Abstract
Hemotropic mycoplasmas (hemoplasmas) are the causative agents of infectious anemia in several mammalian species. Their zoonotic potential has recently been substantiated by the identification of a feline hemoplasma isolate in an immunocompromised human patient. Although species-specific diagnostic molecular methods have been developed, their application as screening tools is limited due to the species diversity of hemoplasmas. The goals of this study were to develop a universal hemoplasma screening assay with broad specificity based on the SYBR green PCR principle, to compare the assay with hemoplasma-specific TaqMan PCR, and to analyze potential tick vectors and human blood samples to address the zoonotic potential. The newly developed PCR assay based on the 16S rRNA gene amplified feline, canine, bovine, porcine, camelid, and murine hemoplasmas, as well as Mycoplasma penetrans and Mycoplasma pneumoniae. The lower detection limit for feline and canine hemoplasmas was 1 to 10 copies/PCR. The assay exhibited 98.2% diagnostic sensitivity and 92.1% diagnostic specificity for feline hemoplasmas. All 1,950 Ixodes ticks were PCR negative, suggesting that Ixodes ticks are not relevant vectors for the above-mentioned hemoplasma species in Switzerland. None of the 414 blood samples derived from anemic or immunocompromised human patients revealed a clear positive result. The SYBR green PCR assay described here is a suitable tool to screen for known and so-far-undiscovered hemoplasma species. Positive results should be confirmed by specific TaqMan PCR or sequencing.
Hemotropic mycoplasmas, also known as hemoplasmas, are small, pleomorphic, cell wall-free bacteria that have been detected in the blood of various mammalian species (17). Originally classified as Haemobartonella and Eperythrozoon species within the order Rickettsiales, these organisms have recently been reclassified within the genus Mycoplasma (17, 19, 21, 27).
Hemotropic mycoplasmas are clinically relevant as causative agents of acute, life-threatening hemolytic anemia in infected animals. Some animals, however, develop only mild clinical signs or remain asymptomatic. Many cofactors, such as gender, age, immune status, or coinfection with other pathogenic agents, have been proposed to be involved in the development of disease (10, 16, 25, 32). It is thought that animals become chronic, asymptomatic carriers after infection, although clearance of the infectious agents from the host blood has been reported (26, 32, 33).
To date, hemoplasmas have been documented in numerous mammalian species (Table 1). The close relationship between the feline hemoplasma “Candidatus Mycoplasma turicensis” and rodent hemoplasmas and the similarity of feline and canine hemoplasmas suggest potential interspecies transmission of these agents (24, 33). This is especially important since hemotropic mycoplasmas are thought to be transmitted by blood-sucking arthropods such as ticks, fleas, and lice, given that these agents have been hypothesized to exhibit zoonotic potential. Some authors have described organisms with morphological similarities to hemotropic mycoplasmas in the blood of human patients (3, 7, 14, 22, 37). Furthermore, a recently published report demonstrating the molecular detection of a feline hemoplasma species in an immunocompromised human patient further substantiated the zoonotic potential of these agents (6).
TABLE 1.
Oligonucleotide sequences of the forward and reverse primers used in the universal SYBR green PCR assay and mismatches of the primer sequences with published hemotropic Mycoplasma 16S rRNA gene sequences
| Namea | Host species | Sequence (5′ → 3′) | |||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SYBR_For | A | G | C | A | A | T | R | C | C | A | T | G | T | G | A | A | C | G | A | T | G | A | A | ||||||||||||||||||||||||||
| SYBR_Rev1 | T | G | G | C | A | C | A | T | A | G | T | T | T | G | C | T | G | T | C | A | C | T | T | ||||||||||||||||||||||||||
| SYBR_Rev2 | G | C | T | G | G | C | A | C | A | T | A | G | T | T | A | G | C | T | G | T | C | A | C | T | |||||||||||||||||||||||||
| M. hemofelis | Felids | A | T | ||||||||||||||||||||||||||||||||||||||||||||||
| “Ca. Mycoplasma hemominutum” | Felids | A | A | ||||||||||||||||||||||||||||||||||||||||||||||
| “Ca. Mycoplasma turicensis” | Felids | G | T | ||||||||||||||||||||||||||||||||||||||||||||||
| M. hemocanis | Dog | A | T | ||||||||||||||||||||||||||||||||||||||||||||||
| “Ca. Mycoplasma hematoparvum” | Dog | A | C | A | |||||||||||||||||||||||||||||||||||||||||||||
| M. suis | Swine | A | C | A | |||||||||||||||||||||||||||||||||||||||||||||
| M. wenyonii | Cow | A | C | A | |||||||||||||||||||||||||||||||||||||||||||||
| “Ca. Mycoplasma hemobovis” | Cow | A | T | ||||||||||||||||||||||||||||||||||||||||||||||
| M. ovis | Sheep | A | C | A | |||||||||||||||||||||||||||||||||||||||||||||
| M. hemomuris | Rat, mouse | T | G | G | T | ||||||||||||||||||||||||||||||||||||||||||||
| M. coccoides | Rat, mouse | G | T | ||||||||||||||||||||||||||||||||||||||||||||||
| M. erythrodidelphis | Opossum | A | C | A | |||||||||||||||||||||||||||||||||||||||||||||
| “Ca. Mycoplasma hemolamae” | Lama, alpaca | A | C | A | |||||||||||||||||||||||||||||||||||||||||||||
| “Ca. Mycoplasma kahanei” | Squirrel monkey | A | C | G | A | ||||||||||||||||||||||||||||||||||||||||||||
For the hemoplasma 16S rRNA GenBank accession number, see Materials and Methods.
