Abstract
Porphyromonas gingivalis is one of the major causative agents of adult periodontitis. One of the features of this periodontal pathogen is its ability to attach to a variety of oral bacterial surfaces and to colonize subgingival dental plaque. We have shown that Streptococcus cristatus CC5A inhibits expression of fimA, a gene encoding the major protein subunit of long fimbriae in P. gingivalis; as a result, S. cristatus interrupts formation of P. gingivalis biofilms. Here we further demonstrate that the inhibitory activity of S. cristatus affects multiple strains of P. gingivalis and that optimal inhibitory activity correlates with levels of arginine deiminase expression in S. cristatus. More strikingly, the impact of S. cristatus on P. gingivalis colonization was revealed by comparing levels of P. gingivalis and S. cristatus in subgingival dental plaque. Spearman correlation analysis indicated a negative correlation between the distributions of S. cristatus and P. gingivalis (r = −0.57; P < 0.05). These data suggest that some early colonizers of dental plaque, such as S. cristatus, may be beneficial to the host by antagonizing the colonization and accumulation of periodontal pathogens such as P. gingivalis.
Periodontitis is a bacterial biofilm (dental plaque)-related infectious disease. Although over 750 oral bacterial taxa can be detected in the oral cavity, three gram-negative species, Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola—known as the red complex—have strong associations with chronic periodontal disease (9, 28). It has been suggested that these gram-negative bacteria are later colonizers of dental plaques and are recruited to microbial communities by earlier colonizers, such as oral streptococci and Actinomyces species, via specific interactions of surface molecules (10). While there is no direct evidence that the earlier colonizers are associated with periodontitis, some of these organisms can provide a favorable environment for periodontal pathogens such as P. gingivalis. A well-studied interaction of earlier colonizers and later colonizers of dental plaque is coadhesion of Streptococcus gordonii and P. gingivalis. Specific adherence of P. gingivalis to S. gordonii strains was demonstrated in in vitro experiments (12). Studies by independent laboratories showed involvement of multiple sets of adhesins in the two bacteria. The first set of adhesins is the P. gingivalis long fimbriae (FimA) and glyceraldehyde-3-phosphate dehydrogenase present on the surfaces of streptococci (17). The second set involves the P. gingivalis short fimbriae (Mfa1) and the streptococcal SspA/B (antigen I/II) adhesins (4, 23). It is likely that these specific protein-protein interactions promote P. gingivalis colonization on existing biofilms consisting of S. gordonii and related oral streptococci (4, 27).
We previously reported an antagonistic relationship between P. gingivalis and Streptococcus cristatus CC5A. P. gingivalis was unable to form microcolonies with S. cristatus, due to repression of fimA expression in the presence of S. cristatus (32). Moreover, the long fimbriae are important for aspects of P. gingivalis colonization. Previous studies have shown that a P. gingivalis strain with a fimA deficiency has a diminished capacity to adhere to human gingival fibroblasts and epithelial cells (6) and is deficient in invasion of epithelial cells (35). The fimA mutant also is less able to induce periodontal bone loss in a gnotobiotic rat model (18). Our recent study identified an S. cristatus surface protein, arginine deiminase (ArcA), responsible for eliciting repression of fimA expression in P. gingivalis 33277 (34). The arcA gene is found in a number of oral bacteria, mainly in streptococci (3). However, arcA is differentially expressed among oral streptococcal strains (16). Higher-level expression of arcA was observed in S. cristatus than in S. gordonii, which may contribute to the ability of the organism to prevent P. gingivalis colonization in the oral cavity. In this study, we tested an antagonistic role of S. cristatus in P. gingivalis colonization. We postulate that inhibition of FimA production in P. gingivalis by S. cristatus requires higher expression of ArcA and that colonization of S. cristatus strains expressing an elevated level of ArcA plays an important role in antagonizing P. gingivalis colonization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. P. gingivalis strains were grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates, supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml), at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Streptococcus strains were grown in Trypticase peptone broth supplemented with 0.5% glucose at 37°C under aerobic conditions.
TABLE 1.
