Abstract
Chagas' disease caused by Trypanosoma cruzi is endemic in Latin America. T. cruzi presents heterogeneous populations and comprises two main genetic lineages, named T. cruzi I and T. cruzi II. Diagnosis in the chronic phase is based on conventional serological tests, including indirect immunofluorescence (IIF) and enzyme-linked immunosorbent assay (ELISA), and diagnosis in the acute phase based on parasitological methods, including hemoculture. The objective of this study was to evaluate the diagnostic procedures of Chagas' disease in adult patients in the chronic phase by using a PCR assay and conventional serological tests, including TESA-blot as the gold standard. Samples were obtained from 240 clinical chronic chagasic patients. The sensitivities, compared to that of TESA-blot, were 70% for PCR using the kinetoplast region, 75% for PCR using the nuclear repetitive region, 99% for IIF, and 95% for ELISA. According to the serological tests results, we recommend that researchers assess the reliability and sensitivity of the commercial kit Chagatest ELISA recombinant, version 3.0 (Chagatest Rec v3.0; Wiener Lab, Rosario, Argentina), due to the lack of sensitivity. Based on our analysis, we concluded that PCR cannot be validated as a conventional diagnostic technique for Chagas' disease. These data have been corroborated by low levels of concordance with serology test results. It is recommended that PCR be used only for alternative diagnostic support. Using the nuclear repetitive region of T. cruzi, PCR could also be applicable for monitoring patients receiving etiologic treatment.
Chagas' disease is a complex zoonosis caused by the parasite Trypanosoma cruzi. This parasite can be genetically classified into two major lineages, namely, T. cruzi I, which is found in northern South American countries, and T. cruzi II, which is found in southern South American countries (46). Chagas' disease is a chronic systemic disease endemic to both South and Central America. T. cruzi is transmitted through the infected dejections of triatomine insects by blood transfusion, congenital infection, laboratory accidents, or oral infection (45). Chagas' disease constitutes a serious public health problem in terms of both social and economic impact. The disease currently affects 15 million people, and about 28 million are at risk of acquiring the infection. In America, nearly 41,200 new cases occur each year, along with an average of 12,500 deaths per year (16).
Chagas' disease presents two distinct clinical stages. The acute phase begins about 1 week after initial infection, and nearly 30% of patients recall having had relevant symptoms and signs during this period. During the chronic disease stage, the parasites are no longer easily detectable in the bloodstream but serological tests remain positive. Diagnosis of Chagas' disease is based on parasitological and serological methods. Infection can usually be detected by microscopic examination or by parasitological tests such as hemoculture or PCR (6, 28). There are several targets for the detection of T. cruzi by PCR. The variable region of the minicircle kinetoplast DNA (kDNA) and a repeat tandem sequence of nuclear DNA (stDNA) of the parasite have been the regions most widely used as target sequences for diagnosis via PCR (2, 6, 13, 32, 44). Serological diagnosis of T. cruzi infection is typically performed by using two of three individual tests, according to availability (45). Enzyme-linked immunosorbent assay (ELISA), indirect immunofluorescence (IIF), and indirect hemagglutination are often used. These three tests, also referred to as the conventional tests, usually employ recombinant and/or crude antigenic T. cruzi preparations (21). The major innovation in Chagas' disease diagnosis with the detection of antibodies against T. cruzi is the TESA-blot (TESA stands for trypomastigote excreted-secreted antigen). This is an immunoblot assay that has been widely used because of its high sensitivity and specificity compared to those of conventional serological methods (38). The isolation and gene cloning of this immunodominant peptide have been intended, and ELISAs based on TESAs have been performed with high-quality results (25). In addition, this test has shown the presence of false negatives when this technique was compared with conventional serology in a cohort from Bolivia (47). These reasons make TESA-blot one of the most feasible and available tests for the diagnosis of Chagas' disease (38, 39, 40). Recently, TESA-blot has shown great usefulness in resolving doubtful serology and cross-antigenicity issues with related protozoan parasites in regions where the disease is endemic (41). Because of these previous reports, TESA-blot has been selected as the gold standard in several different studies due to the high sensitivity and specificity of the test.
Among the conventional techniques used for the serological diagnosis of Chagas' disease are ELISA because of its high sensitivity and IIF due to its specificity. However, it has been observed that these tests can produce a certain number of false positives and false negatives. This makes it necessary to search for diagnostic tests that provide more reliable results (5). As a routine test for the diagnosis of Chagas' disease, the World Health Organization recommends immunological techniques according to the type of diagnosis, and a minimum of two positive serological tests are required for considering a patient to be infected with T. cruzi. Nevertheless, in some cases, there is a need to implement other techniques for the diagnosis of T. cruzi. Because of the number of copies and organization of the kinetoplast and nuclear repetitive DNA of T. cruzi, a PCR assay has been developed (29, 42). Comparative studies of PCR, hemoculture, and serology showed that individuals with positive hemoculture and serology results had a detection rate of 36.5%. When PCR was used, the detection of infection was 83.5%. These results demonstrate the higher sensitivity of PCR compared to hemoculture (13). There is great variability within the results obtained by PCR, xenodiagnosis, and hemoculture that makes PCR a controversial tool of choice for the accurate diagnosis of Chagas' disease.
The specificity of serological techniques has been questioned because of the cross-antigenicity between T. cruzi and parasites of related protozoan diseases, particularly leishmaniasis and infection with T. rangeli (5, 39). This questioning arises because these techniques use crude or partially purified parasite extracts, which can cause false-positive results. In order to avoid false-positive results, recombinant antigens and/or synthetic peptides have been used with success (18, 27, 30, 39, 40). These problems may be overcome by using recombinant antigens containing specific T. cruzi epitopes that elicit an immune response in the majority of chagasic patients (8, 14, 21, 39). Therefore, parasitological tests are still extremely necessary to detect T. cruzi, especially in patients with doubtful serology results and to determine the treatment response.