No in vitro cultivation system has been established to cultivate these organisms outside their hosts, and diagnosis of infection relies mainly upon the molecular detection of hemoplasmal genes in blood or tissue samples. Specific conventional and quantitative real-time TaqMan PCR systems have been established to diagnose hemotropic mycoplasmas (5, 11, 13, 18, 20, 25, 32). These assays provided the initial insight into the epidemiology and pathogenesis of hemoplasmas; however, conventional PCR is laborious and prone to carryover of PCR amplicons. Real-time TaqMan PCR assays, on the other hand, allow quantification, have minimal risk of amplicon carryover (being closed-tube systems), and are highly specific due to the use of a third labeled oligonucleotide. However, because of their high specificity, these assays are unlikely to detect novel hemoplasma species.
Real-time SYBR green PCR assays combine the advantages of conventional and real-time PCR methods. They employ two primers and a dye (SYBR green) in a closed-tube system. The SYBR green principle allows for quantification, and its specificity is less restrictive than that of TaqMan PCR assays. Furthermore, melting curves that differentiate between various PCR amplicons can be generated after PCR amplification.
The goals of this study were to develop a universal hemoplasma screening assay based on the SYBR green PCR principle, to compare the assay to specific TaqMan PCR assays, and to screen potential tick vectors and blood samples from anemic or immunocompromised human patients to address zoonotic potential and elucidate the possible occurrence of human hemotropic mycoplasmas by means of molecular methods.
MATERIALS AND METHODS
Samples.
For optimization of the SYBR green PCR assay and comparison with specific TaqMan PCR assays, nucleic acid (NA) samples from 99 felids (42 uninfected, 15 singly infected, and 42 coinfected with feline hemoplasmas) were included. The samples were obtained from 35 specific-pathogen-free cats (4), 19 Swiss pet cats (32), and 45 African lions (Panthera leo) (35). The pet cats and lions had been analyzed for the presence of feline hemoplasmas by specific real-time TaqMan PCR assays (32, 35). The lions were included in the study because they are commonly coinfected with feline hemoplasmas (35).
To address the zoonotic potential of hemoplasmas, NA samples from a total of 1,950 Ixodes ticks collected from the vegetation in the area around Zurich, Switzerland, by the cloth-dragging method were included (34). The arthropods were mechanically disrupted with sterile scalpel blades and homogenized in a Mixer Mill MM 300 device (Retsch GmbH, Haan, Germany), and NA was extracted with a MagNA Pure LC TNA isolation kit (Roche Diagnostics, Rotkreuz, Switzerland) as described previously (34). After NA extraction, the samples were subjected to an 18S rRNA gene real-time PCR assay as described previously (34) to confirm the presence of amplifiable NA and exclude PCR inhibition.
Furthermore, EDTA-anticoagulated blood samples from 414 anonymous human patients were used, including 200 blood samples collected from immunocompromised patients from Zimbabwe infected with human immunodeficiency virus (HIV) and 214 blood samples from immunocompromised or anemic patients from Switzerland. NA was purified from 100 μl of human blood using the MagNaPure LC TNA isolation external lysis protocol (Roche Diagnostics) with a final elution volume of 100 μl. To monitor cross-contamination, negative controls consisting of 100 μl of phosphate-buffered saline were prepared concurrently with each batch of 15 samples.
SYBR green real-time PCR primer design.
