Eight genotypes (A to H) of hepatitis B virus (HBV) have been described so far (11). Each genotype seems to have a restricted geographical distribution; therefore, it constitutes an invaluable tool in tracing HBV molecular evolution and the pattern and modes in which HBV spreads (5). Genotype H, the focus of the present work, was first considered to be confined to Central America since it was found in Nicaragua and Mexico (1), but it has more recently been reported in the United States and Japan (3, 7, 9). Thus, the natural history and evolution of HBV infection by genotype H are still obscure.
Serum samples corresponding to two Argentinean sisters diagnosed as chronically HBV infected were analyzed. Both tested positive for HBsAg and anti-HBe and negative for HBeAg. Case A, 36 years old, had normal alanine aminotransferase levels throughout the follow-up, and HBV DNA levels were below 5.0 log copies/ml. Needle liver biopsy was performed; grading inflammation was 1/18, and fibrosis was stage 0. Case B, 38 years old, was diagnosed in 2001 at the delivery of her baby. The viral load on the analyzed sample was 5.47 log copies/ml. Unfortunately, no additional information about this case was available.
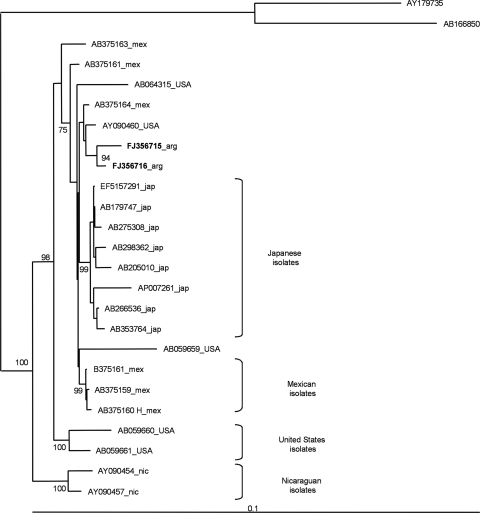
A phylogenetic analysis based on the entire genome sequences of serum samples revealed that sequences were grouped as genotype H (GenBank accession numbers FJ356715 and FJ356716) (not shown). Two clusters and several subclusters of genotype H were supported with high bootstrap values (Fig. 1).
FIG. 1.
Phylogram generated by maximum likelihood analysis of full-length sequences of HBV. Twenty-one full-length sequences of genotype H previously identified were included. The sequences are shown in the phylogram as the GenBank accession numbers followed by the country of origin (mex, Mexico; USA, United States; arg, Argentina; jap, Japan; nic, Nicaragua). The sequences with GenBank accession numbers AY179735 and AB166850 (genotypes F1b and F4, respectively) were used as an outgroup. The numbers at each node correspond to neighbor-joining bootstrap values obtained with 10,000 replicates.
Isolates FJ356715 and FJ356716 showed 35 unique nucleotide substitutions distributed along the four open reading frames (ORF) compared to those in the consensus full-length sequences of genotype H previously identified (Table 1). Substitutions were distributed along the entire genome, and 25 out of 35 substitutions were nonsynonymous, as follows: 8 in the core region, 4 in the S region, 2 in the X region, 10 in the polymerase (Pol) region, and 1 in the precore region. G1816T, affecting the precore initiation codon, abrogates the translation of the HBe antigen, whereas positions 1762/1764 and 1896 retained the wild-type nucleotides.
TABLE 1.
Unique nucleotide and amino acid substitutions in FJ356715 and FJ356716 isolates, in comparison with those in the consensus sequences of genotype Ha
| ORF | Type of regionb | Substitution(s) | Substitutions for: |
|
|---|---|---|---|---|
| Case 1 (FJ356716 isolate) | Case 2 (FJ356715 isolate) | |||
| Nonsynonymous | ||||
| Surface | D | C6T | —d | T125I |
| D | G210T | — | C193F | |
| Dc | A227G | — | I199V | |
| D | T231G | L200R | — | |
| X | S | C1653T | — | H94Y |
| Dc | G1816T | C148F | C148F | |
| Precore | Dc | G1816T | M1I | M1I |
| Core | S | T1961A, C1962A | — | S21N |
| S | T2012C | Y38H | Y38H | |
| S | T2020G | — | D40E | |
| S | A2038T | E46D | E46D | |
| S | T2079C | L60S | L60S | |
| S | C2100A | T67N | — | |
| S | A2171G | T91A | T91A | |
| S | A2200C | L100F | L100F | |
| Pol | D | A2323C | Q6P | — |
| D | A2326G | — | H7R | |
| S | A2736G | — | T144A | |
| S | G2740A | R145K | — | |
| D | T2892C | — | S196P | |
| Dc | A227G | — | N379S | |
| D | T493A | Y468N | — | |
| S | T853A | W588R | — | |
| S | T865A | — | S592T | |
| S | T887G | — | I599R | |
| Synonymous | ||||
| Pol/S | D | C562T | ||
| Pol/S | D | C724T | — | |
| Pol/S | D | C748T | ||
| Pol | S | A1284C | — | |
| Pol | S | C1287A | ||
| Pol | S | C1341T | ||
| Pol/X | D | A1383C | — | |
| Core | S | T1909C | ||
| Core | S | C1987T | — | |
| Core | S | A2032T | — | |
| Core | S | A2035C | ||
| Pol | S | T2529C | ||
Full-length genotype H sequences were retrieved from the 21 HBV strains reported (GenBank accession numbers AY090454, AY090457, AY090460, EF157291, AB179747, AB266536, AB275308, AB298362, AB205010, AP007261, AB353764, AB059659, AB064315, AB059660, AB059661, AB375164, AB375159, AB375163, AB375161, AB375162, and AB375160).
