Abstract
The pathogenesis of gastroduodenal diseases is related to the diversity of Helicobacter pylori strains. CagA-positive strains are more likely to cause gastric cancer than CagA-negative strains. Based on EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs at the carboxyl terminus corresponding to phosphorylation sites, H. pylori CagA is divided into East Asian CagA and Western CagA. The former type prevails in East Asia and is more closely associated with gastric cancer. The present study used full sequences of the cagA gene and CagA protein of 22 H. pylori strains in gastric cancer and peptic ulcer patients from Southern Vietnam to make a comparison of genetic homology among Vietnamese strains and between them and other strains in East Asia. A phylogenetic tree was constructed based on full amino acid sequences of 22 Vietnamese strains in accordance with 54 references from around the world. The cagA gene was found in all Vietnamese H. pylori strains. Twenty-one of 22 (95.5%) strains belonged to the East Asian type and had similar characteristics of amino acid sequence at the carboxyl terminus to other strains from the East Asian region. From evidence of East Asian CagA and epidemiologic cancerous lesions in Vietnam, H. pylori-infected Vietnamese can be classified into a high-risk group for gastric cancer, but further studies on the interaction among environmental and virulence factors should be done. Finally, phylogenetic data support that there is a Japanese subtype in the Western CagA type.
Helicobacter pylori is a gram-negative bacterium that infects about 50% of the world's population. Infections with H. pylori can result in chronic active gastritis and are a risk factor for peptic ulcers, gastric cancer and gastric mucosa-associated lymphoid tissue lymphoma (40, 52, 53, 56). The prevalence of H. pylori infections is not the same in different parts of the world. Recent studies reported that humans actually acquired H. pylori in the early days of their history, long before the migration of modern humans out of Africa, and the diverse distribution of H. pylori today is associated with waves of human migration in the past (19, 32, 36, 61, 62). The rate of H. pylori infections is high in Africa, East Asia, and South Asia; however, the incidence of gastric cancer is high in East Asia but not in South Asia or Africa; this may be explained partly by the diversity of H. pylori strains in these regions (62).
In cases of gastroduodenal diseases, especially gastric cancer, the pathogenesis involves three major factors: H. pylori virulence factors, host factors, and environmental factors (1, 10, 14, 16, 21, 34, 57). Two H. pylori virulence factors that have been focused on in many studies all around the world are VacA and CagA. VacA is encoded by the vacA gene and found in all H. pylori strains. The vacA gene is classified into three major genotypes: s1/m1, s1/m2, and s2/m2. s1/m1 strains produce higher levels of cytotoxin than s1/m2 or s2/m2 strains (2, 3). CagA is encoded by cagA, located in the cagPAI (pathogenicity island) region of the H. pylori genome, and the presence of this protein is a marker of cagPAI. Unlike the vacA gene, the cagA gene has been found in only 50% to 70% of H. pylori strains infecting Western populations: however, 80% to 100% of H. pylori strains from East Asia have the cagA gene, with the cagPAI region, in their genome (17, 49, 55, 61, 64, 65). Patients infected with H. pylori strains possessing CagA were at greater risk of developing gastric adenocarcinoma than those uninfected or infected with CagA-negative strains (11, 41). CagA is injected directly from H. pylori into intragastric epithelial cells via the type IV secretion system. The injected CagA mimics eukaryotic adaptor proteins that recruit multiple host signaling factors into protein complexes that target cellular junctions, cell proliferation, and actin-cytoskeletal rearrangements (54). The translocated CagA undergoes tyrosine phosphorylation by Src and Abl family kinases and binds to SHP-2 in the human gastric mucosa (45, 47, 48). The repeated EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs at the carboxyl-terminal end of CagA are the targets of tyrosine phosphorylation (8, 22, 24, 38, 63). CagA multimerization plays an important role in the pathophysiological activity of CagA in disturbing host cell functions via SHP-2 deregulation, and EPIYA polymorphisms of CagA greatly influence the magnitude and duration of phosphorylation-dependent CagA activity (37, 42). SHP-2, like its Drosophila melanogaster homolog Corkscrew, is known to play an important role in the mitogenic signal transduction that connects receptor tyrosine kinases and ras (20). It is possible that deregulation of SHP-2 by the translocation of CagA plays a role in the acquisition of a cellular-transformed phenotype at a relatively early stage in the carcinogenesis of gastric carcinoma. A recent study on generating CagA in transgenic mice has provided the first direct evidence of the role of CagA as a bacterium-derived oncoprotein that acts in mammals and further indicates the importance of tyrosine phosphorylation, which enables CagA to deregulate SHP-2, in the development of H. pylori-associated neoplasms (39). Based on characteristics of the EPIYA motif, the H. pylori CagA protein could be divided into a Western type and an East Asian type. The East Asian CagA protein exhibits stronger SHP-2-binding activity and so is more pathogenic than the Western CagA protein in H. pylori-infected patients (4, 7, 23, 37). Clinical data from East Asia, Japan, and South Korea indicated that the East Asian form of CagA was more closely related to persistent active inflammation, atrophic gastritis, and a higher risk of gastric cancer than the Western form (5, 6, 26, 44). It is quite clear that by studying the characteristics of H. pylori strains, especially the cagA gene and CagA protein, one can categorize H. pylori-infected patients into those at high risk of developing gastric cancer and those not.
Vietnam is a developing country located in Southeast Asia. However, according to historical and migrational evidence, the Vietnamese are more closely related to people from East Asia than people from South Asia. Gastric cancer is one of the five most common cancers in Vietnam, including lung, stomach, liver, recto-colon and naso-pharynx cancer in males. In females, it ranks third behind breast and cervical-uterine cancer. The prevalence of gastric cancer in Northern Vietnam is as high as that in China or Korea. Its prevalence in Southern Vietnam is lower but still higher than that in Thailand and South Asia (46, 50). A few studies reported that cagA was found in nearly 100% of H. pylori-infected Vietnamese (60, 61), but no studies have examined the type of CagA protein or the full sequence of cagA in Vietnamese patients. The present study reports the diverse characteristics of cagA and classification of CagA in H. pylori-infected patients from Southern Vietnam based on the full genomic cagA sequence.
MATERIALS AND METHODS
H. pylori strains.
Patients had abdominal symptoms and were clinically examined at the Department of Gastroenterology and further underwent upper gastrointestinal endoscopy in the Endoscopic Unit, Cho Ray Hospital, Ho Chi Minh City (formally Saigon), Vietnam. All patients were unrelated and Vietnamese in origin. Patients who had received nonsteroidal anti-inflammatory drugs were excluded from the study, and none of the patients had recently been prescribed antibiotics. Two biopsy specimens from the greater curvature of the gastric antral site and one from the greater curvature of the gastric fundic mucosa were obtained during the endoscopic procedure. Immediately, one specimen from the gastric antral mucosa was used for a rapid urea test. Afterward, one specimen each from the antral and fundic mucosae, respectively, was transferred for H. pylori culture at the Department of Microbiology, Cho Ray Hospital, Ho Chi Minh City, Vietnam. Only positive H. pylori strains were used in the cagA sequencing analysis. For patients who had gastric ulcers, additional biopsy specimens at the margin of the lesions and one at each site of antral and fundic mucosae were obtained for further histological examination. Biopsy specimens were fixed in 10% buffered formalin for the differential diagnosis of ulcers from gastric carcinoma. A total of 22 H. pylori strains from 22 patients were studied: 11 patients with gastric cancer, 2 patients with gastric ulcers, and 9 patients with duodenal ulcers. Fourteen patients were males and 8 were females, and the mean age was 53.09 ± 14.76 years (range, 34 to 84 years). This work was performed according to the principles of the Declaration of Helsinki and approved by the Ethics Committee.