To design primers for a universal hemoplasma SYBR green PCR assay, the 16S rRNA genes of the following hemotropic Mycoplasma species were retrieved from GenBank and aligned using the GCG Wisconsin Package (Accelrys GmbH, Munich, Germany) and ClustalW (29): M. haemofelis (accession no. DQ157160), “Candidatus Mycoplasma haemominutum” (DQ157149), “Candidatus Mycoplasma turicensis” (DQ464421), M. haemocanis (EF416568), “Candidatus Mycoplasma haematoparvum” (EF416569), M. suis (AY492086), M. wenyonii (DQ641256), “Candidatus Mycoplasma haemobovis” (EF616468), M. ovis (AF338268), M. haemomuris (U82963), M. coccoides (AY171918), M. erythrodidelphis (AF178676), “Candidatus Mycoplasma haemolamae” (AF306346), and “Candidatus Mycoplasma kahanei” (AF338269). One forward and two reverse primers were designed using Primer Express software v2.0 (Applied Biosystems, Rotkreuz, Switzerland) (Table 1). The two reverse primers were used as a 1:1 mixture in the SYBR green PCR.
SYBR green real-time PCR primer optimization.
For SYBR green PCR optimization, a primer matrix containing forward or reverse primer concentration combinations of 50 nM, 300 nM, and 900 nM was assessed using plasmids containing the nearly full-length 16S rRNA genes of M. haemofelis, “Candidatus Mycoplasma haemominutum,” and “Candidatus Mycoplasma turicensis” as target templates (32, 33) or no-template controls (NTC). The three target templates (positive controls) were chosen because each represents a member of the three distinct phylogenetic clusters of hemoplasmas (17). The reaction mixture was composed of 12.5 μl of 2× SYBR green PCR master mix (Applied Biosystems), 50 to 900 nM concentrations of the forward and reverse primers, and 5 μl of NA template brought to a total volume of 25 μl with water. Assays were performed using an ABI Prism 7700 sequence detection system (Applied Biosystems). The SYBR green PCR protocol comprised 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After the PCR run, dissociation was performed with the following thermal profile: 95°C for 15 s, 60°C for 20 s, an increase from 60°C to 95°C for 20 min, and finally 95°C for 15 s. The minimum primer concentration demonstrating the maximum difference in fluorescence intensity (ΔRn) was determined. ΔRn was calculated by the ABI Prism 7700 sequence detection system as described previously (2).
SYBR green real-time PCR master mix optimization.
Using the optimized primer concentration and the PCR cycle conditions mentioned above, the SYBR green PCR master mix (Applied Biosystems) was compared to two additional SYBR green PCR master mixes: Power SYBR green PCR master mix (Applied Biosystems) and QuantiTect SYBR green master mix (Qiagen, Hombrechtikon, Switzerland). For this purpose, cats singly infected (n = 8) or coinfected (n = 3) with the three feline hemoplasmas and hemoplasma-uninfected cats (n = 11), based on the results of specific TaqMan PCR, were selected from the above-mentioned samples and run with the three different master mixes. In each PCR run, positive controls and NTC were included. The diagnostic accuracy was calculated (diagnostic accuracy [%] = number of correctly classified samples/number of tested samples) (23), and the master mix demonstrating maximal diagnostic accuracy was selected for further analyses.
SYBR green real-time PCR specificity and sensitivity.
Using the optimized primer concentration and master mix, NA samples from the following bacteria were used to determine the specificity of the SYBR green PCR assay: M. haemofelis, “Candidatus Mycoplasma haemominutum,” “Candidatus Mycoplasma turicensis,” M. haemocanis, “Candidatus Mycoplasma haematoparvum,” M. suis, M. wenyonii, “Candidatus Mycoplasma haemobovis,” “Candidatus Mycoplasma haemolamae,” M. coccoides, M. pneumoniae, M. penetrans, M. equigenitalium, M. argini, M. agalactiae, Chlamydophila felis, Pasteurella multocida, and Cytauxoon felis.
To determine the sensitivity of the assay for canine and feline hemoplasmas, recently published linearized plasmid standards containing the cloned 16S rRNA genes of M. haemofelis, “Candidatus Mycoplasma haemominutum,” “Candidatus Mycoplasma turicensis,” and “Candidatus Mycoplasma haematoparvum” were used (30, 32, 33). The amplification efficiency was calculated as R = 101/−slope − 1 (15).
Comparison of the SYBR green PCR assay with specific real-time TaqMan assays.
For comparison with the universal SYBR green PCR assay, specific real-time TaqMan PCR assays for the detection of M. haemofelis, “Candidatus Mycoplasma haemominutum,” and “Candidatus Mycoplasma turicensis” were performed as previously described (32, 33). Of the above-mentioned NA samples, 93 were used for comparison. They comprised samples from 38 uninfected cats, 15 singly infected cats, and 40 cats coinfected with feline hemoplasmas, as assessed by the specific TaqMan PCR assays.
