Abstract
Metal-responsive transcription factor 1 (MTF-1) mediates both basal and heavy metal-induced transcription of metallothionein genes and also regulates other genes involved in the cell stress response and in metal homeostasis. In resting cells, MTF-1 localizes to both the cytoplasm and the nucleus but quantitatively accumulates in the nucleus upon metal load and under other stress conditions. Here we show that within the DNA-binding domain, a region spanning zinc fingers 1 to 3 (amino acids [aa] 137 to 228 in human MTF-1) harbors a nonconventional nuclear localization signal. This protein segment confers constitutive nuclear localization to a cytoplasmic marker protein. The deletion of the three zinc fingers impairs nuclear localization. The export of MTF-1 to the cytoplasm is controlled by a classical nuclear export signal (NES) embedded in the acidic activation domain. We show that this activation domain confers metal inducibility in distinct cell types when fused to a heterologous DNA-binding domain. Furthermore, the cause of a previously described stronger inducibility of human versus mouse MTF-1 could be narrowed down to a 3-aa difference in the NES; “humanizing” mouse MTF-1 at these three positions enhanced its metal inducibility to the level of human MTF-1.
Cells need to adapt quickly to various forms of stress and adjust their gene expression profile accordingly. Therefore, stress-responsive transcription factors are regulated mostly at the posttranslational level by modulating their activity, stability, or subcellular localization. A number of transcription factors localize predominantly to the cytoplasm but translocate to the nucleus when required. Since the nuclear pore complex typically allows the diffusion of molecules of up to approximately 20 to 40 kDa only (8), larger molecules require active transport to pass through the nuclear pore. Driven by a RanGTP gradient, this cargo transport is facilitated by binding to transport receptors, β-karyopherins (5, 10). Import into the nucleus requires the binding of importin-β to a nuclear localization signal (NLS) in the cargo protein either directly or through an adapter molecule, importin-α. Such NLSs typically consist of mono- or bipartite stretches of basic amino acids. In the nucleus, importin-β binds to RanGTP, which lowers its affinity for the cargo, leading to its release. Classical export from the nucleus to the cytoplasm is controlled by binding to the RanGTP-bound export receptor Crm1 through a leucine-rich nuclear export signal (NES). The cargo is released upon GTP hydrolysis in the cytoplasm. The Ran GTP gradient is maintained by Ran GTPase-activating protein, inducing the Ran-catalyzed hydrolysis of GTP in the cytoplasm, and guanine nucleotide exchange factor, catalyzing the exchange of GDP for GTP in the nucleus (10, 14, 26). Here we study how metal-responsive transcription factor 1 (MTF-1)-regulated transcription is dependent on its subcellular localization and activation status.
MTF-1 is evolutionarily conserved from mammals to insects and has been characterized for various species including human, mouse, capybara, pufferfish, zebrafish, trout, and Drosophila melanogaster (1, 2, 7, 13, 20, 29, 43). This constitutively expressed transcription factor is activated upon diverse stress stimuli, notably metal load but also oxidative stress and hypoxia (6, 24). In resting cells, MTF-1 localizes primarily to the cytoplasm but upon stimulation translocates to the nucleus (32, 37). There, it binds to the enhancers/promoters of target genes that harbor one or multiple copies of a specific recognition sequence, the metal response element (MRE). MREs share the “core” consensus sequence TGCRCNC (29, 38). The best-characterized target genes are those for metallothioneins. Metallothioneins are short, cysteine-rich proteins that bind, and thereby scavenge/detoxify, a variety of heavy metals. In addition, they serve as scavengers of reactive oxygen species (12). Besides its well-characterized role in metallothionein gene transcription, MTF-1 is also involved in the transcriptional regulation of other metal-responsive genes, such as zinc transporter 1 (17), and the recently identified cadmium-responsive target genes N-myc downstream regulated gene 1 (Ndrg1) and glycine-rich protein 1 (42). The loss of MTF-1 in the mouse leads to embryonic lethality at day 14 post coitum due to cell-autonomous hepatocyte degeneration, which shows that MTF-1 also serves a developmental role (11, 39). DNA binding of MTF-1 is accomplished by six tandemly arranged zinc fingers, whereby zinc fingers 1 to 4 are most important (3, 9). The zinc finger region is also implicated in the metal inducibility of MTF-1, since MTF-1 requires elevated zinc concentrations for DNA binding in vitro (13), and mutations in the linker between zinc fingers 1 and 2 lead to constitutive transcriptional activation (19). The C-terminal half of mammalian MTF-1 contains three types of transcriptional activation domains, of which the acidic activation domain is the strongest one (28). In the C-terminal region, there is a cysteine-rich motif (CQCQCAC) that was previously shown to be necessary for metal induction (4). Interestingly, mouse MTF-1 is less responsive to metal than human MTF-1 (hMTF-1) irrespective of whether it is tested with mouse or human cells (2, 23, 29). Activation by heavy metals involves several signal transduction pathways that affect MTF-1. Previously reported kinase inhibitor studies indicated roles for protein kinase C, tyrosine kinase, casein kinase II, and c-Jun N-terminal kinase, whereby no distinction was made between direct or indirect effects (15, 31). A conspicuous cluster of basic amino acids just upstream of zinc finger 1 in hMTF-1 was tested as a candidate NLS. However, a triple-amino-acid substitution in this region only delayed but did not abolish metal-induced nuclear import (32). Antagonizing import, hMTF-1 also contains a determinant for export from the nucleus: a classical leucine-rich NES sequence is embedded in the acidic activation domain. Mutations in this region lead to impaired export and also to impaired transactivation by MTF-1, initially suggesting a link between nuclear export and metal inducibility.
In this study, we identify the major determinant for the nuclear import of MTF-1 to be a nonconventional NLS in the region spanning zinc fingers 1 to 3. Furthermore, we show that the acidic activation domain of hMTF-1 can work as a zinc-inducible activation domain independent of the full-length protein context. The higher metal inducibility of hMTF-1 than mouse MTF-1 can be attributed to a 3-amino-acid (aa) difference in the NES, which is embedded in the acidic activation domain.
MATERIALS AND METHODS
Cell culture conditions and transfection.
