Abstract
We identified 179 cases of acute respiratory illness including 50 cases of radiographically confirmed pneumonia over the course of 4 months on a deployed U.S. Navy vessel. Laboratory tests showed Mycoplasma pneumoniae to be the etiological agent. This report represents the first published description of a shipboard outbreak of this pathogen.
Ships are notorious vectors for respiratory and gastrointestinal pathogens, which can spread readily in crowded onboard populations (1, 13, 20). Despite this, most ships remain isolated from the collection and diagnostic resources needed to identify outbreak pathogens. Military populations, which tend to be of higher density than their civilian counterparts, are particularly susceptible to agents of acute respiratory disease (ARD) and pneumonia (21). A few respiratory disease outbreak investigations with diagnostic laboratory support have been conducted on military ships (6, 13, 24) and cruise ships (2, 3, 4, 19), usually in response to sharp outbreaks with high attack rates, and these have generally identified influenza virus as the disease agent.
Naval Environmental and Preventive Medicine Units can field mobile epidemiological response and investigation teams in response to outbreaks on U.S. Navy vessels. To provide continuous surveillance capability for respiratory pathogens, the Naval Health Research Center (NHRC) began placing diagnostic specimen collection and storage equipment on board vessels in 2002 and offers diagnostically accredited reference laboratory support for the identification of etiological agents in collected specimens. NHRC also supports some ships in maintaining specific PCR capability on board.
Mycoplasma pneumoniae is commonly associated with ARD and pneumonia outbreaks among civilians, usually children and young adults (5), and has been identified in pneumonia outbreaks among crowded military recruit populations (8). M. pneumoniae spreads through close contact with expired respiratory secretions (droplets), like influenza virus, but has a long incubation period (6 to 32 days [10]) and a resulting tendency to generate long, slow-spreading epidemics (7, 22) which may be difficult to recognize and control. M. pneumoniae is challenging and slow to culture (16), and emerging molecular methods such as PCR offer increased sensitivity and more rapid identification from uncultured throat swab specimens or sputum (23). These technologies greatly increase the functional value of testing for M. pneumoniae, as the resulting data can be obtained quickly enough to inform treatment and outbreak response decisions. In the outbreak described here, laboratory-supported identification of the responsible pathogen played a role in guiding the effective treatment of patients and limitation of further transmission.
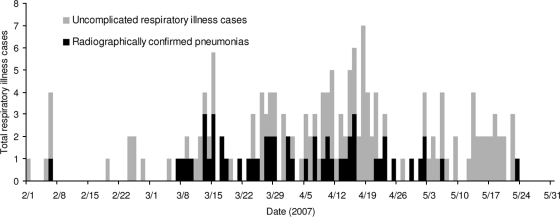
Based upon review of routine syndromic surveillance data, a Navy Environmental Preventive Medicine epidemiological investigation team embarked aboard USS Boxer from 20 to 30 May 2007 to confirm and characterize a suspected respiratory disease outbreak occurring while the ship was at sea. This team conducted chart reviews (the source of the data in Fig. 1) and broad respiratory specimen collection (the source of the samples collected from 24 to 26 May). Samples spanning the breadth of the outbreak had been collected prior to the investigation by crew members using collection and storage equipment provided by NHRC as part of ongoing shipboard respiratory disease surveillance (Institutional Review Board research protocol NHRC.2003.0002).
FIG. 1.
Respiratory disease and pneumonia cases on USS Boxer.
Oropharyngeal (throat) swab samples were collected from patients presenting with respiratory disease. Samples were stored in viral transport medium (Remel, Lenexa, KS), and a cold chain was maintained at −80°C or below from sample collection until extraction.
