Abstract
In the present investigation we molecularly characterized nontypeable rotavirus strains previously identified during surveillance in New Delhi, India. The majority of strains were demonstrated to belong to genotype G1 (54.5%) or P[8] (77.8%) on the basis of nucleotide sequencing of fragments from their VP7 and VP4 genes. The other genotypes detected included G2, G8, G9, G12, and P[4]. A G8P[6] strain, strain DS108, was detected for the first time in northern India. The VP7 gene of DS108 was most homologous with the VP7 gene of a bovine G8 strain, strain A5 (98.9%), indicating its bovine parentage. In contrast, the VP4 gene had a high degree of nucleotide sequence homology (92.9% to 99.1%) with the VP4 genes of human P[6] strains. The VP6 gene and nonstructural genes (NSP1 to NSP3 and NSP5) were most homologous with the VP6 gene and nonstructural genes of human rotaviruses belonging to the DS1 genogroup. Interestingly, the NSP4 gene of DS108 clustered within genotype E6 that until now had only two representative strains, both with G12P[6] specificity (strains RV176-00 and N26-02). Together, these results indicate that G8 strain DS108 belongs to the DS1 genogroup and could be the result of the acquisition of the VP7, VP4, and NSP4 genes by a human G2P[4] strain from more than one donor, similar to the evolution of G12P[6] strain RV176-00. The present study highlights the importance of characterizing multiple genes of nontypeable rotavirus strains to detect novel strains and get a more complete picture of rotavirus evolution.
Rotaviruses (RVs) are the most important etiological agents of severe dehydrating diarrhea in children, claiming approximately 611,000 lives each year (27). Group A RVs belong to the family Reoviridae and possess a genome of 11 segments of double-stranded RNA. The two major surface proteins, the VP7 (G type) glycoprotein and protease-cleaved protein VP4 (P type), form the basis of a dual classification system for RV. To date, 23 G types and 31 P types have been identified (35, 39). Worldwide surveillance studies have reported on the continued predominance of G1 to G4 and G9 RV strains (2, 34). Additionally, G12 RVs were recently reported in neonatal and diarrheal children at high prevalence rates on the Indian subcontinent and may be emerging globally (30, 32, 36, 38). Some RV genotypes may be common in particular regions or countries and may be detected only sporadically elsewhere; for example, G5 is predominant in Brazil and G8 is predominant on the African continent (5, 7, 10, 22).
RV infection in India accounts for both high rates of mortality and high rates of morbidity, leading to approximately 400,000 hospitalizations each year (2, 15). A study recently conducted by Mendelsohn et al. observed that RV is the cause of considerable economic burden in India and suggested that an appropriate effective RV vaccine may provide significant economic savings (24). Two live oral vaccines—Rotateq, a human-bovine pentavalent vaccine, and Rotarix, a G1P[8] monovalent human rotavirus vaccine—have already been licensed (33, 40). In India, 116E, a monovalent vaccine candidate with G9P[11] specificity, has successfully completed a phase II clinical trial; and efficacy studies will be starting shortly (3).
In anticipation of a vaccine program for India, we have conducted surveillance in Delhi, India, to monitor the locally circulating RV strains. We recently reported on the high prevalence of G1, G2, and G9 rotaviruses in Delhi and the emergence of genotype G12 (36). In addition, approximately 9% of the strains were partially or completely nontypeable. To determine the nature of these nontypeable strains, we characterized them molecularly by sequence analysis in the present study. We report on the detection of a variety of strains that were not typeable due to variations in the primer binding regions of the VP4 and VP7 genes as well as the characterization of a novel G8P[6] strain.
MATERIALS AND METHODS
Fecal specimens.
Sixteen RV specimens from our previous study that were nontypeable for either the G or the P genotype or for both the G and the P types and for which a sufficient amount of stool sample remained were characterized (36). Of these, seven specimens were characterized by sequencing of the VP7 gene alone, five specimens were characterized by sequencing of the VP4 gene alone, and four specimens were characterized by sequencing of both the VP7 and the VP4 genes. Additionally, the structural (VP4 and VP6) and nonstructural (NSP1 to NSP5) genes of strain DS108 were also sequenced.
RNA extraction and RT-PCR.
Genomic RNA was extracted from 10% fecal specimens by the Trizol method (Invitrogen Corp, Carlsbad, CA), according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) was performed with a One-Step RT-PCR kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. The VP7 and VP4 genes were amplified by using previously reported consensus primers (6, 8, 31). The amplification of other structural and nonstructural genes was performed with primer pairs reported in earlier studies (13, 23).
Sequence determination.
