Abstract
The hepatitis B virus (HBV) envelope proteins bear two determinants of viral entry: a receptor-binding site (RBS) in the pre-S1 domain of the large envelope protein and a conformation-dependent determinant, of unknown function, in the antigenic loop (AGL) of the small, middle, and large envelope proteins. Using an in vitro infection assay consisting of susceptible HepaRG cells and the hepatitis delta virus (HDV) as a surrogate of HBV, we first investigated whether subelements of the pre-S1 determinant (amino acids 2 to 75), i.e., the N-terminal myristoyl anchor, subdomain 2-48 (RBS), and subdomain 49-75, were functionally separable. In transcomplementation experiments, coexpression of two distinct infectivity-deficient pre-S1 mutants at the surface of HDV virions failed to restore infectivity, indicating that the myristoyl anchor, the 2-48 RBS, and the 49-75 sequence, likely cooperate in cis at viral entry. Furthermore, we showed that as much as 52% of total pre-S1 in the HDV envelope could bear infectivity-deficient lesions without affecting entry, indicating that a small number of pre-S1 polypeptides—estimated at three to four per virion—is sufficient for infectivity. We next investigated the AGL activity in the small or large envelope protein background (S- and L-AGL, respectively) and found that lesions in S-AGL were more deleterious to infectivity than in L-AGL, a difference that reflects the relative stoichiometry of the small and large envelope proteins in the viral envelope. Finally, we showed that C147S, an AGL infectivity-deficient substitution, exerted a dominant-negative effect on infectivity, likely reflecting an involvement of C147 in intermolecular disulfide bonds.
Hepatitis B virus (HBV) remains a major public health concern worldwide, affecting more than 350 millions of chronically infected individuals. Since the discovery of HBV, substantial information has been gathered on the viral replication cycle, but our understanding of the viral entry mechanism remains limited, and the identity of the receptor(s) for HBV is still unknown (15). HBV displays a very narrow host range, which is likely determined at viral entry by a highly specific interaction between the HBV envelope proteins and receptors at the surface of human hepatocytes. The envelope proteins designated large (L-HBsAg), middle (M-HBsAg), and small (S-HBsAg) are membrane-spanning glycoproteins that differ from each other by the size of their N-terminal ectodomain (21). L-HBsAg contains a N-terminal pre-S1, central pre-S2, and C-terminal S domains. M-HBsAg is shorter than L-HBsAg in lacking pre-S1, whereas S-HBsAg consists of the S domain only (Fig. 1). Envelope protein synthesis occurs at the endoplasmic reticulum (ER) membrane. Empty subviral particles (SVPs) assemble from aggregates at a pre-Golgi membrane and exit the cell through the secretory pathway (36). Assembly of mature HBV virions requires, in addition to S-HBsAg, the presence of L-HBsAg as a matrix protein for nucleocapsid envelopment (6). Recent findings indicate that HBV virions and SVPs follow distinct pathways for budding: the late endosomal multivesicular bodies (MVBs) for HBV virions, and the MVB-independent secretory pathway for SVPs (26, 28, 46). The HBV envelope proteins can also package the hepatitis delta virus (HDV) ribonucleoprotein (RNP), in case of HBV/HDV coinfection (5, 45), leading to the formation of HDV virions. Whether HDV uses the SVP secretion pathway rather than an MVB-dependent route is uncertain.
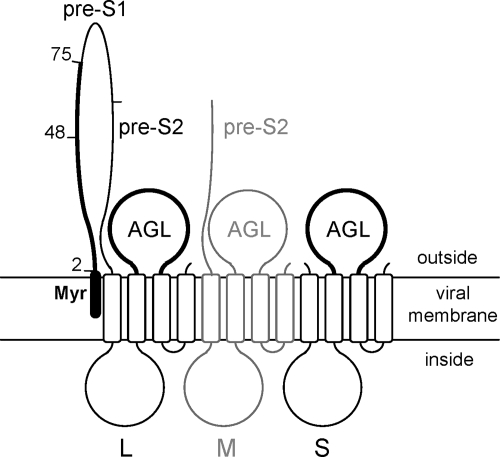
FIG. 1.
Schematic representation of HBV envelope proteins. The topology of the L-, M-, and S-HBsAg proteins at the viral membrane is represented. The pre-S2 domain of L- and M-HBsAg, and the determinants of viral entry, pre-S1 and AGL, are indicated. The M-HBsAg protein, represented in gray, is dispensable for infectivity. The myristic acid (Myr) linked to the L-HBsAg N terminus is indicated (closed box). Subdomains 2-48 and 49-75 of the pre-S1 infectivity determinant are indicated. Open boxes represent transmembrane regions in the S domain.
L-HBsAg, but not M-HBsAg, is crucial to infectivity of both HBV and HDV particles (13, 31, 41, 42). L-HBsAg contains a major infectivity determinant located between amino acid residues 2 and 75 of its N-terminal pre-S1 domain (4, 30), including a myristoyl anchor linked to glycine-2 (1, 8, 18), a putative receptor binding site (RBS) between positions 2 and 48, and a domain of unknown function between amino acids 49 and 75. To date, the most compelling evidence that pre-S1 mediates receptor binding comes from studies demonstrating that myristoylated synthetic peptides specific for the N-terminal 2-to-48 pre-S1 domain can bind to hepatocyte plasma membranes and block infection in vitro (3, 16, 17) and in vivo (37). Beside pre-S1, a second determinant was recently identified in the antigenic loop (AGL) borne by the three HBV envelope proteins (Fig. 1). The AGL participation in viral entry was first established in the HDV model (23) and more recently directly in the HBV model (39). Interestingly, serine substitutions for the AGL cysteine residues, which prove detrimental to the conserved immunodominant “a” determinant, could also block viral entry. Note that the “a” determinant consists in conformational epitopes, which elicit highly neutralizing antibodies (22). Infectivity and the “a” determinant were also lost when virions were treated with membrane-impermeable inhibitors of thiol/disulfide isomerization (2). These findings clearly established a correlation between the AGL cysteine disulfide bonds network, the conformation of the “a” determinant, and infectivity. Hence, the strict conservation of the “a” determinant among all HBV genotypes is related to the AGL function at viral entry. The AGL determinant may operate in association with, or independently of pre-S1, in binding to receptors at the early step of entry and/or in the mechanism of envelope disassembly postentry.