D, double codification region; S, single codification region.
Affecting two overlapped ORF.
—, mutation not detected.
In Argentina, the genotype prevalence varies for the different regions: in Buenos Aires city, genotypes A, D, and F are the most prevalent, whereas in the north region, the most prevalent one is genotype F (2, 10).
Here we report, for the first time in Argentina, two anti-HBe individuals infected with genotype H, a globally rare genotype. The two patients are sisters and have never been to Central America. The phylogenetic analysis showed well-defined clusters and subclusters from different geographical localizations and suggests that the diversification of genotype H is not a consequence of a recent emergence process, as has been recently proposed for genotype E in Africa (6, 8).
The anti-HBe-positive state is a late phase of the HBV chronic infection. There is scarce information about anti-HBe-positive patients infected with genotype H since most of the cases described were HBeAg-positive patients with either acute or chronic infections (3, 7, 9). The two cases described here showed G1816T, a rare substitution (4) and the only one that justified the anti-HBe phenotype at the molecular level.
Further surveys of HBV genotypes in the region are warranted to trace HBV molecular evolution and the pattern and modes in which HBV genotype H spreads.
Nucleotide sequence accession numbers.
The newly determined genotype H sequences are available in GenBank under accession numbers FJ356715 and FJ356716.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. [DOI] [PubMed] [Google Scholar]
- 2.França, P. H., J. E. González, M. S. Munné, L. H. Brandão, V. S. Gouvea, E. Sablon, and B. O. Vanderborght. 2004. Strong association between genotype F and hepatitis B virus (HBV) e antigen-negative variants among HBV-infected Argentinean blood donors. J. Clin. Microbiol. 42:5015-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumagai, I., K. Abe, T. Oikawa, A. Sato, S. Sato, R. Endo, et al. 2007. A male patient with severe acute hepatitis who was domestically infected with a genotype H hepatitis B virus in Iwate, Japan. J. Gastroenterol. 42:168-175. [DOI] [PubMed] [Google Scholar]
- 4.Laras, A., J. Koskinas, K. Avgidis, and S. J. Hadziyannis. 1998. Incidence and clinical significance of hepatitis B virus precore gene translation initiation mutations in e antigen-negative patients. J. Viral Hepat. 5:241-248. [DOI] [PubMed] [Google Scholar]
- 5.Miyakawa, Y., and M. Mizokami. 2003. Classifying hepatitis B virus genotypes. Intervirology 46:329-338. [DOI] [PubMed] [Google Scholar]
- 6.Mulders, M. N., V. Venard, M. Njayou, A. P. Edorh, A. O. Bola Oyefolu, M. O. Kehinde, J. J. Muyembe Tamfum, Y. K. Nebie, I. Maiga, W. Ammerlaan, F. Fack, S. A. Omilabu, A. Le Faou, and C. P. Muller. 2004. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J. Infect. Dis. 190:400-408. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima, A., M. Usui, T. T. Huy, N. K. Hlaing, N. Masaki, T. Sata, et al. 2005. Full-length sequence of hepatitis B virus belonging to genotype H identified in a Japanese patient with chronic hepatitis. Jpn. J. Infect. Dis. 58:244-246. [PubMed] [Google Scholar]
- 8.Odemuyiwa, S. O., M. N. Mulders, O. I. Oyedele, S. O. Ola, G. N. Odaibo, D. O. Olaleye, and C. P. Muller. 2001. Phylogenetic analysis of new hepatitis B virus isolates from Nigeria supports endemicity of genotype E in West Africa. J. Med. Virol. 65:463-469. [PubMed] [Google Scholar]
- 9.Ohnuma, H., A. Yoshikawa, H. Mizoguchi, and H. Okamoto. 2005. Characterization of genotype H hepatitis B virus strain identified for the first time from a Japanese blood donor by nucleic acid amplification test. J. Gen. Virol. 86:595-599. [DOI] [PubMed] [Google Scholar]
- 10.Piñeiro, Y., F. G. Leone, S. C. Pezzano, C. Torres, C. E. Rodríguez, M. Eugenia Garay, H. A. Fainboim, et al. 2008. Hepatitis B virus genetic diversity in Argentina: dissimilar genotype distribution in two different geographical regions; description of hepatitis B surface antigen variants. J. Clin. Virol. 42:381-388. [DOI] [PubMed] [Google Scholar]
- 11.Schaefer, S. 2007. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J. Gastroenterol. 13:14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]