Sequences were determined from the amplicons generated following PCR. The amplification product was purified by the GeneClean purification method (Q-biogene, Cambridge, United Kingdom) and sequenced on an automated sequencer (36). Phylogenetic analysis was performed with Mega software, version 3.1, based on the neighbor-joining method (37). Bootstrapping was statistically supported with 1,000 replicates, and percent bootstrap support was indicated by the value at each node. Values less than 80% were omitted, and the bar indicates the variation scale. All reference sequences were obtained from the GenBank/EMBL database.
Nucleotide sequence accession numbers.
The gene sequences described in the present study have been deposited in the GenBank database under accession numbers FJ827592 to FJ827609 and FJ861654 to FJ861662, inclusive.
RESULTS
In our recent RV surveillance study conducted from 2000 to 2007, we observed a high prevalence of genotypes G1, G2, and G9. However, the most notable finding of the study was the emergence of G12 RVs in combination with various P genotypes, thus confirming their potential to undergo rapid evolution. Additionally, significant numbers of nontypeable RV strains (3.2% G-nontypeable strains, 3.4% P-nontypeable strains, and 2.8% G- and P-nontypeable strains) were also detected. Hence, in the present study we characterized these nontypeable RV by nucleotide sequencing.
Detection of G1 and P[8] genotypes among the nontypeable strains with consistent mismatches in primer binding region.
Among the partially nontypeable RV strains, seven G-nontypeable strains were sequenced; and of these, three were genotype G1 and one each belonged to genotypes G2, G8, G9, and G12. The VP4 gene sequencing data for five P-nontypeable strains revealed three genotype P[8] strains and two strains with P[4] genotype specificity. However, among the four RV strains nontypeable for both the G and the P genotypes, three were G1P[8] and one was G9P[8]. Genotypes G1 and P[8] comprised 54.5% and 77.8% of the G- and P-nontypeable strains, respectively, that were identified by nucleotide sequencing (Table 1). The nucleotide sequence alignment of the six G1 RV strains showed three consistent mismatches at nucleotides (nt) 14 (C-A), 17 (T-T), and 19 (C-A) from the 3′ end of the genotyping primer (Fig. 1A). Additionally, a fourth mismatch at nt 11 (A-C) was observed in five of these six G1 samples, while the sixth one had a mismatch at nt 3 (A-C). Similarly, one G2 strain and two G9 strains also had a single-nucleotide mismatch at nt 19 (T-G) and 5 (T-G), respectively, from the 3′ end of the respective genotyping primers (data not shown). Five of seven partially typed VP4-nontypeable strains identified as P[8] by sequencing had nucleotide mismatches at nt 7 (A-C), 9 (A-A), 10 (G-T), and 12 (T-C) from the 3′ end (Fig. 1B). Additionally, four of these samples also had a mismatch at nt 8 (T-G) and two had a mismatch at nt 1 (C-A).
TABLE 1.
Partial and/or complete nontypeable human rotavirus strains characterized during the study
| Serial no. | Rotavirus strain | G genotypea | P genotypea |
|---|---|---|---|
| 1 | Dan139 | G1 | P[6] |
| 2 | Dan240 | G2 | P[4] |
| 3 | Dan318 | G9 | P[8] |
| 4 | Dan323 | G1 | P[8] |
| 5 | Dan381 | G12 | P[8] |
| 6 | Dan385 | G1 | P[6] |
| 7 | DS108 | G8 | P[6] |
| 8 | Dan092 | G2 | P[4] |
| 9 | Dan157 | G1 | P[8] |
| 10 | Dan370 | G2 | P[4] |
| 11 | Dan480 | G1 | P[8] |
| 12 | Dan481 | G9 | P[8] |
| 13 | Dan103 | G1 | P[8] |
| 14 | Dan114 | G1 | P[8] |
| 15 | Dan250 | G9 | P[8] |
| 16 | Dan279 | G1 | P[8] |
The G and P genotypes assigned during the study following gene sequencing are indicated in boldface.
FIG. 1.
(A) Nucleotide mismatches observed in the VP7 gene of six genotype G1 isolates within the primer (9T1-1) binding region. Reverse complementary sequences of the G1 primer (9T1-1) were used for alignment. Nucleotides that match the primer sequence are denoted by dots. (B) Nucleotide mismatches observed in the VP8* subunit of the VP4 gene of five genotype P[8] isolates within the primer (1T-1) binding region. Reverse complementary sequences of the P[8] primer (1T-1) were used for alignment. Nucleotides that match the primer sequence are denoted by dots.
Detection of a G8P[6] RV strain for the first time in northern India.