Histological analysis.
Biopsy specimens were embedded in paraffin and stained with hematoxylin-eosin. The specimens were evaluated by an experienced histopathologist who had no knowledge of the clinico-endoscopic diagnosis. The histological slides were then examined by an independent expert, who had no information on the patients, for confirmation of the diagnosis.
Isolation and culture of H. pylori.
Gastric biopsy specimens from each patient were inoculated onto a Trypticase soy agar II-5% sheep blood plate and cultured for 3 to 5 days at 37°C under microaerobic conditions (5% O2, 15% CO2, and 80% N2). A single colony was picked from each primary culture plate, inoculated onto a fresh Trypticase soy agar II plate, and cultured under conditions described previously. H. pylori cells were harvested from each plate, transferred into a brucella broth liquid culture medium containing 10% fetal calf serum, and cultured for 3 days under the conditions described above. The liquid culture samples were stored at −80°C in 0.01 M phosphate-buffered saline containing 20% glycerol before being transferred to Japan. At the Department of Gastroenterology, Kobe University Graduate School of Medicine, the H. pylori strains were once again cultured as described above. Part of the liquid-cultured sample was stored at −80°C in 0.01 M phosphate-buffered saline containing 20% glycerol. DNA from each H. pylori isolate was extracted from the pellet of the liquid culture sample by the protease-phenol-chloroform method, suspended in 300 μl of a TE buffer (10 mM Tris-HCl, 1 mM EDTA), and stored at 4°C for the PCR analysis and nucleotide sequencing.
PCR and nucleotide sequencing of the entire cagA gene.
Primers for PCR amplification and direct sequencing of the entire coding region of cagA are listed in Table 1. The full-length cagA gene was amplified by PCR under the following conditions: 94°C for 1 min and 25 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 3 min, followed by 72°C for 10 min. PCR products were determined by 2% agarose gel electrophoresis. The gels were stained with ethidium bromide to determine the size of the cagA gene. PCR products then were purified with Centricon-100 concentrator columns (Amicon, Beverly, MA). H. pylori DNA was sequenced using the dideoxynucleotide chain termination method with a BigDye terminator v.3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) in an ABI Prism 3100-Avant analyzer (Applied Biosystems) according to the manufacturer's recommendations, using the primers marked in Table 1. The sequencing reaction was as follows: 96°C for 5 min and then 25 cycles of 96°C for 10s, 50°C for 5s, and 60°C for 4 min, followed by 4°C for 10 min. The full-length nucleotide sequence of cagA was constructed and translated into the amino acid sequence with the standard code using GENETYX software version 7.0 (Genetyx Corporation, Japan).
TABLE 1.
Oligonucleotide primers used for sequencing of the H. pylori cagA gene
| Primera | Sequence | Corresponding DNA sequenceb |
|---|---|---|
| cagA L2 (+) | 5′-AAGGAGAAACAATGACTAACGAAACTATTG-3′ | −11-19c |
| cagA L2 (−) | 5′-TCCTTTAAGATTTTTGGAAACCACCTTTTG-3′ | +4-3536 |
| cagA B1 (−)* | 5′-CTGCAAAAGATTGTTTGGCAGA-3′ | 520-499 |
| cagA M2 (+)* | 5′-ATACAAGGCTTACCGCCTG-3′ | 754-772 |
| cagA S2* | 5′-GGCAATGGTGGTCCTGGAGCTAGGC-3′ | 976-1000 |
| cagA M1 (−)* | 5′-GTAGCCACATTGTCGCCTTGTTGG-3′ | 1055-1032 |
| cagA C2 (+)* | 5′-GAATTGTCTGATAAACTTGAAA-3′ | 2059-2080 |
| cagA C2 (−)* | 5′-TTTGCTTGCGTTACCTTGCTG-3′ | 2306-2286 |
| cagA C3 (−)* | 5′-GCGTATGTGGCTGTTAGTAGCG-3′ | 3210-3189 |
| cagA 3′F1* | 5′-AAACCCTGAGTGGCTCAAGCTC-3′ | 3273-3294 |
*, primer used for sequencing.
Nucleotide positions in the HP0547 (cagA) gene of H. pylori 26695 (GenBank accession no. AE000569).
Product size of 3,576 bp.
Phylogenetic analysis.
To clarify the phylogenetic relationship between the 22 Vietnamese H. pylori strains of the present study and previously reported H. pylori strains worldwide, sequences of 54 reference strains of the full-length cagA gene in GenBank (accession numbers reported in Fig. 1) were downloaded. Of the 54 references, 32 were from Japan, including 14 strains with the Western type of CagA (Okinawa strains) and 18 strains with the East Asian type (Fukui strains) reported previously (64), 5 were from Hong Kong (China) and China, 2 were from India (South Asia), 4 were from Australia, 4 were from Europe, and 7 were from America. Full-length sequences of the cagA gene of Vietnamese H. pylori strains and references were aligned using the CLUSTAL X program. Genetic distances were estimated by Kimura's two-parameter method (30). Phylogenetic trees were constructed by the neighbor-joining method (43). To confirm the reliability of the phylogenetic trees, bootstrap resampling and reconstruction with 1,000 replicates were used. These analyses were carried out using MEGA software version 3.0 (31).
FIG. 1.
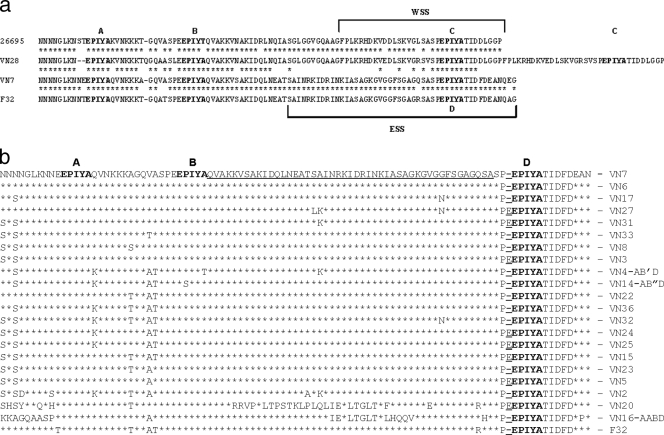
Alignment of the deduced amino acid sequence of the carboxy terminus of CagA in 22 H. pylori strains from Southern Vietnam, the representative Western strain 26695 (accession no. AE000569), and the Japanese strain F32 (accession no. AF202972). (a) Amino acid sequences in strains 26695, VN7 (EPIYA-ABD), VN28 (EPIYA-ABCC), and F32. (b) Eighteen Vietnamese strains had the EPIYA-ABD motif, strain F32 had the EPIYA-ABD motif, strain VN4 had the EPIYA-AB′D motif, strainVN14 had the EPIYA-AB″D motif, and VN16 had the EPIYA-AABD motif. The well-conserved region between motifs EPIYA-B and EPIYA-D is indicated by a double underline, and the completely conserved region is indicated by underlined letters. Strains VN20 and VN16 have marked diversity in amino acid sequence between EPIYA-B and EPIYA-D in comparison to other strains. In the EPIYA-D region, 8 of 21 (38.1%) strains have an additional residue, a glutamine (E), upstream of the EPIYA-D motif (indicated by single underlines).
