
Samples.
All sputum and throat swab samples were cultured by the standard diagnostic laboratory protocols of each hospital. At UMCU, the samples were plated on Trypticase soy agar II with 5% sheep blood (TSA-blood) plates and MacConkey agar plates (Becton, The Netherlands) to detect P. aeruginosa. The plates were incubated at 37°C for 2 days, after which the isolates were identified on the basis of the oxidase reaction and Gram stain. At Haga, sputum samples were washed in phosphate buffer (pH 7.2 to 7.4) and plated on Columbia agar-5% sheep blood-colistin-aztreonam, chocolate, cystine lactose electrolyte-deficient, and cepacia agar plates (Oxoid, The Netherlands). On the second day, all nonfermenters were plated on a cystine lactose electrolyte-deficient plate (Oxoid, The Netherlands) and grown at 37°C in 5% CO2 for 36 h. All lactose-negative colonies were plated on Columbia agar with C-390 and phenanthroline (Oxoid) and were grown at 37°C for a maximum of 3 nights. In addition, oxidase, acetamide, and antibiotic resistance tests (BBL discs; Becton Dickinson, Belgium) were performed. MICs were determined by using the CLSI breakpoints for colistin, tobramycin, ciprofloxacin, ceftazidime, and piperacillin-tazobactam in both hospitals (7). Furthermore, the susceptibilities to piperacillin, amikacin, and meropenem were determined at UMCU and the susceptibility to imipenem was determined at Haga. One P. aeruginosa colony of each different colony morphology (according to rough, smooth, and mucoid characteristics and colony size) per sample was randomly picked and stored at −70°C. These isolates from the first respiratory tract culture that yielded P. aeruginosa from each patient during the study period were genotyped. Multilocus sequence typing (MLST) was used to confirm the correct species identification and to study the genetic relatedness of the P. aeruginosa isolates recovered.
MLST.
MLST was performed according to the protocol of Curran et al. (8). Some adjustments were made, including the use of newly designed primers (Table 1), because the initial protocol did not yield PCR products in all cases. The isolates were recultured on TSA-blood plates (Becton) overnight at 37°C, suspended in 20 μl lysis buffer (0.25% sodium dodecyl sulfate, 0.05 N NaOH), and incubated at 95°C for 20 min. The cell lysate was centrifuged and diluted with 180 μl buffer (10 mM Tris-DCl, pH 8.5). After thorough mixing of the solution, another centrifugation at 16,000 × g for 5 min was performed to remove the cell debris. The supernatants were frozen at −20°C until further use. A total of 2.5 μl of the lysate was used in a touchdown PCR, according to the protocol of Curran et al. (8). Q buffer (Qiagen Benelux B.V., Venlo, The Netherlands) was added to the PCR mix. The PCR was conducted as follows: 10 min at 96°C and then 10 cycles of 30 s at 95°C, 30 s at 65°C with the use of a temperature 1°C lower every cycle, and 1 min at 72°C. This was followed by 25 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C, after which a final step of 7 min at 72°C was used. A check for the presence of PCR products was performed by electrophoresis on a 1% agarose gel. The PCR products were sequenced (BaseClear, Leiden, The Netherlands) with the same primers used for amplification. The sequences were analyzed by using the Bionumerics (version 5.1) program (Applied Maths, St-Martens-Latem, Belgium).
TABLE 1.
Primers used for MLST (both amplification and sequencing)
| Primer | Sequence |
|---|---|
| acsA for | AAGGGCGTGCTGCATACCA |
| acsA rev | CGGCCAGGAGTCGAGGATC |
| aroE for | ATGTCACCGTGCCGTTCAAG |
| aroE rev | GCGCCAGAGGAAGAATGCC |
| guaA for | ACTACGGCGTGCAATTCCAC |
| guaA rev | GAACGGGTGGCGGTAGACC |
| mutL for | AGCCTGGCAGGTGGAAACC |
| mutL rev | CTCTCCAGCACGCTCTCGG |
| nuoD for | GGGACATGTACGGCATCACCT |
| nuoD rev | GCGCAGGATGCTGTTCTTCA |
| ppsA for | CGGTCAAGGTAGTGGACGTCG |
| ppsA rev | TTCTTGCGCACATCGAAACC |
| trpE for | CGCGAGGACTATGAAAACGC |
| trpE rev | CGCTTGTTGATGGTTTCTT |
Sequence types (STs) were compared to those on the P. aeruginosa MLST website (http://pubmlst.org/paeruginosa/) developed by Keith Jolley (17), and new alleles and profiles were sent to the curator (A. Baldwin). MLST data on genetic similarity were also confirmed by PFGE (data not shown).
