Abstract
The genetic background and the presence of several virulence factors of Staphylococcus aureus isolates from intensive care unit (ICU) patients from 14 hospitals in The Netherlands isolated from 1996 until 2006 were investigated. In total, 936 methicillin-susceptible S. aureus (MSSA) and 7 methicillin-resistant S. aureus (MRSA) isolates were collected. The genetic background was determined by spa typing and multilocus sequence typing (MLST). The virulence determinants Panton-Valentine leukocidin (PVL), toxic shock syndrome toxin 1 (TSST-1), and collagen adhesion (CNA) were detected with real-time PCR assays. On the MRSA isolates, mobile resistance staphylococcal cassette chromosome mec (SCCmec) typing was performed. Among the MSSA isolates, 313 different spa types were observed. A genetic background common to MRSA clones, e.g., MLST clonal complex 1 (CC1), CC5, CC8, CC22, CC30, and CC45, was observed among 62% of the isolates. The remaining isolates were associated with MSSA-related MLST CCs. MLST CC1, CC25, and CC30 were continuously present, and other MLST CCs fluctuated over time. Two percent of the MSSA isolates harbored PVL, 21% had TSST-1, and 46% were positive for CNA. There were no changes in the prevalence of the virulence factors over time. Four MRSA isolates were typed as ST8-MRSA-IV (where ST is the MLST sequence type and IV is the SCCmec type), two were ST5-MRSA-II, and one was ST228-MRSA-I. All MRSA isolates were PVL, CNA, and TSST-1 negative except for the two ST5-MRSA-II isolates, which were TSST-1 positive. No changes in the S. aureus genetic background and the prevalence of the virulence factors PVL, CNA, and TSST-1 were observed in ICU patients in The Netherlands over time.
Around 20% of all patients in intensive case units (ICUs) acquire an ICU-related infection as a consequence of frequent use of antibiotics and intensive treatment procedures (1, 31). Of all ICU-related infections, 25% are caused by Staphylococcus aureus (31). Knowledge of the S. aureus population structure and of the prevalence of virulence factors has been proven crucial for the investigation of the epidemiology of S. aureus throughout the world (34).
Methicillin-resistant S. aureus (MRSA) clones can emerge by horizontal transfer of the staphylococcal cassette chromosome mec (SCCmec) between methicillin-resistant coagulase-negative Staphylococcus or MRSA and methicillin-susceptible S. aureus (MSSA) (51). In the event of antibiotic pressure, the MSSA isolates have a high risk of SCCmec transfer and survive. As shown in the literature, MSSA lineages with a MRSA-unrelated background may not provide a stable genomic environment for the integration of SCCmec (4, 23, 30, 32, 36, 43). SCCmec transfer has been found to be stable in MSSA with a MRSA-related genetic background, i.e., multilocus sequence typing (MLST) clonal complex 1 (CC1), CC5, CC8, CC22, CC30, and CC45 (39). The MSSA lineages with a MRSA background possess certain characteristics that favor their persistence in the host as well as the transfer between hosts.
As the highest antibiotic pressure in hospitals is found in ICUs, changes in the genetic background will be the most obvious among isolates from ICU patients. However, little is known about the genetic backgrounds of ICU isolates over time, and, therefore, this study investigates the genetic background and the virulence of S. aureus isolates obtained from 1996 to 2006 from ICU patients from 14 hospitals in The Netherlands.
MATERIALS AND METHODS
Study population.
From 1 January 1996 to 1 July 2006, a total of 943 S. aureus isolates (maximum, 100 isolates per ICU) were collected from blood, wounds, and bronchial aspirate from patients in ICUs of 14 medical centers (two university hospitals and 12 referral hospitals) in The Netherlands. The centers were geographically spread throughout the country. The isolates were identified as S. aureus at the local laboratories, stored at −20°C, and then sent to a central microbiological laboratory for antimicrobial susceptibility testing and genetic characterization. The genetic characterization of all isolates was performed at the department of Medical Microbiology of the Maastricht University Medical Center (MUMC). Only the first S. aureus isolate from each patient was included. Clinical data of the patients were not available.
Typing of the spa locus.
Real-time amplification of the spa locus, followed by sequencing, was performed as described before (45). The spa types were clustered into spa CCs using the algorithm based upon repeat pattern (BURP) with the Ridom StaphType, version 1.5, software package (http://www.ridom.de). The default settings recommended by the manufacturer were used. Since it has been shown that spa typing, together with the algorithm BURP, yields results consistent with typing results obtained by pulsed-field gel electrophoresis and MLST (2, 20, 21), the associated CCs, as determined with MLST, were allocated through the Ridom SpaServer (http://spaserver.ridom.de).
MLST.