Data deposition.
The full sequences of cagA in H. pylori strains from Southern Vietnam have been deposited in GenBank under the accession numbers (FJ798952 to FJ798973) shown in Table 2.
TABLE 2.
Genetic characteristics of cagA in 22 isolates of Vietnamese H. pylori
| Strain (n = 22) | Clinical diagnosisa | Geographic type | EPIYA typeb | Size of full-length cagA gene (bp) | Size of predicted protein (no. of aa) | GenBank accession no. |
|---|---|---|---|---|---|---|
| VN7 | GC | East Asian | ABD | 3,480 | 1,160 | FJ798957 |
| VN6 | GC | East Asian | ABD | 3,546 | 1,182 | FJ798956 |
| VN17 | GC | East Asian | ABD | 3,549 | 1,183 | FJ798962 |
| VN16 | DU | East Asian | AABD | 3,588 | 1,196 | FJ798961 |
| VN4 | DU | East Asian | AB′D | 3,558 | 1,186 | FJ798954 |
| VN23 | DU | East Asian | ABD | 3,525 | 1,175 | FJ798965 |
| VN27 | GC | East Asian | ABD | 3,555 | 1,185 | FJ798968 |
| VN33 | GC | East Asian | ABD | 3,546 | 1,182 | FJ798972 |
| VN2 | GU | East Asian | ABD | 3,579 | 1,193 | FJ798952 |
| VN5 | DU | East Asian | ABD | 3,549 | 1,183 | FJ798955 |
| VN15 | DU | East Asian | ABD | 3,552 | 1,184 | FJ798960 |
| VN31 | DU | East Asian | ABD | 3,549 | 1,183 | FJ798970 |
| VN24 | GC | East Asian | ABD | 3,549 | 1,183 | FJ798966 |
| VN25 | GU | East Asian | ABD | 3,549 | 1,183 | FJ798967 |
| VN20 | GC | East Asian | ABD | 3,549 | 1,183 | FJ798963 |
| VN3 | DU | East Asian | ABD | 3,582 | 1,194 | FJ798953 |
| VN8 | GC | East Asian | ABD | 3,543 | 1,181 | FJ798958 |
| VN36 | DU | East Asian | ABD | 3,549 | 1,183 | FJ798973 |
| VN22 | GC | East Asian | ABD | 3,540 | 1,180 | FJ798964 |
| VN14 | DU | East Asian | AB″D | 3,543 | 1,181 | FJ798959 |
| VN32 | GC | East Asian | ABD | 3,537 | 1,179 | FJ798971 |
| VN28 | GC | Western | ABCC | 3,657 | 1,219 | FJ798969 |
DU, duodenal ulcer; GU, gastric ulcer; GC, gastric cancer.
B′, EPIYT; B″, ESIYA.
RESULTS
Characteristic diversity of CagA.
The alignment of the deduced amino acid sequence in the 3′ region of the cagA gene among strains is shown in Fig. 1. The first and second EPIYA motifs (designated EPIYA-A and EPIYA-B, respectively) are present in almost all CagA proteins, whereas two major subtypes are observed around the third EPIYA motif. Western strains have the WSS (Western CagA-specific, SHP-2 binding sequence) in the third EPIYA motif (designated EPIYA-C) (23). The WSS contains D1, D2, and D3 motifs, as defined by Covacci et al. (15), or R1 and WSR (Western specific region) sequences as defined by Yamaoka et al. (58). Strain 26695 (AE000569) with the representative Western CagA type (EPIYA-ABC) is included in Fig. 1a. VN28 CagA was classified as type “ABCC” since it had two repeats of WSS. The amino acid sequence of CagA from H. pylori isolated in East Asian countries is quite different from that of Western CagA. Predominant East Asia CagA proteins do not have the WSS, but instead possess a distinct sequence which we designated ESS (East Asian CagA-specific, SHP-2-binding sequence) in the corresponding region (23). The ESS contains a JSR (Japanese specific region) sequence previously defined by Yamaoka et al. (58) and also possesses an EPIYA motif, denoted EPIYA-D. F32 (accession no. AF202972), with the representative East Asian type (EPIYA-ABD) is also included in Fig. 1a. VN7-CagA was classified as type “ABD” since it had one ESS. Figure 1b gives amino acid sequences and repeated EPIYA motifs in East Asian CagA. Variations are frequently found between the EPIYA-A motif and EPIYA-B motif. Between the EPIYA-B and EPIYA-D motifs, excluding strain VN20 with the EPIYA-ABD motif and strain VN16 with the EPIYA-AABD motif in Fig. 1b, the sequence of 48 amino acids indicated by double-underlined letters on panel b is quite conserved (QVAKKVSAKIDQLNEATSAINRKIDRINKIASAGKGVGGFSGAGGFSGAGQSAS), with only some variation, except in strains VN16 and VN20. In the EPIYA-D region, 8 of 21 (38.1%) strains have an additional residue, a glutamine (E), upstream of the EPIYA-D motif. This variation is specific for Vietnamese strains; however, there was no association between the variation and clinical outcome.
The Vietnamese H. pylori strains and characteristics of their cagA gene are listed in Table 2. The CagA protein in 21 of 22 (95.5%) strains belonged to the East Asian type. EPIYA-ABD motifs were found in 18 strains, and the EPIYA-AABD motif, EPIYA-AB′D motif, and EPIYA-AB″D motif were each found in one strain. As reported previously (64), B′ represents EPIYT-B, in which the A (alanine) residue is replaced by T (threonine), and B″ represents ESIYA-B, in which the P (proline) residue is replaced by S (serine). Only one H. pylori strain in the study had the Western type of CagA with the EPIYA-ABCC motif, meaning that it has two EPIYA-C motifs. The shortest sequence of the full-length coding region of cagA was 3,480 nucleotides in a strain (VN7) with the EPIYA-ABD motif, and the longest was 3,657 nucleotides in a strain (VN28) with the EPIYA-ABCC motif. The predicted size of CagA in 22 strains ranged from 1,160 to 1,219 amino acids.