Statistics.
Calculations and analyses were performed with the SPSS (version 12.0) program (SPSS, Chicago, IL). Genetic diversity was calculated as described previously by using the EpiCompare (version 1.0) program (14). The eBURST algorithm (10), available at the eBURST (version 3) website (http://eburst.mlst.net), was used to display the P. aeruginosa population snapshot of 591 P. aeruginosa STs available at http://pubmlst.org/paeruginosa/ (September 2009). The following settings for the creation of the eBURST-based snapshot were used: number of loci per isolate, 7; minimum number of identical loci for group definition, 0; minimal single-locus variant count for subgroup definition, 3; and number of resamplings for bootstrap analysis, 1,000.
RESULTS
Patient characteristics.
There are an estimated 1,300 CF patients in The Netherlands, and 596 (46%) of these patients visited UMCU (n = 386) or Haga (n = 210) during the study period. The average ages of the study population were 19.5 years (standard deviation [SD], 11.8 years; range, 1.7 to 55.9 years) for those visiting UMCU and 30.7 years (SD, 14.2 years; range, 0.9 to 69.1 years) for those visiting Haga (P < 0.001). The majority of patients (53%) were male, with the gender distributions in both centers being similar. During the study period, respiratory tract samples (sputum or throat swab samples) were cultured from 551 (92%) patients, and P. aeruginosa was identified in 313, yielding a prevalence in the sampled patients of 57% (55% at UMCU and 60% at Haga). Forty percent of the children (age, <18 years; n = 249) were colonized, as were 70% of the adults (age, ≥18 years; n = 302).
A total of 443 P. aeruginosa isolates from 265 (85%) of 313 patients were genotyped.
Prevalence of P. aeruginosa genotypes.
Among the 443 isolates that were genotyped, 142 different STs were identified, of which 43 were shared by patients. Although 135 (51%) colonized patients were simultaneously colonized with two or more phenotypically different isolates, isolates with different STs were found in only 28 (11%) patients. A separate analysis of throat swab versus sputum specimens did not yield a different interpretation (data not shown).
ST406 was found in 41 patients and ST497 was found in 14 patients, yielding prevalence rates of 15% (41/265) and 5% (14/265), respectively. Other genotypes were found among nine patients (ST274), seven patients (STs 108 and 155), six patients (STs 17, 492, and 511), five patients (STs 27, 170, 395, and 517), four patients (STs 111, 261, 267, 485, 540, and 561), and three patients (STs 179, 244, 277, 481, 508, and 549); and 19 STs were shared by two different patients. The P. aeruginosa population in Dutch CF patients is highly diverse, with the genetic diversity being 97.3% (95% confidence interval [CI], 96.2% to 98.4%). Among 173 patients, 99 different STs were found at UMCU, and 70 different STs were found among 92 patients at Haga. This results in a lower level of genetic diversity among the isolates from patients from UMCU (96.2%; 95% CI, 94.3% to 98.1%) than among the isolates from patients from Haga (98.5%; 95% CI, 97.8% to 99.3%). In total, 70% of all colonized patients harbored P. aeruginosa STs that were also found in other patients and 30% were colonized with isolates with unique STs. Seven percent of all patients were colonized with both isolates with unique STs and isolates with shared STs.
The most prevalent ST, ST406, had a higher prevalence in the UMCU population (20%) than in the Haga population (7%) (risk ratio [RR], 2.6; 95% CI, 1.2 to 5.6). ST497 had a comparable prevalence in both hospital populations (5% and 6% in the UMCU and Haga populations, respectively). The location of the patient's residence was not correlated with the carriage of ST406 or ST497 (Fig. 1).
FIG. 1.
Locations of residences in The Netherlands of CF patients harboring the predominant Dutch P. aeruginosa clones ST406 and ST497.