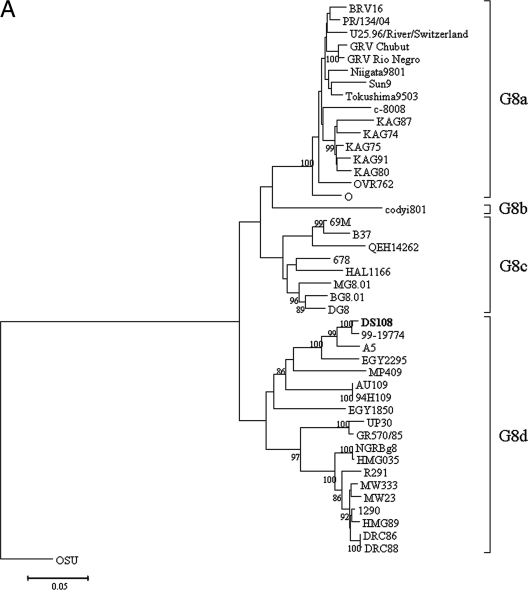
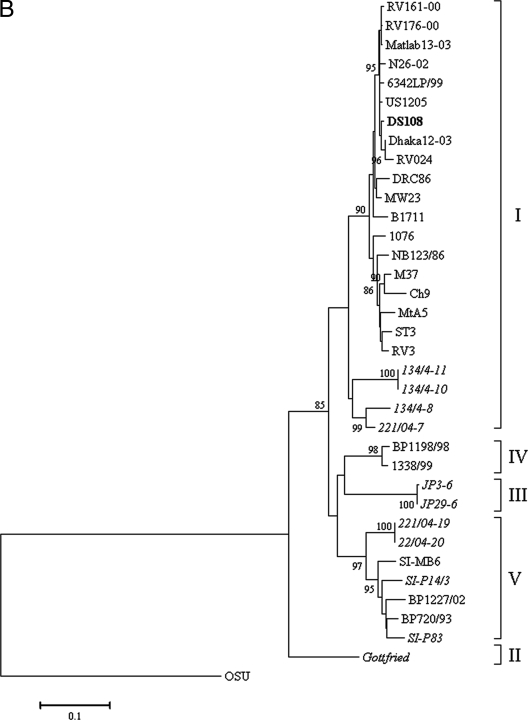
A G8 RV strain (strain DS108) was detected for the first time in the year 2000 from northern India, during the present study. The nucleotide sequence of the VP7 gene of DS108 had a high level of identity (98.9%) with that of strain 99-19774, isolated in 1997 from a diarrheal child in south India. Phylogenetic analysis revealed that strain DS108 was in the same cluster (lineage G8d) as bovine G8 strain A5 and two other human strains, strains EGY2295 and MP409, with 96.6, 93.9, and 90.2% nucleotide sequence identities, respectively (Fig. 2A). The nucleotide sequence similarities with other human and bovine G8 RVs ranged from 82.0% to 89.3%. Phylogenetic analysis of the VP8* subunit of the VP4 gene demonstrated that it clustered with human P[6] RV strains within lineage I (Fig. 2B). The VP4 gene of DS108 demonstrated a high level of nucleotide sequence identity with the G12P[6] strains detected within the Indian subcontinent (98.6% to 99.1%) and with a G9P[6] strain, strain US1205 (99%). However, a relatively low level of identity (80.8% to 91.1%) with porcine P[6] RVs was observed.
FIG. 2.
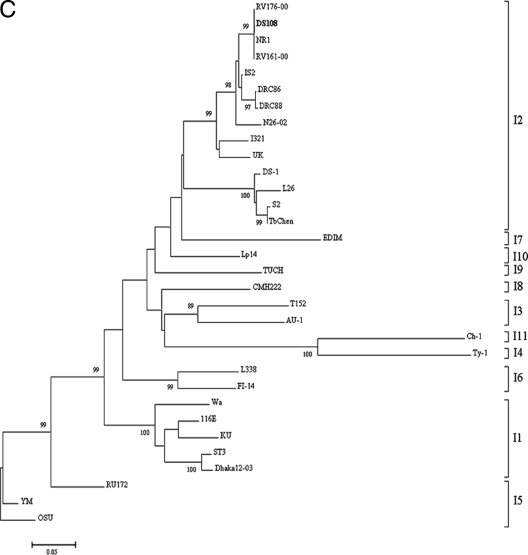
(A) Phylogenetic analysis of VP7 gene (nt 45 to 1015) from human and animal G8 RVs. The tree constructed was rooted with porcine G5 RV strain OSU. (B) Phylogenetic analysis of the VP8* subunit of the VP4 gene (nt 58 to 833) from human and animal RVs with P[6] genotype specificity. Strains in italics represent animal RVs. The tree constructed was rooted with P[7] genotype RV strain OSU. (C) Phylogenetic analysis of the VP6 gene (nt 799 to 1101) of group A RVs. The tree constructed was rooted with RV strain OSU, which has I5 genotype specificity.