The homology of nucleotide sequence and amino acid sequence is shown in Table 3. Within the East Asian CagA group, excluding strain VN32, the highest degree of homology was 99% and the lowest were 93% and 93.1% in full nucleotide sequences and in full amino acid sequences, respectively. Strain VN32 was located in between the East Asian CagA group and Western CagA group in the phylogenetic tree, as reported in Fig. 2, having levels of homology of 92.8% and 92.9% and the lowest degrees of homology—90.7% and 91%—compared with the other strains in the East Asian CagA group for nucleotide sequences and amino acid sequences, respectively. In a comparison between East Asian CagA strains and the Western CagA strain VN28, the highest degrees of homology in nucleotide and amino acid sequences were 90% and 90.3%, respectively, and the lowest were 83.8% for nucleotide sequences and 84.5% for amino acid sequences. A Japanese strain, F32, with the representative East Asian type (EPIYA-ABD) and strain 26695 with the representative Western CagA type (EPIYA-ABC) are included in Table 3. To compare the Japanese strain F32 and strain 26695 with Vietnamese strains, the highest and lowest degrees of homology for nucleotide sequences were 96.3% and 94.4% and 84.9% and 83.8%, respectively; those for amino acid sequences were 96.3% and 94.5% and 85.6% and 84.5%, respectively.
TABLE 3.
Analysis of cagA homology in Vietnamese H. pylori strains
| Strain | % Homology to straina: |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VN7 | VN6 | F32 | VN17 | VN16 | VN4 | VN23 | VN27 | VN33 | VN2 | VN5 | VN15 | VN31 | VN24 | VN25 | VN20 | VN3 | VN8 | VN36 | VN22 | VN14 | VN32 | VN28 | 26695 | |
| VN7 | 98.8 | 95.9 | 96.0 | 95.6 | 95.6 | 95.5 | 95.6 | 95.4 | 94.6 | 94.7 | 95.5 | 95.1 | 95.0 | 94.3 | 94.5 | 94.5 | 95.4 | 95.6 | 94.6 | 93.6 | 91.6 | 85.3 | 85.8 | |
| VN6 | 98.9 | 95.3 | 95.4 | 95.1 | 95.0 | 95.0 | 95.0 | 94.9 | 94.1 | 94.2 | 95.1 | 94.5 | 94.4 | 93.8 | 93.9 | 94.5 | 94.8 | 94.9 | 94.0 | 93.1 | 91.0 | 84.8 | 85.2 | |
| F32 | 95.9 | 95.2 | 95.6 | 95.6 | 95.5 | 95.7 | 95.8 | 96.3 | 95.0 | 95.3 | 95.6 | 95.2 | 95.2 | 94.5 | 94.8 | 94.6 | 95.6 | 95.3 | 95.3 | 94.3 | 91.7 | 85.6 | 84.4 | |
| VN17 | 96.0 | 95.3 | 95.6 | 95.7 | 95.8 | 95.9 | 96.6 | 96.1 | 95.2 | 95.1 | 95.8 | 95.4 | 95.9 | 95.1 | 95.1 | 95.2 | 95.3 | 95.7 | 94.5 | 94.5 | 92.3 | 85.2 | 85.8 | |
| VN16 | 95.6 | 95.0 | 95.5 | 95.7 | 96.3 | 95.8 | 96.1 | 96.3 | 94.9 | 94.8 | 95.7 | 95.2 | 95.6 | 94.9 | 94.7 | 94.8 | 95.6 | 95.8 | 94.3 | 93.9 | 92.6 | 85.7 | 86.2 | |
| VN4 | 95.5 | 94.9 | 95.5 | 95.7 | 96.3 | 96.0 | 96.3 | 96.5 | 95.4 | 94.6 | 96.3 | 95.8 | 96.0 | 95.4 | 95.0 | 94.9 | 95.6 | 95.9 | 94.8 | 94.5 | 92.3 | 86.0 | 86.5 | |
| VN23 | 95.5 | 94.9 | 95.7 | 95.9 | 95.7 | 95.9 | 97.2 | 96.9 | 95.8 | 95.6 | 96.4 | 95.8 | 96.3 | 95.6 | 95.7 | 95.5 | 96.0 | 96.0 | 95.1 | 94.6 | 92.4 | 85.8 | 85.6 | |
| VN27 | 95.6 | 94.9 | 95.8 | 96.6 | 96.1 | 96.4 | 97.2 | 96.9 | 95.9 | 95.7 | 96.2 | 96.3 | 96.3 | 95.6 | 95.6 | 95.8 | 95.7 | 96.1 | 94.9 | 94.5 | 92.9 | 86.1 | 86.6 | |
| VN33 | 95.4 | 94.8 | 96.3 | 96.1 | 96.2 | 96.4 | 96.9 | 96.9 | 95.8 | 95.6 | 96.8 | 96.5 | 96.8 | 96.2 | 95.7 | 95.9 | 96.3 | 96.5 | 95.1 | 95.1 | 92.5 | 85.4 | 85.8 | |
| VN2 | 94.5 | 94.0 | 95.0 | 95.2 | 94.8 | 95.3 | 95.8 | 95.9 | 95.8 | 95.1 | 95.6 | 95.6 | 95.3 | 94.7 | 94.7 | 94.7 | 94.7 | 95.4 | 94.5 | 94.1 | 91.4 | 84.9 | 85.2 | |
| VN5 | 94.7 | 94.1 | 95.2 | 95.1 | 94.7 | 94.5 | 95.6 | 95.6 | 95.6 | 95.0 | 95.1 | 94.9 | 95.5 | 95.0 | 94.7 | 94.5 | 95.2 | 95.0 | 93.9 | 93.7 | 91.2 | 84.8 | 85.1 | |
| VN15 | 95.5 | 95.0 | 95.6 | 95.8 | 95.6 | 96.3 | 96.4 | 96.2 | 96.8 | 95.6 | 95.0 | 96.1 | 95.9 | 95.3 | 95.0 | 95.4 | 96.0 | 96.2 | 95.5 | 94.3 | 92.0 | 85.4 | 85.6 | |
| VN31 | 95.0 | 94.4 | 95.2 | 95.4 | 95.2 | 95.8 | 95.8 | 96.3 | 96.5 | 95.6 | 94.9 | 96.0 | 95.6 | 94.9 | 94.8 | 95.2 | 95.4 | 95.4 | 94.7 | 93.9 | 91.8 | 85.0 | 85.4 | |
| VN24 | 95.0 | 94.3 | 95.2 | 95.8 | 95.6 | 96.0 | 96.2 | 96.3 | 96.9 | 95.3 | 95.4 | 95.9 | 95.5 | 99.0 | 96.3 | 95.8 | 95.9 | 95.9 | 94.4 | 94.4 | 92.4 | 85.5 | 86.0 | |
| VN25 | 94.2 | 93.6 | 94.4 | 95.0 | 94.9 | 95.4 | 95.6 | 95.6 | 96.2 | 94.6 | 94.9 | 95.2 | 94.8 | 99.0 | 95.7 | 95.1 | 95.3 | 95.3 | 93.9 | 93.9 | 91.9 | 84.9 | 85.2 | |
| VN20 | 94.4 | 93.8 | 94.8 | 95.0 | 94.7 | 95.0 | 95.6 | 95.6 | 95.7 | 94.6 | 94.7 | 94.9 | 94.7 | 96.2 | 95.6 | 94.9 | 95.0 | 95.2 | 94.0 | 93.7 | 91.6 | 85.0 | 85.4 | |
| VN3 | 94.5 | 94.5 | 94.5 | 95.1 | 94.7 | 94.8 | 95.4 | 95.8 | 95.9 | 94.7 | 94.5 | 95.4 | 95.2 | 95.7 | 95.0 | 94.8 | 95.1 | 95.0 | 93.8 | 93.5 | 91.5 | 84.6 | 85.1 | |