The present study is a substudy of the BENEFIT (BENznidazol Evaluation For Interrupting Trypanosomiasis) population recruited in Colombia (23), and the objective was to compare serological diagnosis by ELISA, IIF, and TESA-blot to PCR amplification of the variable region of the kDNA and the stDNA of T. cruzi in clinical and serologically ascertained chagasic patients from Colombia. We also aimed to optimize the procedure of DNA parasite amplification by selecting the most suitable DNA extraction method. Similarly, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and the kappa index were calculated. TESA-blot was used as the gold standard due to the above-mentioned characteristics of this test.
MATERIALS AND METHODS
Sample collection.
A total of 240 clinical chronic chagasic patients and 20 negative controls were included in this study. We employed the inclusion and exclusion criteria required for the BENEFIT project (23). Every patient presented clinical heart failure evidenced by echocardiogram and positive serology results. A 10-ml blood sample was collected from all patients and control subjects. Blood samples were mixed with an equal volume of 6 M guanidine HCl-0.2 M EDTA solution immediately after sample collection. The guanidine-EDTA blood mixture was then maintained at room temperature and later stored at 4°C until DNA extraction. A 2-ml blood sample was also collected for serum collection and analysis. The serum aliquots were then stored at 4°C until the serological tests were performed. The 20 individual control subjects were from an area where the disease is not endemic, and they all tested negative for Chagas' disease by IIF, ELISA, PCR, and TESA-blot. All of the control subjects also showed no clinical signs of heart failure.
Comparison of DNA isolation methods.
The phenol-chloroform method for DNA extraction (3) was compared with a commercial DNA isolation kit (Bio-Rad AquaPure Genomics for blood tissue) in 100 positive samples selected randomly, including 10 negative controls. The comparison was conducted by spectrophotometric quantification of 5-μl DNA aliquots and also by estimating the efficiency of amplification of T. cruzi kDNA and stDNA by PCR. A statistical comparison of means (Student one-tailed t test, P < 0.05) was conducted for the selection of the best method of DNA isolation, and a correlation analysis was conducted for both methods of amplification.
DNA isolation.
The samples were immersed in boiling water for 15 min. After cooling, two 200-μl aliquots were taken from each patient blood lysate and successive phenol-chloroform extractions were performed on this material as previously reported (3). The DNA isolation using the commercial kit (Bio-Rad AquaPure Genomics for blood tissue) was performed according to the manufacturer's protocol. The DNA was then stored at −20°C; DNA purity and concentrations were determined with an Eppendorf 6131 biophotometer at wavelengths of 260 and 280 nm.
PCR.
Amplification reactions were performed with a total volume of 21 μl. This reaction mixture consisted of 1× Pfx Taq polymerase amplification buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl; Invitrogen), 100 mM deoxynucleoside triphosphate solution, 25 mM MgCl2 solution, 700 mM KCl solution, 2.5 U/μl of Taq polymerase platinum Pfx (Invitrogen), 56 pM T. cruzi minicircle-specific primers 121 (5′AAATAATGTACGGGKGAGATGCATGA3′) and 122 (5′GGTTCGATTGGGGTTGGTGTAATATA3′), 3 μl of template DNA, and a quantity of water sufficient to give a final volume of 21 μl. The reaction mixture was subjected to 30 cycles of amplification in an automatic thermocycler (Programmable Thermal Controller PTC-100; MJ Research) as reported previously (4). All of the samples were further tested under the same conditions with two oligonucleotides from the human β-globin gene region, PC03 (5′CAACTTCATCCACGTTCACC3′) and PC04 (5′ACACAAACTGTGTTCACTAGC3′), as an internal control for amplification. This was done to check for the possibility that a result showing no amplification could have been due to inhibition of the reaction (33). For amplification of the T. cruzi nuclear repetitive region, the amplification reactions were performed in a total volume of 21 μl. This reaction mixture consisted of 1× Taq polymerase amplification buffer (100 mM Tris-HCl, pH 8.3; Invitrogen), 100 mM deoxynucleoside triphosphate solution, 25 mM MgCl2 solution, 5 U/μl of Taq polymerase platinum (Invitrogen), 50 pM of T. cruzi nuclear repetitive region-specific primers cruzi1 (5′ASTCGGCTGATCGTTTTCGA3′) and cruzi2 (5′AATTCCTCCAAGCAGCGGATA 3′) (31), 3 μl of template DNA, and a quantity of water sufficient to give a final volume of 21 μl. The reaction mixtures were subjected to 40 cycles of amplification in an automatic thermocycler (Bio-Rad iCycler) as reported previously (11). The possibility of contamination of the PCR reagents and of the solutions used to prepare DNA was carefully examined through the use of appropriate controls (DNA from strain VS [T. cruzi IIb] and DNA from strain Dm11 [T. cruzi I] as positive controls and DNA from strain 444 [T. rangeli] and DNA from blood serologically negative as negative controls), and each sample was tested in duplicate. Twenty microliters of each PCR product was analyzed by electrophoresis on a 2% agarose gel and visualized by staining with SYBR green for gel staining (Invitrogen).
Serological methods. (i) IIF.
IIF was carried out as reported elsewhere, with formaldehyde-treated epimastigote forms of T. cruzi I strain X-380 obtained from culture medium as described previously (13). Positive and negative controls, including those positive for anti-Leishmania sp. antibodies, were always included to validate the results obtained.