In the present study, we investigated the pre-S1 determinant by performing transcomplementation experiments between mutants of 3 pre-S1 subelements: the myristoyl anchor, subdomain 2-48, and subdomain 49-75. We analyzed the activity of the AGL determinant in the S- or L-HBsAg background (S- and L-AGL, respectively), and we examined the effect of introducing increasing amounts of infectivity-deficient pre-S1, or AGL, in the virion's envelope on infectivity.
MATERIALS AND METHODS
Plasmids. HDV recombinant plasmid pSVLD3 was used for replication of HDV RNA and production of HDV RNP (27). HBV expression vectors p123 and p124 were used for the production of the wild-type (wt) S- and L-HBsAg proteins, respectively (40). Vectors for the expression of L-HBsAg mutants were derived from p124 by using the PCR technique with two complementary mutagenic oligonucleotides as described previously (25). Mutations in pre-S1 consist in a five-residue deletion and the insertion of Lys-Leu dipeptide as described previously (4). Plasmid p124 Δ115-153 vector encodes a truncated but fully functional L-HBsAg protein, and plasmid p2713 codes for G2A L-HBsAg lacking the myristoylation signal, as previously described (1).
AGL mutations initially created in pTHB2.7 plasmid (2, 23) were introduced in p123 and p124 plasmids (40). A XbaI-NcoI HBV DNA fragment containing the mutation was excised from pTHB2.7-derived mutant plasmid and inserted between the XbaI and NcoI restriction sites of both p123 and p124 plasmids. The mutations are designated by the one-letter code for the targeted amino acid followed by its position in the S domain of the HBV envelope protein (genotype D), and the one-letter code for the substituted amino acid.
To discriminate between wt and mutant S-HBsAg proteins by Western blot analysis, a plasmid coding for wt S-HBsAg bearing a C-terminal influenza virus hemagglutinin (HA) epitope sequence was used as a substitute for wt S-HBsAg (note that the HA tag alters neither HDV assembly nor infectivity).
Production and characterization of HDV particles.
Production of HDV particles was achieved in the Huh-7 human hepatoma cell line as previously described (4). Cells (106/well) were transfected with 1 μg of pSVLD3 and 1 μg each of p123 and p124 (or their derivatives) (40). Transfection was carried out using the Fugene-HD reagent according to the manufacturer's directions (Roche). Culture supernatants were harvested at day 6, 8, 10, and 12 posttransfection and then pooled and analyzed by immunoblotting for the detection of HBV envelope proteins (4). Note that detection of HBV envelope proteins was achieved using a rabbit anti-S antibody (R247) that recognizes a linear epitope in the cytosolic loop-I of the envelope proteins, and a rabbit anti-pre-S2 antibody (R257) for specific detection of L- and M-HBsAg proteins. The relative levels of the immunoblot signals for HBV envelope proteins thus do not reflect the real ratio of L-HBsAg to S-HBsAg. For detection of L-HBsAg proteins bearing deletions in pre-S2 (Δ115-153), a rabbit anti-pre-S1 antibody (R254) specific for the 83-to-106 pre-S sequence was used. Immunoblot signals were quantified by using the NIH ImageJ. 1.41O software. When indicated, PNGase F treatment of envelope proteins was performed as previously reported (4). Supernatants harvested at days 14 and 16 posttransfection served as inocula in infectivity assays after normalization for their HDV RNA content by Northern blot analysis (4).
In vitro infection assay of HepaRG cells.
HepaRG cells were maintained and differentiated as described previously (19). For infection with HDV virions, clarified supernatants from Huh-7 cells were used as inocula after normalization for HDV RNA. HepaRG cells (106 cells/well) were exposed to ∼108 genome equivalents (ge) of HDV virions for 16 h in the presence of 5% polyethylene glycol (PEG) 8000 (Sigma). Cells were harvested at day 7 postinoculation in RLT buffer (Qiagen). Total cellular RNA was purified by using an RNeasy minikit (Qiagen) and tested for the presence of genomic HDV RNA as evidence of infection.
RESULTS
The present analysis of HBV envelope proteins at viral entry was performed using the HDV model as a surrogate for HBV as previously described (43). Production of HDV virions was achieved by transfection of Huh-7 with a mixture of an expression vector for HDV RNP, and two expression vectors for S- and L-HBsAg envelope proteins, respectively (M-HBsAg was omitted because dispensable for infectivity) (7, 13, 29). The resulting particles, designated SL-HDV, bear S- and L-HBsAg proteins only. Infectivity was then assayed by inoculation of Huh-7 cell-derived virions to susceptible HepaRG cells, and measurement of intracellular accumulation of HDV RNA at day 7 postinoculation. HDV infection being nonproductive in the absence of the helper HBV, the level of intracellular viral RNA that accumulates in infected HepaRG cells is proportional to the inoculum titer (4).
Subelements of the pre-S1 infectivity determinant (amino acids 2 to 75) are nonseparable.
It was recently demonstrated that the N-terminal 75 amino acid residues of the L-HBsAg protein constitute the minimum pre-S1 infectivity determinant (4, 30). This sequence can be arbitrarily divided in three subelements: (i) the myristoyl anchor linked to glycine 2, (ii) the sequence from amino acids 2 to 48 likely to contain an RBS, and (iii) the sequence from amino acids 49 to 75 of unknown function. Here, to gain mechanistic insight into the pre-S1 determinant, we addressed the possibility of transcomplementation between infectivity-deficient L-HBsAg mutants of the following types: type 1 was a mutant (G2A) deficient for myristoylation (1); type 2 were mutants bearing deletions in the 2-to-48 sequence (Δ11-15, Δ16-20, Δ21-25, and Δ31-35) and type 3, mutants with deletions in the 49-to-75 sequence (Δ51-55, Δ56-60, Δ61-65, and Δ66-70) (4). All mutations were previously reported as permissive for L-HBsAg synthesis and incorporation into the HDV envelope, but detrimental to infectivity (4). Intra- or intertypic transcomplementations were attempted using SL-HDV particles produced by transfection of Huh-7 cells with a wt S-HBsAg expression vector and equivalent amounts of plasmids for expression of two distinct L-HBsAg mutants as indicated in Fig. 2. As shown in Fig. 2A, wt and mutant SL-HDV particles displayed identical characteristics in terms of envelope proteins and HDV RNA content. Prior to infectivity assays, each preparation of SL-HDV was adjusted to ∼108 ge/ml and inoculated to HepaRG cells in the presence of 5% PEG as described previously (4). As shown in Fig. 2B, none of the transcomplementation combinations was successful in restoring infectivity. This observation suggests that subelements of the pre-S1 infectivity determinant cannot be separated, implying their cooperation in cis in the mechanism of viral entry.