All MRSA isolates were typed with MLST. To confirm the association between spa typing and MLST, MLST was performed on two or three MSSA isolates randomly selected from each of the spa CCs. MLST was performed with primers described by Enright et al. (19), with the modifications described by Deurenberg et al. (11). The amplification was performed on a conventional PCR system (Perkin Elmer GeneAmp 9600), using the following program: 15 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C, and with a final step for 5 min at 72°C. Following amplification, the PCR products were purified using a QIAquick PCR Purification Kit (Qiagen, The Netherlands). Sequencing of the PCR products was performed as described previously (40). The corresponding sequence types (STs) were obtained from the MLST database accessible via http://www.mlst.net. For spa CCs that consisted of fewer than 10 isolates, no MLST was performed.
Other genotypic determinations.
The oxacillin-resistant isolates were analyzed for the presence of the S. aureus-specific femA gene as well as the MRSA-specific mecA gene using a multiplex real-time PCR assay (14). The type of the mobile resistance determinant SCCmec was determined with multiple PCR assays. These PCR assays involved determination of ccr genes and mec complexes as described previously by Ito et al. (29) and Zhang et al. (53).
In all isolates, the presence of the virulence determinants Panton-Valentine leukocidin (PVL), toxic shock syndrome toxin 1 (TSST-1), and collagen adhesion (CNA) (6, 12, 47) was determined with real-time PCR assays (10; R. H., Deurenberg, M. I. A. Rijnders, S. Sebastian, M. A. Welling, P. S. Beisser, and E. E. Stobberingh, submitted for publication).
RESULTS
Of the 943 isolates collected, 7 isolates were oxacillin resistant and mecA positive and were identified as MRSA. The remaining 936 isolates were identified as MSSA.
Distribution of spa types and BURP analysis of the MSSA isolates.
A total of 313 different spa types were found among the 936 MSSA isolates. Seventy-seven spa types (25%) were not described previously. Among the new spa types, three new spa repeats were found (r198, r199, and r202). The 313 spa types were clustered into 19 spa CCs, 4 of which had no founder. Of the 936 isolates, 45 (5%) could not be clustered and were classified as singletons. Seventy-two isolates (8% of the total isolates) were excluded from the analyses because the spa types found were less than five repeats in length (Table 1). The results obtained with spa typing/BURP analyses were in agreement with the results obtained with MLST. With MLST, two new STs, ST282 and ST1302, were found (Table 1).
TABLE 1.
Composition of the spa CCs from isolates of ICU patients for an 11-year period
| spa CC no. or groupj | No. (%) of isolates | No. (%) of spa types | spa type(s)a | MLST CC(s) (ST)k | No. (%) of isolates positive for: |
||
|---|---|---|---|---|---|---|---|
| PVL | CNA | TSST-1 | |||||
| 084 | 181 (19) | 37 (12) | t084,b t085, t091,b t094, t144, t228, t254, t279, t289, t335, t346, t360, t491, t547, t593, t629, t803, t853, t963, t1119, t1204, t1243, t1361, t1685, t2119, t2556, t2636, t2932, t2949, t3003, t3008, t3097, t3099, t3161, t3163, t3190, t3370 | 7 (7), 15 (1036)f | 1 (1) | 18 (10) | 8 (4) |
| 012 | 162 (17) | 42 (13) | t012,b t017,b t018, t019, t021, t046, t122, t137, t275, t318, t338, t342, t382, t631, t685, t707, t710, t822, t840, t871, t974, t1070, t1397, t1504, t1626, t1642, t1654, t1827, t1889, t1945, t2018, t2024, t2271, t2785, t3001, t3007, t3095, t3100, t3110,t3117, t3372, t3373 | 30 (30, 282c) | 7 (4) | 149 (92) | 111 (69) |
| 024 | 87 (9) | 17 (5) | t008,b t024, t064, t104, t121, t190, t304, t334, t430, t451, t656, t701, t711, t723, t1476, t2104, t3052 | 8 (8) | 12 (13) | 5 (12) | |