Strains and plasmids used in this study
| Strains | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. gingivalis | ||
| 33277 | Type strain from ATCC, FimA type I | Lab collection |
| 49417 | Type strain from ATCC, FimA type II | Lab collection |
| BH18/10 | FimA type I | 21 |
| OMZ409 | FimA type II | 21 |
| W83 | FimA type IV | 5 |
| HG564 | FimA type IV | 21 |
| UPF | Derivative of P. gingivalis 33277 containing fimA::lacZ gene fusion in its chromosomal DNA; Emr | 31 |
| S. cristatus | ||
| CC5A | Wild-type strain | 20 |
| ARCE | Derivative of S. cristatus CC5A containing an insertional mutation in the arcA gene; Emr | This study |
| CR3 | Clinical isolate | 7 |
| CR311 | Type strain NCTC 12479 | 8 |
| CH34110 | Clinical isolate | Lab collection |
| PSH1a | Clinical isolate | 7 |
| PSH1b | Clinical isolate | 7 |
| S. gordonii | ||
| DL1 | Wild-type strain | 14 |
Emr, resistance to erythromycin.
Partial purification of the S. cristatus inhibitory protein.
Surface extracts of S. cristatus were collected by sonication and centrifugation (13,000 × g for 30 min) followed by filtration (0.2-μm pore size). The crude extract of CC5A was partially purified by ammonium sulfate fractionation as described earlier (33, 34), and the fraction precipitated with 66% saturated ammonium sulfate was used for the inhibitory activity.
RNA isolation and quantitative RT-PCR.
S. cristatus cells were harvested and resuspended in 300 μl distilled H2O and 900 μl Trizol (Invitrogen, Carlsbad, CA). The cells were disrupted by using a Mini-Beadbeater 3110BX (BioSpec Products, Bartlesville, OK). P. gingivalis was grown on TSB blood agar plates with or without 10 μg of partially purified ArcA, as described before (34). Bacteria were homogenized in Trizol reagent (Invitrogen). The RNA in the supernatant was then purified using an RNeasy minispin column (Qiagen, Valencia, CA). RNA samples were digested on-column with RNase-free DNase. The total RNA was tested using an Agilent 2100 Bioanalyzer to ensure the quality of the samples. Real-time reverse transcriptase PCR (RT-PCR) analysis was performed by using a QuantiTect SYBR green RT-PCR kit (Qiagen) on an iCycler MyiQTM real-time PCR detection system (Bio-Rad Laboratories, Inc., Redmond, WA) according to the manufacturer's instructions. Amplification reactions consisted of a reverse transcription cycle at 50°C for 30 min, an initial activation at 95°C for 15 min, and 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. The melting curve profile was analyzed to verify a single peak for each sample, which indicates primer specificity. The expression levels of the investigated genes for the test sample were determined relative to the untreated calibrator sample by using the comparative cycle threshold (ΔCT) method. The ΔCTs were calculated by subtracting the average CT of the test sample from the average CT of the calibrator sample and were then used to calculate the ratio between the two by assuming 100% amplification efficiency. By loading the same amount of total RNA for any comparable samples, the ΔCT represents the difference in gene expression between the samples. The following primers were used for amplification: CGGAACGAATAACCCAGAGA and CTGACCAACGAGAACCCACT for the fimA gene of 33277, OMZ409, and BH18/10; GGCCTTGACGACTTCTTTGA and ATGCAGTCCCACCAGGATAG for fimA of W83; CTAAAATCGCAGCCCTTGTC and GACGCCTCCAATTCGTATGT for fimA of HG564; and TGAAGTGACGATGAGCCAGT and GCCAATGAAGCACCGAATAG for fimA of 49417. Primers TCCAATGCCAAACCTTTACT and ATACGAGTATCTTCTTCACG were designed to complement the highly conserved regions of arcA. All experiments were performed at least three times.
β-Galactosidase assays.
S. cristatus protein fractions (10 μg) were mixed with 105 cells of P. gingivalis UPF, which contains a chromosomal fimA promoter-lacZ reporter construct (34), and spotted onto a TSB blood agar plate. The ability of the fractions to inhibit fimA expression in P. gingivalis was determined with a β-galactosidase assay. Expression of the lacZ gene under the control of the fimA promoter was measured by the standard spectrophotometric β-galactosidase assay with ONPG (o-nitrophenyl-β-d-galactopyranoside as the substrate, as described previously (31).
Arginine deiminase assay.