Statistical evaluation.
For primer concentration optimization, the ΔRns of the two primer groups (group 1, primer combinations containing 50 nM; group 2, primer combinations containing 300 nM or 900 nM) were compared using the nonparametric Mann-Whitney U test. Differences with a P value of <0.05 were considered significant.
RESULTS
SYBR green PCR assay primer design.
Forward and reverse primers were designed based on published hemotropic Mycoplasma 16S rRNA gene sequences (Table 1). Sequence alignment revealed up to two mismatches in the forward or reverse primer sequence when aligned with those of known hemoplasma species (Table 1). No mismatches were located near the 3′ ends of the primer sequences.
SYBR green PCR assay optimization.
For all three feline hemoplasmas, all primer combinations containing concentrations of 50 nM resulted in significantly lower ΔRn values than for the remaining primer combinations containing only 300 and/or 900 nM (M. haemofelis, P = 0.016; “Candidatus Mycoplasma haemominutum,” P = 0.016; and “Candidatus Mycoplasma turicensis,” P = 0.016). Among the four giving high ΔRn values, the minimum primer concentration was selected; thus, final primer concentrations of 300 and 300 nM were used for further testing. Three master mixes were then evaluated, and the diagnostic accuracy was assessed (Table 2). Based on the highest diagnostic accuracy of the SYBR green PCR master mix (86%), it was used for further testing.
TABLE 2.
Diagnostic accuracy of the three tested SYBR green PCR master mixes compared to feline hemoplasma-specific TaqMan PCR
| Master mix | SYBR green PCR result | No. of samples with TaqMan PCR: |
Diagnostic accuracy (%) | |
|---|---|---|---|---|
| Positive | Negative | |||
| SYBR green PCR master mix | Positive | 9 | 1 | 86 |
| Negative | 2 | 10 | ||
| Power SYBR green PCR master mix | Positive | 10 | 4 | 77 |
| Negative | 1 | 7 | ||
| QuantiTect SYBR green master mix | Positive | 11 | 7 | 68 |
| Negative | 0 | 4 | ||
SYBR green PCR specificity and sensitivity.
All 10 hemotropic mycoplasmas tested were amplified using the SYBR green PCR assay. M. pneumoniae and M. penetrans, two mycoplasmas that are closely related to the hemotropic Mycoplasma group, were also amplified, whereas the remaining agents listed in Materials and Methods revealed threshold cycle (CT) values in the range of those for the NTC.
The lower detection limit for the tested feline and canine standards ranged from 1 to 10 copies/PCR. Amplification efficiencies were calculated using the same threshold and baseline for all four standard curves (threshold, 0.283; baseline, 3 to 10). Amplification efficiencies were ≥92.0%.
Melting curve analysis of SYBR green PCR products.
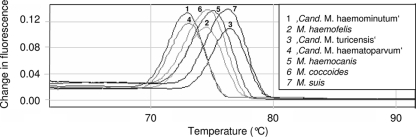
In animals infected with a single hemoplasma species, the melting temperature (Tm) was distinctly different among the three feline, two canine, and two bovine hemoplasmas (Table 3 and Fig. 1). In hemoplasma-coinfected animals, species differentiation was not possible due to Tm variability among the amplification products.
TABLE 3.
Tms of the tested hemotropic Mycoplasma species as assessed by melting curve analyses
| Species | Tm (°C)a |
|---|---|
| M. hemofelis | 74.5-76.0 |
| “Candidatus Mycoplasma hemominutum” | 73.0-74.5 |
| “Candidatus Mycoplasma turicensis” | 76.0-77.5 |
| M. hemocanis | 75.0 |
| “Candidatus Mycoplasma hematoparvum” | 73.0-74.0 |
| M. suis | 76.5 |
| M. wenyonii | 76.5 |
| “Candidatus Mycoplasma hemobovis” | 74.0 |
| M. coccoides | 74.5 |
| “Candidatus Mycoplasma hemolamae” | 73.5-74.0 |
When multiple samples were analyzed, the melting temperature range is specified.
FIG. 1.
Melting curve analysis after hemoplasma SYBR green PCR of samples from animals infected singly with a feline, canine, murine, or porcine hemoplasma species. Each curve represents one PCR amplicon and depicts the change in fluorescence during a continuous increase in temperature. The temperature demonstrating the peak change in fluorescence represents the Tm.
Nonspecific product formation.
When nonspecific product formation was found, it was commonly observed in the NTC rather than in the uninfected samples. The CT values ranged from 34.4 to 39.9, corresponding to <10 copies/PCR. Because the Tm reported for primer dimers (about 75°C) (2) is in the range of the Tms for the different hemoplasmas (Table 3), differentiation of primer dimers and specific product formation by melting curve analysis was not possible.