All samples were shipped by air on dry ice to NHRC for laboratory analysis. Samples were extracted with the QIAamp 96 DNA blood and body fluid kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and tested for common respiratory pathogens by PCR as listed in Table 1. Following PCR analysis, specimens were also tested by a College of American Pathologists-accredited diagnostic protocol to confirm the presence of M. pneumoniae by standard culture methods. A subset of 15 samples were also tested with the TessArray resequencing pathogen microarray kit (RPM v.1; TessArae, LLC, Potomac Falls, VA), a functionally independent method that uses multiplexed PCR and resequencing microarrays to screen for diverse respiratory pathogens (14, 15).
TABLE 1.
PCR results for potential pathogens
| Pathogen (test method reference) | No. (%) positive by PCR |
||
|---|---|---|---|
| Period 1a (n = 2) | Period 2b (n = 31) | Period 3c (n = 12) | |
| Adenovirus (18) | 0 (0) | 0 (0) | 0 (0) |
| Coronavirus (9)d | 0 (0) | 2 (6.5) | 2 (12.5) |
| Influenza virus A (11) | 0 (0) | 0 (0) | 0 (0) |
| Influenza virus B (11) | 0 (0) | 0 (0) | 0 (0) |
| Respiratory syncytial virus (12) | 0 (0) | 0 (0) | 0 (0) |
| Bordetella pertussis (17) | 0 (0) | 0 (0) | 0 (0) |
| Chlamydophila pneumoniae (17) | 0 (0) | 0 (0) | 0 (0) |
| Legionella pneumophila (17) | 0 (0) | 0 (0) | 0 (0) |
| Mycoplasma pneumoniae (17) | 0 (0) | 24 (77.4) | 1 (8.3) |
Samples collected before 31 January 2007.
Samples collected between 1 February and 23 May 2007.
Samples collected between 24 and 26 May 2007.
One of the type-specific coronavirus PCR tests was developed and validated by Kay Holmes and Mary Catherine Smith at the Colorado Health Sciences Center, Denver, and has not been published.
Cases occurred in the chronological sequence illustrated in Fig. 1. Sixty-nine (38.5%) of 179 cases met the clinical case definition used for ARD (including both a fever of ≥38°C and cough or a sore throat or radiologically confirmed pneumonia). All of the samples collected before 24 May met this case definition, while the 12 collected during the investigation (24 to 26 May) were from personnel displaying any respiratory symptom. One hundred fifty-nine (88.8%) of the 179 patients self-presented for respiratory complaints. Of these, 55 (30.7%) had documented fever (≥38°C). Sixty-six patients received a chest X-ray during their evaluation, with 50/66 (75.8%) positive for infiltrate by radiologist-confirmed reading. The average age of the patients was 27.0 years, with 141 (78.8%) males and 38 (21.2%) females. Of the 179 patients, 70 (39%) had a history of smoking.
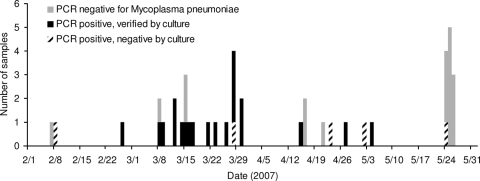
PCR results are shown in Table 1. A time course of M. pneumoniae PCR and culture results is shown in Fig. 2 (only a subset of cases were sampled, primarily on the basis of time available to the ship's medical personnel). Neither of the two samples collected prior to February 2007 (October 2006 and January 2007) was positive for M. pneumoniae (omitted from Fig. 2). PCR testing identified the presence of M. pneumoniae in 24/31 samples collected during the outbreak (1 February to 23 May).
FIG. 2.
Laboratory results for oropharyngeal swab samples collected aboard USS Boxer.
M. pneumoniae was confirmed by tissue culture in 20/24 PCR-positive samples. All PCR-negative samples were also negative by culture. Analysis of 15 samples with the RPM v.1 microarray system also supported the PCR results. Seven were positive for M. pneumoniae by both PCR and microarray analysis, seven were negative by both methods, and one was positive by microarray and negative by PCR.