The arginine deiminase assay was performed in 96-well microplates as previously described (34). Briefly, surface extracts of S. cristatus cells were collected by centrifugation and sonication. A 100-μl (5-μg) volume of sample was added to each well and mixed with 50 μl of 0.1 M l-arginine. The mixtures were allowed to react for 1 h at 37°C and were then terminated by the addition of 50 μl of 20% sulfuric acid. Finally, 1% 2,3-butanedione monoxime (Sigma, St. Louis, MO) was added to each well, and the reaction was developed by incubation in the dark for 1 h at 56°C. The resultant gradients of a peach color were quantitated with a Benchmark plus microplate spectrophotometer (Bio-Rad, Hercules, CA) at 492 nm.
Western blot analysis.
S. cristatus strains were grown in Trypticase peptone broth for 18 h. The surface proteins were collected by sonication and centrifugation as described previously (33). Protein concentrations of the samples were determined by using a Bio-Rad protein assay. The soluble proteins (5 μg) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, along with prestained molecular weight standards (Bio-Rad), and were transferred to nitrocellulose membranes (Gibco BRL, Rockville, MD) with a mini-Transblot electrophoretic transfer cell (Bio-Rad) at 100 V for 1 h. The membrane was treated with 30 ml of blocking solution (3% bovine serum albumin in phosphate-buffered saline [PBS] containing 0.1% Tween 20, pH 7.4) for 1 h and incubated for 1 h with a polyclonal anti-ArcA antibody diluted 1:1,000 in PBS containing 0.1% Tween 20, pH 7.4. The membrane was then rinsed twice and washed three times for 15 min each with 0.1% Tween 20 in PBS. The membrane was incubated with anti-rabbit immunoglobulin horseradish peroxidase-conjugated secondary antibodies for 1 h and rinsed and washed as described above. Antigen-antibody reactivity was visualized by enhanced chemiluminescence (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
Dental plaque sampling.
Twenty patients with chronic periodontitis were recruited among those newly admitted to the clinic of School of Dental Medicine, State University of New York at Buffalo. The study was approved by the Health Science Institutional Review Board. Patients were informed of the protocols and gave informed consent. Subjects with chronic periodontitis presented with radiographic periodontal bone loss of ≥4 mm at multiple teeth. Additional inclusion criteria were an age of ≥21 years, pocket depth of ≥5 mm and concomitant radiographic evidence of bone loss, no systemic conditions that indicate periodontitis as a manifestation of systemic diseases, no use of systemic antibiotics in the previous 6 months, and no prior periodontal treatment. A total of 20 subjects, 12 males and 8 females, with a mean age of 47.8 years, were enrolled.
All samples were obtained by the same dentist to standardize the sampling procedure. Before sampling, the selected teeth were isolated with cotton rolls. Plaque samples were obtained from one of the deepest periodontal sites by inserting sterile endodontic paper points as deep as possible and keeping them in place for 30 s. The paper points were placed in microcentrifuge tubes containing 500 μl Tris-EDTA buffer. The tubes were then vortexed for 60 s and centrifuged at 12,000 × g for 3 min. The pellets in the tubes were stored at −70°C until DNA extraction.
DNA extraction was carried out as follows. The pellets in the microcentrifuge tubes were resuspended in 100 μl Tris-EDTA buffer, boiled for 20 min, and centrifuged at 12,000 × g for 2 min. DNA concentration in the supernatants was determined using a NanoDrop spectrophotometer.
P. gingivalis cells and S. cristatus cells in the plaque were enumerated by using a QuantiTect SYBR green PCR kit with P. gingivalis species-specific 16S rRNA gene primers (TGTAGATGACTGATGGTGAAA and ACTGTTAGCAACTACCGATGT) (29) or CC5A, CR3, and CR311 strain-specific arcA gene primers (CTGACGAAGCGAAAGGTCTG and ATGTGGTTGAGCGATACAGC). Standards used to determine P. gingivalis or S. cristatus cell numbers were prepared using genomic DNAs from the wild-type strain 33277 or CC5A. A fresh culture of bacteria was mildly sonicated to release single cells from the bacterial clumps, and the bacterial culture was then serially diluted in PBS and plated to enumerate CFU at each dilution. DNA was also isolated from the dilutions, and a quantitative PCR assay was performed by using the QuantiTect SYBR green PCR kit (Qiagen) on an iCycler MyiQ real-time PCR detection system (Bio-Rad Laboratories, Inc., Redmond, WA) according to the manufacturer's instructions, to determine cell number. Three trials were performed on three separate cultures.
Statistical analysis.
The Spearman correlation analysis was used to determine the relationship of distributions of P. gingivalis and S. cristatus in subgingival dental plaque. The role of ArcA in fimA expression in P. gingivalis strains was analyzed by two-way analysis of variance.