| VN8 | 95.4 | 94.7 | 95.6 | 95.3 | 95.6 | 95.5 | 96.0 | 95.7 | 96.3 | 94.7 | 95.2 | 96.0 | 95.4 | 95.9 | 95.2 | 95.0 | 95.0 | 95.8 | 95.0 | 94.2 | 91.9 | 85.1 | 85.7 | |
| VN36 | 95.5 | 94.8 | 95.3 | 95.6 | 95.7 | 95.9 | 96.0 | 96.1 | 96.5 | 95.4 | 95.0 | 96.1 | 95.4 | 95.9 | 95.2 | 95.1 | 94.9 | 95.8 | 95.6 | 94.3 | 92.4 | 85.6 | 86.0 | |
| VN22 | 94.5 | 93.9 | 95.2 | 94.4 | 94.2 | 94.7 | 95.0 | 94.8 | 95.1 | 94.4 | 93.8 | 95.4 | 94.7 | 94.3 | 93.8 | 93.9 | 93.8 | 95.0 | 95.6 | 93.2 | 91.2 | 84.5 | 85.2 | |
| VN14 | 93.5 | 93.0 | 94.2 | 94.5 | 93.8 | 94.4 | 94.5 | 94.5 | 95.1 | 94.0 | 93.6 | 94.3 | 93.9 | 94.4 | 93.8 | 93.5 | 93.4 | 94.1 | 94.2 | 93.0 | 91.7 | 84.6 | 84.5 | |
| VN32 | 91.4 | 90.7 | 91.5 | 92.1 | 92.5 | 92.1 | 92.2 | 92.8 | 92.3 | 91.2 | 91.0 | 91.9 | 91.6 | 92.8 | 91.7 | 91.4 | 91.3 | 91.7 | 92.3 | 91.0 | 91.4 | 90.3 | 88.8 | |
| VN28 | 84.7 | 84.1 | 84.9 | 84.6 | 85.1 | 85.4 | 85.2 | 85.5 | 84.7 | 84.3 | 84.1 | 84.8 | 84.4 | 84.9 | 84.2 | 84.3 | 83.9 | 84.4 | 85.0 | 83.8 | 83.9 | 90.0 | 94.5 | |
| 26695 | 85.3 | 84.6 | 85.9 | 85.2 | 85.6 | 85.9 | 85.0 | 86.1 | 85.2 | 84.6 | 84.4 | 85.0 | 84.8 | 85.4 | 84.6 | 84.7 | 84.4 | 85.1 | 85.5 | 84.5 | 83.8 | 88.4 | 94.4 | |
FIG. 2.
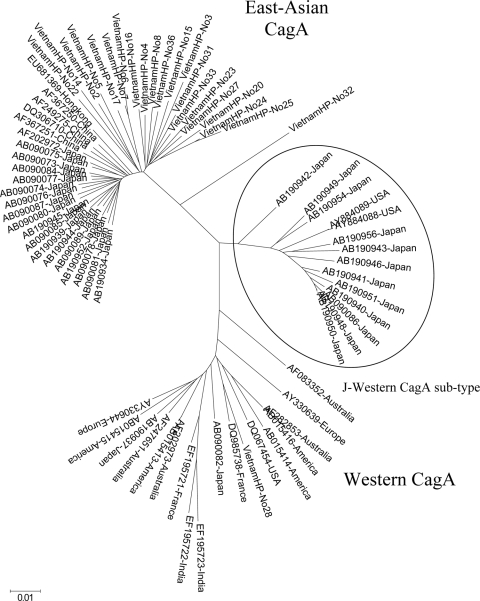
A phylogenetic tree constructed based on the complete amino acid sequence of H. pylori CagA protein in 22 isolates from Southern Vietnam (VietnamHP no. 7, 6, 17, 16, 4, 23, 27, 33, 2, 5, 15, 31, 24, 25, 20, 3, 8, 36, 22, 14, 32, 28 [given in the order shown in Tables 2 and 3]) compared with complete amino acid sequence of H. pylori CagA protein in 54 isolates from the GenBank database, including Japan (32 strains, 14 with the Western CagA type from Okinawa and 18 with the East Asian CagA type from Fukui), China (5 strains), India (2 strains), Australia (4 strains), Europe (4 strains), and America (7 strains). The length of the horizontal bar indicates the number of amino acid substitutions per site.
Phylogenetic tree analysis.
To more precisely determine the genotype of the CagA protein in Vietnamese H. pylori strains, a phylogenetic analysis based on full CagA amino acid sequences in 22 Vietnamese strains plus 54 reference strains from GenBank was carried out (Fig. 2). Twenty of 22 (90.9%) Vietnamese strains belonged to the East Asian type, in accordance with 18 Japanese strains, mainly from Fukui Province, and 5 strains from China. One of 22 (4.5%) Vietnamese strains (VN28) belonged to the Western type. From the results in Fig. 2, it is clear that all East Asian CagA strains from Japan, Vietnam, and China are located in the same cluster, except for one strain from Vietnam. However, all East Asian CagA Vietnamese strains were integrated closely at one site of the cluster and Japanese strains at another site of the cluster. VietnamHP-No32 was located in between the East Asian and Western cluster, though it has the EPIYA-ABD motif. Also even in Western type CagA strains, most strains from Japan (Okinawa strains) formed a different cluster from strains isolated in the West. We therefore believe that there is a Japanese subtype in the Western CagA type (J-Western CagA subtype).
DISCUSSION
H. pylori can survive long term outside of a host, suggesting it has flexible evolutionary strategies in accordance with environmental modifications and enabling bacterial transmission with important clinical manifestations (13). The geographical distribution of H. pylori has been associated with the migration of modern humans, and the prevalence of gastric cancer in the world not only is related to host and environmental factors but also is related to the diversity of H. pylori genomic genotypes (16, 19, 32, 36, 60, 62). H. pylori CagA is an oncoprotein that acts in mammals (39) and has a major role in the pathogenesis of gastroduodenal diseases in H. pylori-infected patients, especially in cases of gastric cancer (11, 14, 17, 38, 40, 41). Today, the so-called “paradox of gastric cancer” is particularly evident in Asian countries (46). The prevalence of H. pylori infections in Japan, Korea, and China is no higher than that in South Asian nations, but the incidence of gastric cancer is significantly higher. This can be partly explained by the diversity of H. pylori strains among Asian nations, especially the polymorphisms of EPIYA motifs in the carboxyl-terminal region of CagA, the Western CagA type (EPIYA-ABC), and East Asian CagA type (EPIYA-ABD) (4, 6, 7, 59-62, 64). East Asian CagA is more pathological than Western CagA (7, 22, 37, 44). H. pylori strains containing many EPIYA-C motifs at the 3′ end of Western CagA could be more likely to cause gastric cancer than strains with fewer EPIYA-C motifs (9).
