Abstract
To study vesicular stomatitis virus (VSV) entry and uncoating, we generated a recombinant VSV encoding a matrix (M) protein containing a C-terminal tetracysteine Lumio tag (rVSV-ML) that could be fluorescently labeled using biarsenical compounds. Quantitative confocal microscopy showed that there is a transient loss of fluorescence at early times after the initiation of endocytosis of rVSV-ML-Green (rVSV-MLG) virions, which did not occur when cells were treated with bafilomycin A1. The reduction in fluorescence occurred 5 to 10 min postentry, followed by a steady increase in fluorescence intensity from 15 to 60 min postentry. A similar loss of fluorescence was observed in vitro when virions were exposed to acidic pH. The reduction in fluorescence required G protein since “bald” ΔG-MLG particles did not show a similar loss of fluorescence at low pH. Based on the pH-dependent fluorescence properties of Lumio Green, we hypothesize that the loss of fluorescence of rVSV-MLG virions during virus entry is due to a G ectodomain-dependent acidification of the virion interior. Biochemical analysis indicated that low pH also resulted in an enhancement of M protein dissociation from partially permeabilized, but otherwise intact, wild-type virions. From these data we propose that low-pH conformational changes in G protein promote acidification of the virus interior, which facilitates the release of M from ribonucleoprotein particles during uncoating.
Vesicular stomatitis virus (VSV) is a prototypic enveloped, nonsegmented, negative-strand RNA virus in the Rhabdoviridae family and is composed of an 11.1-kb genome that contains five genes (3′-N-P-M-G-L-5′). These genes encode the nucleocapsid protein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the large catalytic subunit (L) of the RNA-dependent RNA polymerase. Virions consist of bullet-shaped particles that are approximately 180 nm long and 75 nm wide and are composed of a helical nucleocapsid containing the negative-sense genomic RNA bound by ∼1,200 molecules of N protein such that the phosphodiester backbone of the RNA genome is cradled in a pocket that forms between the N- and C-terminal lobes of the N protein (15). The RNA-dependent RNA polymerase, which consists of the L and P proteins, is associated with the N-RNA complex. The L-P-N-RNA collectively makes up the ribonucleoprotein particle (RNP), which is surrounded by an envelope derived from the host cell membrane and contains approximately 400 trimeric G protein spikes that protrude from the virion surface. During virus assembly at the plasma membrane, M protein associates with the RNP and condenses it into a tightly packed helix, which is referred to as the skeleton (43, 44). M protein-driven condensation results in the characteristic bullet-shaped morphology of mature virions (37).
VSV enters cells through the clathrin-mediated endocytic pathway (24, 39, 51). As virions encounter the decreasing pH within endocytic organelles, G protein undergoes conformational changes that induce fusion of the viral envelope and endosomal membrane, resulting in the release of RNPs into the cytoplasm (19, 54). Either directly after or concomitant with membrane fusion, the RNP core dissociates from M protein, which results in decondensation of the skeleton (43, 44) and completes the uncoating process (46). After uncoating, the decondensed RNP serves as a template for transcription of viral mRNAs by the packaged RNA-dependent RNA polymerase. Without the dissociation of M protein from RNPs, transcription of viral mRNAs (productive infection) cannot proceed since it has been shown that M protein inhibits viral transcription (9, 11, 55). What triggers the dissociation of M protein from RNPs is not well understood. In vitro studies have shown that M reversibly associates and dissociates from RNPs at physiological ionic strength, indicating that M is in a dynamic equilibrium with RNPs (36). This suggested that skeletons dissociate from M protein in a newly infected cell, which has no soluble M in the cytoplasm, and that later in infection when the concentration of M protein is high, binding of M to RNPs is favored, and this results in RNP condensation. The assembly of M into nascent virions occurs only on the inner leaflet of the plasma membrane (12), suggesting the affinity of membrane-bound M is much higher than that of soluble M for RNPs. If true, then it is not clear how RNPs are released from M lining the interior of the virion envelope after membrane fusion within endosomes.