RV strain DS108 belongs to the DS1 genogroup.
In order to assign DS108 to a particular genogroup, the sequencing of various other genes, structural gene VP6 and nonstructural genes NSP1 to NSP5, was undertaken. Phylogenetic analysis demonstrated that the VP6 gene clustered with lineage I2, typical of DS1 genogroup strains, and shared complete nucleotide sequence identity with RV strains NR1, RV161-00, and RV176-00 (Fig. 2C). Comparisons of the nonstructural genes (NSP1 to NSP3 and NSP5) with the corresponding genes of the other RV strains that were obtained from GenBank and sequenced revealed a very high level of identity with G8 strains (strains DRC86 and DRC88) and G12 strains (strains RV161-00 and RV176-00) (Table 2). Altogether, the high degree of nucleotide sequence homology of the structural gene (VP6) and nonstructural genes (NSP1 to NSP3 and NSP5) of DS108 with the RV reference strains belonging to the DS1 genogroup indicate that DS108 belongs to that genogroup.
TABLE 2.
Percent nucleotide sequence identitya of the strain DS108 nonstructural gene segments (NSP1 to NSP3 and NSP5) with that of the respective prototype strain of each genotype
| NSP1 |
NSP2 |
NSP3 |
NSP5 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Strain | % nt sequence homologya | Genotype | Strain | % nt sequence homology | Genotype | Strain | % nt sequence homology | Genotype | Strain | % nt sequence homology |
| A1 | Wa | 75.1 | N1 | Wa | 79.3 | T1 | Wa | 77.8 | H1 | Wa | 83.9 |
| A1 | KU | 74.5 | N1 | KU | 79.3 | T1 | KU | 77.6 | H1 | KU | 84.7 |
| A2 | DS1 | 92.1 | N2 | DS1 | 85.2 | T2 | DS1 | 94.8 | H2 | DS1 | 95.2 |
| A2 | DRC86 | 97.4 | N2 | DRC86 | 97.2 | T2 | DRC86 | 98.5 | H2 | DRC86 | 97.9 |
| A2 | DRC88 | 97.4 | N2 | DRC88 | 96.9 | T2 | DRC88 | 98.1 | H2 | DRC88 | 97.9 |
| A2 | RV176-00 | 98.6 | N2 | RV176-00 | 99.7 | T2 | RV176-00 | 98.7 | H2 | RV176-00 | 98.7 |
| A2 | RV161-00 | 98.6 | N2 | RV161-00 | 99.7 | T2 | RV161-00 | 98.6 | H2 | RV161-00 | 98.8 |
| A3 | Au-1 | 59.2 | N3 | Au-1 | 80.9 | T3 | Au-1 | 81.0 | H3 | Au-1 | 83.7 |
| A4 | Po-13 | 36.6 | N4 | Po-13 | 66.0 | T4 | Po-13 | 55.5 | H4 | Po-13 | 66.2 |
| A5 | SA11g4“O” (5N) | 46.5 | N5 | SA11g4“O” (5N) | 79.8 | T5 | SA11g4“O” (5N) | 75.5 | H5 | SA11g4“O” (5N) | 84.8 |
| A6 | L338 | 43.9 | T6 | OVR762 | 77.2 | H6 | RRV | 84.5 | |||
| A7 | EW | 43.4 | T7 | UK | 76.7 | H6 | T152 | 84.8 | |||
| A8 | Gottfried | 69.8 | |||||||||
| A9 | ALA | 46.0 | |||||||||
| A10 | FI14 | 46.3 | |||||||||
| A11 | OVR762 | 60.3 | |||||||||
| A12 | T152 | 53.9 | |||||||||
| A13 | B223 | 62.1 | |||||||||
| A14 | A5-13 | 61.8 | |||||||||
A high degree of nucleotide sequence identity is indicated in boldface.
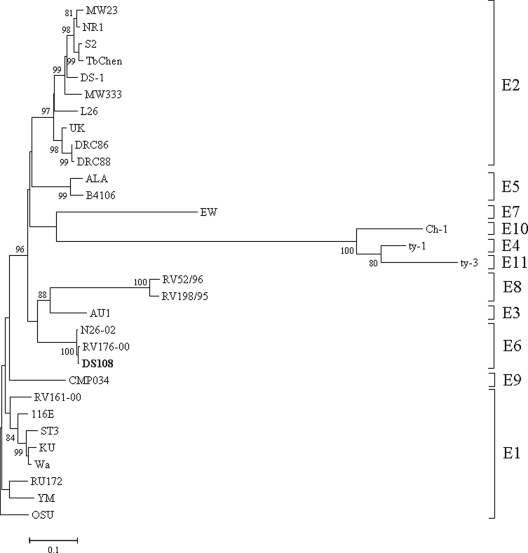
E6 genotype specificity of NSP4 gene of strain DS108.