RESULTS
Inhibitory spectrum of S. cristatus ArcA on fimA expression in P. gingivalis.
We have previously demonstrated the ability of S. cristatus CC5A ArcA to repress expression of the fimA gene in P. gingivalis 33277 (34). Since several variants of FimA have been identified according to their deduced amino acid sequences (21), we tested the inhibitory activity of S. cristatus ArcA in a group of P. gingivalis strains representing different ATCC type strains and clinical isolates, including strains displaying type I, II, and IV fimbriae. As shown in Table 2, although the greatest ArcA action on fimA expression was detected in P. gingivalis 33277, expression of fimA was inhibited at least twofold in P. gingivalis strains carrying type I and II fimbriae (33277, BH18/10, OMZ409, and ATCC 49417). However, S. cristatus ArcA had a modest effect on fimA expression in type IV FimA strains (W83 and HG567). These results suggest that the inhibitory activity of S. cristatus ArcA on fimA expression is fimbrial-type specific and that the inhibitory activity of ArcA on fimA expression is greater in type I and II fimA genotypes of P. gingivalis.
TABLE 2.
Quantification of fimA expression in P. gingivalis strains
| P. gingivalis strain (FimA type) | Fold decrease of fimA expression in the presence of ArcAa |
|---|---|
| ATCC33277 (I) | 12.8 ± 0.7 (a) |
| BH18/10 (I) | 3.1 ± 0.2 (bc) |
| OMZ409 (II) | 5.2 ± 1.5 (b) |
| ATCC 49417 (II) | 2.3 ± 0.5 (c) |
| HG564 (IV) | 1.4 ± 0.1 (c) |
| W83 (IV) | 1.3 ± 0.2 (c) |
Levels of fimA transcripts were measured by real-time PCR, and the change in expression levels was calculated by dividing the copy number of the gene transcript in the P. gingivalis strains grown in the absence of S. cristatus CC5A ArcA by that in the strains grown in the presence of ArcA. Results are means and standard deviations from three independent experiments. Two-way analysis of variance revealed a significant difference in changes among the groups (P < 0.001). Means with different letters are significantly different by the post hoc test (Student-Newman-Keuls method, P < 0.05).
Expression and inhibitory function of arcA in S. cristatus.
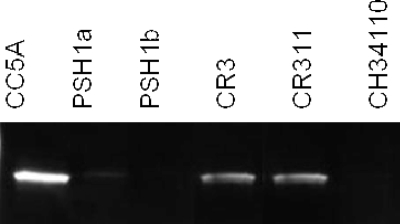
Since we observed both a higher expression of ArcA and fimA inhibitory activity in S. cristatus CC5A than in S. gordonii DL1, we sought to determine if all S. cristatus strains produced the same level of ArcA. Using PCR analysis, we showed that all six strains of our S. cristatus collection carried the arcA gene (data not shown). However, differential expression of arcA in these strains was observed by using real-time PCR. The expression level of arcA in CC5A, CR3, and CR311 was at least 10-fold higher than that in PSH1a, PSH1b, and CH34110 (Table 3). As much as a 75-fold difference in expression of arcA was observed between S. cristatus strains CR311 and PSH1a. To determine differential expression of ArcA in S. cristatus at the protein level, we performed Western blot analysis with anti-CC5A antibodies as a probe. The expression pattern of mRNA corresponded with the observed ArcA protein levels. Higher expression of ArcA was found in CC5A, CR3, and CR311 than in PSH1a, PSH1b, and CH34110 (Fig. 1). These results suggest a differential expression of arcA among S. cristatus strains that may result in a difference in their ability to repress fimA expression in P. gingivalis.
TABLE 3.
Quantification of arcA expression in S. cristatus strainsa
| Strain | arcA expression |
|---|---|
| S. cristatus CC5A | 1.00 ± 0.06 |
| S. cristatus PSH1a | 0.04 ± 0.05 |
| S. cristatus PSH1b | 0.05 ± 0.01 |
| S. cristatus CR3 | 1.73 ± 0.01 |
| S. cristatus CR311 | 3.70 ± 0.40 |
| S. cristatus CH34110 | 0.09 ± 0.01 |
| S. cristatus ARCE (arcA mutant) | 0.00 ± 0.00 |
| S. gordonii DL1 | 0.09 ± 0.01 |
Transcript levels were measured by real-time PCR, and the relative expression of arcA was normalized by 23S rRNA to the expression level in S. cristatus CC5A. Results are means and standard deviations from four independent experiments.