To study the release of RNPs from M protein during uncoating, we developed and characterized a recombinant VSV encoding M proteins with N- or C-terminal tetracysteine Lumio tags (rVSV-LM or rVSV-ML, respectively) which can be fluorescently labeled using biarsenical compounds (16). This allowed us to follow the fate of incoming M protein during VSV entry into cells. A similar rVSV was recently described by others (10). Here, we report that there is a transient loss of fluorescence at early times after Lumio Green-labeled rVSV-ML (rVSV-MLG) is endocytosed, and at later times the fluorescent signal subsequently recovers and then increases above the initial input fluorescence levels. The loss of fluorescence was recapitulated in vitro when rVSV-MLG particles were exposed to decreasing pH. Based on the pH-dependent fluorescence properties of Lumio Green (16), we propose that the loss of fluorescence of rVSV-MLG virions during the initial stages of virus entry is due to acidification of the virion interior. The effect of pH on the fluorescence of MLG within intact virus was dependent on G protein, and biochemical analysis indicated that low pH resulted in an enhancement of M dissociation from partially permeabilized, but otherwise intact, virions. These results suggest that the previously described viroporin activity of G protein at low pH (7, 26) may allow acidification of the virus interior, which reduces the interaction of M protein with RNPs, thereby facilitating the release of RNPs into the cytoplasm during uncoating. This would be similar to the well-characterized uncoating mechanism of influenza virus RNPs in which the M1 protein is released following the M2-dependent acidification of the interior of influenza virions (20, 45, 49).
MATERIALS AND METHODS
Plasmid design and construction.
pBS-M-Lumio was constructed using pBluescript-SK+ (Stratagene) by annealing two overlapping oligonucleotides (CEM6 and CEM7) that encoded the C-terminal tetracysteine (in boldface) Lumio tag (AEAAAREACCRECCARA). The oligonucleotides were phosphorylated with T4 polynucleotide kinase, annealed to form a linker with NheI and EagI overhangs, and ligated to pBS-M-Flag-C that was digested with NheI and EagI.
The pBS-Lumio-M plasmid was constructed by using PCR amplification of pBS-MΦT as a template with a forward primer (CEM8), which had 5′ KpnI and AscI restriction sites, a Kozak sequence (in boldface) (TTCATCATGG) followed by the sequence for the Lumio tag (encoded as MAEAAAREACCRECCARA; the tag is shown in boldface), and the first 24 bp of the coding sequence of M, and with a 3′ reverse primer that annealed to the T3 promoter in pBS. The product was digested with KpnI and EagI, gel purified, and ligated into pBS-MΦT digested with the same enzymes. Sequences of all constructed cDNAs were determined by the Molecular Resources Center at the University of Tennessee Health Sciences Center, and no changes from predicted sequences were found.
Generation of full-length VSV cDNAs encoding ML and LM proteins.
The ML and LM chimera sequences were cloned into the polylinker of a modified version of the VSV anti-genome pΔM-PLF (21) except that there was no green fluorescent protein between the G and L coding regions. The resulting plasmids were called pVSV-LM and pVSV-ML.
Recovery and characterization of rVSV.
Recombinant viruses were recovered using reverse genetics (32) with some modifications, as described previously (21). Single-step growth curves were performed by adsorbing virus to baby hamster kidney (BHK-21) cells at a multiplicity of infection (MOI) of 10 for 1 h at 31°C with constant rocking. The inoculum was removed, the cells were washed four times with serum-free Dulbecco's minimal Eagle's medium (SF-DMEM) to remove unbound virus, DMEM containing 5% fetal bovine serum (D-5) was added, and the cells were placed at 37°C. Every 2 h postinfection (hpi), a 5% aliquot of medium was collected and replaced with the same volume of fresh medium, and then virus titers were determined in duplicate by plaque assay on BHK-21 cells. Growth curves were performed in triplicate for each virus.