| 015 | 86 (9) | 43 (14) | t004, t015,b t029, t031, t040, t050, t065, t073, t095, t116, t230, t302, t330, t331, t340, t445, t505, t620, t630, t671, t715, t737, t772, t1081, t1402, t1510, t1574, t1575, t1601, t1801, t1996, t2056, t2334, t2338,bt2733, t3011, t3054, t3062, t3103, t3157, t3164, t3189, t3493 | 45 (45) | 76 (88) | 7 (8) | |
| 002 | 71 (8) | 16 (5) | t001, t002,b t010, t105, t179, t311, t447, t450, t494, t548, t688, t954, t1815, t1818, t3012, t3371,b | 5 (5) | 6 (8) | 17 (24) | |
| 127 | 70 (7) | 17 (5) | t098,b t114, t127,b t174, t177, t189, t224, t231, t267, t345, t405, t591, t922, t3051, t3158, t3159, t3192, | 1 (1) | 1 (1) | 38 (54) | 1 (14) |
| 078 | 40 (4) | 20 (6) | t056. t078,b t167,b t280, t349, t353, t469, t660, t762, t814, t1315, t1350, t1439, t1541, t2523, t2992, t3006, t3111, t3116,bt3188 | 25/101 (25) | 2 (5) | 5 (13) | 3 (8) |
| 166 | 27 (3) | 9 (3) | t089, t136,b t153, t166,b t240, t369, t3101,bt3187, t3368 | 30 (938,g 30) | 1 (4) | 21 (78) | |
| 005 | 22 (2) | 9 (3) | t005b, t474, t1132, t1276, t1629, t1869, t3002,bt3162, t3366 | 22 (217,h 22) | 1 (4) | 20 (80) | 3 (12) |
| 160/156 | 19 (2) | 8 (3) | t156, t160,b t213, t771, t909, t1137, t1693, t3118,b | 12 (12) | 19 (100) | 2 (11) | |
| 159 | 14 (1) | 9 (3) | t159, t162, t272, t408, t645,b t659, t741, t1077, t1596 | 121 (121, 1302d) | 2 (14) | 14 (100) | 1 (7) |
| 100 | 14 (1) | 8 (3) | t099, t100,b t587, t733, t1045, t3064,bt3094, t3102 | 9 (9, 27i) | 1 (7) | 1 (7) | |
| 164 | 5 (1) | 3 (1) | t164, t731, t3004 | 20e | |||
| 273 | 5 (1) | 3 (1) | t273, t1491, t1778 | 1e | 5 (100) | ||
| 800 | 4 (0) | 3 (1) | t209, t337, t800 | 9e | 1 (25) | 1 (25) | |
| No founder | 6 (1) | 2 (1) | t216, t3093 | 59e | |||
| No founder | 2 (0) | 2 (1) | t246, t705 | 2 (100) | |||
| No founder | 2 (0) | 2 (1) | t3017, t3112 | 2 (100) | 1 (50) | ||
| No founder | 2 (0) | 2 (1) | t257, t3091 | ||||
| Singleton | 28 (3) | 19 (6) | t287, t332, t525, t576, t1039, t1096, t1531, t2050, t2559, t2834, t2998, t2999, t3010, t3049, t3063,t3096, t3160, t3166, t3191, t3360,t3367 | 1 (2) | 3 (7) | ||
| 2 | 1 | t034 | 398e | 1 (2) | |||
| 3 | 1 | t148 | 8e | ||||
| 4 | 1 | t185 | 50e | 3 (7) | |||
| 2 | 1 | t186 | 88e | ||||
| 1 | 1 | t409 | 1e | 1 (2) | |||
| 1 | 1 | t436 | 25e | ||||
| 2 | 1 | t493 | 182e | 1 (2) | |||
| 1 | 1 | t1191 | (445)e | ||||
| 1 | 1 | t1194 | 1021e | 1 (2) | |||
| Excluded | 26 | 23 | t129, t282, t287, t643, t693, t779, t929, t1040, t1050, t1509, t1552, t2353, t2413, t2524, t2853, t2887, t3018, t3019, t3050, t3098, t3104, t3165, t3369 | 2 (3) | 14 | 1 | |
| 32 | 1 | t026 | 45e | 30 (42) | 2 (3) | ||
| 1 | 1 | t059 | 8e | ||||
| 2 | 2 | t111, t777 | 5e | ||||
| 1 | 1 | t233 | 30/36 (39)e | 1 (1) | 1 (1) | ||
| 2 | 1 | t227 | 25e | ||||
| 5 | 2 | t528, t605 | 1e | 1 (1) | 1 (1) | ||
| 3 | 1 | t2383 | 398e | 1 (1) | - | ||
Underlined spa types are new spa types.
Isolates on which MLST was performed.
This is a new ST with MLST profile 2-4-2-2-6-3-2 (spa type t012).
This is a new ST with MLST profile 6-2-6-98-7-5-5 (spa type t645).
These MLST CCs are associated through the spa server.
Single-locus variant of ST15 at locus 7 (yqiL).
Single-locus variant of ST 30 at locus 4 (gmk).
Single-locus variant of ST 22 at the locus 6 (tpi).
Single-locus variant of ST9 at the locus 6 (tpi).
The 313 spa types were clustered into 19 spa CCs, four which had no founder; 45 isolates could not be clustered and were classified as singletons, and 72 isolates were excluded from the analyses because the spa types found were less than five repeats in length.
Data represent the results of MLST. Most of the time, only the ST of the main cluster was present. In some cases, locus variants were observed.