FIG. 1.
Western blot analysis of ArcA production in S. cristatus. Streptococcal strains were grown in Trypticase peptone broth for 18 h. Cell extracts (5 μg) from streptococcal cells were used.
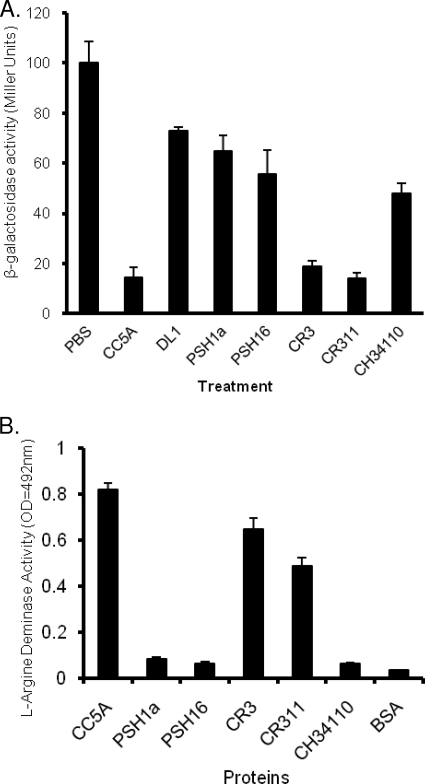
Our recent study indicated that ArcA is a dual-function protein (34). Besides the ability to repress fimA expression in P. gingivalis, the enzymatic activity of ArcA catalyzes the hydrolysis of l-arginine to l-citrulline and ammonia. To determine whether S. cristatus strains producing more ArcA have higher enzymatic and inhibitory activities, we compared both activities in six strains of S. cristatus. The ability to repress fimA expression in P. gingivalis was measured by a β-galactosidase assay. As shown in Fig. 2A, higher arcA expression levels in CC5A, CR3, and CR311 corresponded to a significantly stronger inhibition of fimA expression. Expression of fimA in P. gingivalis was decreased more than 10-fold in the presence of surface extracts isolated from CC5A, CR3, and CR311. However, only 30 to 50% inhibitory activity was observed in PSH1a, PSH1b, and CH34110, similar to that found in S. gordonii DL1. We next examined arginine hydrolytic activity in S. cristatus strains. As shown in Fig. 2B, high hydrolytic activity was found in CC5A, CR3, and CR311, while much lower hydrolytic activity was observed in PSH1a, PSH1b, and CH34110.
FIG. 2.
Evaluation of ArcA function among S. cristatus strains. (A) Expression of fimA in P. gingivalis in response to streptococcal strains. P. gingivalis UPF carrying a fimA promoter-lacZ fusion gene was tested for β-galactosidase activity in the presence and absence of surface extracts (50 μg) isolated from S. cristatus or S. gordonii strains. The β-galactosidase activity of P. gingivalis grown in the presence of S. cristatus is indicated relative to the activity level in P. gingivalis grown in the absence of S. cristatus surface extract (set at 100). Standard errors are indicated (n = 3). (B) Comparison of arginase activity in protein fractions of S. cristatus strains. Arginine deiminase levels are means (n = 3) ± standard deviations.
Distribution of S. cristatus strains expressing higher levels of ArcA and P. gingivalis in dental plaque.
Our previous studies indicated that S. cristatus CC5A prevents P. gingivalis colonization in in vitro studies (32). Thus, we postulated that the distribution of P. gingivalis in subgingival bacterial communities is negatively correlated with the distribution of S. cristatus, especially strains carrying an arcA gene with a high degree of homology with the CC5A arcA gene. To test this hypothesis, we collected dental plaques from the deepest periodontal pocket in 20 periodontitis patients prior to periodontal treatment. P. gingivalis cells and S. cristatus cells in the plaque were enumerated by quantitative PCR with P. gingivalis species-specific 16S rRNA gene primers (29) or strain-specific primers for the arcA gene for CC5A, CR3, and CR311. The P. gingivalis 16S rRNA gene was detected in 13 samples (65%), while the S. cristatus arcA gene was detected in 12 samples (55%). Seven samples were negative for both S. cristatus arcA and P. gingivalis 16S rRNA; therefore, they were not used for further correlation analysis due to the possibility of a false-negative result. As shown in Table 4, there was a statistically significant negative correlation between P. gingivalis and S. cristatus in the 13 positive samples, by the Spearman correlation analysis (r = −0.57222, P < 0.05).