The mean age of the patients colonized with ST406 strains was 19.8 years (SD, 3.6 years), whereas the mean ages were 32.8 years (SD, 8.6 years) for those colonized with ST497 strains and 26.8 years (SD, 14.1 years) for those colonized with non-ST406 and non-ST497 strains (P = 0.001). The prevalence of ST406 was 40 to 53% among colonized patients between 15 and 24 years of age (Fig. 2), yielding an RR of 17.8 (95% CI, 7.2 to 43.6) for ST406 carriage in this age group compared to the other age groups. The prevalence of ST497 was as high as 14% among colonized patients between 25 and 35 years of age (Fig. 2), and the RR for the carriage of ST497 in CF patients older than 25 years was 6.0 (95% CI, 1.4 to 26.1). Similar to the two highly prevalent clones, other genotypes shared by at least three patients also appeared to be associated with age, with some STs (STs 27, 108, 540, 17, 481, and 492) being almost exclusively present in patients younger than 25 years of age and others (STs 549, 274, 511, and 508) being almost exclusively present in those older than 25 years of age (Fig. 3). Most of these age-associated STs were present in patients from both hospitals. The unique STs were found in patients of all ages.
FIG. 2.
Patients colonized with P. aeruginosa ST406 and ST497 strains as a percentage of all patients colonized with P. aeruginosa by age group (n = total number of patients who were positive for P. aeruginosa [PA] in each group). The age categories are indicated in brackets.
FIG. 3.
Age per sequence type. Black line, mean age; box, 25 and 75 percentile; ○, outliers between 1.5 and 3 box lengths from either end of the box; *, cases more than 3 box lengths from either end of the box. The number of patients colonized by each ST is noted in parentheses. STs 27, 261, 108, 540, 17, 492, 170, and 406 were almost exclusively present in patients younger than 25 years of age; and STs 497, 549, 274, and 511 were almost exclusively present in those older than 25 years.
Compared to strains of all other STs, strains of ST406 and ST497 could not be characterized by a specific antibiotic resistance profile. In general, ST406 strains were more frequently resistant to carbapenems and susceptible to ciprofloxacin. Both ST406 and ST497 strains were more often resistant to amikacin (data not shown).
Among our patient population, there were 28 sibling pairs and 5 sibling trios. Of these 33 sibling sets, both or all three siblings were colonized with P. aeruginosa in 13 instances, and in 10 cases all siblings shared strains of identical STs. In another 13 cases, all siblings had negative cultures. Thus, 79% of siblings had identical (or concordant) colonization status (negative for carriage or carriage of strains of the same genotypes).
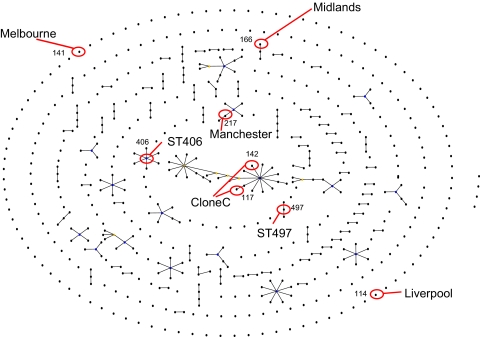
A total of 126 STs from the present study had not been described before and were added to the existing 524 genotypes in the international P. aeruginosa MLST database. The eBURST picture of 591 STs from the available isolates in the MLST database revealed a high number of singletons, with only a few, small clonal complexes typical for species with a panmictic population structure being detected (Fig. 4). ST406 and ST497 appeared to be genotypically different from all known STs in the international database, indicating that they are most likely unique for the Dutch CF population. Moreover, they are not genetically linked to each other, nor are they linked to any of the known epidemic strains, such as the Midlands, the Liverpool, the Melbourne, and the Manchester clones and clone C. Furthermore, none of these clones were detected among this cohort of Dutch CF patients (Fig. 4).
FIG. 4.
Snapshot of population of 591 P. aeruginosa STs on the basis of MLST allelic profiles using the eBURST algorithm (10). Numbers and dots represent STs. Lines connect single-locus variants, which are STs that differ in only one of the seven housekeeping genes. The snapshot shows all clonal complexes (connected STs), singletons (unconnected STs), and patterns of evolutionary descent. The placement of singletons as well as the length of the lines is arbitrary. Blue dots indicate probable ancestors of clonal complexes. The two highly prevalent Dutch clones as well as the previous described epidemic clones are indicated.
DISCUSSION
In this cross-sectional study, which included individuals representing about 45% of the Dutch CF population, the prevalence of P. aeruginosa colonization was 57%, and the individuals were colonized with a genetically highly diverse bacterial population. In all, 70% of the patients were colonized with strains that had genotypes that were also found in other patients, and strains with two highly prevalent STs, ST406 and ST497, were identified. Colonization with STs shared by at least three patients, including ST406 and ST497, was age dependent. ST406 and ST497 appeared to be genotypically different from all known STs in the international database and are not genetically closely linked to each other or to any of the known epidemic strains, such as the Midlands, the Liverpool, the Melbourne, and the Manchester clones and clone C. Besides, none of these clones were detected among the cohort of Dutch CF patients evaluated in this study.