The prevalence of gastric cancer in Vietnam is higher than that in other countries in the region such as Thailand, Indonesia, and the Philippines and also higher than that in South Asian nations such as India, Pakistan, and Nepal (7, 46, 50). The prevalence rates of gastric cancer in males and females in Northern Vietnam are 34.5/100,000 and 16.4/100,000, respectively. The prevalence rates in Southern Vietnam are 16.5/100,000 and 7.5/100,000, respectively (50). This means that the prevalence of gastric cancer in Northern Vietnam may be as high as that in China, Korea, and some regions of Japan. The prevalence of gastric cancer in Southern Vietnam is lower than that in Northern Vietnam, but higher than that in other Southeast Asian nations and in South Asia. According to studies of cancerous lesions among Asian-Pacific ethnicities in the United States, Vietnamese Americans have a high prevalence of gastric cancer (33, 35). Previous studies reported that the cagA gene was common in H. pylori from Vietnamese patients both in Vietnam and overseas (60, 61). However, to our knowledge, the present study is the first not only to detail polymorphisms at the carboxyl terminal end of CagA but also to fully sequence the cagA gene and CagA protein in H. pylori strains from Vietnamese patients. It was found that all of the H. pylori strains having cagA in the genome and most others (95.5%) as well had the East Asian form of CagA. As in East Asian countries such as Japan, Korea, and China, the data from the present study and previous studies (60, 61) indicate that most CagA proteins in Vietnamese H. pylori strains are of the East Asian type. The full-length open reading frame of cagA in all Vietnamese H. pylori strains is over 3,400 (3,480 to 3,657) nucleotides long, with a predicted protein of 1,160 to 1,219 amino acids. In the present study, in the East Asian CagA group, except for one strain located between the East Asian type and Western type in the phylogenetic analysis, Vietnamese H. pylori strains had a high degree of homology in both the nucleotide sequence of cagA and the amino acid sequence of CagA (93 to 99%).
Consistent with previous studies of populations from East Asia (26, 27, 51), the present study found that the amino acid sequences of the EPIYA-B and EPIYA-D motifs are quite conserved. A sequence of 18 amino acid sequences (AINRKIDRINKIASAGKG) in the conserved region was designated East Asian-specific immunogens for immunohistochemical genotyping of cagA in previous studies (27, 51). Most of the Vietnamese H. pylori strains with the East Asian form of CagA have similar characteristics of amino acid sequences in the region to other strains from the East Asian region (Japan and Korea), so the immunohistochemical genotyping for CagA protein reported in previous studies (27, 51) can be applied to H. pylori strains from Vietnam. However, in the present study, CagA in Vietnamese H. pylori strains was classified based on the full sequence of the cagA gene. This method not only is a very good way of genotyping but also can count how many EPIYA motifs are present at the carboxyl-terminal end of CagA, since the number of EPIYA motifs is associated with clinical outcome in H. pylori-infected patients (9, 23).
An increasing number of studies have carried out a phylogenetic analysis based on nucleotide sequences of H. pylori strains in different parts of the world. However, when examining the cagA gene, most phylogenetic studies have analyzed the 3′ region or 5′ region only, with or without nucleotide sequences of the vacA gene or other housekeeping genes (12, 18, 28, 29, 61). Some studies from Japan that carried out a phylogenetic analysis based on full genomic sequences of the cagA gene or cagPAI region were reported, but unfortunately they focused mainly on intergroup examinations of Japanese H. pylori strains (5, 64) or had very few H. pylori reference strains from other countries (25). The phylogenetic tree in the present study was based on full nucleotide sequences of cagA in 22 strains from Vietnam in comparison with 54 reference strains from East Asia and many parts of the world. So the present study has analyzed the largest number of full nucleotide sequences of cagA published to date. The data show that except for one East Asian CagA strain from Vietnam located between East Asian CagA and Western CagA, all East Asian CagA strains (Japan, Vietnam, China, and Hong Kong) are found in the same cluster with a slightly different location of Vietnamese strains and Japanese strains in the cluster. Vietnamese strains are very closely located at one site, and Japanese strains are very closely located at another site. As the number of H. pylori-infected patients is huge, we hope that in the future, more full-nucleotide sequences of the East Asian CagA type will be published, and more diverse characteristics of East Asian CagA will be found. It is interesting that in Western type CagA strains, most strains from Japan (Okinawa strains) formed a different cluster from strains isolated in the West. We therefore believe that there is a Japanese subtype in the Western CagA type (J-Western CagA). Since H. pylori with the Western CagA type is found in many regions of the world, it is better to carry out further studies on different features of intergroups of Western CagA protein involved in the pathogenesis of gastroduodenal diseases and gastric cancer.
Evidently, East Asian CagA binds more strongly to SHP-2 than Western CagA (6, 23, 37), and H. pylori-infected patients with East Asian CagA have a higher risk of gastric cancer (4, 7, 26, 44). The present study found that CagA is present in all H. pylori strains from Southern Vietnam and mostly (95.5%) of the East Asian type; this observation supports the notion that H. pylori-infected Southern Vietnamese can be considered at high risk of developing gastric cancer. However, the prevalence rates of H. pylori infections are ∼50% in Southern Vietnam and 70% in Northern Vietnam, yet the prevalence rates of gastric cancer are different between these two regions (50). This cannot simply be explained by the prevalence of H. pylori infection. There are about 54 ethnic groups in Vietnam, but more than 84% of the population are of the majority King's group. Most data in studies of H. pylori-infected Vietnamese are from King's people. Further studies examining environmental factors in H. pylori-infected Vietnamese are important. Weather and humidity, lifestyle, and dietary factors such as kinds of vegetables and fruits are quite different between Northern and Southern Vietnam.
In conclusion, the present study found that cagA is present in all H. pylori strains from Southern Vietnam. Most of the CagA proteins in Vietnamese strains of H. pylori are of the East Asian type and have three EPIYA motifs (A, B, and D) in the carboxyl-terminal region. Vietnamese strains contain the same characteristic diversity of amino acid sequence between EPIYA-B and EPIYA-D motifs as H. pylori strains from East Asia, excluding a very small proportion with very diverse amino acid sequences in this region. Finally, the phylogenetic analysis supported that there is a Japanese subtype in the Western CagA type (J-Western CagA).
Acknowledgments
We thank Sueyoshi Kumiko for technical assistance.