TABLE 4.
Distribution of P. gingivalis and S. cristatus in subgingival plaques
| Patient | No. of bacterial cells/100 ng DNA/samplea |
|
|---|---|---|
| S. cristatus | P. gingivalis | |
| 1 | 53 | 2,626 |
| 2 | 462 | 1 |
| 3 | 207 | 7 |
| 4 | 0 | 1,862 |
| 5 | 660 | 2 |
| 6 | 12 | 220 |
| 7 | 518 | 12 |
| 8 | 105 | 1,302 |
| 9 | 707 | 160 |
| 10 | 1,239 | 7 |
| 11 | 103 | 20 |
| 12 | 3,157 | 100 |
| 13 | 289 | 1,220 |
The numbers of P. gingivalis and S. cristatus cells were determined by using quantitative PCR with 16S rRNA and arcA-specific primers, respectively.
DISCUSSION
Periodontal diseases such as chronic periodontitis are among the most common chronic infectious diseases occurring in humans. Because of the chronic nature of periodontitis and concerns of emerging bacterial resistance to antibiotics, development of novel therapeutic agents for the management of periodontitis is warranted. Therapeutic agents that can locally interfere with the colonization of periodontal pathogens would be desirable, since the virulence of the periodontal pathogens is dependent on their initial colonization in the oral cavity. FimA is one of major surface adhesions of P. gingivalis and is responsible for bacterial attachment to a number of host and bacterial substrates (13, 30, 31). P. gingivalis is classified into at least five types based on the variation of the fimA nucleotide sequence (2, 21). We have demonstrated here that ArcA from S. cristatus inhibits expression of fimA in type I and II genotypes of P. gingivalis. Studies of FimA genotypes of P. gingivalis showed that the majority of periodontitis patients carried suggested type II FimA genotype, suggesting a strong association with adult periodontitis (1, 5, 19). Although the function and mechanism of FimA variants are not clear, it appears that recombinant type II FimA has higher adhesive activity to human epithelial cells than other types of FimA (22). Therefore, the ability of ArcA to repress type II FimA production may make ArcA a good candidate for a novel therapeutic agent for periodontitis prevention.
Our data demonstrate that not all S. cristatus strains are able to inhibit FimA production in P. gingivalis. Only strains producing elevated levels of ArcA may be considered antagonistic to P. gingivalis. Therefore, based on the expression level of arcA and the ability to repress fimA expression in P. gingivalis, S. cristatus strains may be divided into two groups. The promoter regions of arcA of CC5A, CR3, and CR311 were sequenced, and DNA alignments show 98% homology among these strains (data not shown). It appears, however, that there is a low degree of homology between the arcA promoter regions for the two groups, since amplification of the promoter regions of PSH1a, PSH1b, and CH34110 by PCR using several sets of CC5A arcA primers was unsuccessful. It is likely that differential expression of arcA in S. cristatus strains is due to variation of their promoter structures.
Several recent studies indicated that the prevalence of P. gingivalis as detected with PCR ranges from 61% to 79% in patients without any periodontal treatment (15, 24, 25). We report here a 65% detection rate of P. gingivalis in previously untreated periodontitis patients. Using quantitative PCR analysis, the numbers of P. gingivalis organisms in dental plaque samples were determined and compared with the numbers of S. cristatus organisms detected in the same samples. Interestingly, more P. gingivalis cells were detected in the plaque samples where the number of S. cristatus cells was low. Conversely, fewer P. gingivalis cells were discovered in the samples harboring more S. cristatus. This negative correlation between the P. gingivalis and S. cristatus distributions provides evidence of an antagonistic relationship between these oral bacteria. It is likely that lower numbers of P. gingivalis detected in dental plaque are due to repression of FimA production induced by the presence of S. cristatus. Further studies are required to investigate if prevalence of S. cristatus in subgingival plaque is associated with the severity and prognosis of periodontitis. Apparently, the numbers of S. cristatus organisms in the subgingival dental plaques are not sufficient to eliminate P. gingivalis. It is well known that the oral streptococci are major colonizers in supragingival plaque and can constitute up to 80% of the plaque (26). However, the numbers of oral streptococci and gram-positive bacteria decrease in subgingival plaque in which gram-negative anaerobic bacteria such as P. gingivalis are proportionally increased (11, 26). Our observation of the negative correlation between P. gingivalis and S. cristatus distribution provides opportunities to develop therapeutics specifically against P. gingivalis colonization in subgingival plaque.