Transient expression of ML and LM proteins.
BHK-21 cells at ∼90% confluence on glass coverslips in 35-mm dishes were infected with a recombinant vaccinia virus (vTF7-3) encoding T7 RNA polymerase (13) at an MOI of 5 for 1 h at 31°C in SF-DMEM. The inoculum was removed, and the cells were transfected with 5 μg of pBS-MΦT (wild-type M protein [M-wt], Indiana serotype), pBS-M-Lumio, or pBS-Lumio-M using TransfectACE (48) in SF-DMEM. At 5 h posttransfection (p.t.) the DNA-lipid complex was removed and replaced with D-5 containing antibiotics (100 U/ml streptomycin and penicillin), and at 18 h p.t. the cells were either fixed with 4% paraformaldehyde in phosphate-buffered saline, pH 7.4, and processed for immunofluorescence or used in a budding assay as described below.
Budding assays.
Cells transiently expressing the indicated M proteins were washed twice with methionine-free, SF-DMEM and then incubated in this medium for 15 min to deplete the intracellular methionine pools at 5 h p.t. After depletion, a medium composed of one part D-5 and nine parts methionine-free medium and supplemented with 50 μCi of 35S-Express Protein Labeling Mix (Perkin-Elmer) per ml, was added, and 18 h later medium and cells were collected separately and processed for immunoprecipitation.
To examine M protein released by budding, the medium was centrifuged at 170 × g for 10 min to remove dislodged cells, and the supernatants were placed on ice after the addition of 200 U of aprotinin (U.S. Biochemicals). The supernatants were made to contain 50 mM Tris-HCl, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 20 mM NaN3, and 0.3% sodium dodecyl sulfate (SDS); radiolabeled M proteins were immunoprecipitated with an anti-M monoclonal antibody (MAb 23H12) (34) overnight at 4°C, and immune complexes were collected using Pansorbin (catalog number 507861; Calbiochem).
Cell extracts were prepared by washing the cells twice with phosphate-buffered saline, and then cells were subjected to lysis with 800 μl of detergent solution (10 mM Tris-HCl [pH 7.4], 66 mM EDTA, 0.4% sodium deoxycholate, 1% Triton X-100, 0.05% NaN3) containing 200 U/ml of aprotinin for 5 min at ambient temperature on a rocker. The cell extract was collected, nuclei and insoluble material were removed by centrifugation at 10,000 × g for 5 min, and a 200-μl aliquot of the cell extract was made to 0.3% SDS and immunoprecipitated with M MAb 23H12 overnight at 4°C, followed by Pansorbin retrieval. One-quarter of the radiolabeled M protein in the cell extracts was compared to the amount released into the supernatant by SDS-polyacrylamide gel electrophoresis (PAGE) on a 9% polyacrylamide gel and visualized by fluorography.
Lumio Green labeling and quantification of rVSV.
BHK-21 cells on 100-mm plates at ∼95% confluence were infected with wt rVSV (rVSV-wt), rVSV-ML, or G-complemented ΔG-ML at an MOI of 1. The inoculum was removed after 1 h, the cells were washed twice with SF-DMEM, and then 10 ml of D-5 was added. For labeling with Lumio Green reagent (Invitrogen), the cells were washed twice with reduced-serum Opti-MEM I medium, and then 10 ml of Opti-MEM I (Invitrogen) containing 200 nM Lumio Green was added at 4 to 5 hpi. At 18 hpi the supernatant was collected, and labeled virus was concentrated by centrifugation over a 20% sucrose cushion at 38,000 rpm in an SW41 swinging bucket rotor (Beckman) for 1 h and 15 min. The viral pellet was resuspended on ice in 1 ml of 10% sucrose-TN buffer (10 mM Tris [pH 7.4]-150 mM NaCl). Residual, unbound Lumio reagent was removed from the resuspended virus preparation by adding 100-μl aliquots to Sephadex G-50 (Fine) columns (Roche). The void volume containing the virus was collected and stored at −80°C, except for “bald” ΔG virions which were used immediately (e.g., on the same day of preparation without freezing) for experiments since the pseudotyped viruses were more sensitive to cycles of freezing and thawing. In contrast, the Lumio Green-labeled “wt” rVSV-ML virus (rVSV-MLG) was stable for two to three freeze-thaw cycles without loss of infectivity or fluorescence. Titers were determined by standard plaque assay on BHK-21 cells, and protein concentration was determined by a bicinchoninic acid protein assay (Pierce) according to the manufacturer's directions.