This investigation confirms an outbreak of respiratory illness aboard a U.S. Navy ship lasting from 1 February to 23 May 2007. The ship experienced roughly 16 cases of respiratory illness per week during this period in a population of 1,074 personnel. Additionally, USS Boxer experienced 48 cases of radiographically confirmed pneumonias over a period of 9 weeks (5.4 cases per week).
The epidemiological and laboratory data implicate M. pneumoniae as the most likely causative organism. By late May, disease transmission had experienced a decrease, resulting in a downslope in the disease curve, as shown in Fig. 1. Only 1 of the 12 oropharyngeal specimens collected from 24 to 26 May was positive for M. pneumoniae by PCR, as shown in Table 1 and Fig. 2. It should be reiterated that these 12 samples were collected with a broader case definition (any respiratory symptom) than previous samples, as described above. In this investigation, the observed respiratory illness attack rate was approximately 17% (179 cases/1,074 at-risk personnel).
This is the first documented shipboard outbreak of M. pneumoniae. The shape of the epidemiological curve (multiple peaks, propagative pattern) is inconsistent with transmission patterns of typical seasonal influenzas, which generally create a much steeper singular curve. The high rate of pneumonias relative to ARD cases is also different than would be expected for influenza among healthy young adults.
Based on the identification of the source of this outbreak, the ship's medical personnel implemented screening, case recognition, treatment, and isolation procedures that quickly brought the outbreak under control. While these measures are not the subject of this report, they were dependent on the investigation and laboratory findings reported here.
Acknowledgments
For the permissions, access, and assistance necessary to conduct this study, we acknowledge the commanding officer and the medical staff of USS Boxer. We acknowledge the administrative support of the Henry M. Jackson Foundation for Military Medicine.
The views expressed in this report are ours and do not reflect the official policy or position of the Department of the Navy or the U.S. Government.
This research has been conducted in compliance with all applicable federal and international regulations governing the protection of human subjects in research.
We declare that no conflict of interest exists.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Barry, J. M. 2004. The great influenza: the epic story of the deadliest plague in history. Viking Books, New York, NY.
- 2.Brotherton, J. M. L., V. C. Delpech, G. L. Gilbert, S. Hatzi, P. D. Paraskevopoulos, J. M. McAnulty, and the Cruise Ship Outbreak Investigation Team. 2003. A large outbreak of influenza A and B on a cruise ship causing widespread morbidity. Epidemiol. Infect. 130:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1988. Acute respiratory illness among cruise-ship passengers—Asia. MMWR Morb. Mortal. Wkly. Rep. 37:63-66. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1997. Update: influenza activity—United States, 1997-98 season. MMWR Morb. Mortal. Wkly. Rep. 46:1094-1098. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1993. Outbreaks of Mycoplasma pneumoniae respiratory infection—Ohio, Texas and New York, 1993. MMWR Morb. Mortal. Wkly. Rep. 42:931, 937-939. [PubMed] [Google Scholar]
- 6.Earhart, K. C., C. Beadle, L. K. Miller, M. W. Pruss, G. C. Gray, E. K. Ledbetter, and M. R. Wallace. 2001. Outbreak of influenza in highly vaccinated crew of U.S. Navy ship. Emerg. Infect. Dis. 7:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernald, G. W., and W. A. Clyde, Jr. 1989. Epidemic pneumonia in university students. J. Adolesc. Health Care 10:520-526. [DOI] [PubMed] [Google Scholar]
- 8.Forsyth, B. R., H. H. Bloom, K. M. Johnson, and R. M. Chanock. 1965. Etiology of primary atypical pneumonia in a military population. JAMA 191:364-368. [DOI] [PubMed] [Google Scholar]