Comparison of the universal SYBR green PCR assay with specific TaqMan PCR assays.
NA samples extracted from 93 felines were used for comparison. Using SYBR green PCR, 54 out of 55 infected and 35 out of 38 uninfected samples were correctly identified (Table 4). The SYBR green assay exhibited 98.2% diagnostic sensitivity and 92.1% diagnostic specificity compared to feline hemoplasma-specific TaqMan PCR assays. The one “Candidatus Mycoplasma haemominutum”-infected sample that was not detected by the SYBR green assay was obtained from a Swiss pet cat. By quantitative real-time TaqMan PCR, this animal had a hemoplasma blood load of 1,960 copies/ml of blood. The three hemoplasma-uninfected samples that displayed a positive result in SYBR green PCR but a negative result in specific TaqMan PCR assays were obtained from two specific-pathogen-free cats and one African lion.
TABLE 4.
Comparison of the results obtained by universal hemoplasma SYBR green PCR and feline hemoplasma-specific TaqMan PCR
| TaqMan PCR-positive hemoplasma species | Total no. of samplesa | No. SYBR green PCRb: |
|
|---|---|---|---|
| Positive | Negative | ||
| M. hemofelis | 5 | 5 | 0 |
| “Candidatus Mycoplasma hemominutum” | 8 | 7 | 1 |
| “Candidatus Mycoplasma turicensis” | 2 | 2 | 0 |
| M. hemofelis-“Candidatus Mycoplasma hemominutum” | 5 | 5 | 0 |
| “Candidatus Mycoplasma hemominutum”-“Candidatus Mycoplasma turicensis” | 10 | 10 | 0 |
| M. hemofelis-“Candidatus Mycoplasma hemominutum”-“Candidatus Mycoplasma turicensis” | 25 | 25 | 0 |
| Total | |||
| Positive | 55 | 54 | 1 |
| Negative | 38 | 3 | 35 |
| Total no. | 93 | 57 | 36 |
Number of samples uninfected, singly infected, or coinfected with feline hemoplasmas, as assessed by specific TaqMan PCR assays.
Results based on melting curve analysis.
Ticks.
In order to assess whether hemoplasmas could be found in the most common tick in Switzerland, Ixodes ricinus, and pose a potential zoonotic risk, 1,950 unfed Ixodes ticks were analyzed. All NA samples extracted from ticks tested 18S rRNA PCR positive (CT values of <27), confirming the presence of amplifiable NA and the absence of relevant PCR inhibition. In SYBR green PCR, all 1,950 samples revealed PCR-negative results for hemoplasma species.
Human blood samples.
None of the 414 human blood samples revealed clear positive results. Five blood samples from immunocompromised patients from Switzerland exhibited CT values that were slightly below 40 (range from 39.2 to 39.9, corresponding to <1 copy/PCR); however, CT values in this range were also observed for some NTC. The high CT values did not allow for further analysis, e.g., by sequencing.
DISCUSSION
The universal SYBR green PCR assay described here was designed as a quantitative, closed-tube method for inexpensive and rapid screening of samples for hemotropic Mycoplasma species. The assay was shown to amplify all 10 hemoplasma species tested, including feline, canine, bovine, porcine, camelid, and murine hemoplasmas. To the best of our knowledge, no other hemoplasma PCR assay published thus far has been able to amplify that many different hemoplasma species. For feline hemoplasmas, the assay exhibited 98% diagnostic sensitivity. Because high assay sensitivity is a prerequisite for the detection of infections at low prevalence, our assay represents an excellent hemoplasma screening method. Recently, the number of reported hemoplasma species in mammals has steadily increased, and the universal SYBR green PCR assay, with its broad specificity, represents an important tool to simplify and boost the search for other, thus-far-unknown hemoplasma species.
The recent identification of the feline hemoplasma M. haemofelis in an immunocompromised HIV-positive patient in Brazil represents the first report of human hemoplasma infection based on molecular methods (6). This discovery supports the hypothesis of the zoonotic potential of these agents and underscores the importance of searching for hemoplasma species in humans using assays with broad specificity. Through testing of human blood samples from Switzerland and Zimbabwe, patients living in different climate zones and social environments were included in the present study. All human patients were either anemic or immunocompromised and therefore were assumed to be at risk of hemoplasma infection. Furthermore, a remarkably high hemoplasma prevalence was recently reported in cats in South Africa and dogs in Sudan (12, 28, 36), and hemoplasmas were detected in lions and in ticks collected from lions in Tanzania (8, 35). Nonetheless, none of the human blood samples tested here were PCR positive.