(ii) ELISA.
ELISA was performed with the commercial kit Chagatest ELISA recombinant, version 3.0 (Chagatest Rec v3.0; Wiener Lab, Rosario, Argentina), which had a mixture of recombinant proteins. Each serum was analyzed in duplicate, and the positive and negative controls were analyzed in triplicate. A sample was considered positive if the optical density (OD) at 450 nm was ≥0.345 and negative if the OD was ≤0.344. Positive and negative controls, including sera positive for anti-Leishmania sp. antibodies, which were used for specificity control, were always included to validate the results obtained.
(iii) TESA-blot.
TESAs from T. cruzi II strain Y were obtained from the supernatant of infected LLC-MK2 cells and used for immunoblotting as described before (38). Membrane strips (5 mm) were later incubated with serum diluted 1:200 in Tris-buffered saline (TBS)-1% milk for 2 h with mechanical agitation. After four 5-min washes in TBS, the bound antibodies were detected by using peroxidase-conjugated anti-human immunoglobulin G (Sigma) diluted 1:2,500 in TBS-1% milk for 2 h. After new cycles of washes, the immune complexes were revealed by addition of H2O2 and 4-chloro-1-naphthol. The reaction was stopped with deionized water.
Statistical analyses.
To validate the reliability of the results obtained, the parameters used were sensitivity, specificity, PPV, NPV, and the kappa index (27). TESA-blot was used as the gold standard due to its high sensitivity and specificity and also due to the high level of accurate results for doubtful serology samples and cross-reaction samples that have been previously reported (25, 38, 41, 47).
RESULTS
Comparison of DNA isolation methods.
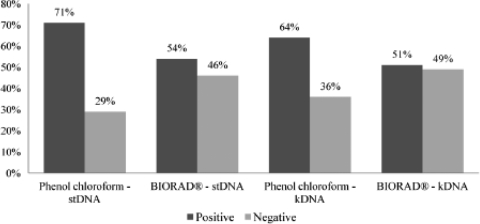
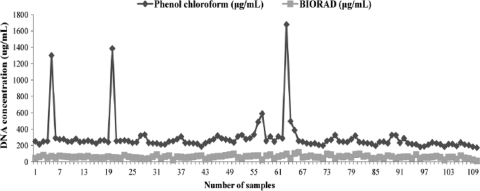
A comparison of the proposed DNA isolation methods was performed based on the efficiency of PCR amplification using the kDNA and stDNA genomic regions when testing 100 positive samples that were ELISA, IIF, and TESA-blot positive as well as 10 negative control samples. Likewise, a comparison of DNA concentrations was also performed by spectrophotometry. A one-tailed paired Student's t test (P < 0.05) was performed. Results of PCR efficiency amplification showed that the phenol-chloroform extraction method was 17% more sensitive than the AquaPure Genomics extraction for blood tissue kit from Bio-Rad for the stDNA PCR and 13% more sensitive for the kDNA PCR. Our analysis indicated that the differences between the two extraction methods using both PCR detection targets (Fig. 1) were statistically significant (P = 0.040 and P = 0.047, respectively). All of the negative controls showed an absence of amplification by PCR by both methods. A statistically significant difference was observed (P = 0.017) when evaluating the DNA concentration, demonstrating that the final DNA concentration obtained by the phenol-chloroform method was much higher than that obtained with the commercial kit (Fig. 2). Additionally, there was a positive correlation (P = 0.002, P < 0.05) between the results observed by amplification with stDNA and kDNA. We have therefore concluded that the ideal method for the detection of amplified DNA from T. cruzi is the phenol-chloroform DNA extraction method.
FIG. 1.
Results comparing the phenol-chloroform DNA isolation method and Bio-Rad AquaPure Genomics for blood tissue based on 100 positive samples. The differences between the results obtained with stDNA and kDNA were statistically significant (P =0.040 and P = 0.047, respectively).
FIG. 2.
Results comparing the phenol-chloroform DNA isolation method and Bio-Rad AquaPure Genomics for blood tissue based on DNA concentration measurement by spectrophotometry. The difference between the results obtained was statistically significant (P =0.017).
T. cruzi DNA detection.
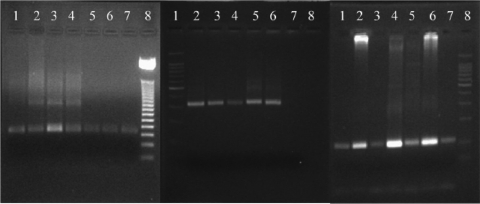
A representative PCR result from both the stDNA and kDNA reactions is shown in Fig. 3A and B. The 20 individual controls from an area where the disease is not endemic which were negative by IIF and ELISA were all negative by PCR. The expected 330-bp corresponding kDNA region of the T. cruzi amplified product was observed in 178 (70%) of the 240 patients, and the 166-bp amplified product corresponding to stDNA was seen in 180 (75%) patients; however, this difference was not statistically significant (P = 0.125, P < 0.05). Although there was no statistically significant difference, the stDNA target was more sensitive than the kDNA target for PCR amplification in the samples from chronic chagasic patients. T. cruzi strain VS (T. cruzi IIb) DNA and T. cruzi strain Dm11 (T. cruzi I) DNA were used as positive controls, and T. rangeli strain 444 DNA and DNA from negative blood samples were used as negative controls. The PCRs were performed in duplicate. For a sample to be considered positive, both reactions had to be positive. Also, the results were validated with the amplification of the 110-bp human β-globin gene region to ensure that the absence of amplification was not attributable to PCR inhibitors. All of the human positive control samples were positive (Fig. 3C). The detection limit of the PCR procedure was established as 0.5 fg for kDNA and 0.1 fg for stDNA (data not shown) according to the protocol proposed (19).