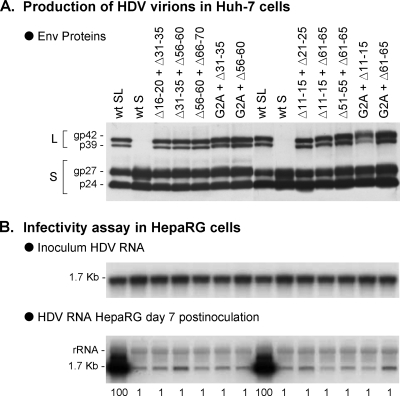
FIG. 2.
Relative infectivity of HDV particles bearing two distinct infectivity-deficient pre-S1 mutants. SL-HDV particles bearing wt S-HBsAg proteins and two distinct infectivity-deficient L-HBsAg proteins in equivalent proportions were produced in Huh-7 cells. Infectivity-deficient L-HBsAg proteins included G2A, a protein lacking a myristoylation signal, and proteins with small deletions in the 2-to-48 (Δ11-15, Δ16-20, Δ21-25, and Δ31-35) or the 49-to-75 pre-S1 sequence (Δ51-55, Δ56-60, Δ61-65, and Δ66-70). (A) Particles from 0.5 ml of cell culture supernatant were analyzed for S- and L-HBsAg proteins by immunoblotting. The glycosylated (gp) and nonglycosylated (p) forms of S- and L-HBsAg are indicated. (B) Genomic HDV RNA from 140 μl of supernatant was analyzed by Northern blotting. After normalization of the different preparations of HDV virions for their HDV RNA content, they were assayed for infectivity in cultures of HepaRG cells. Infectivity was evaluated by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV. The experiment was repeated with consistent results.
Dose-dependent effect of an infectivity-deficient pre-S1 at viral entry.
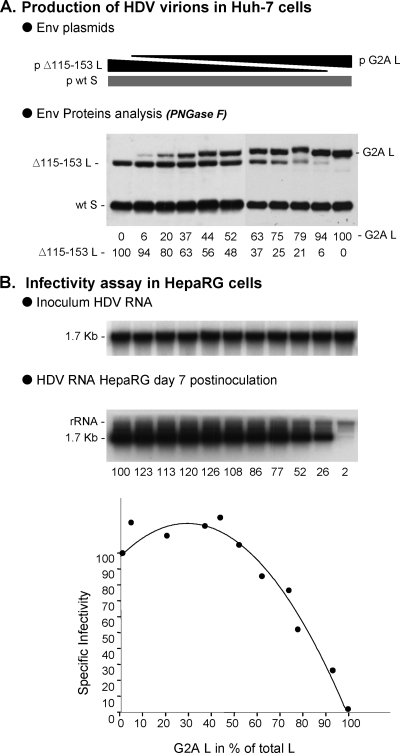
Assuming that pre-S1 plays a role in receptor recognition, a minimum number of pre-S1 molecules may be required at the surface of an infectious virion. We addressed this question indirectly by measuring the dose-dependent effect of incorporating an infectivity-deficient pre-S1 mutant in the HDV envelope. We produced SL-HDV particles bearing various ratios of G2A/wt pre-S1: the G2A substitution leads to the synthesis of a nonmyristoylated, infectivity-deficient pre-S1 (1). SL-HDV production was achieved as described above, except that in place of the wt L-HBsAg expression vector, a mixture of wt and G2A L-HBsAg encoding plasmids was used in the transfection procedure. The different ratios of G2A/wt L- HBsAg vectors are indicated in the legend to Fig. 3. To monitor for a dose-dependent incorporation of G2A L-HBsAg in the SL-HDV envelope, we modified our experimental procedure by substituting to wt L-HBsAg expression vector, a plasmid for expression of Δ115-153 L-HBsAg, a protein bearing a deletion previously demonstrated as fully permissive to infectivity (4). The Δ115-153 and G2A L-HBsAg proteins having different molecular weights, we could quantify their relative incorporation in the viral envelope by performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis with an anti-pre-S1 antibody (R254) that equally recognizes the two proteins. To further facilitate quantification, envelope protein samples were treated with PNGase F prior to SDS-PAGE to resolve each Δ115-153 and G2A L-HBsAg (ca. 50% of each are glycosylated at Asn-146 in the S domain) as a single form of unglycosylated protein. As shown in Fig. 3A, L-HBsAg proteins were detected at the expected G2A/Δ115-153 ratios in the different preparations of HDV particles.
FIG. 3.
Relative infectivity of HDV particles bearing increasing ratios of G2A infectivity-deficient to wt pre-S1. Production of SL-HDV particles was achieved by transfection of 106 Huh-7 cells with 1 μg of pSVLD3 plasmid for production of HDV RNP, 1 μg of p123 for expression of wt S-HBsAg, together with 1 μg of a mixture of Δ115-153 L-HBsAg (as a surrogate for wt L-HBsAg) and G2A L-HBsAg expression vectors. The ratios of G2A to wt plasmids used for transfection were 0/100, 10/90, 20/80, 30/70, 40/60, 50/50, 60/40, 70/30, 80/20, 90/10, and 100/0. (A) Particles from 0.5 ml of supernatant from transfected cells were analyzed for S- and L-HBsAg proteins by immunoblotting after PNGase F treatment. Quantification of G2A and Δ115-153 proteins signals is indicated as percentages of total L-HBsAg. (B) After normalization of the different preparations of HDV particles for their viral RNA content, infectivity was evaluated in cultures of HepaRG cells by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of intracellular HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV. Specific infectivity (y axis) is indicated as a function of the G2A/total L-HBsAg ratio in the viral envelope (x axis).