This work was supported by a Grant-in-Aid for G-COE Research and by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Amieva, M. R., and E. M. El-Omar. 2008. Host-bacterial interaction in Helicobacter pylori infection. Gastroenterology 134:306-323. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C., P. Cao, R. M. Peek, M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., R. M. Peek, K. T. Tham, T. L. Cover, and M. J. Blaser. 1997. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology 112:92-99. [DOI] [PubMed] [Google Scholar]
- 4.Azuma, T., A. Yamakawa, S. Yamazaki, K. Fukuta, M. Ohtani, Y. Ito, M. Dojo, Y. Yamazaki, and M. Kuriyama. 2002. Correlation between variation of the 3′ region of the cagA gene in Helicobacter pylori and diseases outcome in Japan. J. Infect. Dis. 186:1621-1630. [DOI] [PubMed] [Google Scholar]
- 5.Azuma, T., A. Yamakawa, S. Yamazaki, M. Ohtani, Y. Ito, A. Muramatsu, H. Suto, Y. Yamazaki, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Distinct diversity of the cag pathogenicity island among Helicobacter pylori strains in Japan. J. Clin. Microbiol. 42:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azuma, T., S. Yamazaki, A. Yamakawa, M. Ohtani, A. Muramatsu, H. Suto, Y. Ito, M. Dojo, Y. Yamazaki, M. Kuriyama, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Association between diversity in the Src homology 2 domain-containing tyrosine phosphatase biding site of Helicbacter pylori CagA protein and gastric atrophy and cancer. J. Infect. Dis. 189:820-827. [DOI] [PubMed] [Google Scholar]
- 7.Azuma, T. 2004. Helicobacter pylori CagA protein variation associated with gastric cancer in Asia. J. Gastroenterol. 39:97-103. [DOI] [PubMed] [Google Scholar]
- 8.Backert, S., S. Moese, M. Selbach, V. Brinkmann, and T. F. Meyer. 2001. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol. Microbiol. 42:631-644. [DOI] [PubMed] [Google Scholar]
- 9.Basso, D., C. F. Zambom, D. P. Letley, A. Stranges, A. Marchet, J. L. Rhead, S. Schiavon, G. Guariso, M. Ceroti, D. Nitti, M. Rugge, M. Plebani, and J. C. Atherton. 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135:91-99. [DOI] [PubMed] [Google Scholar]
- 10.Beevers, D. G., G. Y. H. Lip, and A. Blann. 2004. Salt intake and Helicobacter pylori infection. J. Hypertens. 22:1475-1477. [DOI] [PubMed] [Google Scholar]
- 11.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 12.Caroll, I. M., N. Ahmed, S. M. Beesley, A. A. Khan, S. Ghousunnissa, C. A. Morain, C. M. Habibullah, and C. J. Smyth. 2004. Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients. J. Med. Microbiol. 53:669-677. [DOI] [PubMed] [Google Scholar]
- 13.Cellini, L., R. Grande, E. Di Campli, S. Di Bartolomeo, M. Di Giulio, T. Traini, and O. Trubiani. 2008. Characterization of an Helicobacter pylori environmental strain. J. Appl. Microbiol. 105:761-769. [DOI] [PubMed] [Google Scholar]
- 14.Correa, P. 1992. Human gastric carcinogenesis: a multistep and multifactoral process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 52:6735-6740. [PubMed] [Google Scholar]
- 15.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, and N. Figura. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 17.Cover, T. L., Y. Glupczynski, A. P. Lage, A. Burette, M. K. R. Tummuru, G. I. Perez-Perez, and M. J. Blaser. 1995. Serologic detection of infection with cagA+ Helicobacter pylori strains. J. Clin. Microbiol. 33:1496-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devi, S. M., I. Ahmed, P. Francalacci, M. A. Hussain, Y. Akhter, A. Alvi, L. A. Sechi, F. Megraud, and N. Ahmed. 2007. Ancestral European roots of Helicobacter pylori in India. BMC Genomics 8:184-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Kazowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 229:1582-1585. [DOI] [PubMed] [Google Scholar]
- 20.Freeman, R., J. Plutzky, and B. G. Neel. 1992. Identification of a human src homology 2-containing protein-tyrosine-phosphatase: a putative homolog of Drosophila corkscrew. Proc. Natl. Acad. Sci. USA 89:11239-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halminton, J. P., and S. J. Meltzer. 2006. A review of the genomics of gastric cancer. Clin. Gastroenterol. Hepatol. 4:416-425. [DOI] [PubMed] [Google Scholar]
- 22.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama. 2002. SHP-2 tyrosine phosphatase as intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 23.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA 29:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higashi, H., K. Yokoyama, Y. Fujii, S. Ren, H. Yuasa, I. Saadat, N. Murata-Kamiya, T. Azuma, and M. Hatakeyama. 2005. EPIYA motif is a membrance-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J. Biol. Chem. 280:23130-23137. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino, F. B., K. Katayama, K. Watanabe, S. Takahashi, H. Uchimura, and T. Ando. 2000. Heterogeneity found in the cagA gene of Helicobacter pylori from Japanese and non-Japanese isolates. J. Gastroenterol. 35:890-897. [DOI] [PubMed] [Google Scholar]
- 26.Jones, K. R., Y. M. Joo, S. Jang, Y. J. Yoo, H. S. Lee, I. S. Chung, C. H. Olsen, J. M. Whitmire, D. S. Merrell, and J. H. Cha. 2009. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J. Clin. Microbiol. [Epub ahead of print.] doi: 10.1128/JCM.02330-08. [DOI] [PMC free article] [PubMed]
- 27.Kamada, R., T. Uchida, Y. Tsukamoto, L. T. Nguyen, N. Hijiya, K. Matsuura, M. Kodama, T. Okomoto, K. Murakami, T. Fujioka, S. Yanagisawa, and M. Moriyama. 2008. Genotyping of the cagA gene of Helicobacter pylori on immunohistochemistry with East Asian CagA-specific antibody. Pathol. Int. 58:218-225. [DOI] [PubMed] [Google Scholar]
- 28.Kauser, F., M. A. Hussain, I. Ahmed, S. Srinivas, S. M. Devi, A. A. Majeed, K. R. Rao, A. A. Khan, L. A. Cechi, and N. Ahmed. 2005. Comparative genomics of Helicobacter pylori isolates recovered from ulcer disease patients in England. BMS Microbiol. 5:32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauser, F., M. A. Hussain, I. Ahmed, N. Ahmad, A. Habeeb, A. A. Khan, and N. Ahmed. 2005. Comparing genomes of Helicobacter pylori strains from the high-altitude desert of Ladakh, India. J. Clin. Microbiol. 43:1538-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, S., T. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 32.Linz, B., F. Balloux, Y. Moodley, A. Manica, H. Liu, P. Roumagnac, D. Falush, C. Stamer, F. Prugnolle, S. W. van der Merwe, Y. Yamaoka, D. Y. Graham, E. Perez-Trallero, T. Wadstrom, S. Suerbaum, and M. Achtman. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken, M., M. Olsen, M. S. Chen, A. Jemal, M. Thun, V. Cokkinides, D. Deapen, and E. Ward. 2007. Cancer incidence, mortality, and associated risk factors among Asian Americans of Chinese, Filipino, Vietnamese, Korean, and Japanese ethnicities. CA Cancer J. Clin. 57:190-205. [DOI] [PubMed] [Google Scholar]