Interestingly, the NSP4 gene of RV strain DS108 was highly homologous to the cognate gene of recently detected G12P[6] RV strains RV176-00 (99.5%) and N26-02 (99.1%). The NSP4 genes of both RV176-00 and N26-02 were previously considered to belong to NSP4 genotype C; however, according to the new classification system for all 11 RV gene segments, they represent a separate genotype, genotype E6, in which the NSP4 gene of strain DS108 also clustered (Fig. 3). The amino acid sequences of these three representative strains of genotype E6 were compared with those of other NSP4 genotype strains. Certain amino acid residues were found to be conserved within this genotype. They were the presence of methionine (at amino acid [aa] 10) and asparagine (at aa 11) within the H1 hydrophobic domain, arginine (at aa 115) in the enterotoxin domain, isoleucine (at aa 139) and glutamic acid (at aa 140) in the VP4 binding domain, and aspartic acid (at aa 161) in the single-shelled particle binding domain (data not shown).
FIG. 3.
Phylogenetic analysis of the NSP4 gene (nt 52 to 686) of group A RVs. The tree constructed was rooted with E1 genotype RV strain OSU.
DISCUSSION
RV surveillance studies regularly conducted worldwide aim to monitor the prevalence of predominant genotypes and identify rare and emerging strains that may become important in the future. Such studies are particularly important in countries such as India that are considering the introduction of an RV vaccine program (3). In this regard, we recently reported a high prevalence of genotype G1, G2, and G9 RV strains with the emergence of the G12 genotype in Delhi, India (36). However, during that study, low but significant numbers of nontypeable RV strains were detected in comparison to the numbers of nontypeable RV strains detected on previous studies (2). This prompted us to characterize these nontypeable strains to identify novel strains and further ascertain the diversity of the RVs within the local population.
The VP7 and VP4 gene sequencing data for the nontypeable RVs identified common G genotypes (G1, G2, G9, and G12) and P genotypes (P[4] and P[8]). Nucleotide sequence analysis of these partially sequenced genes clearly demonstrated consistent multiple-nucleotide mismatches with the type-specific primer. The nucleotide mismatches observed between the G1 VP7 gene and type-specific primer 9T1-1 were identical to those observed in a previous study; however, the percentage of genotype G1 strains that could not be typed was much lower (5, 28, 29). Similarly, five of seven samples initially nontypeable for the VP4 gene that turned out to have the P[8] genotype had identical mismatches in the type-specific primer (primer 1T-1), as was observed in other studies (5, 12, 19). However, for the two remaining samples of genotype P[8], the partial VP4 gene sequences obtained did not yield the sequence of the primer binding region. Altogether, this finding suggests that the use of alternative sets of G1 and P[8] type-specific primers in the primer pool is essential for future RV surveillance studies. Additionally, the sequences of the other nontypeable strains that could not be assigned a genotype revealed either a single-nucleotide mismatch (G2 and G9) or were completely homologous to the sequence of the type-specific primer (G12). Their unsuccessful typing previously could be attributed to a failed PCR.
The highlight of the study was the detection of a novel G8P[6] RV strain with a DS1-like genogroup and an atypical NSP4 gene from Delhi, India. G8 RVs were first detected in diarrheal children in Indonesia and were subsequently detected in cattle (21, 25). A retrospective analysis reported in 1995 demonstrated that the bovine Cody strain of neonatal calf diarrhea virus (NCDV-Cody) isolated in Nebraska in 1968, which was initially thought to be the virulent counterpart of the attenuated NCDV strain (NCDV-Lincoln, G6P[1]), bore a G8P[1] specificity (20). That report as well as a recent report indicates that the bovine G8 serotype has been circulating among cattle in the United States for more than three decades (4). The G8 genotype has also been sporadically detected in several countries in combination with genotypes P[1], P[4], P[6], P[8], and P[14] (1, 5, 11, 22, 26, 41). In addition, G8 RVs have been associated with both of the widely prevalent genogroups, genogroups Wa and DS1 (7, 23). Sporadic cases of G8 RVs have been reported among children with RV gastroenteritis in India since 1990 (9, 14, 16-18).