Cells were maintained in Dulbecco's modified Eagle medium (Gibco) supplemented with 8% fetal bovine serum (Biochrom AG, Berlin, Germany), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Transfections were performed using the standard calcium phosphate coprecipitation method. Dko7 cells were derived from MTF-1−/− mouse embryonic stem cells (13) that were allowed to differentiate into fibroblast-type cells and immortalized by transformation with simian virus 40 (SV40) large T antigen (28).
Plasmid constructions.
The vector expressing myc-tagged chicken pyruvate kinase (myc-PK) was a generous gift from Gideon Dreyfuss (Howard Hughes Medical Institute, Philadelphia, PA) and was described previously (36). The plasmids expressing myc-PK fusion proteins were generated by cloning PCR-amplified fragments with either KpnI and NotI sites or Acc65I and NotI sites (for aa 110 to 228 and 137 to 228 because of an endogenous KpnI site) into the KpnI-NotI- or Acc65I-NotI-digested myc-PK vector. Wild-type plasmid pChMTF-1-VSV; pChMTF-1-VSV with the mutation H185Y, 135KRK→LLI, or 4L→A (32); or pEGFP-hMTF-1(Δ133-139) was used as a template. Primer sequences are available upon request. The myc-PK_aa130-140 fusion construct was generated by annealing the oligonucleotides 5′-GGCCGCTCAGTAGCGCTTTACTTCTTTACGTTTTGTTTCCGGGTAC-3′and 5′-CCGGAAACAAAACGTAAAGAAGTAAAGCGCTACTGAGC-3′ and subsequent cloning into the KpnI-NotI-digested myc-PK vector. The myc-PK_SV40NLS fusion construct was generated in the same way using the following oligonucleotides: 5′-CCGAAGAAGAAGCGAAAGGTATGAGC-3′ and 5′-GCGGCCGCTCATACCTTTCGCTTCTTCTTCGGGTAC-3′. Reporter plasmid 4×MREd-OVEC and reference plasmid OVEC-Ref were described previously (41). Plasmids expressing hMTF-1-vesicular stomatitis virus (VSV) (29), mouse MTF-1-VSV (2), and hMTF-1-VSV(4L→A) (32) were described previously. Plasmids with the mutations H185Y, C337Y/D340E/S342G, C337Y, and S342G in hMTF-1 and Y336C/E339D/G341S, Y336C, and G341S in mouse MTF-1 were generated by using site-directed mutagenesis (QuikChange; Stratagene) according to the manufacturer's instructions. Primer sequences are available upon request. To generate plasmid pEGFP-hMTF-1, plasmid pC-hMTF-1 (13) was digested with NcoI, blunt ended with Klenow polymerase, and subsequently XhoI digested. The resulting 3,254-bp fragment was cloned into BspEI-digested, Klenow-treated, and subsequently SalI-digested vector pEGFP-C1 (Clontech). The pEGFP-hMTF-1 deletion constructs were generated by PCR using primers, containing restriction sites, flanking the regions to be deleted. Detailed instructions and primer sequences are available upon request. The plasmid expressing the reporter gene under the control of five Gal4 binding sites, 5×GOVEC, was described previously (34). The vector expressing the Gal4 DNA binding domain (DBD) (aa 1 to 93) fused to amino acids 327 to 410 of mouse MTF-1 was described previously (28). Vector pSCTGal(1-93), expressing only aa 1 to 93 of the Gal4 DBD (aa 1 to 93), was described previously (35). The plasmids expressing the Gal4 DBD fused to fragments of hMTF-1 or the control activation domain of TFE3 were generated either by PCR amplification or by restriction digestion and subsequent cloning into vector pSCTGal(1-93). All sequences were confirmed by sequencing. Detailed information about the primers and the cloning procedure are available upon request.
S1 nuclease protection assay.
For testing the transcriptional activity of wild-type and mutant MTF-1, dko7 cells were seeded into 10-cm dishes and transfected with 10 μg of 4×MREd reporter plasmid, 4 μg OVEC-Ref reference plasmid, and 1 μg of the respective MTF-1 plasmid. To test the transcriptional activity of the acidic activation domain, cells were transfected with 10 μg 5×GOVEC reporter plasmid, 4 μg OVEC-Ref reference plasmid, and 0.5 μg of the respective effector plasmid. Herring sperm DNA was added to each sample to a total amount of 20 μg DNA. Sixteen hours after transfection, the precipitate was washed off with Tris-buffered saline. Twenty-four hours after washing, cells were treated as indicated. For metal induction, tissue culture medium was supplemented with zinc sulfate (final concentration, 100 μM) or cadmium chloride (final concentration, 50 μM) for 4 h. Leptomycin B (LMB) (final concentration, 10 ng/ml) (catalog no. 431050; Calbiochem) dissolved in ethanol or solvent alone was added to the cells 2 h prior to zinc supplementation. RNA isolation and S1 nuclease protection assays were performed as described previously (40, 41). The samples were analyzed by use of a 10% polyacrylamide (acrylamide-bisacrylamide, 19:1)-7.5 M urea gel. Bands were visualized using an FLA-7000 fluorescent image analyzer and quantified using ImageGauge software (Fujifilm Life Science). Reporter bands were normalized to the reference bands and compared to the sample value with noninduced hMTF-1, which was set to 1.
Indirect immunofluorescence.
U2OS or dko7 cells were plated onto coverslips in 12-well tissue culture dishes and transfected with 1 μg expression vector per well as indicated. Cells were either left untreated or treated with either 100 μM zinc sulfate or 10 ng/ml LMB for 2 h. Immunofluorescent staining was done as described previously (32). Each experiment was repeated three times. The following antibodies were used: mouse anti-VSV at a 1:500 dilution (V5507; Sigma), fluorescein-coupled rabbit anti-mouse immunoglobulin G at a 1:1,000 dilution (Molecular Probes), mouse anti-c-myc at a 1:1,000 dilution (catalog no. OP10; Oncogene), and Alexa Fluor 594 goat anti-mouse immunoglobulin G at a 1:1,000 dilution of the 2-mg/ml stock (Invitrogen). Nuclei were visualized by staining with 4′,6′-diamidino-2-phenylindole (DAPI) at a concentration of 1 μg/ml.
Preparation of nuclear extracts and EMSA.