The amount of fluorescence in the virus suspensions was determined by adding 1 μg of virus to a cuvette that contained 3 ml of HMTN buffer (10 mM HEPES, 10 mM morpholineethanesulfonic acid, 10 mM Tris-HCl [pH 8.0], 150 mM NaCl) using a Perkin-Elmer Luminescence Spectrometer (LS 50B). Protein-specific fluorescence was examined by dissociating virus in SDS-PAGE sample buffer containing 1/10 the normal concentration of β-mercaptoethanol and examining the protein profile on an SDS-10% polyacrylamide gel. Fluorescent protein bands were detected using a Storm 860 fluorescence imager (480 nm) and ImageQuant software (Molecular Dynamics). The same gel that was scanned for fluorescence was processed for immunoblot analysis using the anti-M MAb 23H12, and proteins were visualized as described above.
ΔG-ML was pseudotyped with G-wt or GSHA (where GS is the G-stem and HA is an N-terminal hemagglutinin tag) as described previously (22) with the following modifications for labeling with Lumio Green. BHK cells at ∼80% confluence were transfected with 2 μg of pCAGGS-G or pCAGGS-GSHA plasmid and infected with G-complemented ΔG-ML at an MOI of 10 at 18 h p.t. Virus was labeled in cells grown in Opti-MEM I containing 200 nM Lumio Green beginning at 4 hpi and processed as described above.
To label virus with 35S-methionine, ∼1 × 106 BHK-21 cells in 35-mm dishes were infected with rVSV-wt at an MOI of 5. At 4 hpi the cells were starved of methionine for 30 min in methionine-free DMEM, and then methionine-free DMEM containing 50 μCi of 35S-Express Protein Labeling Mix (Perkin-Elmer) per ml was added. At 14 hpi the culture medium was collected, and virus was concentrated by ultracentrifugation (45,000 rpm for 35 min in a Sorvall AH-650 rotor) through a 20% sucrose cushion (10 mM Tris, pH 7.4, 150 mM NaCl, 20% sucrose). The amount of protein in the viral preparation was determined using a bicinchoninic acid protein assay (Pierce) according to the manufacturer's instructions. Aliquots of 35S-labeled rVSV-wt were stored at −80°C and used within 2 weeks of freezing.
Live-cell synchronized entry assay.
To examine the entry of Lumio Green-labeled virus into live cells, BHK-21 cells in 35-mm glass-bottomed culture dishes (MatTek Corporation) at 90% confluence were washed twice with 2 ml of ice-cold Opti-MEM I (with or without 1 μM bafilomycin A1) and placed at 4°C for 10 min. For the bafilomycin A1-treated cells, the cultures were preincubated in D-5 containing 1 μM bafilomycin A1 for 30 min at 37°C and then chilled as described above with Opti-MEM I containing 1 μM bafilomycin A1. rVSV-MLG and 50 μg of transferrin conjugated to Texas Red (Tfn-TR) were adsorbed to cells for 90 min in 100 μl of ice-cold Opti-MEM I in the dark with rocking every 15 min. After a 90-min adsorption, the inoculum was removed, and medium at 37°C with or without bafilomycin A1 was added to initiate entry. For the nonentry time point (time zero, or t-0), the inoculum was replaced with 2 ml of ice-cold Opti-MEM I, and images of the cells were collected using a laser scanning confocal microscope ([LSCM] Zeiss LSM 510) on an ambient temperature stage using 488-nm excitation from an argon ion laser for Lumio Green fluorescence and 543-nm excitation from a 1 mW helium-neon laser for Tfn-TR fluorescence. Separate plates were used for each of the other time points to examine rVSV-MLG entry after the addition of 37°C medium by confocal microscopy using a heated stage and objective heater set to 37°C. At least four individual fields were collected for each time point. Lumio Green fluorescence was quantified by determining pixel intensity using the Profile function in the physiology package of the LSM 510 software (version 3.2). Only Lumio Green fluorescence that was contained within Tfn-TR-positive endosomes was used for quantification. Pixel intensity values were imported into Excel and averaged, and then GraphPad Prism 5 software was used to calculate the P values for the statistical analysis of the mean pixel intensity for each time point. One plate of cells (with or without bafilomycin A1) was allowed to incubate for 4 h, and then the cells were fixed and stained for VSV N protein using N MAb 10G4 (34) and rabbit anti-mouse conjugated to Alexa Fluor 633 after permeabilization with 1% Triton X-100. These cells were imaged by LSCM using the 488-nm, 543-nm, and 633-nm lasers.