The main spa CC was spa CC 084 (19% of the isolates). This spa CC consisted of two MLST CCs, i.e., MLST CC7 (n = 65; founder t091) and MLST CC15 (n = 116; founder t084). The two MLST CCs are “connected” through spa type t2636. Other MLST CCs observed were MLST CC30 (spa CC012; n = 162; 17%), MLST CC8 (spa CC024; n = 87; 9%), MLST CC45 (spa CC15; n = 86; 9%), MLST CC5 (spa CC002; n = 71; 8%), and MLST CC1 (spa CC127; n = 70; 7%). Among the remaining MLST CCs, 5% or less of the total number of MSSA isolates were represented.
One of the four spa CCs with no founder was associated with MLST CC59. The others were associated with an undetermined MLST CC. Of the 72 excluded MSSA isolates, 32 were associated with MLST CC45.
Sixty-two percent of the MSSA isolates had a genetic background common to the MRSA lineages MLST CC1, CC5, CC8, CC22, CC30, CC45, CC59, and CC398. The genetic background of 17% of the MSSA isolates was associated with MSSA-associated lineages, i.e., MLST CC7, CC9, CC12, CC15, CC20, CC25, CC121, CC182, and CC1021. Of the remaining isolates (6%), the associated MLST CC could not be determined.
Distribution of S. aureus lineages over time.
The prevalence of the isolates with a MRSA-related genetic background, i.e., MLST CC1, CC5, CC8, CC22, CC30, CC45, CC59, and CC398, remained relatively stable during the study period, with the smallest amount in 1999 (53%) and the largest amount in 2004 (71%). The prevalence of isolates with a MSSA-related genetic background, i.e., MLST CC7, CC9, CC12, CC15, CC20, CC25, CC121, CC182, and CC1021, remained relatively stable during the years, with the smallest amount in the 1998 (23%) and the largest amount in 1999 (37%).
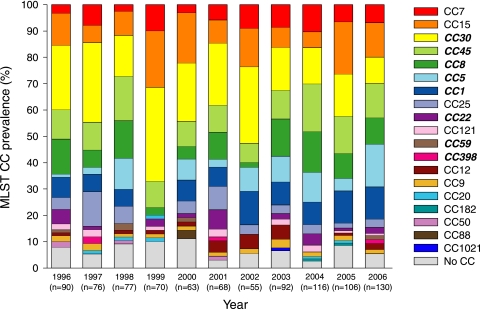
Several MRSA-related MLST CCs (MLST CC30 and 45) were continuously present between 1996 and 2006. MLST CC398 was present only in 1997, 2001, and 2006. The prevalence of MLST CC1, CC5, CC22, and CC8 fluctuated during the study period. Some MSSA-related MLST CCs fluctuated over time, i.e., MLST CC7, CC9, CC12, CC15, CC20, and CC25, while other MSSA-related MLST CCs, i.e., MLST CC88, MLST CC1021, MLST CC182, and MLST CC50, were observed only sporadically (Fig. 1).
FIG. 1.
Distribution of the MLST CCs between 1996 and 2006. The italic and bold MLST CCs in the legend are the MRSA-related MLST CCs. The remaining MLST CCs in the legend are MSSA-related MLST CCs.
Prevalence of virulence factors PVL, CNA, and TSST-1.
Of the 943 isolates, 17 (2%) were PVL positive. Seven of these isolates were associated with MLST CC30; the others were associated with MLST CC15 (n = 1), MLST CC1 (n = 1), MLST CC25/101 (n = 2), MLST CC22 (n = 1), and MLST CC121 (n = 2). The three other PVL-positive isolates were related to undetermined MLST CCs. Fourteen of the PVL-positive strains were also positive for the cna gene, six of which also harbored the tst gene.
A total of 433 isolates (46%) were positive for the cna gene. These isolates were predominantly associated with MLST CC30 (n = 150; 35%) and MLST CC45 (n = 76; 18%). The remaining CNA-positive isolates were associated with different MLST CCs, i.e., MLST CC1 (n = 43; 10%), MLST CC22 (n = 20; 5%), MLST CC7/15 (n = 18; 4%), MLST CC12 (n = 18; 4%), MLST CC121 (n = 14; 3%), MLST CC5 (n = 6; 1%), MLST CC25/101 (n = 5; 1%), and MLST CC9 (n = 2; <1%). Furthermore, 4 of the 433 (<1%) CNA-positive isolates were associated with a spa CC with no founders, 17 (4%) were associated with singletons, and 47 (11%) CNA-positive isolates were excluded from the BURP analyses.
A total of 194 (21%) S. aureus isolates were positive for the tst gene, of which 132 isolates (89%) were associated with MLST CC30 (spa CC012, n = 111; spa CC166, n = 21). The remaining tst-positive isolates were present among multiple MLST CCs (Table 1).
MRSA.
Seven of the 943 S. aureus isolates carried the mecA gene and were classified as MRSA. Four isolates were typed as ST8-MRSA-IV ([UK MRSA-2/-6] the roman numeral indicates the SCCmec type), two were ST5-MRSA-II (New York/Japan clone), and one was ST228-MRSA-I (Southern Germany clone). All MRSA isolates were PVL negative. The two ST5-MRSA-II isolates were TSST-1 positive.