HEK293 cells were transfected with 2 μg of either pChMTF-1-GFP wild-type or deletion constructs as indicated. Herring sperm DNA was added to a final amount of 20 μg DNA per 10-cm dish. Sixteen hours after transfection, the precipitate was washed away with Tris-buffered saline, and fresh medium was added. Forty hours after transfection, cells were treated with 100 μM zinc sulfate for 2 h or left untreated as indicated. Nuclear extracts were prepared as described previously (33). The electrophoretic mobility shift assay (EMSA) was performed as described previously (29) by using, for each sample, 10 μg of nuclear protein extract and 25 fmol of end-labeled, 31-bp-long oligonucleotide containing the core MRE sequence (MRE-s), TGCACAC. The samples were analyzed on a 4% polyacrylamide gel (acrylamide-bisacrylamide, 29:1). The bands were visualized using an FLA-7000 fluorescent image analyzer and ImageGauge software (Fujifilm Life Science).
RESULTS
Zinc fingers 1 to 3 are necessary and sufficient for nuclear localization.
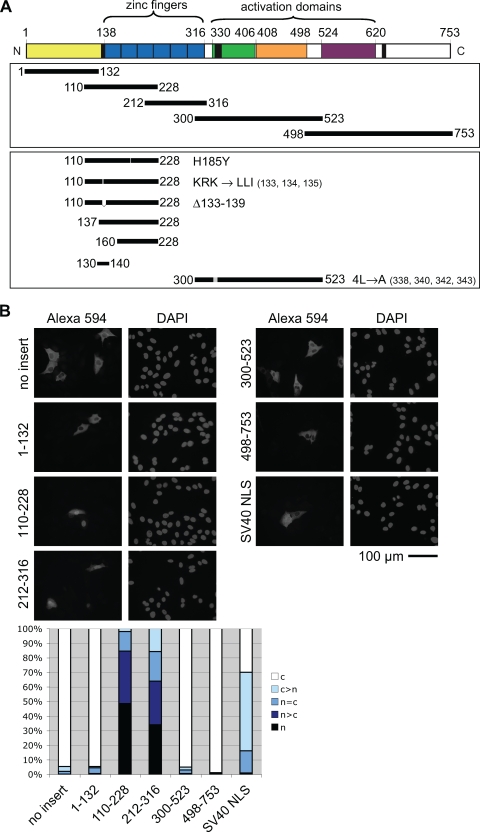
MTF-1 translocates to the nucleus in response to metal exposure, but the only segment resembling classical NLS motifs, a cluster of basic amino acids N terminal to the zinc finger domain, was previously shown not to be essential for nuclear import (32), suggesting that an additional part of MTF-1 was required. To search for such an NLS function in hMTF-1, overlapping fragments of the cDNA sequence (Fig. 1A) were fused in frame into a vector expressing myc-PK, a protein that was previously shown to be localized exclusively to the cytoplasm (36). The subcellular distribution of these fusion proteins was examined (Fig. 1B and C). To exclude confounding effects due to nuclear export, we also tested a mutant fragment where four leucines were replaced by alanines, thereby destroying export function (aa 300 to 523 [4L→A]) (Fig. 1A and C) (32). Also included was an MTF-1 segment in which the histidine at position 185 was replaced by tyrosine. An His185-containing variant of hMTF-1 was described previously by us (2) but represents a rare polymorphism or mutation since other hMTF-1 isolates contain a tyrosine at position 185 (25; our unpublished observations). Both the His185 and the Tyr185 versions display the same metal inducibility (see Fig. S1 in the supplemental material) and import activity of the fragment at aa 110 to 228 (Fig. 1B and C). As depicted in Fig. 1B, only the segments spanning the zinc fingers conferred nuclear localization to myc-PK. Nuclear import was efficient and clearly better than that with the SV40-NLS construct with a minimal NLS, as defined previously by Kalderon et al. (16). Further dissection of the zinc finger region revealed that the region spanning aa 137 to 228 had almost exclusive nuclear localization (Fig. 1C). An N-terminally extended fragment, aa 110 to 228 (Fig. 1B), was less efficiently imported, suggesting some inhibitory function in the extension. A mutation (LLI) or deletion (Δ133-139) of the string of basic amino acids whose mutation had been shown to delay nuclear import (32) impaired the nuclear localization of the fragment at aa 110 to 228. However, a minimal fragment containing this basic stretch (aa 130 to 140) fused to the marker protein was insufficient to confer nuclear localization (Fig. 1C). We therefore dubbed this basic string an auxiliary NLS (aNLS). Unlike full-length MTF-1, whose nuclear localization is stress regulated, treatment of the cells with 100 μM zinc for 2 h did not change the localization pattern of any of the constructs (data not shown).
FIG. 1.
Zinc fingers 1 to 3 of hMTF-1 are sufficient to direct nuclear localization of a cytoplasmic marker protein, pyruvate kinase (myc-PK). (A) Scheme of the hMTF-1 domains and overview on the fragments tested for NLS function. Yellow, potentiation domain; blue, zinc fingers; green, acidic activation domain; orange, proline-rich activation domain; purple, serine/threonine-rich activation domain; black box preceding zinc fingers, arginine/lysine-rich motif; black box following zinc fingers, NES; black box at the C terminus, cysteine cluster. Fragments of MTF-1 were fused into the vector with the myc-tagged marker (myc-PK); the first and last amino acid positions and mutations/deletions are indicated. (B and C) General (B) and refined (C) searches for an NLS in MTF-1. U2OS (human osteosarcoma) cells were transfected with myc-PK with or without fragments of MTF-1 or the SV40 NLS inserted and were analyzed for their subcellular localization by anti-myc immunostaining. For quantification, 200 cells each were classified into the indicated categories (c, cytoplasm; n, nucleus) and counted. Nuclei were visualized by DAPI staining.