FIG. 3.
(A) Results of PCR amplification of a 166-bp fragment with T. cruzi nuclear repetitive region-specific primers cruzi1 and cruzi2 extracted from DNA. Lane 1, patient sample 1; lane 2, patient sample 2; lane 3, patient sample 123; lane 4, patient sample 220; lane 5, patient sample 72; lane 6, positive control T. cruzi I strain Dm11; lane 7, positive control T. cruzi II strain VS; lane 8, molecular weight marker. (B) Results of PCR amplification of a 330-bp fragment with T. cruzi minicircle-specific primers 121 and 122 extracted from DNA. Lane 1, molecular weight marker; lane 2, patient sample 1; lane 3, patient sample 2; lane 4, patient sample 123; lane 5, positive control T. cruzi II strain VS; lane 6, positive control T. cruzi I strain Dm11; lane 7, negative control 1; lane 8, negative control 2. (C) Results of PCR amplification of a 110-bp fragment of the human β-globin region as an internal control. Lane 1, patient sample 1; lane 2, patient sample 2; lane 3, patient sample 123; lane 4, patient sample 200; lane 5, patient sample 127; lane 6, patient sample 122; lane 7, patient sample 256; lane 8, molecular weight marker.
Serological tests.
The 20 samples from healthy individuals were negative by IIF and ELISA, and of the 240 samples from chronic chagasic patients 99.2% (n = 238) were positive by IIF, while 95% (n = 228) were positive by ELISA. Likewise, the gold standard TESA-blot, because of its high sensitivity and specificity, showed a positivity of 99.2% (n = 238) (Table 1). All samples were tested in duplicate. A sample was considered seroreactive in IIF when both reaction mixtures had a titer of ≥1/32. For ELISA, a sample was considered seroreactive when both reactions showed ODs greater than 0.345, and with TESA-blot, a sample was considered seroreactive when the 150-kDa band was observed in the membrane strips. These results were previously validated with positive and negative controls for ELISA, IIF, and TESA-blot. Similarly, all of the six anti-Leishmania sp. antibody-positive sera tested were negative by all of the methods used, resulting in 100% specificity (Table 1).
TABLE 1.
Sensitivities, specificities, PPVs, NPVs, and kappa indexes calculated according to PCR results and serological tests (IIF and ELISA) with TESA-blot as the gold standard
| Comparison | % Sensitivity | % Specificity | % PPV | % NPV | Kappa indexc |
|---|---|---|---|---|---|
| IIF vs TESA | 100 | 100 | 100 | 100 | 1 |
| ELISA vs TESA-blota | 95 | 100 | 95 | 70 | 0.8 |
| ELISA vs TESA-blotb | 100 | 100 | 100 | 100 | 1 |
| stDNA PCR vs TESA-blot | 75 | 100 | 100 | 27 | 0.3 |
| kDNA PCR vs TESA-blot | 70 | 100 | 100 | 24 | 0.2 |
Values calculated without the results of corrected discrepant samples.
Values calculated with the results of corrected discrepant samples.
Less than 0.40, poor agreement; 0.40 to 0.60, fair agreement; 0 61 to 0.80, good agreement; more than 0.80, excellent agreement.
Statistical analyses.
Sensitivity, specificity, PPV, NPV, and the kappa index were calculated. TESA-blot was used as the gold standard, and the values mentioned above were obtained (27) (Table 1). Interestingly, there were some discrepancies in the ELISA results obtained. Due to these discrepancies, the 12 samples that were discrepant based on ELISA were submitted double blind to the reference center in Colombia, where an ELISA based on a crude antigenic preparation from local strains is performed. These samples were also processed by a Stat-Pak immunochromatographic assay (21) according to the manufacturer's recommendations. The parameters were then recalculated with these results (Table 1).
DISCUSSION
A considerable number of studies that examined the sensitivity of PCR for the detection of T. cruzi in the blood of patients with chronic Chagas' disease have been published. Avila et al. described 100% sensitivity for samples from chagasic patients when serological techniques and xenodiagnosis were compared, but not all of the patients were in the chronic phase, which is evidenced by an absence of tissue failure; therefore, their probability of detecting parasites was much higher (2). In previous reports, the sensitivity of PCR for samples from chronic patients ranged from 45 to 60% (4, 6, 13, 29, 44) when the number of parasites was relatively low. The PCR sensitivity obtained in our study was 70% by kDNA and 75% by stDNA. This can be explained by the low number of parasites in the chronic phase of the disease, thus preventing efficient amplification by PCR (1). Although some authors have demonstrated that PCR is more sensitive than hemoculture and xenodiagnosis (6), its sensitivity is not 100% due to the fact that detecting a parasite in 10 ml of blood is slightly complicated in terms of steric hindrance and the availability of template DNA within the same reaction mixture. Some authors have also reported the possibility of using a nested PCR in order to increase the amount of parasite template DNA (22).