Melting curve analysis is commonly used to differentiate amplicons after SYBR green PCR. The Tm depends on both the PCR amplicon size and the GC content. In the present study, melting curve analysis revealed different Tm values for the three feline, two canine, and two bovine hemoplasmas. The Tm value was unpredictable, however, in samples containing two or more hemoplasma species, suggesting that species differentiation in coinfected animals is not feasible. Similar results have recently been reported for other SYBR green PCR assays (9). Since coinfection with different hemoplasma species is very common (20, 32, 35, 36), species-specific TaqMan PCR assays or sequencing of PCR products remains a prerequisite for specification of the hemoplasma species present in coinfected animals.
In some instances, low-level nonspecific product formation was observed. According to the manufacturer's instructions (2), the weakly positive signals probably represent primer dimer formation or elongation of primers nonspecifically bound to genomic DNA. Because the Tms for the tested hemoplasma species ranged from 73.0°C to 77.5°C, discrimination of primer dimer formation (with a reported Tm of 75°C) and specific product formation by melting curve analysis was not feasible.
All 1,950 Ixodes ticks investigated in the present study tested negative by SYBR green PCR, suggesting that these ticks do not play a role in the transmission of the hemoplasma species detected by our assay in Switzerland. This extends our recent results showing that Ixodes ticks are not a relevant vector for feline hemoplasmas in Switzerland (34). Since Ixodes ricinus represents the most common tick species in Switzerland (1, 31), vector-borne transmission of hemoplasmas via ticks from animals to humans in Switzerland seems unlikely.
In conclusion, the present study demonstrates that the SYBR green PCR assay described here is suitable to screen for known and so-far-undiscovered hemoplasma species. Positive results should be confirmed by species-specific TaqMan PCR assays or sequencing of the PCR products. Despite the recent molecular detection of M. haemofelis infection in an HIV-positive patient, in this study no hemoplasma infections were detected in blood samples derived from anemic or immunocompromised humans. The question of whether undiscovered hemoplasma species could play a role in human health should be addressed in subsequent studies. The assay described here will be an important tool in the pursuit of this issue.
Acknowledgments
We thank V. Cattori, R. Tandon, C. Brunner, B. Weibel, T. Meili Prodan, and E. Gönczi for helpful contributions. Laboratory work was performed using the logistics of the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich, Switzerland.
This study was supported by the UBS AG. B. Willi was supported by the Roche Research Foundation, Basel. R.H.-L. is the recipient of a professorship from the Swiss National Science Foundation (grants PP00B-102866/1 and PP00B-119136/1).
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Aeschlimann, A. 1972. Ixodes ricinus, Limmeus, 1758 (Ixodoidea: Ixodidae). Preliminary study of the biology of the species in Switzerland. Acta Trop. 29:321-340. [PubMed] [Google Scholar]
- 2.Applied Biosystems. 2006. SYBR® Green PCR master mix and RT-PCR reagents, protocol. Applied Biosystems, Foster City, CA.
- 3.Archer, G. L., P. H. Coleman, R. M. Cole, R. J. Duma, and C. L. Johnston, Jr. 1980. Hemotropic bacteria. N. Engl. J. Med. 302:1151-1152. [DOI] [PubMed] [Google Scholar]
- 4.Brunner, C., T. Kanellos, M. L. Meli, D. J. Sutton, R. Gisler, M. A. Gomes-Keller, R. Hofmann-Lehmann, and H. Lutz. 2006. Antibody induction after combined application of an adjuvanted recombinant FeLV vaccine and a multivalent modified live virus vaccine with a chlamydial component. Vaccine 24:1838-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Criado-Fornelio, A., A. Martinez-Marcos, A. Buling-Sarana, and J. C. Barba-Carretero. 2003. Presence of Mycoplasma haemofelis, Mycoplasma haemominutum and piroplasmids in cats from southern Europe: a molecular study. Vet. Microbiol. 93:307-317. [DOI] [PubMed] [Google Scholar]