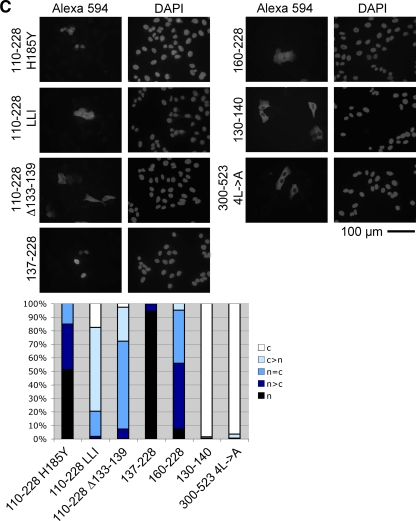
To further test whether the segment spanning zinc fingers 1 to 3 is necessary for the nuclear localization of MTF-1, we analyzed the localization of GFP-tagged hMTF-1 with various deletions in the zinc finger region (Fig. 2A and B). A complete deletion of the zinc finger domain as well as the deletion of only zinc fingers 1 to 3, including the aNLS, abolished import. A smaller deletion of only zinc fingers 2 and 3 showed reduced import. Outside of zinc fingers 1 to 3, there was another, albeit weak, activity for nuclear import: zinc fingers 4 to 6 of MTF-1 (aa 212 to 228) conferred nuclear localization to the pyruvate kinase marker protein (Fig. 1B) but were not sufficient to direct efficient nuclear import within the context of MTF-1 lacking zinc fingers 1 to 3 (Fig. 2B). MTF-1 lacking all zinc fingers still showed partial nuclear localization in cells treated with the export inhibitor LMB (Fig. 2B). These are, however, minor activities that are overridden by the presence of a functional export machinery. Furthermore, the nucleocytoplasmic distribution of the deletion constructs was no longer affected by zinc status (Fig. 2B). Unexpectedly, the elimination of the aNLS either by amino acid substitutions or by deletion gave different results. The replacement of basic amino acids (K135L, R136L, and K137I) delayed import (32), while a complete deletion (Δ133-139) led to constitutive nuclear localization (with a concomitant loss of activity, possibly due to steric alterations) (see below and Fig. 4C). Altogether, these results show that the region spanning zinc fingers 1 to 3 is necessary and sufficient for the nuclear localization of MTF-1. This region does not resemble the classical mono- or bipartite NLSs (Fig. 2C).
FIG. 2.
Amino acids 133 to 226 are essential for nuclear localization of hMTF-1. (A and B) MTF-1 fused to GFP, with or without deletions in the zinc finger region, was transfected into U2OS (human osteosarcoma) cells. Thirty-six hours after transfection cells were treated with 100 μM zinc sulfate or 10 ng/ml LMB for 2 h or left untreated. Cells were fixed, and subcellular localization was monitored by GFP fluorescence. For quantification, 200 cells each were classified into the indicated categories (c, cytoplasm; n, nucleus). Nuclei were visualized by DAPI staining. nt, not treated; wt, wild type. (C) Sequence of the region necessary and sufficient for the nuclear localization of hMTF-1. Basic amino acids are highlighted in yellow. ZF1, zinc finger 1.
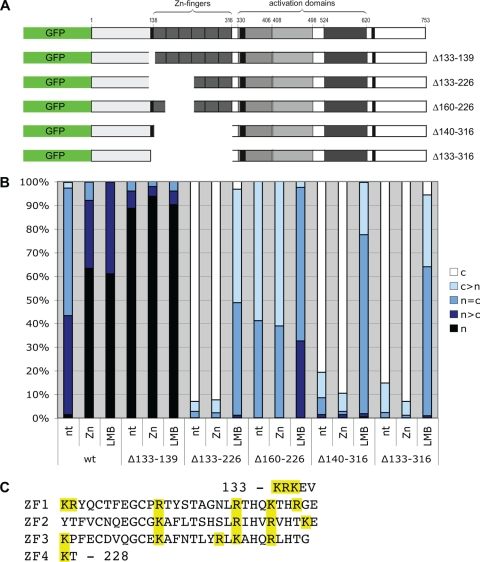
FIG. 4.
Mutations in MTF-1 suggest a link between export and metal inducibility. (A) To analyze the subcellular localization of the chimeric constructs, MTF-1 null cells (dko7) were transfected with wild-type or mutated MTF-1-VSV of mouse or human. Cells were stained using an anti-VSV antibody. Two hundred cells each were counted and classified into the indicated categories (c, cytoplasm; n, nucleus; n and c, equally distributed). The mutant with impaired metal induction showed increased nuclear localization, and the mutant with high metal inducibility showed increased cytoplasmic localization. mMTF-1, mouse MTF-1; nt, not treated. (B) Sequence comparison of the MTF-1 NESs of different species (corresponding to aa 335 to 346 of hMTF-1). The human sequence differs from the rodent sequences in 2 and 3 aa, respectively. The leucine residues, characteristic for NESs, are well conserved. (C) The transcriptional activity of MTF-1 with a mutation outside the NES that is impaired in nuclear export was analyzed by an S1 nuclease protection assay. This mutant is not metal inducible. Error bars indicate standard deviations of data from three independent experiments. wt, wild type. (D) Test for DNA binding by EMSA. HEK293 cells were transfected with MTF-1-GFP constructs as indicated and treated with 100 μM zinc sulfate or left untreated. MTF-1-GFP and MTF-1-GFP(Δ133-139) bind to the MRE-containing oligonucleotide, while the constructs with larger deletions did not. The band visible in the latter case is the band shift from endogenous (endog.) MTF-1 from HEK293 cells, which migrates faster because it lacks the GFP moiety.
Three amino acids in the acidic activation domain specify metal inducibility.