The nuclear repetitive region (stDNA) was a more suitable region than the variable region of minicircle kDNA for the detection of T. cruzi in chronic patients. These regions have been previously selected because of the high number of copies (5,000 to 10,000 copies of kDNA per cell and 10% of stDNA in the T. cruzi genome) (2, 4, 24). PCR methodology for direct detection of parasite DNA in blood was standardized in this study. In addition, the limit of detection for amplification was determined to be 0.5 fg of parasite DNA per ml of blood for kDNA and 0.1 fg for stDNA (data not shown). In regard to primers used, many authors report the variable region of minicircle kDNA to amplify T. cruzi DNA (2, 4) and the nuclear repetitive DNA region (9, 20, 29). Likewise, many authors have used other regions for the detection of T. cruzi in blood. Silber et al. (35) reported the use of primers to amplify the region that encodes flagellar protein F29, where the sensitivity was 95%. This study was carried out with samples from chronic chagasic patients and also with samples with a high number of parasites, such as the feces of infected vectors and acute-phase patients. Lastly, Chiurillo et al. (7) reported the use of telomeric sequences with a sensitivity of 100% where T. cruzi was detected in artificially infected blood of mice and in triatomine feces. In these studies, we highlight the great variability of the primers used to detect T. cruzi in blood; similarly, it can be inferred that, according to the sensitivity ranges, the best primers for T. cruzi detection in blood might correspond to the nuclear repetitive region of stDNA and the variable region of kDNA.
The primers used to amplify the variable region of T. cruzi kDNA have been designed by sequencing T. cruzi II strains from Brazil. Similarly, all reports on the detection and amplification of DNA from T. cruzi are based on the kDNA of T. cruzi II strains. According to some authors (17, 34, 36), the genetic variability of strains of T. cruzi I can lead to the inference that a difference may exist between the kDNA sequences of strains belonging to the T. cruzi I lineage and those belonging to the T. cruzi II lineage. Phylogenetic analysis based on the variable region of T. cruzi kDNA demonstrated genetic variability when isolates belonging to the T. cruzi I and II lineages were compared (37). Also, the influence of the stDNA copy number difference between the T. cruzi lineages has been established (12, 15, 24), where the number of stDNA copies in T. cruzi I is lower than the number in T. cruzi II. It is important to mention that parasites circulating in Colombia belong to T. cruzi I but there is evidence of patients infected with T. cruzi II parasites (26, 46). This fact could affect the sensitivity of the PCR due to changes in the sequence and copy number seen in the parasites according to the primers used. This may be a factor that can explain the lower sensitivity obtained when using the PCR method. Virreira et al. (43) observed that the intensity of DNA bands might vary according to the genetic lineage of T. cruzi when amplification was carried out with primers used for the detection of T. cruzi kDNA. These facts corroborate the importance of considering genetic variation and virulence factors in the detection of the parasite.
Although previous reports suggest that the use of recombinant antigens is the best choice for the serodiagnosis of Chagas' disease (8, 40), the sensitivity of the commercial kit Chagatest ELISA recombinant, version 3.0, was 95% when samples from Colombian chagasic patients were tested. The origin of recombinant protein is always controversial, since the majority of theses peptides are obtained from T. cruzi II and the T. cruzi strains that circulate in Colombia and the other countries of northern South America are predominantly T. cruzi I (26). Likewise, it would be ideal to clone, express, and purify recombinant antigens from T. cruzi I strains to assess their sensitivity in the diagnosis of chronic chagasic Colombian patients. In a multicenter study performed with 53 chronic chagasic patients from an area of Brazil where the disease is endemic, where it is known that infections are reported to be due to T. cruzi II, the Chagatest ELISA recombinant showed 100% sensitivity (5). Chagatest ELISA recombinant was also evaluated in other study in which five commercially available ELISAs were evaluated to determine their diagnostic accuracy for Chagas' disease in Brazilian and Panamanian chagasic patients. A sensitivity of 100% was obtained with Brazilian patients, but a sensitivity of 81.25% was obtained with chagasic patients in Panama, where T. cruzi I is predominant. Interestingly, in this study, the samples from Panamanian chronic chagasic patients showed lower titers in all five ELISAs than Brazilian samples (5). Likewise, it has already been reported that there is a difference in immunoglobulin profiles between the different genotypes of T. cruzi, another factor that can explain the low sensitivity of the commercial kit used (10). These developments have a great epidemiological and social impact due to blood bank screening, where these kits are often used to determine if a sample of blood for transfusion is positive or negative for anti-T. cruzi antibodies.
Sensitivities, specificities, PPVs, NPVs, and kappa indexes were considered (Table 1). TESA-blot was chosen as the gold standard test based on its sensitivity and specificity. IIF results showed a kappa index of 1, but with the discrepant ELISA results obtained the sensitivity dropped to 95%. In relation to the 12 samples with discrepant ELISA results, it was observed that the sensitivities and specificities were 100% in the ELISA results provided by the reference center, highlighting the limitations of the commercially available ELISA kits and the high potential of Stat-Pak as a rapid diagnostic test in the chronic phase of Chagas' disease (21). However, it is also necessary to consider the usefulness of PCR in doubtful or negative serology, because two samples that were negative by IIF, ELISA, and TESA-blot were positive by PCRs with stDNA and kDNA. In the PCR comparison, the 70% sensitivity obtained with kDNA and the 75% sensitivity obtained with stDNA highlight the disadvantages of using this test in the diagnosis of chronic Chagas' disease. In addition, the kappa indexes showed a very low concordance of PCR with serological methods, although the stDNA PCR showed a higher concordance than did the kDNA PCR, suggesting that PCR may not be a reliable diagnostic test for Chagas' disease but, alternatively, may be useful as a diagnostic tool. The commercial kit Chagatest ELISA recombinant, version 3.0 (Chagatest Rec v3.0; Wiener Lab, Rosario, Argentina) provided false-negative results in the diagnosis of chronic patients. It is important to consider that the reference center in Colombia uses crude antigenic extracts from T. cruzi I strains to perform ELISAs. The use of commercial kits must be evaluated due to the high proportion of false-negative results that may lead to serious public health implications, given the fact that these kits are used to screen for anti-T. cruzi antibody in blood banks (5).