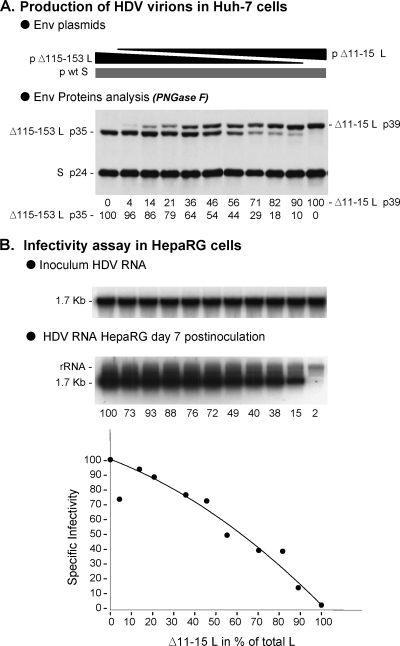
When particles were assayed for infectivity in HepaRG cells after normalization for HDV RNA, a nonlinear, dose-dependent effect of G2A L-HBsAg was observed. It is remarkable that G2A L-HBsAg could account for as much as 52% of total L-HBsAg proteins without affecting infectivity (Fig. 3), clearly indicating that only a limited number of pre-S1 polypeptides at the surface of HDV virion suffices to confer infectivity. We attempted to calculate this number by first separating cell culture-derived HDV virions from SVPs by two successive rate-zonal sedimentation procedures, followed by quantification of the S- and L-HBsAg proteins in the HDV RNA-positive fractions by Western blotting with an anti-HBsAg antibody that equally recognizes both proteins (24). The percentage of L-HBsAg relative to L- + S-HBsAg was at 5 to 6% (data not shown). Note that this estimation is very close to the one obtained for in vitro-produced, HDV particles (10), but slightly different from that of serum-derived virions in which L-HBsAg was reported to account for only 1% of total envelope proteins (5). Then, we estimated the number of total envelope proteins at the surface of an HDV particle (36 nm in diameter) to be 130, based on the measurement by Gilbert et al. (14) of 48 S-HBsAg polypeptides per 22 nm-diameter SVP. An average HDV virion would thus bear seven to eight L-HBsAg proteins. If, as shown here, infectivity-deficient L-HBsAg mutants can account for 52% of the total L-HBsAg proteins without consequence at viral entry, as few as three to four L-HBsAg molecules per virion would suffice to confer infectivity. Using a different infectivity-deficient L-HBsAg protein, Δ11-15 (4), similar results were obtained whereby SL-HDV particles in which Δ11-15 L-HBsAg accounted for 46% of total L-HBsAg, maintained infectivity at 72% of that of wt particles (Fig. 4).
FIG. 4.
Relative infectivity of HDV particles bearing increasing ratio of Δ11-15 infectivity-deficient/wt pre-S1. Production of SL-HDV particles was achieved by transfection of 106 Huh-7 cells with 1 μg of pSVLD3 plasmid for production of HDV RNP, 1 μg of p123 for expression of wt S-HBsAg, together with 1 μg of a mixture of Δ115-153 L-HBsAg (as a surrogate for wt L-HBsAg) and Δ11-15 L-HBsAg expression vectors. The ratios of Δ11-15 to Δ115-153 plasmids used for transfection were 0/100, 10/90, 20/80, 30/70, 40/60, 50/50, 60/40, 70/30, 80/20, 90/10, and 100/0. (A) Particles from 0.5 ml of supernatant from transfected cells were analyzed for S- and L-HBsAg proteins by immunoblotting. Quantification of Δ11-15 and Δ115-153 proteins signals is indicated as percentages of total L-HBsAg. (B) After normalization of the different preparations of HDV particles for their viral RNA content, infectivity was evaluated in cultures of HepaRG cells by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of intracellular HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV. Specific infectivity (y axis) is indicated as a function of the Δ11-15/total L-HBsAg ratio in the viral envelope (x axis).
S-AGL is critical for infectivity.
It was shown recently that the AGL bears a determinant of infectivity, which is closely related to the immunodominant “a” determinant. This was demonstrated using HDV or HBV particles bearing AGL substitutions in L-, M-, and S-HBsAg proteins (2, 39). Since M-HBsAg—and therefore M-AGL—is dispensable at viral entry, the functional AGL can be L-AGL and/or S-AGL.
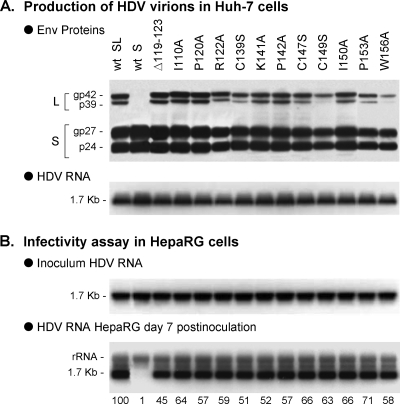
To sort between L- and S-AGL activity, we first produced SL-HDV bearing lesions in L-AGL. Such lesions consisted in a small deletion (Δ119-123) or single amino acid substitutions (I110A, P120A, R122A, C139S, K141A, P142A, C147S, C149S, I150A, P153A, and W156A), which have been described previously as drastically reducing HDV infectivity when present in all three S-, M-, and L-HBsAg proteins (2, 39). SL-HDV particles production was achieved as described above, except that in place of wt L-HBsAg expression vector, derivative plasmids bearing the above-mentioned mutations were used in the transfection procedure. Supernatants from transfected Huh-7 cells were characterized as indicated in the legend to Fig. 2. The results show that SL-HDV mutants displayed the characteristics of wt virions in terms of envelope proteins and HDV RNA content (Fig. 5A). When assayed for infectivity in HepaRG cell cultures, all SL-HDV mutants demonstrated a limited loss of infectivity relative to wt HDV ranging between 45% (Δ119-123) and 71% (P153A). (Fig. 5B).