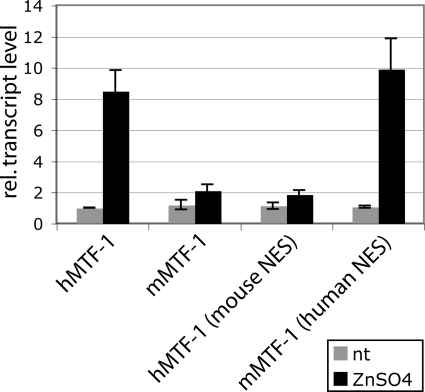
In transfection experiments using various cell lines and promoter constructs, we previously observed that hMTF-1 exhibited a much higher metal inducibility than MTF-1 of rodents (mouse and capybara) (20, 23; this paper). The substitution of a 65-aa segment within the murine acidic activation domain with the corresponding human sequence conferred high metal inducibility (23). A sequence comparison between the mouse and human proteins showed that this 65-aa segment differs in 9 aa. Three of these are clustered at the NES motif, and we considered them to be likely candidates for the effect. To test this, we converted the 3 aa in mouse MTF-1 to the human type and vice versa (see Fig. 4B). MTF-1 null mutant cells (dko7) were transfected with the respective mutant or wild-type MTF-1 constructs, a reference plasmid, and a reporter plasmid under the control of four tandem MREs. This reporter offers the advantage of being dependent exclusively on MTF-1; i.e., it is completely inactive unless an MTF-1 expression clone is cotransfected. In line with the above-described results, mouse MTF-1 displayed about twofold metal inducibility, and hMTF-1 displayed about eightfold inducibility (Fig. 3). The metal inducibility of mouse MTF-1 with the “humanized” NES was indeed as strong as that with the human wild-type protein, whereas the reciprocal substitution construct, hMTF-1 with the mouse NES, behaved like wild-type mouse MTF-1. For this effect, the simultaneous exchange of all three amino acids was required: single-amino-acid substitutions did decrease the metal inducibility of hMTF-1, but in the converse situation, they did not improve metal induction in mouse MTF-1 (data not shown). Taken together, these results show that the 3-aa difference in the NES region between the mouse MTF-1 and hMTF-1 proteins accounts for the different metal inducibilities.
FIG. 3.
A 3-aa change in the NES of mouse MTF-1 (mMTF-1) to the corresponding human residues confers high metal inducibility. MTF-1 null cells (dko7) were transfected with a reporter plasmid under the control of four tandem MREs, a reference plasmid, and the indicated wild-type or mutated MTF-1 construct of mouse or human. Thirty-six hours after transfection cells were treated with 100 μM zinc sulfate for 4 h. RNA was isolated from the cells and quantified by the S1 nuclease protection assay. Error bars indicate the standard deviations of data from three independent experiments. rel., relative; nt, not treated.
A link between nuclear export and MTF-1 activity?
Analysis of the subcellular localization of the chimeric constructs described above revealed that the triple-amino-acid substitution also changed the subcellular distribution of MTF-1 (Fig. 4A). hMTF-1 with the mouse NES showed enhanced nuclear localization, whereas “humanized” mouse MTF-1 showed increased cytoplasmic localization in comparison to wild-type mouse MTF-1. Another mutant MTF-1 with a change of four leucines to alanines (aa 338, 341, 343, and 344) in the NES was also previously shown to exhibit impaired export together with impaired zinc inducibility (32). Since the NES is embedded within the acidic activation domain, this correlation might be explained by a simple spatial overlap of the two functional domains. To investigate this further, we examined the transcriptional activity of export-impaired MTF-1 with a mutation outside the NES. As shown above, the deletion of aa 133 to 139 led to constitutive nuclear localization (Fig. 2B). This construct also showed impaired metal inducibility (Fig. 4C), even though it retained DNA-binding ability, as shown by EMSA (Fig. 4D). These results suggested that nuclear export might be functionally linked to the metal inducibility of MTF-1. As expected, none of the mutant constructs with a partial or complete deletion of the zinc fingers was able to bind DNA (Fig. 4D).
Nuclear export is not essential for metal inducibility.
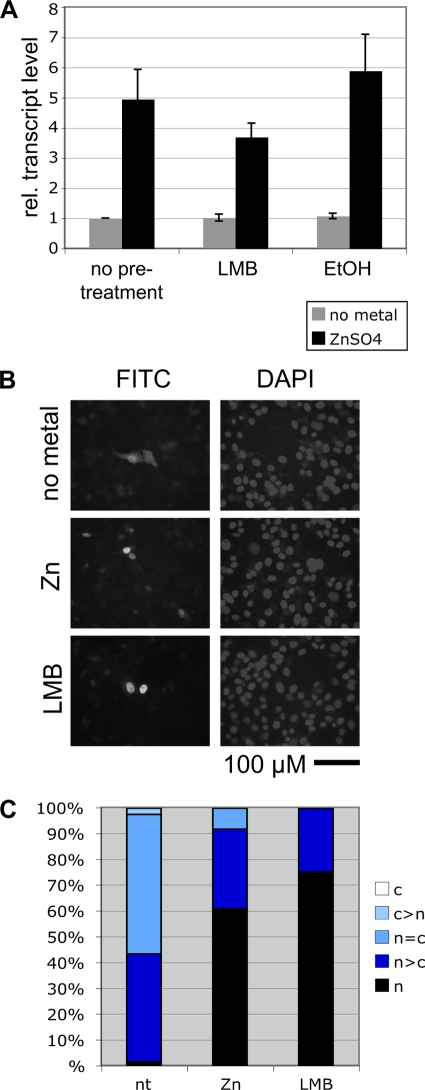
To clarify this issue, i.e., whether the nuclear export of MTF-1 is necessary for metal induction, we made use of the export inhibitor LMB. We observed that LMB pretreatment for 2 h followed by zinc treatment for 4 h led to only a mild reduction in levels of MTF-1-dependent transcription (Fig. 5A). As expected, under conditions of export inhibition, immunofluorescent staining confirmed the nuclear accumulation of MTF-1 (Fig. 5B and C). We conclude that export, while it might support MTF-1 function, is not an essential step in the process of metal induction. The loss-of-function mutations in the NES are thus compromising primarily an activation function rather than affecting metal response indirectly via the inhibition of nuclear export.
FIG. 5.
Inhibition of nuclear export does not abolish metal inducibility. (A) MTF-1 null cells (dko7) were transfected with a reporter plasmid under the control of four tandem MREs, a reference plasmid, and hMTF-1-VSV. Thirty-six hours after transfection cells were treated with either LMB or solvent alone or left untreated. After 2 h, zinc sulfate (final concentration, 100 μM) was added to the cells as indicated (while LMB remained present). RNA was isolated from the cells and quantified by an S1 nuclease protection assay. Export inhibition by LMB treatment does not abolish induction by zinc (center). Error bars indicate the standard deviations of data from three independent experiments. EtOH, ethyl alcohol; rel., relative. (B and C) To test whether export inhibition by LMB was efficient, MTF-1 null cells (dko7) were transfected and treated with zinc or LMB as described above, and the subcellular localization of MTF-1-VSV was analyzed by anti-VSV immunofluorescent staining. For quantification, 200 cells each were counted and classified into the indicated categories (c, cytoplasm; n, nucleus). FITC, fluorescein isothiocyanate; nt, not treated.