The observed prevalence of P. aeruginosa carriage in the Dutch CF population of 57% (the prevalence is 40% in children and 70% in adults) is similar to prevalence rates reported among CF populations in the United Kingdom, the United States, and Australia, which range from 40 to 85% (9, 13; United Kingdom CF database, 2008).
In the Dutch CF population, two P. aeruginosa clones are present in 20% of all patients colonized. Although CF patients mainly harbored unique P. aeruginosa strains, with infrequent transmission events occurring in some studies, others reported that specific P. aeruginosa strains were shared by from 14% to 60% of CF patients, with the rates of patient-to-patient transmission being presumed to be high (3, 5, 6, 11, 19, 25, 28, 29). In a cohort of 849 CF patients colonized with P. aeruginosa from 31 CF centers in the United Kingdom, 11% and 10% carried the so-called Liverpool and Midlands 1 genotypes, respectively (24). In an Australian cohort of 100 CF patients from two clinics, 39 patients shared strains of one common genotype (23). This indicates that the presence of specific predominant clones circulating in the CF patient population is not unique to the Dutch situation.
The reported proportions of CF patients colonized with genotypes also found in other CF patients were 28%, 30%, and 59% in studies from the United Kingdom, Belgium, and Australia, respectively (23, 24, 29). The fact that 70% of our patients were colonized with genotypes also detected in other CF patients, indeed, suggests an important role for patient-to-patient transmission. The high level of congruence of colonization status and Pseudomonas genotype between tested siblings, which has also been found in other studies, is also an indication that patient-to-patient transmission plays an important role (20, 25, 29).
The number of shared STs might be related to the typing technique used, as the use of a detection method with a higher discriminatory power will result in more unique types. In the current study, we used MLST to infer the clonality of strains, which was shown to be slightly less discriminatory than pulsed-field gel electrophoresis (PFGE) (16). Considering the small difference in discriminatory power between PFGE and MLST, it is unlikely that the high proportion of patients carrying shared genotypes results from a discriminatory power of MLST that is too low. We have chosen MLST because of its high discriminatory index, its use in revealing clonal relatedness between isolates when this is not readily apparent by PFGE, and its unambiguous, reproducible results that are easily electronically portable between laboratories, allowing the international comparison of genotypes (12).
Colonization with ST406 and ST497 was associated with the patient's age. ST406 was the dominant genotype in patients between 15 and 24 years of age, while ST497 was more prevalent in older patients. In retrospect, our findings confirm those from a previous study of 80 CF patients between 6 and 20 years old attending four Dutch CF summer camps in 2001. MLST of the isolates in that study revealed that the most prevalent amplified fragment length polymorphism (AFLP) type represented ST406 and was found equally frequently among those between 6 and 15 years of age but less frequently among the oldest CF patients (ages, 15 to 19 years). An AFLP type representing ST497 was present only in these older patients (4). Furthermore, in a study involving 157 pediatric patients in The Netherlands, a clone detected in CF patients on the basis of PFGE (the strain was ST406 when it was typed by MLST; C. van der Ent, personal communication) was not found in other non-CF patient groups (27).
The observed association with age, as determined in this study, is much stronger than that detected in previous studies of epidemic clones. For instance, both the Melbourne clone (pulsotype I) and the Liverpool epidemic strain were first identified in pediatric clinics (3, 6), but later they were also identified in adult patients (2, 22, 23). The Australian pulsotype 2 (PT2) strain was also found both in pediatric patients and in adults. However, almost all adults who harbored PT2 strains could be linked to the pediatric clinic where PT2 was found in abundance. In retrospect, this might also indicate an association of PT2 to a specific age group (23).
The absence of ST406 in patients older than 31 years of age is, as yet, unexplained, but several mechanisms could be hypothesized. Initial P. aeruginosa colonization in usually young CF patients is mostly transient and is followed by sustained colonization with a particular genotype in later years (21, 29). ST406 is possibly lost after a certain age, perhaps due to competition with other strains like the clonal strain of ST497, which is found almost exclusively in older patients. Superinfections with a transmissible P. aeruginosa strain have been demonstrated to occur in CF patients who were already colonized by other strains (15, 22). It is also possible that ST406 has only recently (in the last one or two decades) been introduced into the CF population and is therefore linked to the younger cohort of patients who may have participated in activities with other CF patients. Finally, the carriage of ST406 in younger patients could be an effect of host tropism, in which ST406 favors the colonization of young patients above the colonization of older patients. The fact that ST406 was more prevalent in patients from UMCU than those from Haga might be related to differences in the patient population, such as age (with the UMCU patients being younger; 19.4 years versus 30.6 years for the Haga patients), which might have led to more frequent and more intense contacts with each other and thus an increased risk of transmission. In fact, host tropism, competition between strains, and age-related association patterns between patients could all interact. Quantification of these different aspects is needed to elucidate the underlying mechanisms of our observations. In addition, longitudinal studies are needed to determine the clinical consequences of colonization with certain genotypes.