Based on the evaluation and comparison of the five tests, we concluded that the serological tests have higher sensitivity and specificity than PCR for the diagnosis of Chagas' disease. Thus, it is advisable to evaluate serological techniques for the diagnosis of Chagas' disease based on crude extracts of T. cruzi strains that are circulating at the site where transmission occurs. Others studies, as reported by Caballero et al. (5), are recommended to assess available commercial diagnostic kits for Chagas' disease due to their tendency to increase false-negative results. PCR may be an alternative diagnostic technique and particularly valuable for the confirmation of doubtful results by serology and possibly screening in blood banks. Some evidence also suggests that PCR may be a useful method to determine the response to etiologic treatment, as reversion of seropositivity may take several decades (11, 28). Our study made a maximum optimization of the conventional PCR method, obtaining a sensitivity of 75% by stDNA and 70% by kDNA and a specificity of 100%. The phenol-chloroform method was shown to be the most reliable DNA extraction method for the detection of parasite DNA by PCR. Finally, it is recommended to use the available serological tests, and we recommend corroborating these results with PCR using the nuclear repetitive region of T. cruzi in order to validate the parasitological and serological diagnosis of patients with chronic Chagas' disease.
Acknowledgments
Financial support was obtained from the Universidad de los Andes Faculty of Sciences Research Fund and the BENEFIT project, supported by Canadian Institute for Health Research (CIHR) grant no. 150896 and TDR/WHO grant no. A30755.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Andersson, J. 2004. Molecular diagnosis of experimental Chagas disease. Trends Parasitol. 20:52-53. [DOI] [PubMed] [Google Scholar]
- 2.Avila, H., D. Sigman, L. Cohen, R. Millikan, and L. Simpson. 1991. Polymerase chain reaction amplification of Trypanosoma cruzi kinetoplast minicirlce DNA isolated from whole blood lysates: diagnosis of chronic Chagas' disease. Mol. Biochem. Parasitol. 48:211-221. [DOI] [PubMed] [Google Scholar]
- 3.Britto, C., M. A. Cardoso, P. Wincker, and C. M. Morel. 1993. A simple protocol for the physical cleavage of Trypanosoma cruzi kinetoplast DNA present in blood samples and its use in polymerase chain reaction (PCR)-based diagnosis of chronic Chagas disease. Mem. Inst. Oswaldo Cruz 88:171-172. [DOI] [PubMed] [Google Scholar]
- 4.Britto, C., A. Cardoso, C. Ravel, A. Santoro, J. Borges-Pereira, J. R. Coura, C. M. Morel, and P. Wincker. 1995. Trypanosoma cruzi: parasite detection and strain discrimination in chronic chagasic patients from northeastern Brazil using PCR amplification of kinetoplast DNA and nonradioactive hybridization. Exp. Parasitol. 81:462-471. [DOI] [PubMed] [Google Scholar]
- 5.Caballero, Z., O. Sousa, W. Marques, A. Saez-Alquezar, and E. S. Umesawa. 2007. Evaluation of serological tests to identify Trypanosoma cruzi infection in humans and determine cross-reactivity with Trypanosoma rangeli and Leishmania spp. Clin. Vaccine Immunol. 14:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro, A. M., A. O. Luquetti, A. Rassi, G. G. Rassi, E. Chiari, and L. M. C. Galvao. 2002. Blood culture and polymerase chain reaction for the diagnosis of the chronic phase of human infection with Trypanosoma cruzi. Parasitol. Res. 88:894-900. [DOI] [PubMed] [Google Scholar]
- 7.Chiurillo, M. A., G. Crisante, A. Rojas, A. Peralta, M. Dias, P. Guevara, N. Añez, and J. L. Ramírez. 2003. Detection of Trypanosoma cruzi and Trypanosoma rangeli infection by duplex PCR assay based on telomeric sequences. Clin. Diagn. Lab. Immunol. 10:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silveira, J. F., E. S. Umezawa, and A. O. Luquetti. 2001. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 17:286-291. [DOI] [PubMed] [Google Scholar]
- 9.Diaz, C., V. Nussenzweig, and A. Gonzalez. 1992. An improved polymerase chain reaction assay to detect Trypanosoma cruzi in blood. Am. J. Trop. Med. Hyg. 46:616-623. [DOI] [PubMed] [Google Scholar]
- 10.dos Santos, D. M., A. Talvani, P. M. Da Mata Guedes, G. L. L. Machado-Coelho, M. De Lana, and M. T. Bahía. 2009. Trypanosoma cruzi: genetic diversity influences the profile of immunoglobulins during experimental infection. Exp. Parasitol. 121:8-14. [DOI] [PubMed] [Google Scholar]
- 11.Duffy, T., M. Bisio, J. Altcheh, J. M. Burgos, M. Diez, M. J. Levin, R. R. Favaloro, H. Freilij, and A. G. Schijman. 2009. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in Chagas disease patients. PLoS Negl. Trop. Dis. 3:e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias, A. C., N. S. Vargas, B. Zingales, and S. Schenkman. 2003. Organization of satellite DNA in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 129:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, M. L., L. M. Galvao, A. M. Macedo, S. Pena, and E. Chiari. 1999. Chagas' disease diagnosis: comparative analysis of parasitologic, molecular, and serological methods. Am. J. Trop. Med. Hyg. 60:205-210. [DOI] [PubMed] [Google Scholar]