DISCUSSION
No changes in the S. aureus population structure of ICU patients in The Netherlands over time were observed. The prevalence of the virulence factors PVL, CNA, and TSST-1 in S. aureus isolated in ICUs was stable over the years and associated with the several MLST CCs. However, MLST CC30 was the most frequently observed CC.
Distribution of MSSA clones.
MRSA clones can arise through horizontal gene transfer between patients. ICU patients have the highest risk of acquiring MRSA (46) because of the high selective pressure caused by the high antibiotic usage and because SCCmec transfers only in MSSA isolates with an MRSA-related background (39), whereas in this study 62% of the MSSA isolates had a MRSA-related background. This percentage is higher than the 50% described by Nulens et al. (40) and Hallin et al. among hospital isolates (25), probably due to the high antibiotic usage in ICUs.
The genetic background of the MSSA isolates observed in the present study was heterogeneous. However, the main MLST CC was CC30, as has been described previously in studies from in- and outpatients in The Netherlands (9, 37, 40).
Distribution of S. aureus lineages over time.
The prevalence of MLST CC1, CC25, and CC30 was stable and present throughout the study period. These MLST CCs were also found in studies from in- and outpatients in The Netherlands (37, 40). The prevalence of MLST CC8, CC15, and CC45 fluctuated over time. The change in prevalence might be due to import from abroad or from cross-border healthcare (8, 52), but further studies to confirm this hypothesis are needed.
MLST CC398, associated with a pig MRSA, was found among the MSSA isolates (33). These isolates probably derived from pigs or from MRSA from pigs that lost their SCCmec (15, 16). Huijsdens et al. reported the first ST398 MRSA in 2004 (27), which was later isolated in several countries worldwide, for example, France and China, and in several regions in The Netherlands (5, 28, 48). The first association between pigs and ST398 was observed by Armand Lefevre et al. (5). This study observed both the first MSSA and MRSA associated with ST398, isolated around 1998 (5). In the present study, two MSSA isolates with MLST CC398 were observed for the first time in 1997 in the southern region of The Netherlands, which is an agricultural region (48). It is possible that the MSSA isolates with this genetic background were the ancestors of the MRSA isolates found later in multiple Dutch studies.
Prevalence of virulence factors.
Two percent of the isolates were PVL positive, which was lower than described by Monecke et al. (38), in which 30% of the clinical MSSA isolates were PVL positive. This difference in PVL prevalence is most likely due to the origin of the isolates. Isolates derived from skin infections, as in the study of Monecke et al., have a higher prevalence of PVL because of the association of PVL with skin infections (6). In this study various MLST CCs, mainly MLST CC30, were associated with the PVL-positive strains in this study. Aires-de-Sousa et al. and Diep et al. also found that several MLST CCs, i.e., MLST CC1, CC8, CC30, CC59, CC80, and CC121, were associated with PVL-positive MSSA isolates (3, 13). This diversity in PVL-positive strains might be due to the fact that the PVL genes are located on several different phages and can thus spread into different S. aureus lineages (38).
In a Spanish study, a progressive increase in PVL-positive S. aureus isolates was observed in invasive isolates from 21 hospitals in the year 2006 (42). Almost 50% of the MSSA isolates were PVL positive whereas 2% were found in earlier studies in Spain (7, 42). This observation was not seen in our study, where the PVL prevalence remains the same over time.
In previous studies, CNA was found among S. aureus lineages MLST CC1, CC12, CC22, CC30, CC45, and CC51 (12, 35). The CNA-positive isolates in this study were predominantly associated with MLST CC30 and CC45. In the present study, S. aureus lineages, associated with MLST CC5, CC7, CC15, and CC25, were found to be CNA positive. Thus, the virulence factor CNA is not limited to the MLST CCs mentioned above. This can be explained by the notion that CNA is encoded within a genetic element and could be associated with a lysogenic bacteriophage (22).
About 16% of the isolates were positive for TSST-1. In a German study a TSST-1 prevalence of 40% among the MSSA isolates was found (32). As in other studies, in The Netherlands, Germany, and Japan a prevalence of TSST-1 between 5 and 20% was found (9, 40, 41, 44). TSST-1 has already been observed in MLST CC5, CC6, CC8, CC30, and CC45 (17, 26, 32, 41). In this study, several MLST lineages were associated with tst-positive strains, some of which were not described before, i.e., MLST CC1, CC7, CC9, and CC25. As the tst gene is situated on SaPI2, a mobile genomic island, it can be transferred horizontally between the different S. aureus lineages (32).
MRSA.
