Hospital-acquired infections caused by vancomycin-resistant enterococci (VRE) are increasing. The resistance of VRE to multiple antibiotics makes them clinically challenging. There are at least six phenotypes of VRE, including VanA, VanB, VanC, VanD, VanE, and VanG (2, 8). These phenotypes correspond with the genotypes vanA, vanB, vanC, vanD, vanE, and vanG. VanA and VanB phenotypes are common among VRE isolates. In general, vanA genotype VRE shows VanA phenotype with high-level resistance to both vancomycin and teicoplanin, whereas vanB genotype VRE shows VanB phenotype characterized by various levels of resistance to vancomycin but susceptibility to teicoplanin (2). Interestingly, vanA genotype VRE strains were recently found to be susceptible to teicoplanin in East Asia (Japan, South Korea, Taiwan, and even mainland China) and named VanB phenotype-vanA genotype VRE (3, 4, 6, 7, 11). Until now, the mechanism for this phenomenon has not been clear. Among these strains, point mutations in the key region of the vanS gene or impairment of vanY or vanZ in the Tn1546-like element were discovered (3, 4, 7, 11). Also, several vancomycin-heteroresistant Enterococcus faecium isolates with the vanA genotype have been reported since 2001 (1, 5). Several novel point mutations in the vanR, vanS, vanH, vanA, vanX, and vanY genes of Tn1546 were also discovered, resulting in amino acid replacements of VanR, VanS, and VanH. It was concluded that independent novel mutations can give rise to polymorphism, heteroresistance, and clonal diversity among VRE strains.
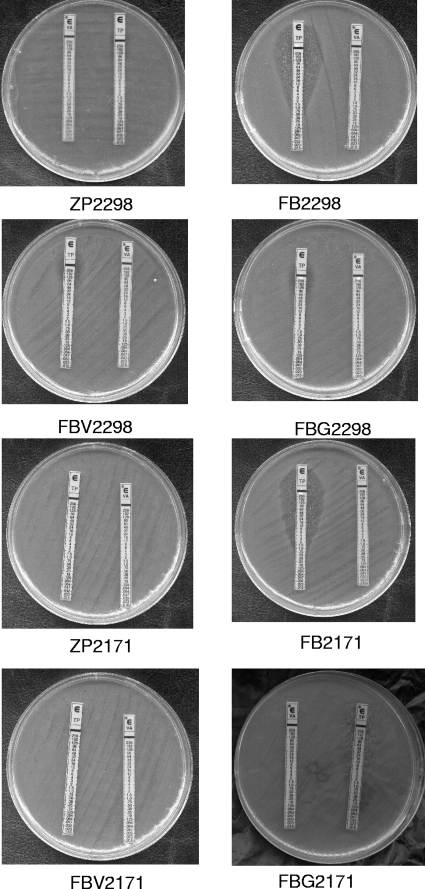
In the present study, we isolated two clinical VRE strains of E. faecium, ZP2298 and ZP2171, with a high level of resistance to vancomycin (MICs > 256 μg/ml) and teicoplanin (MICs of 96 and >256 μg/ml, respectively) from sputum samples from patients in a teaching hospital in mainland China. No prior exposure to glycopeptides was found in the isolates. Filter mating was performed using the two clinical VRE strains of E. faecium ZP2298 and ZP2171 as donors and E. faecium BM4105RF (Fusr Rifr) as the recipient as previously described (9). Transconjugants were selected on brain heart infusion (BHI) agar plates containing vancomycin (32 μg/ml) and rifampin (rifampicin) (256 μg/ml). After conjugation, the transconjugants of E. faecium ZP2298 and ZP2171, named E. faecium FB2298 and FB2171, respectively, exhibit high-level resistance to vancomycin (MICs > 256 μg/ml), but subcolonies were within the zone of inhibition around the teicoplanin Etest strip described for teicoplanin-heteroresistant isolates with teicoplanin MICs of 96 and >256 μg/ml, respectively (Fig. 1). The subcolonies, named E. faecium FBV2298 and FBV2171, were selected and showed high-level resistance to both vancomycin and teicoplanin. E. faecium FBV2171 maintained high level of resistance to vancomycin (MIC > 256 μg/ml) and was heteroresistant to teicoplanin after three serial passages on the BHI agar without vancomycin (named FBG2171). However, the resistance profile of E. faecium FBV2298 did not change after three serial passages on the BHI agar without vancomycin (named FBG2298). This is the first report of a teicoplanin-heteroresistant E. faecium isolate.
FIG. 1.