ST406 and ST497 are different from other known (international) clones, and neither of these STs has been found in other countries so far, indicating that these clones are specifically linked to Dutch CF patients. Until now, there has been only limited evidence of the international spread of the known CF-related epidemic strains. In one study, a number of major P. aeruginosa clones appeared to be widespread in clinical and environmental habitats and were also isolated from CF patients in different countries (30). Although this observation could have been related to the global environmental spread of these strains and the local acquisition by CF patients, some of the CF-related epidemic strains were found in other habitats as well. On the basis of strain typing by use of an ArrayTube chip, the Midlands epidemic strain was assigned to the same clonal group as strains from a German intensive care unit and one strain found in a German river, and the Liverpool epidemic strain belonged to the same type as an isolate from a patient with bacteremia in a Swiss clinic (30).
Rapid evolution of P. aeruginosa clones might decrease their genetic relatedness and obscure their epidemiological linkage, since the timed spread, genome plasticity, and discriminatory power of the typing method determine whether one will find a local, regional, or global spread of clones.
In this report, we describe a population structure of P. aeruginosa in Dutch CF patients that is characterized by the presence of two prevalent STs that are associated with certain age groups and that are not genetically linked to previously described epidemic clones.
In 2006, a segregation policy for CF patients was implemented in The Netherlands. Many CF patients find it difficult to adhere to these strict rules. The population structure of P. aeruginosa and the prevalence rates of genotypes in the Dutch CF patient population in 2007 to 2008 may now serve as a reference point for the future evaluation of the efficacy of this segregation policy. If it is successful, such a policy should reduce, or at least stabilize, the prevalence of dominant P. aeruginosa genotypes.
Acknowledgments
This publication made use of the Pseudomonas aeruginosa MLST website (http://pubmlst.org/paeruginosa/) developed by Keith Jolley (17) and sited at the University of Oxford.
The development of the Pseudomonas aeruginosa MLST website has been funded by the Wellcome Trust. M.B. is supported by The Netherlands Organization for Scientific Research (VICI NWO grant 918.76.611).
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Al-Aloul, M., J. Crawley, C. Winstanley, C. A. Hart, M. J. Ledson, and M. J. Walshaw. 2004. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax 59:334-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, D., S. Bell, M. Robinson, P. Bye, B. Rose, C. Harbour, C. Lee, H. Service, M. Nissen, M. Syrmis, and C. Wainwright. 2003. Evidence for spread of a clonal strain of Pseudomonas aeruginosa among cystic fibrosis clinics. J. Clin. Microbiol. 41:2266-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, D. S., G. M. Nixon, R. Carzino, A. Bigham, J. B. Carlin, R. M. Robins-Browne, and K. Grimwood. 2002. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am. J. Respir. Crit. Care Med. 166:983-987. [DOI] [PubMed] [Google Scholar]
- 4.Brimicombe, R. W., L. Dijkshoorn, T. J. van der Reijden, I. Kardoes, T. L. Pitt, P. J. van den Broek, and H. G. Heijerman. 2008. Transmission of Pseudomonas aeruginosa in children with cystic fibrosis attending summer camps in The Netherlands. J. Cyst. Fibros. 7:30-36. [DOI] [PubMed] [Google Scholar]
- 5.Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444-452. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, K., R. L. Smyth, J. R. Govan, C. Doherty, C. Winstanley, N. Denning, D. P. Heaf, H. van Saene, and C. A. Hart. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639-642. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. M100-S16. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Curran, B., D. Jonas, H. Grundmann, T. Pitt, and C. G. Dowson. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doring, G., S. P. Conway, H. G. Heijerman, M. E. Hodson, N. Hoiby, A. Smyth, and D. J. Touw. 2000. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur. Respir. J. 16:749-767. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluge, G., B. Ojeniyi, N. Hoiby, A. Digranes, O. Ciofu, E. Hunstad, O. C. Haanaes, and O. T. Storrosten. 2001. Typing of Pseudomonas aeruginosa strains in Norwegian cystic fibrosis patients. Clin. Microbiol. Infect. 7:238-243. [DOI] [PubMed] [Google Scholar]