Four MRSA isolates were associated with ST8-MRSA-IV (UK MRSA-2/-6), which was earlier found in Dutch hospitals and in multiple countries worldwide (9, 50). Two MRSA isolates were associated with ST5-MRSA-II (New York/Japan clone), which was identified as the dominant MRSA in hospitals in New York and Japan (9) and was recently observed to be a dominant clone in the hospitals in The Netherlands as well (8). The other MRSA isolate was associated with ST228-MRSA-I (southern Germany clone) (9). The clones from southern Germany have been found sporadically in The Netherlands (9). Despite the high number of isolates with a MRSA-related background, the prevalence of MRSA in The Netherlands is still low. This probably results from (i) the national search-and-destroy policy implemented in 1988 (18, 49) and (ii) the low use of antibiotics, almost the lowest in Europe and globally (24).
In summary, the prevalence of MSSA with a genetic background common to MRSA clones, e.g., MLST CC1, CC5, CC8, CC22, CC30, and CC45, fluctuated between 53 to 72%, which suggests that there is a relatively high chance that ICU patients will be infected with a MSSA with a MRSA-related genetic background. Despite the unique patient population in ICUs, no differences were observed in the genetic background and prevalences of virulence factors of S. aureus in ICUs in The Netherlands over time. Furthermore, no differences in the genetic background of S. aureus strains were observed between in- and outpatient studies and the ICU.
Acknowledgments
The members of the Antibiotic Resistance Surveillance Group in The Netherlands were the following: H. van Dessel, MUMC; P. Bloembergen, Laboratory, Zwolle; M. G. R. Hendrix, Laboratory of Microbiology, Twente/Achterhoek; W. H. M. Vogels, Martini Hospital, Groningen; W. D. H. Hendriks, Medical Centre Rotterdam, location Clara; P. J. G. M. Rietra, Onze Lieve Vrouwe Gasthuis, Amsterdam; B. M. Dejongh, Sint Antonius Hospital, Nieuwegein; A. G. M. Buiting, St. Elisabeth Hospital, Tilburg; A. J. Beunders, Public Health Laboratory, Kennemerland Haarlem; K. Waar, Regional Public Health Laboratory, Leeuwarden; L. J. M. Sabbe, Regional Laboratory, Zeeland Goes; P. Sturm, University Medical Centre, Nijmegen; T. A. M. Trienekens, VieCuri Medical Centre, Venlo; and H. A. Bijlmer, Bronovo Hospital, the Hague, for the collection of the isolates.
Furthermore, we thank the colleagues from the research laboratory of the Department of Medical Microbiology of the MUMC for their laboratory support.
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Aarts, M. A., C. Brun-Buisson, D. J. Cook, A. Kumar, S. Opal, G. Rocker, T. Smith, J. L. Vincent, and J. C. Marshall. 2007. Antibiotic management of suspected nosocomial ICU-acquired infection: does prolonged empiric therapy improve outcome? Intensive Care Med. 33:1369-1378. [DOI] [PubMed] [Google Scholar]
- 2.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires-de-Sousa, M., T. Conceicao, and H. de Lencastre. 2006. Unusually high prevalence of nosocomial Panton-Valentine leukocidin-positive Staphylococcus aureus isolates in Cape Verde Islands. J. Clin. Microbiol. 44:3790-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aires-de-Sousa, M., and H. de Lencastre. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 40:101-111. [DOI] [PubMed] [Google Scholar]
- 5.Armand-Lefevre, L., R. Ruimy, and A. Andremont. 2005. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11:711-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., and R. S. Daum. 2007. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab. Investig. 87:3-9. [DOI] [PubMed] [Google Scholar]
- 7.Cuevas, O., E. Cercenado, A. Vindel, C. Castellares, J. Guinea, and E. Bouza. 2006. Molecular characterization of Staphylococcus aureus isolated in a nationwide prevalence study in Spain. Abstr. 16th Eur. Cong. Clin. Microbiol. Infect. Dis., Nice, France, abstr. P465. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland.