- 14.Gomes, Y. M., V. R. A. Pereira, M. Nakazawa, D. S. Rosa, M. D. D. S. Barros, A. G. P. Ferreira, E. D. Silva, S. F. Yamada Ogata, M. A. Krieger, and S. Goldenberg. 2001. Serodiagnosis of chronic Chagas infection by using EIE-recombinant-Chagas-Biomanguinhos kit. Mem. Inst. Oswaldo Cruz 96:497-501. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, A., E. Prediger, M. E. Huecas, N. Nogueira, and P. M. Lizardi. 1984. Minichromosomal repetitive DNA in Trypanosoma cruzi: its use in a high sensitivity parasite detection assay. Proc. Natl. Acad. Sci. USA 81:3356-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guhl, F. 2007. Chagas disease in Andean countries. Mem. Inst. Oswaldo Cruz 102(Suppl. 1):29-38. [DOI] [PubMed] [Google Scholar]
- 17.Herrera, C., M. D. Bargues, A. Fajardo, M. Montilla, O. Triana, G. A. Vallejo, and F. Guhl. 2007. Identifying four Trypanosoma cruzi I isolate haplotypes from different geographic regions in Colombia. Infect. Genet. Evol. 7:535-539. [DOI] [PubMed] [Google Scholar]
- 18.Houghton, R. L., D. R. Benson, L. D. Reynolds, P. D. McNeill, P. R. Sleath, M. J. Lodes, Y. A. Skeiky, D. A. Leiby, R. Badaro, and S. G. Reed. 1999. A multi-epitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruzi in radioimmunoprecipitation-confirmed and consensus-positive sera. J. Infect. Dis. 179:1226-1234. [DOI] [PubMed] [Google Scholar]
- 19.Jones, E. M., D. G. Colley, S. Tostes, E. Reis, C. L. Vnecak-Jones, and T. M. McCurley. 1993. Amplification of Trypanosoma cruzi DNA sequence from inflammatory lesions in human chagasic cardiomyopathy. Am. J. Trop. Med. Hyg. 48:348-357. [DOI] [PubMed] [Google Scholar]
- 20.Kirchhoff, L. V., J. R. Votava, D. E. Ochs, and D. R. Moser. 1996. Comparision of PCR and microscopic methods for detecting Trypanosoma cruzi. J. Clin. Microbiol. 34:1171-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luquetti, A. O., C. Ponce, E. Ponce, J. Esfandiari, A. Schijman, S. Revollo, N. Añez, B. Zingales, R. Ramgel-Aldao, A. Gonzalez, M. J. Levin, E. S. Umezawa, and J. F. Da Silveira. 2003. Chagas' disease diagnosis: a multicentric evaluation of Chagas Stat-Pal, a rapid immunochromatographic assay with recombinant proteins of Trypanosoma cruzi. Diagn. Microbiol. Infect. Dis. 46:265-271. [DOI] [PubMed] [Google Scholar]
- 22.Marcon, G. E. B., P. D. Andrade, D. M. Alburquerque, J. D. S. Wanderley, E. A. de Almeida, M. E. Gauriento, and S. C. Botelho Costa. 2002. Use of a nested polymerase chain reaction (N-PCR) to detect Trypanosoma cruzi in blood samples from chronic chagasic patients and patients with doubtful serologies. Diagn. Microbiol. Infect. Dis. 43:39-43. [DOI] [PubMed] [Google Scholar]
- 23.Marin-Neto, J. A., A. Rassi, Jr., C. A. Morillo, A. Avezum, S. J. Connolly, S. Sosa-Estani, F. Rosas, and S. Yusuf on behalf of BENEFIT Investigators. 2008. Rationale and design of a randomized placebo-controlled trial assessing the effects of etiologic treatment in Chagas' cardiomyopathy: the BENznidazole Evaluation For Interrupting Trypanosomiasis (BENEFIT). Am. Heart J. 156:37-43. [DOI] [PubMed] [Google Scholar]
- 24.Martins, C., C. S. Baptista, S. Ienne, G. C. Cerqueira, D. C. Bartholomeu, and B. Zingales. 2008. Genomic organization and transcription analysis of the 195-bp satellite DNA in Trypanosoma cruzi. Mol. Biochem. Parasitol. 160:60-64. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto, T. K., P. C. Cotrim, J. F. Da Silveira, A. M. S. Stolf, and E. S. Umezawa. 2002. Trypanosoma cruzi: isolation of an immunodominant peptide of TESA (trypomastigote excreted-secreted antigens) by gene cloning. Diagn. Microbiol. Infect. Dis. 42:187-192. [DOI] [PubMed] [Google Scholar]
- 26.Mejía-Jaramillo, A. M., V. H. Hugo-Peña, and O. Triana-Chávez. 2009. Trypanosoma cruzi: biological characterization of lineages I and II supports the predominance of lineage I in Colombia. Exp. Parasitol. 121:83-91. [DOI] [PubMed] [Google Scholar]
- 27.Moncayo, A., and A. O. Luquetti. 1990. Multicentre double blind study for evaluation of Trypanosoma cruzi defined antigens as diagnostic reagents. Mem. Inst. Oswaldo Cruz 85:489-495. [DOI] [PubMed] [Google Scholar]
- 28.Moncayo, A., and M. Ortiz Yanine. 2006. An update on Chagas disease (human American trypanosomiasis). Ann. Trop. Med. Parasitol. 100:663-677. [DOI] [PubMed] [Google Scholar]