However, studies involving the in-depth characterization of these G8 RVs from India are limited. A previous study from south India involving the sequencing of some genes (VP4, VP7, NSP1, NSP3, and NSP4) of two G8P[1] strains detected indicated that the strains were a result of reassortment between human and bovine RV strains (14). In another study from south India that detected a G8P[8] strain (strain 99-19774), only the VP7 gene was sequenced, and it was found to be highly homologous to that of bovine strain A5 (16). Therefore, to update the knowledge on the evolution of G8 RV and to determine the extent of diversity within the circulating G8 RVs in India, sequencing of the structural genes (VP4, VP6, and VP7) and nonstructural genes (NSP1 to NSP5) of strain DS108 was done.
The VP7 gene sequence of strain DS108 was highly homologous to that of strain 99-19774, detected in south India (16). The phylogram generated from the sequences obtained clustered all the Indian G8 RVs together with bovine strain A5, indicating that the VP7 gene of DS108 had a bovine donor (14). However, the high percentage of nucleotide sequence identity with a human strain of the P[6] genotype suggested that DS108 may have undergone further reassortment. Thus, sequencing of other genes became more important, as it was essential to understand the genetic makeup of DS108 because strains with similar G-P combinations had become predominant in the Democratic Republic of Congo (22). They were considered to have evolved as a result of reassortment between the human G2P[4] strains and strains donating their VP7 and VP4 gene segments (22). This notion was strengthened by the sequencing data for other structural gene VP6 and nonstructural genes NSP1 to NSP3 and NSP5. High degrees of homology were observed with the strains belonging to the DS1 genogroup, specifically, with Bangladeshi G12P[6] strains RV176-00 and RV161-00 and G8P[6] strains DRC86 and DRC88 from the Democratic Republic of Congo (22, 30).
However, on the contrary, the NSP4 gene of strain DS108 clustered in the E6 lineage instead of the E2 lineage, which is represented by DS1-like strains, such as DRC86 (22). This indicated that DS108 may have a different evolutionary path than DRC86. The E6 lineage was previously considered to be distantly related to NSP4 genotype C, and to date, only two RV strains with G12P[6] specificity (strains RV176-00 and N26-02) represent this lineage (30). This is the first report of the association of a G8 RV strain with the E6 lineage. Therefore, altogether the evolution of DS108 seems to be somewhat similar to the evolution of first-generation G12P[6] strain RV176-00 from Bangladesh, with which all its genes except VP7 share maximum homology, indicating that both strains have evolved from a common parent with different VP7 donors.
In conclusion, the present study documents the first detection of a G8 strain in Delhi along with the frequent detection of sequence variations in strains with the same G and P genotypes. This highlights the need for continued surveillance to detect such novel RV strains that may emerge in the local community and also regularly update our typing method with addition of new primers.
Acknowledgments
The financial support from the Department of Biotechnology, Government of India, is greatly acknowledged. Sumit Sharma received a senior research fellowship from the Council of Scientific and Industrial Research, India.
We thank Jon R. Genstch for critical review of the manuscript and Roger I. Glass for helpful comments.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Adah, M. I., S. Nagashima, M. Wakuda, and K. Taniguchi. 2003. Close relationship between G8-serotype bovine and human rotaviruses isolated in Nigeria. J. Clin. Microbiol. 41:3945-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahl, R., P. Ray, S. Subodh, P. Shambharkar, M. Saxena, U. Parashar, J. Gentsch, R. Glass, and M. K. Bhan. 2005. Incidence of severe rotavirus diarrhea in New Delhi, India, and G and P types of the infecting rotavirus strains. J. Infect. Dis. 192(Suppl. 1):S114-S119. [DOI] [PubMed] [Google Scholar]
- 3.Bhandari, N., P. Sharma, S. Taneja, T. Kumar, T. Rongsen-Chandola, M. B. Appaiahgari, A. Mishra, S. Singh, and S. Vrati. 2009. A dose-escalation safety and immunogenicity study of live attenuated oral rotavirus vaccine 116E in infants: a randomized, double-blind, placebo-controlled trial. J. Infect. Dis. 200:421-429. [DOI] [PubMed] [Google Scholar]