FIG. 5.
Effect of L-AGL mutations on HDV infectivity. SL-HDV particles bearing mutations in L-AGL were produced in Huh-7 cells. Mutations included a deletion of residues 119 to 123 (Δ119-123) in the S domain of L-HBsAg, or single amino acid substitutions as indicated. (A) Particles from 0.5 ml of cell culture supernatant were analyzed for S- and L-HBsAg proteins by immunoblotting. The glycosylated (gp) and nonglycosylated (p) forms of S- and L-HBsAg are indicated. Genomic HDV RNA from 140 μl of supernatant was analyzed by Northern blotting. (B) After normalization of the different preparations of HDV particles for their viral RNA content, they were assayed for infectivity in cultures of HepaRG cells. Infectivity was evaluated by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV. The experiment was repeated with consistent results.
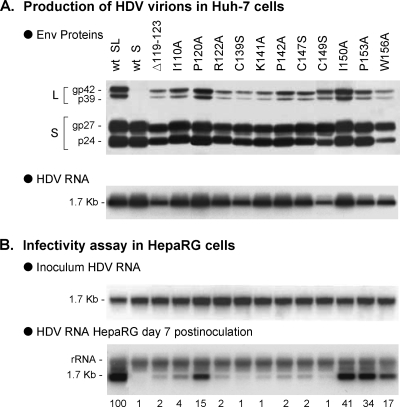
When mutations were borne by S-AGL (Fig. 6), infectivity was more severely affected. For most mutants, e.g., the Δ119-123, I110A, R122A, C139S, K141A, P142A, C147S, and C149S mutants, infectivity was measured at 1 to 4% of that of wt HDV. For P120A, I150A, P153A, and W156A, a smaller effect was observed: 15, 41, 34, and 17%, respectively of that of wt HDV.
FIG. 6.
Effect of S-AGL mutations on HDV infectivity. SL-HDV particles bearing mutations in S-AGL were produced in Huh-7 cells. (A) Particles from 0.5 ml of cell culture supernatant were analyzed for S- and L-HBsAg proteins by immunoblotting. The glycosylated (gp) and nonglycosylated (p) forms of S- and L-HBsAg are indicated. Genomic HDV RNA from 140 μl of supernatant was analyzed by Northern blotting. (B) After normalization of the different preparations of HDV virions for their HDV RNA content, infectivity was evaluated in cultures of HepaRG cells by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV. The experiment was repeated with consistent results.
Taken together, these results show that L-AGL is not critical to HDV infectivity, suggesting that it does not function in cis with pre-S1. Infectivity is most affected when S-AGL is deficient. It may pertain to a particular conformation, or surface accessibility of S-AGL, or the relative stoichiometry of S- and L-HBsAg. If S-HBsAg accounts for 95 to 99% of the HBV envelope proteins at the surface of HDV (5), infectivity-deficient lesions in S-AGL should have much greater impact on infectivity than should the same lesions in L-AGL (see discussion).
Dose-dependent effect of infectivity-deficient AGL at viral entry.
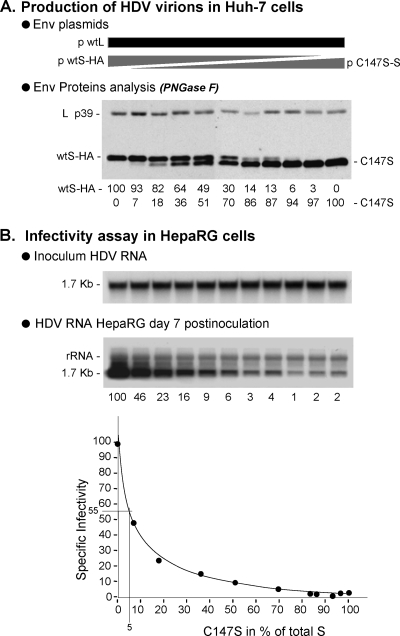
To measure the dose-dependent effect of an infectivity-deficient AGL substitution, we selected C147S, one of the most deleterious substitutions in the 3-envelope-protein background (2), to produce SL-HDV particles bearing various ratios of C147S/wt S-HBsAg proteins. SL-HDV particles production was achieved as described above, except that wt S-HBsAg expression plasmid was replaced with a mixture of wt and C147S S-HBsAg expression vectors in the transfection procedure. The different ratios of C147S/wt S-HBsAg encoding plasmids are indicated in the legend to Fig. 7. To monitor for a dose-dependent incorporation of C147S S-HBsAg in SL-HDV envelope, we substituted a HA-tagged wt S-HBsAg for wt S-HBsAg expression vector in the transfection procedure. Note that the HA-tag has no significant impact on HDV assembly or infectivity (data not shown). As a consequence, the incorporation of C147S S-HBsAg in the envelope of secreted particles is easily monitored by SDS-PAGE and immunoblotting because C147S and wt HA-tagged proteins have different molecular weights. As above, prior to SDS-PAGE, samples were treated with PNGase F to resolve each type of envelope protein as a single band of unglycosylated form. As depicted in Fig. 7A, C147S and HA-tagged wt S-HBsAg were detected at the expected ratios in the different preparations of HDV particles, indicating that the C147S substitution does not affect the capacity of the mutant to coassemble with HA-tagged wt S-HBsAg. When particles were assayed for infectivity in HepaRG cell cultures after normalization for HDV RNA, a dominant-negative effect of C147S was observed. As shown in Fig. 7B, when C147S was measured at 51% of total S-HBsAg in the viral envelope, infectivity was down to 9% of that of the wt HDV.
FIG. 7.
Relative infectivity of HDV particles bearing increasing ratio of C147S infectivity-deficient/wt AGL. Production of SL-HDV particles was achieved by transfection of 106 Huh-7 cells with 1 μg of pSVLD3 plasmid for production of HDV RNP, 1 μg of p124 for expression of wt L-HBsAg, together with 1 μg of a mixture of HA-tagged wt S-HBsAg and C147S S-HBsAg expression vectors. Ratios of C147S/wt plasmids were 0/100, 10/90, 20/80, 30/70, 40/60, 50/50, 60/40, 70/30, 80/20, 90/10, and 100/0. (A) Particles from 0.5 ml of supernatant from transfected cells were analyzed for S- and L-HBsAg proteins by immunoblotting after PNGase F treatment. Quantification of HA-tagged (wtS-HA) and C147S S-HBsAg signals is indicated as percentages of total S-HBsAg proteins. (B) After normalization of the different preparations of HDV virions for HDV RNA, infectivity was evaluated in cultures of HepaRG cells by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of intracellular HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV. Specific infectivity (y axis) is indicated as a function of the C147S/total S-HBsAg ratio in the viral envelope (x axis).