The acidic activation domain can work as an autonomous metal-inducible unit.
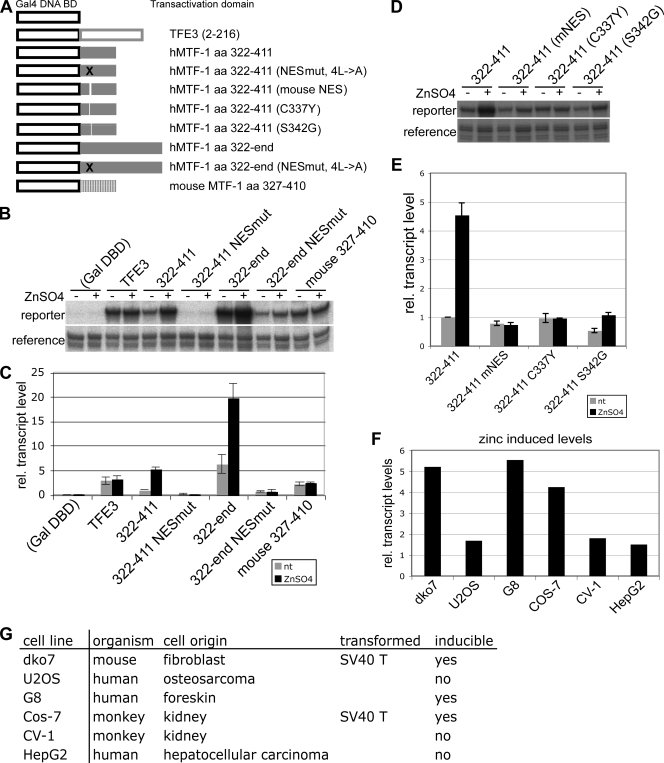
In the course of dissecting metal inducibility and the nuclear export of hMTF-1, we decided to test the transactivation activity of wild-type and NES mutant MTF-1 proteins in a heterologous context. To this end, we fused the C terminus of MTF-1 (aa 322 to 753) or the acidic activation domain only (aa 322 to 411) to the DBD derived from the yeast transcription factor Gal4 (Fig. 6A). The NES motif was mutated either by the substitution of four leucines by alanines (32) or by replacing human-specific amino acids by the ones from mouse, and the metal inducibility of the wild-type and NES mutant constructs was then tested in an S1 assay with a reporter gene under the control of five Gal4-binding sites (Fig. 6B to E). The respective construct containing the mouse acidic activation domain was also included. A metal-independent activation domain of an unrelated transcription factor (TFE3) was used as a control (Fig. 6A to C). As expected, the latter was not zinc responsive, whereas the constructs containing the acidic activation domain or the full C terminus of hMTF-1 were inducible upon zinc supplementation of the cell culture medium (fivefold and threefold, respectively). The corresponding mutant constructs carrying the four leucine-to-alanine mutations displayed a reduced basal activity and, more importantly, were completely nonresponsive to zinc. Also, the construct containing the mouse acidic activation domain was not metal inducible (Fig. 6B and C). This is in line with our observations that wild-type hMTF-1 shows higher metal inducibility than mouse MTF-1, as mentioned above. Accordingly, “mousifying” the NES in of the Gal_322-411 construct by triple- or single-amino-acid substitutions abolished metal inducibility (Fig. 6D and E), as described above for hMTF-1 (Fig. 3). The Gal_322-411 construct was also tested in different cell lines, namely, U2OS, G8, COS7, CV-1, and HepG2. The results described above could be reproduced in G8 cells (human foreskin cells) and COS7 cells (SV40 T-antigen-transformed CV-1 cells), but no zinc induction was observed in CV-1 (African green monkey cells), HepG2 (human hepatoma cells), and U2OS (human osteosarcoma cells) cell lines (Fig. 6F and G). Note that the metal-responsive fusion construct Gal_322-411 did not contain the C-terminal cysteine cluster, which was shown previously to be essential for MTF-1 activity (4). In mammalian MTF-1, this cluster seems to fulfill primarily a structural role in the context of the complete protein (V. Günther and W. Schaffner, unpublished results).
FIG. 6.
The acidic activation domain of hMTF-1 works as an independent zinc-inducible unit in some, but not all, cells. (A) To dissect the transactivation function of hMTF-1, mutant or wild-type fragments of human or mouse MTF-1, or the activation domain of an unrelated factor (TFE3) as a control, were fused to the GAL4 DBD as indicated. NESmut, NES mutant. (B to E) The indicated fusion constructs were tested for the transcriptional activation of a Gal4-responsive reporter gene (5×GOVEC) in MTF-1 null cells (dko7) by the S1 nuclease protection assay. The reporter signal was normalized to the reference signal of a cotransfected plasmid. The metal inducibility of the acidic activation domain of hMTF-1 is fivefold, and that of the full C terminus is threefold (due to the saturation of the signal, induction might have been more than threefold). Neither the acidic activation domain of mouse MTF-1 nor mutant versions of the human NES/acidic activation domain display any metal response. Error bars indicate the standard deviations of data from three independent experiments. nt, not treated; rel. relative. (F) The zinc responsiveness of a Gal4_322-411 fusion protein was tested by use of various cell lines. Zinc-induced levels normalized to the uninduced signal are shown. Zinc inducibility was observed in dko7 cells, G8 cells, and COS-7 cells but not in U2OS cells, CV-1 cells, and HepG2 cells. (G) Tested cell types and their properties. No common characteristics can be assigned to the cells that show inducibility of the isolated acidic activation domain (except that two of the three lines were transformed by SV40).