- 9.Fouchier, R. A. M., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. M. E. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foy, H. M., J. T. Grayston, G. E. Kenny, E. R. Alexander, and R. McMahan. 1966. Epidemiology of Mycoplasma pneumoniae infection in families. JAMA 197:859-866. [PubMed] [Google Scholar]
- 11.Freed, N. E., C. A. Myers, K. L. Russell, E. A. Walter, M. Irvine, R. G. Coon, and D. Metzgar. 2007. Diagnostic discrimination of live attenuated influenza vaccine strains and community-acquired pathogenic strains in clinical samples. Mol. Cell. Probes 21:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grondahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 37:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ksiazek, T. G., J. G. Olson, G. S. Irving, C. S. Settle, R. White, and R. Petrusso. 1980. An influenza outbreak due to A/USSR/77-like (H1N1) virus aboard a US Navy ship. Am. J. Epidemiol. 112:487-494. [DOI] [PubMed] [Google Scholar]
- 14.Lin, B., K. M. Blaney, A. P. Malanoski, A. G. Ligler, C. E. Meador, J. M. Schnur, D. Metzgar, K. L. Russell, and D. A. Stenger. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J. Clin. Microbiol. 45:443-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, B., Z. Wang, G. J. Vora, J. A. Thornton, J. M. Schnur, D. C. Thach, K. M. Mueller, A. G. Ligler, A. P. Malanoski, J. Santiago, E. A. Walter, B. K. Agan, D. Metzgar, D. Seto, L. T. Daum, R. Kruzelock, R. K. Rowley, E. H. Hanson, C. Tibbetts, and D. A. Stenger. 2006. Broad-spectrum respiratory tract pathogen identification using re-sequencing DNA microarrays. Genome Res. 16:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marmion, B. P., J. Williamson, D. A. Worswick, T.-W. Kok, and R. J. Harris. 1993. Experience with newer techniques for the laboratory detection of Mycoplasma pneumoniae infection: Adelaide, 1978-1992. Clin. Infect. Dis. 17(Suppl. 1):S90-S99. [DOI] [PubMed] [Google Scholar]
- 17.McDonough, E. A., C. P. Barrozo, K. L. Russell, and D. Metzgar. 2005. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis in clinical specimens. Mol. Cell. Probes 19:314-322. [DOI] [PubMed] [Google Scholar]
- 18.Metzgar, D., M. Osuna, A. E. Kajon, A. W. Hawksworth, M. Irvine, and K. L. Russell. 2007. Abrupt emergence of diverse species B adenoviruses in US military recruit training centers. J. Infect. Dis. 196:1465-1473. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. M., T. W. S. Tam, S. Maloney, K. Fukuda, N. Cox, J. Hockin, D. Kertesz, A. Klimov, and M. Cetron. 2000. Cruise ships: high-risk passengers and the global spread of new influenza. Clin. Infect. Dis. 31:433-438. [DOI] [PubMed] [Google Scholar]
- 20.Minooee, A., and L. S. Rickman. 1999. Infectious diseases on cruise ships. Clin. Infect. Dis. 29:737-744. [DOI] [PubMed] [Google Scholar]
- 21.Russell, K. L. 2006. Respiratory infections in military recruits, p. 227-253. In M. K. Lenhart, D. E. Lounsbury, and R. B. North, Jr., (ed.), Textbooks of military medicine: recruit medicine, 1st ed. Borden Institute, Walter Reed Army Medical Center, Washington, DC.
- 22.Steinberg, P., R. J. White, S. L. Fuld, R. R. Gutekunst, R. M. Chanock, and L. B. Senterfit. 1969. Ecology of Mycoplasma pneumoniae infections in Marine recruits at Parris Island, South Carolina. Am. J. Epidemiol. 89:62-73. [DOI] [PubMed] [Google Scholar]
- 23.Templeton, K. E., S. A. Scheltinga, A. W. Graffelman, J. M. Van Schie, J. W. Crielaard, P. Sillekens, P. J. Van Den Broek, H. Goossens, M. F. Beersma, and E. C. Claas. 2003. Comparison and evaluation of real-time PCR, real-time nucleic acid sequence-based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae. J. Clin. Microbiol. 41:4366-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White, D. C. 1957. A shipboard epidemic of Asian influenza. U.S. Armed Forces Med. J. 8:1717-1725. [PubMed] [Google Scholar]