- 4.Cao, D., B. Igboeli, L. Yuan, A. Z. Kapikian, J. L. Ayers, F. R. Abinanti, and Y. Hoshino. 2009. A longitudinal cohort study in calves evaluated for rotavirus infections from 1 to 12 months of age by sequential serological assays. Arch. Virol. 154:755-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esona, M. D., A. Geyer, N. Page, A. Trabelsi, I. Fodha, M. Aminu, V. A. Agbaya, B. Tsion, T. K. Kerin, G. E. Armah, A. D. Steele, R. I. Glass, and J. R. Gentsch. 2009. Genomic characterization of human rotavirus G8 strains from the African rotavirus network: relationship to animal rotaviruses. J. Med. Virol. 81:937-951. [DOI] [PubMed] [Google Scholar]
- 8.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gladstone, B. P., M. Iturriza-Gomara, S. Ramani, B. Monica, I. Banerjee, D. W. Brown, J. J. Gray, J. Muliyil, and G. Kang. 2008. Polymerase chain reaction in the detection of an ‘outbreak’ of asymptomatic viral infections in a community birth cohort in south India. Epidemiol. Infect. 136:399-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouvea, V., L. de Castro, M. C. Timenetsky, H. Greenberg, and N. Santos. 1994. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 32:1408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes, J. L., C. D. Kirkwood, G. Gerna, J. D. Clemens, M. R. Rao, A. B. Naficy, R. Abu-Elyazeed, S. J. Savarino, R. I. Glass, and J. R. Gentsch. 1999. Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Arch. Virol. 144:1381-1396. [DOI] [PubMed] [Google Scholar]
- 12.Iturriza-Gomara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38:898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iturriza Gomara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J. Virol. 76:6596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagannath, M. R., R. R. Vethanayagam, B. S. Reddy, S. Raman, and C. D. Rao. 2000. Characterization of human symptomatic rotavirus isolates MP409 and MP480 having ‘long’ RNA electropherotype and subgroup I specificity, highly related to the P6[1],G8 type bovine rotavirus A5, from Mysore, India. Arch. Virol. 145:1339-1357. [DOI] [PubMed] [Google Scholar]
- 15.Jain, V., U. D. Parashar, R. I. Glass, and M. K. Bhan. 2001. Epidemiology of rotavirus in India. Indian J. Pediatr. 68:855-862. [DOI] [PubMed] [Google Scholar]
- 16.Kang, G., J. Green, C. I. Gallimore, and D. W. Brown. 2002. Molecular epidemiology of rotaviral infection in south Indian children with acute diarrhea from 1995-1996 to 1998-1999. J. Med. Virol. 67:101-105. [DOI] [PubMed] [Google Scholar]
- 17.Kelkar, S. D., P. G. Ray, and S. S. Bedekar. 1996. Assay of neutralizing antibodies to animal rotavirus strains and human rotavirus serotype G8 by a modified method in the residents of Pune, India. J. Diarrhoeal Dis. Res. 14:101-106. [PubMed] [Google Scholar]
- 18.Kelkar, S. D., and V. L. Ayachit. 2000. Circulation of group A rotavirus subgroups and serotypes in Pune, India, 1990-1997. J. Health Popul. Nutr. 18:163-170. [PubMed] [Google Scholar]
- 19.Lee, J. I., M. O. Song, J. Y. Chung, T. H. Han, Y. M. Ahn, J. W. Seo, M. S. Kim, M. Y. Kim, W. Y. Kim, and C. H. Lee. 2008. Outbreak of rotavirus variant P[8] in Seoul, South Korea. J. Med. Virol. 80:1661-1665. [DOI] [PubMed] [Google Scholar]
- 20.Lu, W., G. E. Duhamel, Y. Hoshino, D. A. Benfield, E. A. Nelson, and R. A. Hesse. 1995. Characterization of the bovine group A rotavirus strain neonatal calf diarrhea virus-Cody (NCDV-Cody). J. Clin. Microbiol. 33:990-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuno, S., A. Hasegawa, A. Mukoyama, and S. Inouye. 1985. A candidate for a new serotype of human rotavirus. J. Virol. 54:623-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthijnssens, J., M. Rahman, X. Yang, T. Delbeke, I. Arijs, J. P. Kabue, J. J. Muyembe, and M. Van Ranst. 2006. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 44:1801-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthijnssens, J., M. Ciarlet, E. Heiman, I. Arijs, T. Delbeke, S. M. McDonald, E. A. Palombo, M. Iturriza-Gomara, P. Maes, J. T. Patton, M. Rahman, and M. Van Ranst. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82:3204-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendelsohn, A. S., J. R. Asirvatham, D. Mkaya Mwamburi, T. V. Sowmynarayanan, V. Malik, J. Muliyil, and G. Kang. 2008. Estimates of the economic burden of rotavirus-associated and all-cause diarrhoea in Vellore, India. Trop. Med. Int. Health 13:934-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ojeh, C. K., D. R. Snodgrass, and A. J. Herring. 1984. Evidence for serotypic variation among bovine rotaviruses. Arch. Virol. 79:161-171. [DOI] [PubMed] [Google Scholar]