Acknowledgments
This work was supported by Public Health Service grants DE014699 (H.X.), DE017708 (B.Y.W.), and DE12505 (R.L.) from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Amano, A., M. Kuboniwa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79:1664-1668. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 37:1426-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Daep, C. A., R. J. Lamont, and D. R. Demuth. 2008. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect. Immun. 76:3273-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enersen, M., I. Olsen, O. Kvalheim, and D. A. Caugant. 2008. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J. Clin. Microbiol. 46:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamada, N., K. Watanabe, C. Sasakawa, M. Yoshikawa, F. Yoshimura, and T. Umemoto. 1994. Construction and characterization of a fimA mutant of Porphyromonas gingivalis. Infect. Immun. 62:1696-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handley, P. S., P. L. Carter, J. E. Wyatt, and L. M. Hesketh. 1985. Surface structures (peritrichous fibrils and tufts of fibrils) found on Streptococcus sanguis strains may be related to their ability to coaggregate with other oral genera. Infect. Immun. 47:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handley, P. S., F. F. Correia, K. Russell, B. Rosan, and J. M. DiRienzo. 2005. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol. Immunol. 20:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkinson, H. F., and R. J. Lamont. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589-595. [DOI] [PubMed] [Google Scholar]
- 10.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamont, R. J., S. G. Hersey, and B. Rosan. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7:193-197. [DOI] [PubMed] [Google Scholar]
- 13.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc, D. J., and F. P. Hassell. 1976. Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J. Bacteriol. 128:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leys, E. J., S. R. Lyons, M. L. Moeschberger, R. W. Rumpf, and A. L. Griffen. 2002. Association of Bacteroides forsythus and a novel Bacteroides phylotype with periodontitis. J. Clin. Microbiol. 40:821-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, X., R. J. Lamont, J. Wu, and H. Xie. 2008. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J. Bacteriol. 190:4367-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malek, R., J. G. Fisher, A. Caleca, M. Stinson, C. J. van Oss, J. Y. Lee, M. I. Cho, R. J. Genco, R. T. Evans, and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Missailidis, C. G., J. E. Umeda, C. Ota-Tsuzuki, D. Anzai, and M. P. Mayer. 2004. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol. Immunol. 19:224-229. [DOI] [PubMed] [Google Scholar]
- 20.Mouton, C., H. S. Reynolds, and R. J. Genco. 1980. Characterization of tufted streptococci isolated from the “corn cob” configuration of human dental plaque. Infect. Immun. 27:235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa, I., A. Amano, R. K. Kimura, T. Nakamura, S. Kawabata, and S. Hamada. 2000. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J. Clin. Microbiol. 38:1909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa, I., A. Amano, M. Kuboniwa, T. Nakamura, S. Kawabata, and S. Hamada. 2002. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 70:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, Y., M. R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ready, D., F. D'Aiuto, D. A. Spratt, J. Suvan, M. S. Tonetti, and M. Wilson. 2008. Disease severity associated with presence in subgingival plaque of Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Tannerella forsythia, singly or in combination, as detected by nested multiplex PCR. J. Clin. Microbiol. 46:3380-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romano, F., A. Barbui, and M. Aimetti. 2007. Periodontal pathogens in periodontal pockets and in carotid atheromatous plaques. Minerva Stomatol. 56:169-179. [PubMed] [Google Scholar]
- 26.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 27.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19:254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 29.Tran, S. D., and J. D. Rudney. 1999. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J. Clin. Microbiol. 37:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, J., X. Lin, and H. Xie. 2007. Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS Microbiol. Lett. 271:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie, H., S. Cai, and R. J. Lamont. 1997. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect. Immun. 65:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie, H., W. O. Chung, Y. Park, and R. J. Lamont. 2000. Regulation of the Porphyromonas gingivalis fimA (fimbrillin) gene. Infect. Immun. 68:6574-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie, H., N. Kozlova, and R. J. Lamont. 2004. Porphyromonas gingivalis genes involved in fimA regulation. Infect. Immun. 72:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie, H., X. Lin, B. Y. Wang, J. Wu, and R. J. Lamont. 2007. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 153:3228-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yilmaz, O., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 4:305-314. [DOI] [PubMed] [Google Scholar]