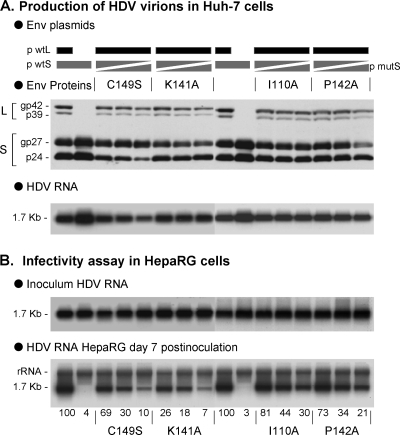
We conducted a similar analysis using different infectivity-deficient AGL mutations: I110A, K141A, P142A, and C149S. In each case, we prepared SL-HDV particles bearing 0, 25, 50, and 75% of mutant relative to total S-HBsAg. As shown in Fig. 8, increasing the proportion of mutant S-HBsAg in SL-HDV envelope translated into a dose-dependent loss of infectivity. However, unlike C147S, a dominant-negative effect was not as clear, since the substitution of 50% of total S-HBsAg envelope proteins with, C149S, K141A, I110A, or P142A, resulted in a lesser loss of infectivity, recorded at 30, 18, 44, and 34%, relative to wt HDV, respectively. The difference between C149S, K141A, I110A, P142A, and C147S substitutions may correspond to different functions of the AGL at viral entry.
FIG. 8.
Relative infectivity of HDV particles bearing increasing ratio of various infectivity-deficient S-AGL. Production of SL-HDV particles was achieved by transfection of 106 Huh-7 cells with 1 μg of pSVLD3 plasmid for production of HDV RNP, 1 μg of p124 for expression of wt L-HBsAg, together with 1 μg of a mixture of wt S-HBsAg and either C149S, K141A, I110A, and P142A S-HBsAg expression vectors. For each assay, the ratios of mutant to wt plasmids were 25/75, 50/50, and 75/25. (A) Particles from 0.5 ml of supernatant from transfected cells were analyzed for S- and L-HBsAg proteins by immunoblotting. The glycosylated (gp) and nonglycosylated (p) forms of S- and L-HBsAg are indicated. (B) After normalization of the different preparations of HDV virions for viral RNA, infectivity was evaluated in cultures of HepaRG cells by measuring the accumulation of HDV RNA in cells harvested at day 7 postinoculation. Quantification of intracellular HDV RNA signals by phosphorimager is indicated as percentages of the value for wt SL-HDV.
DISCUSSION
One major impediment in the study of the HBV entry pathway remains the lack of information on cellular receptor(s) for the virus. The HBV envelope proteins bear at least two determinants of viral entry: (i) the 75 first residues of pre-S1 (4, 30) and (ii) the conformational AGL determinant (2, 39). The pre-S1 2-75 sequence is thought to contain all of the necessary information for receptor recognition as shown by the potent neutralization activity of anti-preS1 antibodies (35) and preS1-specific lipopeptides (3, 16, 17), but in the absence of information on the cell receptor engaged by pre-S1, the receptor-binding mechanism remains hypothetical. Here, to gain insights into the role of pre-S1, we first addressed the possibility of assigning separable functions to 3 pre-S1 subelements: (i) the N-terminal myristoyl moiety and recognition sequence for N-myristoyltransferase (amino acids 1 to 8), (ii) the downstream domain including a putative RBS (2-47), 48 and (iii) the C-terminal amino acids 49 to 75. Clearly, our results are not in agreement with pre-S1 subelements acting independently of each other, but rather in support of their cooperation in cis during entry. Such a mechanism was proposed by Urban et al. (44) for the duck hepatitis B virus pre-S infectivity determinant. In that model, virus attachment to its receptor, carboxypeptidase D, would initiate with a low-affinity binding mediated by a pre-S C-terminal α-helix, followed by the stabilization of the complex by a pre-S N-terminal structure. HBV entry may involve the coordinated functions of distinct subdomains within pre-S1 2-75: amino acids 2 to 48, possibly with the assistance of the myristoyl anchor, could provide the receptor binding function that would be predefined, or stabilized, by the 49-75 domain acting as an allosteric effector. Recent analysis indicate that pre-S1 is a natively unstructured polypeptide containing multiple prestructured motifs within the N-terminal 40 residues that could serve as “recognition antennae” for receptor interactions (11).
Whatever mechanism is engaged by pre-S1 for receptor binding, a rather small number of these molecules, estimated here at three to four per virion, appears to suffice for infectivity. However, we feel that these numbers should be confirmed by additional experiments, because (i) they are based on the assumption that the preparations of SL-HDV virions that were used for our analysis were free of SVPs and homogeneous with regard to envelope proteins composition (i.e., assuming that every single HDV virion bears 5% of L-HBsAg) and (ii) the presence of PEG in the infectivity assay may create aggregates of particles and, thereby, introduce a bias into our calculations. Experiments are being conducted to obtain a more precise quantification using preparations of SVP-free virions in PEG-free infection assays. In addition, further information is needed on the three-dimensional structure of the functional pre-S1 determinant to establish a precise stoichiometry of HBV envelope protein in the entry process. This type of analysis has been achieved recently for the human immunodeficiency virus type 1 entry, which may require a single viral envelope protein trimer to achieve virus entry (32, 47).