Vancomycin and teicoplanin Etest results for VREF isolates. The two clinical VREF strains of E. faecium, ZP2298 and ZP2171, had a high level of resistance to vancomycin and teicoplanin (MICs > 256 μg/ml). After conjugation, the transconjugants of these two strains of E. faecium, FB2298 and FB2171, exhibit high-level resistance to vancomycin (MICs > 256 μg/ml), with resistant subcolonies present in the clear zone of inhibition around the teicoplanin Etest strip, described for teicoplanin-heteroresistant isolates. The resistant subcolonies, E. faecium FBV2298 and FBV2171, had teicoplanin MICs of 96 and >256 μg/ml, respectively. E. faecium FBG2298 and FBG2171 strains were generated after three serial passages of E. faecium FBV2298 and FBV2171, respectively.
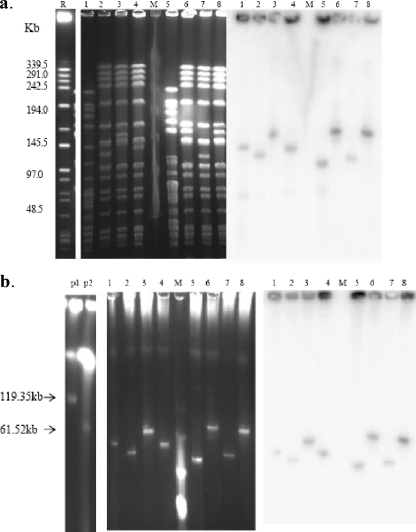
Pulsed-field gel electrophoresis (PFGE) of SmaI-digested genomic material from the recipient strain (E. faecium BM4145), transconjugant colonies, and variants (E. faecium FB2298, FB2171, FBV2298, FBV2171, FBG2298, and FBG2171) produced patterns with significant similarity, suggesting a genetic relationship (Fig. 2a). Minor differences (only one or two bands) were observed (Fig. 2a). It eliminated the possibility that mixed cultures were responsible for the findings observed. These patterns were distinct from those for the two clinical vancomycin-resistant E. faecium (VREF) strains (E. faecium ZP2298 and ZP2171) identified so far (Fig. 2a).
FIG. 2.
(a) PFGE of genomic DNA and hybridization profiles of the vanA gene. (b) Undigested PFGE to identify plasmids and hybridization profiles of the vanA gene. Lanes: R, the recipient strain, E. faecium BM4105RF (Fusr Rifr); 1, E. faecium ZP2298 (clinical VRE isolate); 2, E. faecium FBV2298, which showed teicoplanin-resistant subcolonies present in the clear zone of inhibition; 3, transconjugant E. faecium FB2298 (teicoplanin heteroresistant); 4, FBG2298, strain after three serial passages on the BHI agar without vancomycin from FBV2298; M, lambda ladder; 5, E. faecium ZP2171 (clinical VRE isolate); 6, E. faecium FBV2171, which showed teicoplanin-resistant subcolonies present in the clear zone of inhibition; 7, transconjugant E. faecium FB2171 (teicoplanin heteroresistant); 8, FBG2171, strain after three serial passages on the BHI agar without vancomycin from FBV2171; p1, plasmid R448 (119.35 kb); p2, plasmid R626 (61.52 kb). Preparations were run in 1% agarose under 6.0 V/cm for 22 h with a pulsing time linearly ramped from 3 to 20 s.
Undigested PFGE plugs were used to identify low- and high-molecular-weight plasmids. The vanA gene was the glycopeptide resistance determinant found in all of the VRE isolates by PCR. The vanA hybridization results of SmaI-digested PFGE absolutely corresponded with the plasmids identified by PFGE analysis of undigested DNA (Fig. 2a and b). Interestingly, the vanA-containing plasmids with different sizes were identified by undigested PFGE among these VREF strains, including E. faecium ZP2298, ZP2171, FB2298, FB2171, FBV2298, FBV2171, FBG2298, and FBG2171 (Fig. 2b). No plasmids were identified in E. faecium BM4145. However, it is difficult to find the different sizes of plasmids extracted by Qiagen kit by conventional electrophoresis (data not shown). It is suggested that the differences in the PFGE and undigested PFGE profiles were due to the change in size of plasmids containing the vanA gene.