DISCUSSION
In this study we investigated distinct domains of MTF-1 and their role in metal induction and subcellular localization. A subsegment of the DBD, encompassing zinc fingers 1 to 3, contains the main determinant for nuclear import (NLS) of hMTF-1. The NLS identified here does not conform to the classical stretch of basic amino acids but instead resembles the recently characterized NLS of the zinc finger transcription factor SNAIL1. This novel NLS is composed of six basic amino acids situated at distinct positions in three consecutive zinc fingers. Single mutations of these basic residues in SNAIL1 did not affect its subcellular localization (22). Such a delocalized arrangement of basic residues has also been seen for other zinc finger proteins (such as Egr1, EKLF, RFLAT, SP1, and FEZ1) (22). We note that four of the six basic residues are conserved in zinc fingers 1 to 3 of hMTF-1, and five of six are conserved in Drosophila MTF-1. Thus, we consider it likely that the NLS of MTF-1 is of the same category, with zinc finger folding bringing the basic residues into close proximity, as in a contiguous classical NLS. Although MTF-1's intracellular localization is regulated by the response to stress, notably metal stress, the isolated NLS and NES motifs fused to marker proteins are not zinc responsive but rather are constitutively active, suggesting that metal-dependent nuclear accumulation is mediated by conformational changes that rely on an intact MTF-1 (see also below).
The most elusive feature of MTF-1 function remains its metal inducibility. Previously, we and others showed that MTF-1 requires a higher zinc concentration for DNA binding than other factors, such as Oct-1 and the zinc finger factor Sp1 (13). Moreover, in a cell-free assay system, we demonstrated MTF-1-dependent transcription in response to zinc and even in response to cadmium and copper upon the liberation of zinc from zinc-saturated metallothionein (44). These data suggested an important role of zinc fingers in metal inducibility but did not explain why transfected hMTF-1 is much more metal inducible than mouse MTF-1. Here we show that a crucial region for metal inducibility is the NES/acidic activation domain and that the difference in metal responsiveness between the human and mouse MTF-1 proteins is due to a 3-aa difference in a segment that resembles a recently identified motif, the “9-aa transactivation domain” (27). This motif is characterized by a distinct pattern of hydrophobic patches around positions 3 and 7 and an embedded hydrophilic region around position 5 (see Fig. S2 in the supplemental material). It was identified in the transcription factors Gal4, Oaf1, Pip2, and Sox18 and also occurs within the transactivation domains of VP16, p53, HSF1, NFAT1, and NF-κB, all of which interact with TAF9 (27, 30). Since the above-mentioned factors are not metal inducible, such an arrangement is probably necessary but not sufficient for the metal response. Similarly, interactions of the acidic activation domain of mouse MTF-1 with the cofactor p300 in a zinc-dependent manner (18) and of Drosophila MTF-1 with TFIID and Mediator, two members of regulatory coactivator complexes (21), are probably a prerequisite for metal inducibility. For example, the interaction of the NES with the export receptor Crm1 in nonstressed cells is probably lost upon metal stress, with the concomitant recruitment of coactivators to this domain.
In another attempt to dissect metal inducibility, segments of MTF-1 were fused to the Gal4 DBD. We found that the acidic activation domain of hMTF-1 can act as an autonomous zinc-inducible unit in a Gal4-DBD fusion construct. However, the metal response was not as robust as that with intact hMTF-1, which readily works in every cell line tested so far irrespective of primate or rodent origin. In contrast, metal induction with Gal4-MTF-1 fusions was seen in only a subset of cells tested, probably due to differences in signaling pathways prevalent in particular cell lines. In spite of the caveat that further components are required to bring about a strong, ubiquitous metal inducibility, it is worth pointing out that this is the first demonstration of zinc-responsive transcriptional activation for an isolated MTF-1 activation domain. Since the target genes of MTF-1 are involved in fundamental processes such as metal homeostasis, neurodegeneration, and angiogenesis, advances in the knowledge of MTF-1 regulation will have an impact on these fields.
Supplementary Material
Acknowledgments
We are indebted to Gideon Dreyfuss for the kind gift of Myc-tagged plasmid pcDNA3-PK and to George Hausmann for critical reading of the manuscript.
This work was supported by the Swiss National Science Foundation and the Kanton Zürich.
Footnotes
Published ahead of print on 21 September 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Auf der Maur, A., T. Belser, G. Elgar, O. Georgiev, and W. Schaffner. 1999. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol. Chem. 380:175-185. [DOI] [PubMed] [Google Scholar]
- 2.Brugnera, E., O. Georgiev, F. Radtke, R. Heuchel, E. Baker, G. R. Sutherland, and W. Schaffner. 1994. Cloning, chromosomal mapping and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res. 22:3167-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, X., M. Chu, and D. P. Giedroc. 1999. MRE-binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity, and specificity of the metal-response element complex. Biochemistry 38:12915-12925. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., B. Zhang, P. M. Harmon, W. Schaffner, D. O. Peterson, and D. P. Giedroc. 2004. A novel cysteine cluster in human metal-responsive transcription factor 1 is required for heavy metal-induced transcriptional activation in vivo. J. Biol. Chem. 279:4515-4522. [DOI] [PubMed] [Google Scholar]
- 5.Cook, A., F. Bono, M. Jinek, and E. Conti. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76:647-671. [DOI] [PubMed] [Google Scholar]
- 6.Dalton, T. P., Q. Li, D. Bittel, L. Liang, and G. K. Andrews. 1996. Oxidative stress activates metal-responsive transcription factor-1 binding activity. Occupancy in vivo of metal response elements in the metallothionein-I gene promoter. J. Biol. Chem. 271:26233-26241. [DOI] [PubMed] [Google Scholar]
- 7.Dalton, T. P., W. A. Solis, D. W. Nebert, and M. J. Carvan III. 2000. Characterization of the MTF-1 transcription factor from zebrafish and trout cells. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126:325-335. [DOI] [PubMed] [Google Scholar]
- 8.Fried, H., and U. Kutay. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giedroc, D. P., X. Chen, M. A. Pennella, and A. C. LiWang. 2001. Conformational heterogeneity in the C-terminal zinc fingers of human MTF-1: an NMR and zinc-binding study. J. Biol. Chem. 276:42322-42332. [DOI] [PubMed] [Google Scholar]
- 10.Gorlich, D., N. Pante, U. Kutay, U. Aebi, and F. R. Bischoff. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584-5594. [PMC free article] [PubMed] [Google Scholar]