With respect to the AGL, its role is as yet unclear, and we cannot exclude that it be engaged in multiple functions, including (i) an initial binding to cell surface-exposed glycosaminoglycans, (ii) a specific attachment to a receptor (or coreceptor at a postattachment step), and (iii) a role in the envelope disassembly process postentry. Furthermore, entry may require cooperation between pre-S1 and AGL, whereby the latter could, for instance, assist pre-S1 in the formation of a functional RBS. In view of our results, the AGL is more likely to act as an allosteric activator in trans (S-AGL) rather than in cis (L-AGL) on pre-S1 to promote receptor binding or to engage a postbinding event. In addition, or alternatively, the AGL could act independently of pre-S1, in which case lesions in S-AGL would be expected to have a strong impact on infectivity as a consequence of the relative stoichiometry of S- and L-HBsAg at the surface of HDV particles (5, 9, 21).
The AGL infectivity determinant is conserved in the sequences of all human HBV genotypes, the woolly monkey hepatitis B virus and, to a lesser extent, the woodchuck hepatitis virus (WHV), suggesting that it is not species specific. A recent study by Gudima et al. (20), showed that HDV particles bearing the WHV envelope proteins could infect human hepatocytes in vitro, whereas HDV virions bearing the HBV L- and S-HBsAg proteins were noninfectious in woodchuck hepatocyte cultures. In addition, substituting the small WHV envelope protein for S-HBsAg in the later particles failed to confer infectivity in woodchuck hepatocyte cultures, suggesting also that the S-AGL is not sufficient for species specificity at viral entry.
In the present study, we observed that the introduction of infectivity-deficient substitutions in the sole L-AGL did not abolish infectivity, as opposed to the effect of the same substitutions in the three-envelope-protein background, suggesting that pre-S1 does not require L-AGL for function. If the contrary were true, any infectivity-deficient substitution in L-AGL would abrogate infectivity. In contrast, most of the infectivity-deficient substitutions (I110A, R122A, C139S, K141A, P142A, C147S, and C149S) demonstrated a strong inhibitory effect when present in S-HBsAg only (Fig. 6), which was often as potent as that observed in the three-envelope-protein background (2, 39). There are two possible explanations to these results: (i) S- and L-AGL are functionally indistinguishable, and therefore substitutions introduced in L-AGL lead to a small number of modified AGLs per virion, and (ii) S-AGL is more active than L-AGL as the result of differences of structure or accessibility at the virion's surface. Our results are clearly in support of the former explanation, whereby AGL infectivity-deficient substitutions have a greater impact in the S-HBsAg background, solely because S-AGLs are more numerous than L-AGLs in the viral envelope. In fact, virions bearing as little as 7% of S-AGLs with a C147S substitution, demonstrate a greater than twofold reduction of infectivity (Fig. 7). Assuming that the HDV envelope consists of 95% S-HBsAg and 5% L-HBsAg (5) and that S- and L-AGL are identical, C147S would be expected to have equivalent impact on infectivity when present on 100% of the L-AGLs or 5% of the S-AGLs. As shown here, C147S in all L-AGLs reduced HDV infectivity to 66% of that of the wt (Fig. 5), whereas according to the dose-response curve shown in Fig. 7, the same substitution in 5% of S-AGLs leads to an infectivity down to 55% of that of wt HDV. These results are thus consistent with the assumption that S- and L-AGL are functionally indistinguishable. However, it is important to mention that the detrimental effect of a given AGL substitution may vary according to the nature of the modified amino acid residue (Fig. 8). As investigated here, AGL substitutions targeting cysteines, positively charged or uncharged hydrophobic residues, may impact different functions of the AGL determinant. For instance, the I150A substitution that reduced infectivity to only 41% relative to wt, may pertain to an AGL function other than the one affected by the C139S, K141A, P142A, C147S, or C149S substitutions (Fig. 6).
Finally, the peculiar dominant-negative impact observed with C147S is in agreement with an implication of cysteine-147 in intermolecular disulfide bonds as previously reported (33, 34). Substitution of one cysteine residue in proteins, such as S-HBsAg, which are cysteine-rich and known to be highly cross-linked, may lead to the formation of numerous illegitimate disulfides bonds. The introduction of a small number of C147S AGLs in the HDV envelope could act in trans on the disulfide bonds that structure the coexisting wt AGLs, leading to a dominant-negative effect on infectivity.
Overall, our results further support the existence of two functionally distinct infectivity determinants in the HBV envelope proteins. These determinants, pre-S1 and AGL, differ from each other by their relative stoichiometry in the viral envelope and by their response to infectivity-deficient mutations (Fig. 3 and 7). These characteristics may offer the possibility to develop at least two classes of entry-specific antiviral drugs, including pre-S1 specific lipopeptides to block receptor binding (37) and agents capable of interfering with the AGL disulfide cross-linking (12, 38).