- 8.Deurenberg, R. H., E. Nulens, H. Valvatne, S. Sebastian, C. Driessen, J. Craeghs, E. De Brauwer, B. Heising, Y. J. Kraat, J. Riebe, F. S. Stals, T. A. Trienekens, J. Scheres, A. W. Friedrich, F. van Tiel, P. S. Beisser, and E. E. Stobberingh. 2009. Cross-border dissemination of methicillin-resistant Staphylococcus aureus, Euregio Meuse-Rhin Region. Emerg. Infect. Dis. 15:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deurenberg, R. H., and E. E. Stobberingh. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8:747-763. [DOI] [PubMed] [Google Scholar]
- 10.Deurenberg, R. H., C. Vink, C. Driessen, M. Bes, N. London, J. Etienne, and E. E. Stobberingh. 2004. Rapid detection of Panton-Valentine leukocidin from clinical isolates of Staphylococcus aureus strains by real-time PCR. FEMS Microbiol. Lett. 240:225-228. [DOI] [PubMed] [Google Scholar]
- 11.Deurenberg, R. H., C. Vink, G. J. Oudhuis, J. E. Mooij, C. Driessen, G. Coppens, J. Craeghs, E. De Brauwer, S. Lemmen, H. Wagenvoort, A. W. Friedrich, J. Scheres, and E. E. Stobberingh. 2005. Different clonal complexes of methicillin-resistant Staphylococcus aureus are disseminated in the Euregio Meuse-Rhine Region. Antimicrob. Agents Chemother. 49:4263-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep, B. A., H. A. Carleton, R. F. Chang, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 193:1495-1503. [DOI] [PubMed] [Google Scholar]
- 13.Diep, B. A., and M. Otto. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donker, G. A., R. H. Deurenberg, C. Driessen, S. Sebastian, S. Nys, and E. E. Stobberingh. 2009. The population structure of Staphylococcus aureus among general practice patients from The Netherlands. Clin. Microbiol. Infect. 15:137-143. [DOI] [PubMed] [Google Scholar]
- 15.Donnio, P. Y., F. Fevrier, P. Bifani, M. Dehem, C. Kervegant, N. Wilhelm, A. L. Gautier-Lerestif, N. Lafforgue, M. Cormier, and A. Le Coustumier. 2007. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob. Agents Chemother. 51:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnio, P. Y., L. Louvet, L. Preney, D. Nicolas, J. L. Avril, and L. Desbordes. 2002. Nine-year surveillance of methicillin-resistant Staphylococcus aureus in a hospital suggests instability of mecA DNA region in an epidemic strain. J. Clin. Microbiol. 40:1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand, G., M. Bes, H. Meugnier, M. C. Enright, F. Forey, N. Liassine, A. Wenger, K. Kikuchi, G. Lina, F. Vandenesch, and J. Etienne. 2006. Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France. J. Clin. Microbiol. 44:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutch Workingparty on Infection Prevention. 2004. Policy for methicillin-resistant Staphylococcus aureus. http://www.wip.nl/UK/free_content/Richtlijnen/MRSA(1).pdf. Dutch Workingparty on Infection Prevention, Leiden, The Netherlands. Accessed 5 August 2009.
- 19.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fossum, A. E., and G. Bukholm. 2006. Increased incidence of methicillin-resistant Staphylococcus aureus ST80, novel ST125 and SCCmecIV in the south-eastern part of Norway during a 12-year period. Clin. Microbiol. Infect. 12:627-633. [DOI] [PubMed] [Google Scholar]
- 21.Frenay, H. M., A. E. Bunschoten, L. M. Schouls, W. J. van Leeuwen, C. M. Vandenbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15:60-64. [DOI] [PubMed] [Google Scholar]
- 22.Gillaspy, A. F., J. M. Patti, F. L. Pratt, Jr., J. J. Iandolo, and M. S. Smeltzer. 1997. The Staphylococcus aureus collagen adhesin-encoding gene (cna) is within a discrete genetic element. Gene 196:239-248. [DOI] [PubMed] [Google Scholar]
- 23.Gomes, A. R., H. Westh, and H. de Lencastre. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 50:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goossens, H., M. Ferech, R. Vander Stichele, and M. Elseviers. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579-587. [DOI] [PubMed] [Google Scholar]
- 25.Hallin, M., O. Denis, A. Deplano, R. De Mendonca, R. De Ryck, S. Rottiers, and M. J. Struelens. 2007. Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey. J. Antimicrob. Chemother. 59:465-472. [DOI] [PubMed] [Google Scholar]
- 26.Holtfreter, S., D. Grumann, M. Schmudde, H. T. Nguyen, P. Eichler, B. Strommenger, K. Kopron, J. Kolata, S. Giedrys-Kalemba, I. Steinmetz, W. Witte, and B. M. Broker. 2007. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 45:2669-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huijsdens, X. W., B. J. van Dijke, E. Spalburg, M. G. van Santen-Verheuvel, M. E. Heck, G. N. Pluister, A. Voss, W. J. Wannet, and A. J. de Neeling. 2006. Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ip, M., R. W. Yung, T. K. Ng, W. K. Luk, C. Tse, P. Hung, M. Enright, and D. J. Lyon. 2005. Contemporary methicillin-resistant Staphylococcus aureus clones in Hong Kong. J. Clin. Microbiol. 43:5069-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 6:41-52. [DOI] [PubMed] [Google Scholar]
- 30.Katayama, Y., D. A. Robinson, M. C. Enright, and H. F. Chambers. 2005. Genetic background affects stability of mecA in Staphylococcus aureus. J. Clin. Microbiol. 43:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollef, M. H., and V. J. Fraser. 2001. Antibiotic resistance in the intensive care unit. Ann. Intern. Med. 134:298-314. [DOI] [PubMed] [Google Scholar]
- 32.Layer, F., B. Ghebremedhin, W. Konig, and B. Konig. 2006. Heterogeneity of methicillin-susceptible Staphylococcus aureus strains at a German University Hospital implicates the circulating-strain pool as a potential source of emerging methicillin-resistant S. aureus clones. J. Clin. Microbiol. 44:2179-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis, H. C., K. Molbak, C. Reese, F. M. Aarestrup, M. Selchau, M. Sorum, and R. L. Skov. 2008. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14:1383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsay, J. A. (ed.). 2008. Staphylococcus molecular genetics.