- 6.dos Santos, A. P., R. P. dos Santos, A. W. Biondo, J. M. Dora, L. Z. Goldani, S. T. de Oliveira, A. M. de Sa Guimaraes, J. Timenetsky, H. A. de Morais, F. H. Gonzalez, and J. B. Messick. 2008. Hemoplasma infection in HIV-positive patient, Brazil. Emerg. Infect. Dis. 14:1922-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte, M. I., M. S. Oliveira, M. A. Shikanai-Yasuda, O. N. Mariano, C. F. Takakura, C. Pagliari, and C. E. Corbett. 1992. Haemobartonella-like microorganism infection in AIDS patients: ultrastructural pathology. J. Infect. Dis. 165:976-977. [PubMed] [Google Scholar]
- 8.Fyumagwa, R. D., S. Simmler, B. Willi, M. L. Meli, A. Sutter, R. Hoare, G. Dasen, R. Hofmann-Lehmann, and H. Lutz. 2007. Molecular detection of hemotropic mycoplasmas in Rhipicephalus sanguineus tick species collected on lions (Panthera leo) from Ngorongoro Crater, Tanzania. S. Afr. J. Wildl. Res. 38:117-122. [Google Scholar]
- 9.Giglio, S., P. T. Monis, and C. P. Saint. 2003. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 31:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grindem, C. B., W. T. Corbett, and M. T. Tomkins. 1990. Risk factors for Haemobartonella felis infection in cats. J. Am. Vet. Med. Assoc. 196:96-99. [PubMed] [Google Scholar]
- 11.Hoelzle, L. E., M. Helbling, K. Hoelzle, M. Ritzmann, K. Heinritzi, and M. M. Wittenbrink. 2007. First LightCycler real-time PCR assay for the quantitative detection of Mycoplasma suis in clinical samples. J. Microbiol. Methods 70:346-354. [DOI] [PubMed] [Google Scholar]
- 12.Inokuma, H., M. Oyamada, B. Davoust, M. Boni, J. Dereure, B. Bucheton, A. Hammad, M. Watanabe, K. Itamoto, M. Okuda, and P. Brouqui. 2006. Epidemiological survey of Ehrlichia canis and related species infection in dogs in eastern Sudan. Ann. N. Y. Acad. Sci. 1078:461-463. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, W. A., M. R. Lappin, S. Kamkar, and W. J. Reagan. 2001. Use of a polymerase chain reaction assay to detect and differentiate two strains of Haemobartonella felis in naturally infected cats. Am. J. Vet. Res. 62:604-608. [DOI] [PubMed] [Google Scholar]
- 14.Kallick, C. A., S. Levin, K. T. Reddi, and W. L. Landau. 1972. Systemic lupus erythematosus associated with haemobartonella-like organisms. Nat. New Biol. 236:145-146. [DOI] [PubMed] [Google Scholar]
- 15.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 16.Luria, B. J., J. K. Levy, M. R. Lappin, E. B. Breitschwerdt, A. M. Legendre, J. A. Hernandez, S. P. Gorman, and I. T. Lee. 2004. Prevalence of infectious diseases in feral cats in Northern Florida. J. Feline Med. Surg. 6:287-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messick, J. B. 2003. New perspectives about hemotrophic mycoplasma (formerly, Haemobartonella and Eperythrozoon species) infections in dogs and cats. Vet. Clin. N. Am. Small Anim. Pract. 33:1453-1465. [DOI] [PubMed] [Google Scholar]
- 18.Messick, J. B., L. M. Berent, and S. K. Cooper. 1998. Development and evaluation of a PCR-based assay for detection of Haemobartonella felis in cats and differentiation of H. felis from related bacteria by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 36:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neimark, H., K. E. Johansson, Y. Rikihisa, and J. G. Tully. 2002. Revision of haemotrophic Mycoplasma species names. Int. J. Syst. Evol. Microbiol. 52:683. [DOI] [PubMed] [Google Scholar]
- 20.Peters, I. R., C. R. Helps, B. Willi, R. Hofmann-Lehmann, and S. Tasker. 2008. The prevalence of three species of feline haemoplasmas in samples submitted to a diagnostics service as determined by three novel real-time duplex PCR assays. Vet. Microbiol. 126:142-150. [DOI] [PubMed] [Google Scholar]
- 21.Rikihisa, Y., M. Kawahara, B. Wen, G. Kociba, P. Fuerst, F. Kawamori, C. Suto, S. Shibata, and M. Futohashi. 1997. Western immunoblot analysis of Haemobartonella muris and comparison of 16S rRNA gene sequences of H. muris, H. felis, and Eperythrozoon suis. J. Clin. Microbiol. 35:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ristic, M., and J. P. Kreier. 1979. Hemotropic bacteria. N. Engl. J. Med. 301:937-939. [DOI] [PubMed] [Google Scholar]
- 23.Stockham, S. L., and A. M. Scott. 2008. Fundamentals of veterinary clinical pathology, 2nd ed. Iowa State University Press, Ames, IA.