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Abou-Jaoude, G., S. Molina, P. Maurel, and C. Sureau. 2007. Myristoylation signal transfer from the large to the middle or the small HBV envelope protein leads to a loss of HDV particles infectivity. Virology 365:204-209. [DOI] [PubMed] [Google Scholar]
- 2.Abou-Jaoude, G., and C. Sureau. 2007. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J. Virol. 81:13057-13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera, A., B. Guerra, L. Notvall, and R. E. Lanford. 2005. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J. Virol. 79:9786-9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchet, M., and C. Sureau. 2007. Infectivity determinants of the hepatitis B virus pre-S domain are confined to the N-terminal 75 amino acid residues. J. Virol. 81:5841-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonino, F., K. H. Heermann, M. Rizzetto, and W. H. Gerlich. 1986. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 13:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruss, V., J. Hagelstein, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. [DOI] [PubMed] [Google Scholar]
- 9.Chai, N., H. E. Chang, E. Nicolas, S. Gudima, J. Chang, and J. Taylor. 2007. Assembly of hepatitis B virus envelope proteins onto a lentivirus pseudotype that infects primary human hepatocytes. J. Virol. 81:10897-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai, N., H. E. Chang, E. Nicolas, Z. Han, M. Jarnik, and J. Taylor. 2008. Properties of subviral particles of hepatitis B virus. J. Virol. 82:7812-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi, S. W., D. H. Kim, S. H. Lee, I. Chang, and K. H. Han. 2007. Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus. Protein Sci. 16:2108-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenouillet, E., R. Barbouche, and I. M. Jones. 2007. Cell entry by enveloped viruses: redox considerations for HIV and SARS-coronavirus. Antioxid. Redox Signal. 9:1009-1034. [DOI] [PubMed] [Google Scholar]
- 13.Fernholz, D., P. R. Galle, M. Stemler, M. Brunetto, F. Bonino, and H. Will. 1993. Infectious hepatitis B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology 194:137-148. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, R. J., L. Beales, D. Blond, M. N. Simon, B. Y. Lin, F. V. Chisari, D. I. Stuart, and D. J. Rowlands. 2005. Hepatitis B small surface antigen particles are octahedral. Proc. Natl. Acad. Sci. USA 102:14783-14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glebe, D., and S. Urban. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 13:22-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glebe, D., S. Urban, E. V. Knoop, N. Cag, P. Krass, S. Grun, A. Bulavaite, K. Sasnauskas, and W. H. Gerlich. 2005. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology 129:234-245. [DOI] [PubMed] [Google Scholar]
- 17.Gripon, P., I. Cannie, and S. Urban. 2005. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 79:1613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gripon, P., J. Le Seyec, S. Rumin, and C. Guguen-Guillouzo. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213:292-299. [DOI] [PubMed] [Google Scholar]
- 19.Gripon, P., S. Rumin, S. Urban, J. Le Seyec, D. Glaise, I. Cannie, C. Guyomard, J. Lucas, C. Trepo, and C. Guguen-Guillouzo. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 99:15655-15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudima, S., Y. He, N. Chai, V. Bruss, S. Urban, W. Mason, and J. Taylor. 2008. Primary human hepatocytes are susceptible to infection by hepatitis delta virus assembled with envelope proteins of woodchuck hepatitis virus. J. Virol. 82:7276-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heermann, K. H., and W. H. Gerlich. 1991. Surface proteins of hepatitis B viruses, p. 109-144. In A. Maclachlan (ed.), Molecular biology of the hepatitis B virus. CRC Press, Inc., Boca Raton, FL.
- 22.Hollinger, F. B. 2007. Hepatitis B virus genetic diversity and its impact on diagnostic assays. J. Viral Hepat. 14(Suppl. 1):11-15. [DOI] [PubMed] [Google Scholar]
- 23.Jaoude, G. A., and C. Sureau. 2005. Role of the antigenic loop of the hepatitis B virus envelope proteins in infectivity of hepatitis delta virus. J. Virol. 79:10460-10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenna, S., and C. Sureau. 1998. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology 251:176-186. [DOI] [PubMed] [Google Scholar]
- 25.Jenna, S., and C. Sureau. 1999. Mutations in the carboxyl-terminal domain of the small hepatitis B virus envelope protein impair the assembly of hepatitis delta virus particles. J. Virol. 73:3351-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kian Chua, P., M. H. Lin, and C. Shih. 2006. Potent inhibition of human hepatitis B virus replication by a host factor Vps4. Virology 354:1-6. [DOI] [PubMed] [Google Scholar]
- 27.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert, C., T. Doring, and R. Prange. 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J. Virol. 81:9050-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanford, R. E., C. Sureau, J. R. Jacob, R. White, and T. R. Fuerst. 1994. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology 202:606-614. [DOI] [PubMed] [Google Scholar]
- 30.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magnus, C., P. Rusert, S. Bonhoeffer, A. Trkola, and R. R. Regoes. 2009. Estimating the stoichiometry of human immunodeficiency virus entry. J. Virol. 83:1523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangold, C. M., and R. E. Streeck. 1993. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J. Virol. 67:4588-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangold, C. M., F. Unckell, M. Werr, and R. E. Streeck. 1995. Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology 211:535-543. [DOI] [PubMed] [Google Scholar]
- 35.Neurath, A. R., B. Seto, and N. Strick. 1989. Antibodies to synthetic peptides from the preS1 region of the hepatitis B virus (HBV) envelope (env) protein are virus-neutralizing and protective. Vaccine 7:234-236. [DOI] [PubMed] [Google Scholar]
- 36.Patient, R., C. Hourioux, P. Y. Sizaret, S. Trassard, C. Sureau, and P. Roingeard. 2007. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 81:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen, J., M. Dandri, W. Mier, M. Lutgehetmann, T. Volz, F. von Weizsacker, U. Haberkorn, L. Fischer, J. M. Pollok, B. Erbes, S. Seitz, and S. Urban. 2008. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat. Biotechnol. 26:335-341. [DOI] [PubMed] [Google Scholar]
- 38.Ryser, H. J., and R. Fluckiger. 2005. Progress in targeting HIV-1 entry. Drug Discov. Today 10:1085-1094. [DOI] [PubMed] [Google Scholar]
- 39.Salisse, J., and C. Sureau. 2009. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J. Virol. 83:9321-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sureau, C., C. Fournier-Wirth, and P. Maurel. 2003. Role of N glycosylation of hepatitis B virus envelope proteins in morphogenesis and infectivity of hepatitis delta virus. J. Virol. 77:5519-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sureau, C., B. Guerra, and R. E. Lanford. 1993. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sureau, C., B. Guerra, and H. Lee. 1994. The middle hepatitis B virus envelope protein is not necessary for infectivity of hepatitis delta virus. J. Virol. 68:4063-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sureau, C., J. R. Jacob, J. W. Eichberg, and R. E. Lanford. 1991. Tissue culture system for infection with human hepatitis delta virus. J. Virol. 65:3443-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urban, S., C. Schwarz, U. C. Marx, H. Zentgraf, H. Schaller, and G. Multhaup. 2000. Receptor recognition by a hepatitis B virus reveals a novel mode of high-affinity virus-receptor interaction. EMBO J. 19:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C. J., P. J. Chen, J. C. Wu, D. Patel, and D. S. Chen. 1991. Small-form hepatitis B surface antigen is sufficient to help in the assembly of hepatitis delta virus-like particles. J. Virol. 65:6630-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, T., E. M. Sorensen, A. Naito, M. Schott, S. Kim, and P. Ahlquist. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. USA 104:10205-10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, X., S. Kurteva, X. Ren, S. Lee, and J. Sodroski. 2005. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 79:12132-12147. [DOI] [PMC free article] [PubMed] [Google Scholar]