- 35.Lindsay, J. A., and M. T. Holden. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6:186-201. [DOI] [PubMed] [Google Scholar]
- 36.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 114:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melles, D. C., F. C. Tenover, M. J. Kuehnert, H. Witsenboer, J. K. Peeters, H. A. Verbrugh, and A. van Belkum. 2008. Overlapping population structures of nasal isolates of Staphylococcus aureus from healthy Dutch and American individuals. J. Clin. Microbiol. 46:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monecke, S., P. Slickers, M. J. Ellington, A. M. Kearns, and R. Ehricht. 2007. High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus. Clin. Microbiol. Infect. 13:1157-1164. [DOI] [PubMed] [Google Scholar]
- 39.Nubel, U., P. Roumagnac, M. Feldkamp, J. H. Song, K. S. Ko, Y. C. Huang, G. Coombs, M. Ip, H. Westh, R. Skov, M. J. Struelens, R. V. Goering, B. Strommenger, A. Weller, W. Witte, and M. Achtman. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 105:14130-14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nulens, E., E. E. Stobberingh, H. van Dessel, S. Sebastian, F. H. van Tiel, P. S. Beisser, and R. H. Deurenberg. 2008. Molecular characterization of Staphylococcus aureus bloodstream isolates collected in a Dutch University Hospital between 1999 and 2006. J. Clin. Microbiol. 46:2438-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsonnet, J., R. V. Goering, M. A. Hansmann, M. B. Jones, K. Ohtagaki, C. C. Davis, and K. Totsuka. 2008. Prevalence of toxic shock syndrome toxin 1 (TSST-1)-producing strains of Staphylococcus aureus and antibody to TSST-1 among healthy Japanese women. J. Clin. Microbiol. 46:2731-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Vazquez, M., A. Vindel, C. Marcos, J. Oteo, O. Cuevas, P. Trincado, V. Bautista, H. Grundmann, and J. Campos. 2009. Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of the aminoglycoside-modifying enzyme gene ant(4′)-Ia and the efflux pump genes msrA/msrB. J. Antimicrob. Chemother. 63:21-31. [DOI] [PubMed] [Google Scholar]
- 43.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz, F. J., C. R. MacKenzie, R. Geisel, S. Wagner, H. Idel, J. Verhoef, U. Hadding, and H. P. Heinz. 1997. Enterotoxin and toxic shock syndrome toxin-1 production of methicillin resistant and methicillin sensitive Staphylococcus aureus strains. Eur. J. Epidemiol. 13:699-708. [DOI] [PubMed] [Google Scholar]
- 45.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, D. S. 2004. Methicillin-resistant Staphylococcus aureus in a general intensive care unit. J. R Soc Med. 97:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tristan, A., T. Ferry, G. Durand, O. Dauwalder, M. Bes, G. Lina, F. Vandenesch, and J. Etienne. 2007. Virulence determinants in community and hospital methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 65(Suppl. 2):105-109. [DOI] [PubMed] [Google Scholar]
- 48.van Belkum, A., D. C. Melles, J. K. Peeters, W. B. van Leeuwen, E. van Duijkeren, X. W. Huijsdens, E. Spalburg, A. J. de Neeling, H. A. Verbrugh, and Dutch Working Party on Surveillance and Research of MRSA-SOM. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verhoef, J., D. Beaujean, H. Blok, A. Baars, A. Meyler, C. van der Werken, and A. Weersink. 1999. A Dutch approach to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:461-466. [DOI] [PubMed] [Google Scholar]
- 50.Wannet, W. J. B., X. W. Huijsdens, M. E. O. C. Heck, G. N. Pluister, M. G. van Santen-Verheuve, E. Spalburg, M. van Luit, T. Bosch, A. Haenen, E. W. Tiemersma, and A. J. de Neeling. 2007. MRSA in Nederlandse ziekenhuizen: surveillance-resultaten 2005-2006. Inf. Bull. 18:347-351. [Google Scholar]
- 51.Wielders, C. L., M. R. Vriens, S. Brisse, L. A. de Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2001. In-vivo transfer of mecA DNA to Staphylococcus aureus [corrected]. Lancet 357:1674-1675. [DOI] [PubMed] [Google Scholar]
- 52.Witte, W. 2004. International dissemination of antibiotic resistant strains of bacterial pathogens. Infect. Genet. Evol. 4:187-191. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]