- 34.Miki, K., and Y. Urita. 2007. Using serum pepsinogens wisely in a clinical practice. J. Dig. Dis. 8:8-14. [DOI] [PubMed] [Google Scholar]
- 35.Miller, B. A., K. C. Chu, B. F. Hankey, and L. A. Ries. 2008. Cancer incidence and mortality pattern among specific Asian and Pacific Islander populations in the US. Cancer Causes Control 19:227-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moodley, Y., B. Linz, Y. Yamaoka, H. M. Windsor, S. Breurec, J. Y. Wu, A. Maady, S. Bernhoft, J. M. Thiberge, S. Phuanukoonnon, G. Jobb, P. Siba, D. Y. Graham, B. J. Marshall, and M. Achtman. 2009. The peopling of the Pacific from a bacterial perspective. Science 323:527-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naito, M., T. Yamazaki, R. Tsutsumi, H. Higashi, K. Onoe, S. Yamazaki, T. Azuma, and M. Hatakeyama. 2006. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 130:1181-1190. [DOI] [PubMed] [Google Scholar]
- 38.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 39.Ohnishi, N., H. Yuasa, S. Tanaka, H. Sawa, M. Miura, A. Matsui, H. Higashi, M. Musashi, K. Iwabuchi, M. Suzuki, G. Yamada, T. Azuma, and M. Hatakeyama. 2008. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoetic neoplasms in mouse. Proc. Natl. Acad. Sci. USA 105:1003-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1170-1171. [DOI] [PubMed] [Google Scholar]
- 41.Parsonnet, J., G. D. Friedman, N. Orentreich, and H. Vogelman. 1997. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 40:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren, S., H. Higashi, H. Lu, T. Azuma, and M. Hatakeyama. 2006. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J. Biol. Chem. 281:32344-32352. [DOI] [PubMed] [Google Scholar]
- 43.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 44.Satomi, S., A. Yamakawa, S. Matsunaga, R. Masaki, T. Inagaki, T. Okuda, H. Suto, Y. Ito, Y. Yamazaki, M. Kuriyama, Y. Keida, H. Kutsumi, and T. Azuma. 2006. Relationship between the diversity of the cagA gene of Helicobacter pylori and gastric cancer in Okinawa, Japan. J. Gastroenterol. 41:668-673. [DOI] [PubMed] [Google Scholar]
- 45.Selbach, M., S. Moese, C. R. Hauck, T. F. Meyers, and S. Backert. 2002. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277:6775-6778. [DOI] [PubMed] [Google Scholar]
- 46.Singh, K., and U. C. Ghoshal. 2006. Causal role of Helicobacter pylori infection in gastric cancer: an Asian enigma. World J. Gastroenterol. 12:1346-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein, M., F. Bagnoli, R. Halenbeck, R. Rappuoli, W. J. Fantl, and A. Covacci. 2002. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43:971-980. [DOI] [PubMed] [Google Scholar]
- 48.Tammer, I., S. Brandt, R. Hartig, W. Konig, and S. Backert. 2007. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology 132:1309-1319. [DOI] [PubMed] [Google Scholar]
- 49.Tan, H. J., A. M. Rizal, M. Y. Rosmadi, and K. L. Goh. 2005. Distribution of Helicobacter pylori cagA, cagE and vacA in different ethnic groups in Kuala Lumpur, Malaysia. J. Gastroenterol. Hepatol. 20:589-594. [DOI] [PubMed] [Google Scholar]
- 50.Truong, B. X., V. T. C. Mai, H. H. Hai, T. Long, N. K. Trach, and T. Azuma. 2008. Helicobacter pylori infection and gastric cancer situation in Northern and Southern Vietnam. Vietnamese J. Gastroenterol. 12:5-11. [Google Scholar]
- 51.Uchida, T., R. Kanada, Y. Tsukamoto, N. Hijiya, K. Matsuura, S. Yano, S. Yokoyama, T. Kishida, M. Kodama, K. Murakami, T. Fujioka, and M. Moriyama. 2007. Immunohistochemical diagnosis of the cagA-gene genotype of Helicobacter pylori with anti-East Asian CagA-specific antibody. Cancer Sci. 98:521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]
- 53.Warren, J. R., and B. J. Marshall. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1273-1275. [PubMed] [Google Scholar]
- 54.Wessler, S., and S. Backert. 2008. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 16:397-405. [DOI] [PubMed] [Google Scholar]
- 55.Wong, B. C. Y., Y. Yin, D. E. Berg, H. H. X. Xia, J. Z. Zhang, W. H. Wang, W. M. Wong, X. S. Y. Tang, and S. K. Lam. 2001. Distribution of distinct vacA, cagA and iceA alleles in Helicobacter pylori in Hong Kong. Helicobacter 6:317-324. [DOI] [PubMed] [Google Scholar]
- 56.Wotherspoon, A. C., C. Ortiz-Hidalgo, M. R. Falzon, and P. G. Isaacson. 1991. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 338:1175-1176. [DOI] [PubMed] [Google Scholar]
- 57.Yamada, S., T. Matsuhisa, L. Makonkawkeyoon, S. Chaidath, S. Kato, and N. Matsukura. 2006. Helicobacter pylori infection in combination with the serum pepsinogen I/II ratio and interleukin-1β-511 polymorphisms are independent risk factors for gastric cancer in Thais. J. Gastroenterol. 41:1169-1177. [DOI] [PubMed] [Google Scholar]
- 58.Yamaoka, Y., H. M. El-Zimaity, O. Gutierrez, N. Figura, J. G. Kim, T. Kodama, K. Kashima, and D. Y. Graham. 1999. Relationship between the cagA 3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology 117:342-349. [DOI] [PubMed] [Google Scholar]
- 59.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaoka, Y., H. M. Malaty, M. O. Osato, and D. Y. Graham. 2000. Conservation of Helicobacter pylori genotypes in different ethnic groups in Houston, Texas. J. Infect. Dis. 181:2083-2086. [DOI] [PubMed] [Google Scholar]
- 61.Yamaoka, Y., E. Orito, M. Mizokami, O. Gutierrez, N. Saitou, T. Kodama, M. S. Osato, J. G. Kim, F. C. Ramirez, V. Mahachai, and D. Y. Graham. 2002. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 517:180-184. [DOI] [PubMed] [Google Scholar]
- 62.Yamaoka, Y., M. Kato, and M. Asaka. 2008. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern. Med. 47:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamazaki, S., A. Yamakawa, Y. Ito, M. Ohtani, H. Higashi, M. Hatakeyama, and T. Azuma. 2003. The CagA protein of Helicobacter pylori is translocated into epithelial cells and binds to SHP-2 in human gastric mucosa. J. Infect. Dis. 187:334-337. [DOI] [PubMed] [Google Scholar]
- 64.Yamazaki, S., A. Yamakawa, T. Okuda, M. Ohtani, H. Suto, Y. Ito, Y. Yamazaki, Y. Keida, H. Higashi, M. Hatakeyama, and T. Azuma. 2005. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J. Clin. Microbiol. 43:3906-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou, W., S. Yamazaki, A. Yamakawa, M. Ohtani, Y. Ito, Y. Keida, H. Higashi, M. Hatakeyama, J. Si, and T. Azuma. 2004. The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS. Immunol. Med. Microbiol. 40:81-87. [DOI] [PubMed] [Google Scholar]