- 12.Giske, C. G., B. Libisch, C. Colinon, E. Scoulica, L. Pagani, M. Fuzi, G. Kronvall, and G. M. Rossolini. 2006. Establishing clonal relationships between VIM-1-like metallo-beta-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J. Clin. Microbiol. 44:4309-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths, A. L., K. Jamsen, J. B. Carlin, K. Grimwood, R. Carzino, P. J. Robinson, J. Massie, and D. S. Armstrong. 2005. Effects of segregation on an epidemic Pseudomonas aeruginosa strain in a cystic fibrosis clinic. Am. J. Respir. Crit. Care Med. 171:1020-1025. [DOI] [PubMed] [Google Scholar]
- 14.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelsbak, L., H. K. Johansen, A. L. Frost, R. Thogersen, L. E. Thomsen, O. Ciofu, L. Yang, J. A. Haagensen, N. Hoiby, and S. Molin. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 75:2214-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J. K., S. M. Arduino, O. C. Stine, J. A. Johnson, and A. D. Harris. 2007. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 45:3707-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, A. M., M. E. Dodd, J. R. Govan, C. J. Doherty, C. M. Smith, B. J. Isalska, and A. K. Webb. 2005. Prospective surveillance for Pseudomonas aeruginosa cross-infection at a cystic fibrosis center. Am. J. Respir. Crit. Care Med. 171:257-260. [DOI] [PubMed] [Google Scholar]
- 19.Jones, A. M., J. R. Govan, C. J. Doherty, M. E. Dodd, B. J. Isalska, T. N. Stanbridge, and A. K. Webb. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557-558. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, N. M., M. X. Fitzgerald, E. Tempany, C. O'Boyle, F. R. Falkiner, and C. T. Keane. 1982. Does pseudomonas cross-infection occur between cystic-fibrosis patients. Lancet ii:688-690. [DOI] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCallum, S. J., J. Corkill, M. Gallagher, M. J. Ledson, C. A. Hart, and M. J. Walshaw. 2001. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet 358:558-560. [DOI] [PubMed] [Google Scholar]
- 23.O'Carroll, M. R., M. W. Syrmis, C. E. Wainwright, R. M. Greer, P. Mitchell, C. Coulter, T. P. Sloots, M. D. Nissen, and S. C. Bell. 2004. Clonal strains of Pseudomonas aeruginosa in paediatric and adult cystic fibrosis units. Eur. Respir. J. 24:101-106. [DOI] [PubMed] [Google Scholar]
- 24.Scott, F. W., and T. L. Pitt. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53:609-615. [DOI] [PubMed] [Google Scholar]
- 25.Speert, D. P., M. E. Campbell, D. A. Henry, R. Milner, F. Taha, A. Gravelle, A. G. Davidson, L. T. Wong, and E. Mahenthiralingam. 2002. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am. J. Respir. Crit. Care Med. 166:988-993. [DOI] [PubMed] [Google Scholar]
- 26.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 27.Tramper-Stranders, G. A., C. K. van der Ent, T. F. Wolfs, J. L. Kimpen, A. Fleer, U. Johansen, H. K. Johansen, and N. Hoiby. 2008. Pseudomonas aeruginosa diversity in distinct paediatric patient groups. Clin. Microbiol. Infect. 14:935-941. [DOI] [PubMed] [Google Scholar]
- 28.Tubbs, D., W. Lenney, P. Alcock, C. A. Campbell, J. Gray, and C. Pantin. 2001. Pseudomonas aeruginosa in cystic fibrosis: cross-infection and the need for segregation. Respir. Med. 95:147-152. [DOI] [PubMed] [Google Scholar]
- 29.Van Deale, S., M. Vaneechoutte, K. De Boeck, C. Knoop, A. Malfroot, P. Lebecque, J. Leclercq-Foucart, L. Van Schil, K. Desager, and F. De Baets. 2006. Survey of Pseudomonas aeruginosa genotypes in colonised cystic fibrosis patients. Eur. Respir. J. 28:740-747. [DOI] [PubMed] [Google Scholar]
- 30.Wiehlmann, L., G. Wagner, N. Cramer, B. Siebert, P. Gudowius, G. Morales, T. Kohler, C. van Delden, C. Weinel, P. Slickers, and B. Tummler. 2007. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 104:8101-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]