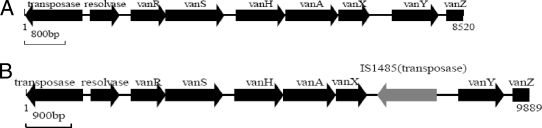
A more thorough investigation of the Tn1546-like elements harboring vanA in these isolates was performed by using PCR mapping and DNA sequencing (10). Sequence analysis of the van operon revealed a 1.5-kb IS1485 insertion element between the vanX and vanY genes in all the original and transconjugant VRE strains (Fig. 3). The locations of the IS1485 insertion in this region were identical, corresponding to bp 8698 of Tn1546 (GenBank accession no. M97297) with a 3-bp duplication of the target sequence (CTT). This is the first time the presence of IS1485 in the vanX-vanY intergenic region of the vanA gene cluster has been demonstrated. No point mutations in the vanR, vanS, vanH, vanA, vanX, and vanY genes were discovered. The nucleotide sequences of the Tn1546-like element were submitted to GenBank (GenBank accession no. EU599211). The vanA-containing transposons in the teicoplanin-heteroresistant isolates had no detectable changes compared to the teicoplanin-resistant isolates. However, it was difficult to amplify the vanZ genes from the teicoplanin-heteroresistant transconjugant VREF strains by PCR. Also, it was shown that the sizes of the plasmids containing the vanA gene changed after conjugation and serial passages. It is presumed that the resistance phenotypic changes may be due to the size changes of plasmids containing the van operon. However, we could not determine what change happened among these plasmids, which needs to be studied further. No independent novel mutations in the van operon previously reported in the polymorphism, heteroresistance, and clonal diversity of VRE strains were found (3, 4, 7, 11). These teicoplanin resistance phenotypic changes were observed during the conjugation experiment and serial passages. It is concluded that the teicoplanin resistance phenotype of VRE may change during natural conjugation and serial passages. The emergence of VanB phenotype-vanA genotype VRE isolates may be related to conjugation and serial passages. However, we yet do not know the mechanism for the emergence of teicoplanin-heteroresistant vanA genotype VREF isolates. Further investigation to elucidate other molecular mechanisms of these strains is under way.
FIG. 3.
Genetic maps of Tn1546 types of E. faecium isolates in this study. (A) Typical Tn1546. (B) Tn1546-like element with IS1485 inserted between vanXY. VRE isolates in this study included E. faecium ZP2298 and ZP2171, FB2298, FB2171, FBV2298, FBV2171, FBG2298, and FBG2171. The open reading frames are shown by boxes with arrows, with the arrow indicating the direction of transcription. The arrow on the gray box indicates the transcriptional orientation of the inserted IS1485 element.
Acknowledgments
This work was supported by a research grant from National Basic Research Program 973 of China (grant 2005CB523101) and by a research grant from the National Natural Science Foundation of China (grant NSFC30800035).
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Alam, M. R., S. Donabedian, W. Brown, J. Gordon, J. W. Chow, M. J. Zervos, and E. Hershberger. 2001. Heteroresistance to vancomycin in Enterococcus faecium. J. Clin. Microbiol. 39:3379-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu, L., B. Cao, Y. Liu, P. Guo, S. Song, R. Li, H. Dai, and C. Wang. 2009. A new Tn1546 type of VanB phenotype-vanA genotype vancomycin-resistant Enterococcus faecium isolates in mainland China. Diagn. Microbiol. Infect. Dis. 63:70-75. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto, Y., K. Tanimoto, Y. Ozawa, T. Murata, and Y. Ike. 2000. Amino acid substitutions in the VanS sensor of the VanA-type vancomycin-resistant Enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol. Lett. 185:247-254. [DOI] [PubMed] [Google Scholar]
- 5.Khan, S. A., K. Sung, S. Layton, and M. S. Nawaz. 2008. Heteroresistance to vancomycin and novel point mutations in Tn1546 of Enterococcus faecium ATCC 51559. Int. J. Antimicrob. Agents 31:27-36. [DOI] [PubMed] [Google Scholar]
- 6.Lauderdale, T. L., L. C. McDonald, Y. R. Shiau, P. C. Chen, H. Y. Wang, J. F. Lai, and M. Ho. 2002. Vancomycin-resistant enterococci from humans and retail chickens in Taiwan with unique VanB phenotype-vanA genotype incongruence. Antimicrob. Agents Chemother. 46:525-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, W. G., J. Y. Huh, S. R. Cho, and Y. A. Lim. 2004. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangements. Antimicrob. Agents Chemother. 48:1379-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKessar, S. J., A. M. Berry, J. M. Bell, J. D. Turnidge, and J. C. Paton. 2000. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob. Agents Chemother. 44:3224-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simjee, S., S. E. Manzoor, A. P. Fraise, and M. J. Gill. 2000. Nature of transposon-mediated high-level gentamicin resistance in Enterococcus faecalis isolated in the United Kingdom. J. Antimicrob. Chemother. 45:565-575. [DOI] [PubMed] [Google Scholar]
- 10.Simonsen, G. S., M. R. Myhre, K. H. Dahl, O. Olsvik, and A. Sundsfjord. 2000. Typeability of Tn1546-like elements in vancomycin-resistant enterococci using long-range PCRs and specific analysis of polymorphic regions. Microb. Drug Resist. 6:49-57. [DOI] [PubMed] [Google Scholar]
- 11.Song, J. H., K. S. Ko, W. S. Oh, S. Park, S. T. Heo, K. T. Kwon, S. Y. Ryu, K. R. Peck, and N. Y. Lee. 2006. High frequency of vancomycin-resistant Enterococcus faecium isolates with VanB phenotype and vanA genotype in Korean hospitals. Diagn. Microbiol. Infect. Dis. 56:401-406. [DOI] [PubMed] [Google Scholar]