- 11.Gunes, C., R. Heuchel, O. Georgiev, K. H. Muller, P. Lichtlen, H. Bluthmann, S. Marino, A. Aguzzi, and W. Schaffner. 1998. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 17:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haq, F., M. Mahoney, and J. Koropatnick. 2003. Signaling events for metallothionein induction. Mutat. Res. 533:211-226. [DOI] [PubMed] [Google Scholar]
- 13.Heuchel, R., F. Radtke, O. Georgiev, G. Stark, M. Aguet, and W. Schaffner. 1994. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 13:2870-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izaurralde, E., U. Kutay, C. von Kobbe, I. W. Mattaj, and D. Gorlich. 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16:6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, H., K. Fu, and G. K. Andrews. 2004. Gene- and cell-type-specific effects of signal transduction cascades on metal-regulated gene transcription appear to be independent of changes in the phosphorylation of metal-response-element-binding transcription factor-1. Biochem. J. 382:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 17.Langmade, S. J., R. Ravindra, P. J. Daniels, and G. K. Andrews. 2000. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem. 275:34803-34809. [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., T. Kimura, R. W. Huyck, J. H. Laity, and G. K. Andrews. 2008. Zinc-induced formation of a coactivator complex containing the zinc-sensing transcription factor MTF-1, p300/CBP, and Sp1. Mol. Cell. Biol. 28:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y., T. Kimura, J. H. Laity, and G. K. Andrews. 2006. The zinc-sensing mechanism of mouse MTF-1 involves linker peptides between the zinc fingers. Mol. Cell. Biol. 26:5580-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindert, U., L. Leuzinger, K. Steiner, O. Georgiev, and W. Schaffner. 2008. Characterization of metal-responsive transcription factor (MTF-1) from the giant rodent capybara reveals features in common with human as well as with small rodents (mouse, rat). Chem. Biodivers. 5:1485-1494. [DOI] [PubMed] [Google Scholar]
- 21.Marr, M. T., II, Y. Isogai, K. J. Wright, and R. Tjian. 2006. Coactivator cross-talk specifies transcriptional output. Genes Dev. 20:1458-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mingot, J. M., S. Vega, B. Maestro, J. M. Sanz, and M. A. Nieto. 2009. Characterization of snail nuclear import pathways as representatives of C2H2 zinc finger transcription factors. J. Cell Sci. 122:1452-1460. [DOI] [PubMed] [Google Scholar]
- 23.Muller, H. P., E. Brungnera, O. Georgiev, M. Badzong, K. H. Muller, and W. Schaffner. 1995. Analysis of the heavy metal-responsive transcription factor MTF-1 from human and mouse. Somat. Cell Mol. Genet. 21:289-297. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, B. J., G. K. Andrews, D. Bittel, D. J. Discher, J. McCue, C. J. Green, M. Yanovsky, A. Giaccia, R. M. Sutherland, K. R. Laderoute, and K. A. Webster. 1999. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 59:1315-1322. [PubMed] [Google Scholar]
- 25.Otsuka, F., I. Okugaito, M. Ohsawa, A. Iwamatsu, K. Suzuki, and S. Koizumi. 2000. Novel responses of ZRF, a variant of human MTF-1, to in vivo treatment with heavy metals. Biochim. Biophys. Acta 1492:330-340. [DOI] [PubMed] [Google Scholar]
- 26.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187-198. [DOI] [PubMed] [Google Scholar]
- 27.Piskacek, S., M. Gregor, M. Nemethova, M. Grabner, P. Kovarik, and M. Piskacek. 2007. Nine-amino-acid transactivation domain: establishment and prediction utilities. Genomics 89:756-768. [DOI] [PubMed] [Google Scholar]
- 28.Radtke, F., O. Georgiev, H. P. Muller, E. Brugnera, and W. Schaffner. 1995. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 23:2277-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radtke, F., R. Heuchel, O. Georgiev, M. Hergersberg, M. Gariglio, Z. Dembic, and W. Schaffner. 1993. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 12:1355-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandholzer, J., M. Hoeth, M. Piskacek, H. Mayer, and R. de Martin. 2007. A novel 9-amino-acid transactivation domain in the C-terminal part of Sox18. Biochem. Biophys. Res. Commun. 360:370-374. [DOI] [PubMed] [Google Scholar]
- 31.Saydam, N., T. K. Adams, F. Steiner, W. Schaffner, and J. H. Freedman. 2002. Regulation of metallothionein transcription by the metal-responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J. Biol. Chem. 277:20438-20445. [DOI] [PubMed] [Google Scholar]
- 32.Saydam, N., O. Georgiev, M. Y. Nakano, U. F. Greber, and W. Schaffner. 2001. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 276:25487-25495. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seipel, K., O. Georgiev, H. P. Gerber, and W. Schaffner. 1993. C-terminal domain (CTD) of RNA-polymerase II and N-terminal segment of the human TATA binding protein (TBP) can mediate remote and proximal transcriptional activation, respectively. Nucleic Acids Res. 21:5609-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 11:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smirnova, I. V., D. C. Bittel, R. Ravindra, H. Jiang, and G. K. Andrews. 2000. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 275:9377-9384. [DOI] [PubMed] [Google Scholar]
- 38.Stuart, G. W., P. F. Searle, H. Y. Chen, R. L. Brinster, and R. D. Palmiter. 1984. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc. Natl. Acad. Sci. USA 81:7318-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., U. Wimmer, P. Lichtlen, D. Inderbitzin, B. Stieger, P. J. Meier, L. Hunziker, T. Stallmach, R. Forrer, T. Rulicke, O. Georgiev, and W. Schaffner. 2004. Metal-responsive transcription factor-1 (MTF-1) is essential for embryonic liver development and heavy metal detoxification in the adult liver. FASEB J. 18:1071-1079. [DOI] [PubMed] [Google Scholar]
- 40.Weaver, R. F., and C. Weissmann. 1979. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5′ termini of 15 S beta-globin mRNA precursor and mature 10 S beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 7:1175-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westin, G., T. Gerster, M. M. Muller, G. Schaffner, and W. Schaffner. 1987. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 15:6787-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wimmer, U., Y. Wang, O. Georgiev, and W. Schaffner. 2005. Two major branches of anti-cadmium defense in the mouse: MTF-1/metallothioneins and glutathione. Nucleic Acids Res. 33:5715-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, B., D. Egli, O. Georgiev, and W. Schaffner. 2001. The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell. Biol. 21:4505-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, B., O. Georgiev, M. Hagmann, C. Gunes, M. Cramer, P. Faller, M. Vasak, and W. Schaffner. 2003. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol. Cell. Biol. 23:8471-8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.