- 24.Sykes, J. E., N. L. Bailiff, L. M. Ball, O. Foreman, J. W. George, and M. M. Fry. 2004. Identification of a novel hemotropic mycoplasma in a splenectomized dog with hemic neoplasia. J. Am. Vet. Med. Assoc. 224:1946-1951, 1930-1931. [DOI] [PubMed] [Google Scholar]
- 25.Tasker, S., S. H. Binns, M. J. Day, T. J. Gruffydd-Jones, D. A. Harbour, C. R. Helps, W. A. Jensen, C. S. Olver, and M. R. Lappin. 2003. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in cats in the United Kingdom. Vet. Rec. 152:193-198. [DOI] [PubMed] [Google Scholar]
- 26.Tasker, S., S. M. Caney, M. J. Day, R. S. Dean, C. R. Helps, T. G. Knowles, P. J. Lait, M. D. Pinches, and T. J. Gruffydd-Jones. 2006. Effect of chronic FIV infection, and efficacy of marbofloxacin treatment, on Mycoplasma haemofelis infection. Vet. Microbiol. 117:169-179. [DOI] [PubMed] [Google Scholar]
- 27.Tasker, S., C. R. Helps, C. J. Belford, R. J. Birtles, M. J. Day, A. H. Sparkes, T. J. Gruffydd-Jones, and D. A. Harbour. 2001. 16S rDNA comparison demonstrates near identity between an United Kingdom Haemobartonella felis strain and the American California strain. Vet. Microbiol. 81:73-78. [DOI] [PubMed] [Google Scholar]
- 28.Tasker, S., C. R. Helps, M. J. Day, D. A. Harbour, S. E. Shaw, S. Harrus, G. Baneth, R. G. Lobetti, R. Malik, J. P. Beaufils, C. R. Belford, and T. J. Gruffydd-Jones. 2003. Phylogenetic analysis of hemoplasma species: an international study. J. Clin. Microbiol. 41:3877-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wengi, N., B. Willi, F. S. Boretti, V. Cattori, B. Riond, M. L. Meli, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2008. Real-time PCR-based prevalence study, infection follow-up and molecular characterization of canine hemotropic mycoplasmas. Vet. Microbiol. 126:132-141. [DOI] [PubMed] [Google Scholar]
- 31.Wicki, R., P. Sauter, C. Mettler, A. Natsch, T. Enzler, N. Pusterla, P. Kuhnert, G. Egli, M. Bernasconi, R. Lienhard, H. Lutz, and C. M. Leutenegger. 2000. Swiss Army survey in Switzerland to determine the prevalence of Francisella tularensis, members of the Ehrlichia phagocytophila genogroup, Borrelia burgdorferi sensu lato, and tick-borne encephalitis virus in ticks. Eur. J. Clin. Microbiol. Infect. Dis. 19:427-432. [DOI] [PubMed] [Google Scholar]
- 32.Willi, B., F. S. Boretti, C. Baumgartner, S. Tasker, B. Wenger, V. Cattori, M. L. Meli, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2006. Prevalence, risk factor analysis, and follow-up of infections caused by three feline hemoplasma species in cats in Switzerland. J. Clin. Microbiol. 44:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willi, B., F. S. Boretti, V. Cattori, S. Tasker, M. L. Meli, C. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2005. Identification, molecular characterization, and experimental transmission of a new hemoplasma isolate from a cat with hemolytic anemia in Switzerland. J. Clin. Microbiol. 43:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willi, B., F. S. Boretti, M. L. Meli, M. V. Bernasconi, S. Casati, D. Hegglin, M. Puorger, H. Neimark, V. Cattori, N. Wengi, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2007. Real-time PCR investigation of potential vectors, reservoirs, and shedding patterns of feline hemotropic mycoplasmas. Appl. Environ. Microbiol. 73:3798-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willi, B., C. Filoni, J. L. Catao-Dias, V. Cattori, M. L. Meli, A. Vargas, F. Martinez, M. E. Roelke, M. P. Ryser-Degiorgis, C. M. Leutenegger, H. Lutz, and R. Hofmann-Lehmann. 2007. Worldwide occurrence of feline hemoplasma infections in wild felid species. J. Clin. Microbiol. 45:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willi, B., S. Tasker, F. S. Boretti, M. G. Doherr, V. Cattori, M. L. Meli, R. G. Lobetti, R. Malik, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2006. Phylogenetic analysis of “Candidatus Mycoplasma turicensis” isolates from pet cats in the United Kingdom, Australia, and South Africa, with analysis of risk factors for infection. J. Clin. Microbiol. 44:4430-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, D., X. Tai, Y. Qiu, and S. Yun. 2000. Prevalence of Eperythrozoon spp. infection and congenital eperythrozoonosis in humans in Inner Mongolia, China. Epidemiol. Infect. 125:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]