- 29.Moser, D. R., L. Kirchhoff, and J. E. Donelson. 1989. Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J. Clin. Microbiol. 27:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastini, A. C., S. R. Iglesias, V. C. Carricarte, M. E. Guerin, D. O. Sanchez, and A. A. C. Frasch. 1994. Immunoassay with recombinant Trypanosoma cruzi antigens potentially useful for screening donated blood and diagnosing Chagas' disease. Clin. Chem. 40:1893-1897. [PubMed] [Google Scholar]
- 31.Piron, M., R. Fisa, N. Casamitjana, P. López-Chejade, L. Puig, M. Vergés, J. Gascón, J. Gomez i Prat, M. Portús, and S. Sauleda. 2007. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 103:195-200. [DOI] [PubMed] [Google Scholar]
- 32.Russomando, G., A. Figueredo, M. Almirón, M. Sakamoto, and K. Morita. 1992. Polymerase chain reaction-based detection of Trypanosoma cruzi DNA in serum. J. Clin. Microbiol. 30:2864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saiki, R. K., S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich, and N. Arnheim. 1985. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 34.Salazar, A., A. Schijman, and O. Triana-Chávez. 2006. High variability of Colombian Trypanosoma cruzi lineage I stocks as revealed by low-stringency single primer-PCR minicircle signatures. Acta Trop. 100:110-118. [DOI] [PubMed] [Google Scholar]
- 35.Silber, A. M., J. Búa, B. Porcel, E. Segura, and A. Ruiz. 1997. Trypanosoma cruzi: specific detection of parasites by PCR in infected humans and vectors using a set of primers (BP1/BP2) targeted to a nuclear DNA sequence. Exp. Parasitol. 85:225-232. [DOI] [PubMed] [Google Scholar]
- 36.Spotorno, A. E., L. Córdova, and A. Solari. 2008. Differentiation of Trypanosoma cruzi I subgroups through characterization of cytochrome b gene sequences. Infect. Genet. Evol. 8:898-900. [DOI] [PubMed] [Google Scholar]
- 37.Telleria, J., B. Lafay, M. Virreira, C. Barbané, M. Tibayrenc, and M. Svoboda. 2006. Trypanosoma cruzi: sequence analysis of the variable region of kinetoplast minicircles. Exp. Parasitol. 10:1016-1026. [DOI] [PubMed] [Google Scholar]
- 38.Umezawa, E. S., M. S. Nascimento, N. Kesper, J. R. Coura, J. Borges-Pereira, A. C. Junqueira, and M. E. Camargo. 1996. Immunoblot assay using excreted-secreted antigens of Trypanosoma cruzi in serodiagnosis of congenital, acute and chronic Chagas' disease. J. Clin. Microbiol. 34:2143-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umezawa, E. S., S. F. Bastos, M. E. Camargo, L. M. Yamauchi, M. R. Santos, A. Gonzalez, B. Zingales, M. J. Levin, O. Sousa, R. Rangel-Aldao, and J. F. Da Silveira. 1999. Evaluation of recombinant antigens for Chagas' disease serodiagnosis in South and Central America. J. Clin. Microbiol. 37:1554-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umezawa, E. S., and J. F. Da Silveria. 1999. Serological diagnosis of Chagas disease with purified and defined Trypanosoma cruzi antigens. Mem. Inst. Oswaldo Cruz 94(Suppl. 1):285-288. [DOI] [PubMed] [Google Scholar]
- 41.Umezawa, E. S., A. I. Souza, V. Pinedo-Cancino, M. Marcondes, A. Marcili, L. M. A. Camargo, A. A. Camacho, A. M. S. Stolf, and M. M. G. Teixeira. 2009. TESA-blot for the diagnosis of Chagas disease in dogs from co-endemic regions for Trypanosoma cruzi, Trypanosoma evansi and Leishmania chagasi. Acta Trop. 111:15-20. [DOI] [PubMed] [Google Scholar]
- 42.Vallejo, G., F. Guhl, E. Chiari, and A. M. Macedo. 1999. Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammalian hosts by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop. 72:203-212. [DOI] [PubMed] [Google Scholar]
- 43.Virreira, M., F. Torrico, C. Truyens, C. Alonso-Veja, M. Solano, Y. Carlier, and M. Svoboda. 2003. Comparison of polymerase chain reaction methods for reliable and easy detection of congenital Trypanosoma cruzi infection. Am. J. Trop. Med. Hyg. 68:574-582. [DOI] [PubMed] [Google Scholar]
- 44.Wincker, P., M. F. Bosseno, C. Britto, N. Yaksic, M. A. Cardoso, C. M. Morel, and S. F. Breniere. 1994. High correlation between Chagas' disease serology and PCR based detection of Trypanosoma cruzi kinetoplast DNA in Bolivian children living in an endemic area. FEMS Microbiol. Lett. 124:419-423. [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization. 2007. Report of the Scientific Working Group on Chagas Disease. Buenos Aires, Argentina, 17-20 April 2005. Update July 2007. World Health Organization, Geneva, Switzerland. http://www.who.int/tdrold/diseases/chagas/swg_chagas.pdf.
- 46.Zafra, G., J. C. Mantilla, H. M. Valadares, A. M. Macedo, and C. I. González. 2008. Evidence of Trypanosoma cruzi II infection in Colombian chagasic patients. Parasitol. Res. 103:731-734. [DOI] [PubMed] [Google Scholar]
- 47.Zarate-Blades, C. R., N. Bladés, M. S. Nascimiento, J. F. Da Silveira, and E. S. Umezawa. 2007. Diagnostic performance of tests base don Trypanosoma cruzi excreted-secreted antigens in an endemic area for Chagas' disease in Bolivia. Diagn. Microbiol. Infect. Dis. 57:229-232. [DOI] [PubMed] [Google Scholar]