- 26.Palombo, E. A., R. Clark, and R. F. Bishop. 2000. Characterisation of a “European-like” serotype G8 human rotavirus isolated in Australia. J. Med. Virol. 60:56-62. [PubMed] [Google Scholar]
- 27.Parashar, U. D., C. J. Gibson, J. S. Bresse, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra, G. I., and E. E. Espinola. 2006. Nucleotide mismatches between the VP7 gene and the primer are associated with genotyping failure of a specific lineage from G1 rotavirus strains. Virol. J. 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman, M., R. Sultana, G. Podder, A. S. Faruque, J. Matthijnssens, K. Zaman, R. F. Breiman, D. A. Sack, M. Van Ranst, and T. Azim. 2005. Typing of human rotaviruses: nucleotide mismatches between the VP7 gene and primer are associated with genotyping failure. Virol. J. 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman, M., J. Matthijnssens, X. Yang, T. Delbeke, I. Arijs, K. Taniguchi, M. Iturriza-Gomara, N. Iftekharuddin, T. Azim, and M. Van Ranst. 2007. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 81:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray, P., M. Fenaux, S. Sharma, J. Malik, S. Subodh, S. Bhatnagar, H. Greenberg, R. I. Glass, J. Gentsch, and M. K. Bhan. 2006. Quantitative evaluation of rotaviral antigenemia in children with acute rotaviral diarrhea. J. Infect. Dis. 194:588-593. [DOI] [PubMed] [Google Scholar]
- 32.Ray, P., S. Sharma, R. K. Agarwal, K. Longmei, J. R. Gentsch, V. K. Paul, R. I. Glass, and M. K. Bhan. 2007. First detection of G12 rotaviruses in newborns with neonatal rotavirus infection at all India Institute of Medical Sciences, New Delhi, India. J. Clin. Microbiol. 45:3824-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Palacios, G. M., I. Perez-Schael, F. R. Velazquez, H. Abate, T. Breuer, S. C. Clemens, B. Cheuvart, F. Espinoza, P. Gillard, B. L. Innis, Y. Cervantes, A. C. Linhares, P. Lopez, M. Macias-Parra, E. Ortega-Barria, V. Richardson, D. M. Rivera-Medina, L. Rivera, B. Salinas, N. Pavia-Ruz, J. Salmeron, R. Ruttimann, J. C. Tinoco, P. Rubio, E. Nunez, M. L. Guerrero, J. P. Yarzabal, S. Damaso, N. Tornieporth, X. Saez-Llorens, R. F. Vergara, T. Vesikari, A. Bouckenooghe, R. Clemens, B. De Vos, and M. O'Ryan. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 354:11-22. [DOI] [PubMed] [Google Scholar]
- 34.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 35.Schumann, T., H. Hotzel, P. Otto, and R. Johne. 2009. Evidence of interspecies transmission and reassortment among avian group A rotaviruses. Virology 386:334-343. [DOI] [PubMed] [Google Scholar]
- 36.Sharma, S., P. Ray, J. R. Gentsch, R. I. Glass, V. Kalra, and M. K. Bhan. 2008. Emergence of G12 rotavirus strains in Delhi, India, in 2000 to 2007. J. Clin. Microbiol. 46:1343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subodh, S., M. K. Bhan, and P. Ray. 2006. Genetic characterization of VP3 gene of group A rotaviruses. Virus Genes 33:143-145. [DOI] [PubMed] [Google Scholar]
- 38.Uchida, R., B. D. Pandey, J. B. Sherchand, K. Ahmed, M. Yokoo, T. Nakagomi, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2006. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J. Clin. Microbiol. 44:3499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ursu, S., P. Kisfali, D. Rigo, E. Ivanics, K. Erdelyi, A. Dan, B. Melegh, V. Martella, and K. Banyai. 2009. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch. Virol. 154:1365-1369. [DOI] [PubMed] [Google Scholar]
- 40.Vesikari, T., D. O. Matson, P. Dennehy, P. Van Damme, M. Santosham, Z. Rodriguez, M. J. Dallas, J. F. Heyse, M. G. Goveia, S. B. Black, H. R. Shinefield, C. D. Christie, S. Ylitalo, R. F. Itzler, M. L. Coia, M. T. Onorato, B. A. Adeyi, G. S. Marshall, L. Gothefors, D. Campens, A. Karvonen, J. P. Watt, K. L. O'Brien, M. J. DiNubile, H. F. Clark, J. W. Boslego, P. A. Offit, and P. M. Heaton. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 354:23-33. [DOI] [PubMed] [Google Scholar]
- 41.Volotão, E. M., C. C. Soares, A. G. Maranhão, L. N. Rocha, Y. Hoshino, and N. Santos. 2006. Rotavirus surveillance in the city of Rio de Janeiro-Brazil during 2000-2004: detection of unusual strains with G8P[4] or G10P[9] specificities. J. Med. Virol. 78:263-272. [DOI] [PubMed] [Google Scholar]