Fluorometric assay.
To measure the fluorescence intensity of rVSV-MLG virions in vitro, we added 1 μg of virus to HMN buffer (10 mM HEPES, 10 mM morpholineethanesulfonic acid, 150 mM NaCl, pH 7) in a cuvette with constant mixing using a magnetic stir bar and measured fluorescence at 1-s intervals with a Perkin Elmer LS 50B fluorometer using 508-nm excitation and 528-nm emission. To determine the effect of pH on fluorescence, we recorded the fluorescence signal at pH 7 and then added 1 M NaOH to raise the pH to 7.5 and then to pH 8.0, taking fluorescence readings after each adjustment of pH. Similarly, we reduced the pH by adding 1 M HCl to reach pH values of 6.5, 6.0, 5.5, and 5.0 and recorded the fluorescence signal at each pH. The percent change in fluorescence intensity from pH 7 was then calculated. The same procedure was used with the bald ΔG-MLG and GSHA-complemented ΔG-MLG virions. For these experiments the amount of virus that was used was normalized to equivalent fluorescence signals at pH 7.0 rather than by the number of PFU since these viruses are not infectious. After fluorescence values were recorded at the lowest pH, virions were solubilized by the addition of Triton X-100 to a 0.1% final concentration, and the percent fluorescence change was calculated as described above.
Lysolecithin treatment of rVSV-wt.
Approximately 1 μg of 35S-labeled rVSV-wt was treated with various concentrations of lysolecithin at pH 7.0 or pH 5.5 for 10 min on ice. The suspensions were layered over a 20% sucrose-TN buffer cushion, and centrifuged for 35 min at 45,000 rpm (Sorvall AH-650) to separate proteins that were released into the supernatant from proteins that remained virion-associated, which were pelleted. The supernatant was trichloroacetic acid (TCA) precipitated, resuspended in reducing Laemmli sample buffer, and compared to the pellet, which was also resuspended in reducing Laemmli sample buffer, by SDS-PAGE. The amount of M protein released into the supernatant was quantified and compared to the amount of N protein in the pellet by phosphorimager analysis (Molecular Dynamics).
Electron microscopy.
Virions were treated with lysolecithin, pelleted through 20% sucrose-TN buffer, and gently resuspended in 10% sucrose-TN buffer on ice. To prevent caramelization of samples during exposure to the electron beam, the sucrose concentration was reduced by diluting the virus preparations 1:10 in filtered, deionized water, and then the samples were immediately prepared for transmission electron microscopy (TEM) by adsorbing virus to carbon-coated grids using the drop-to-drop method, followed by negative staining with 2% phosphotungstic acid (pH 5.9). Images were collected using a JEOL JEM1200EX II TEM with an ATM 2K bottom mount digital camera.
RESULTS
Recovery and characterization of rVSV encoding fluorescent M proteins.
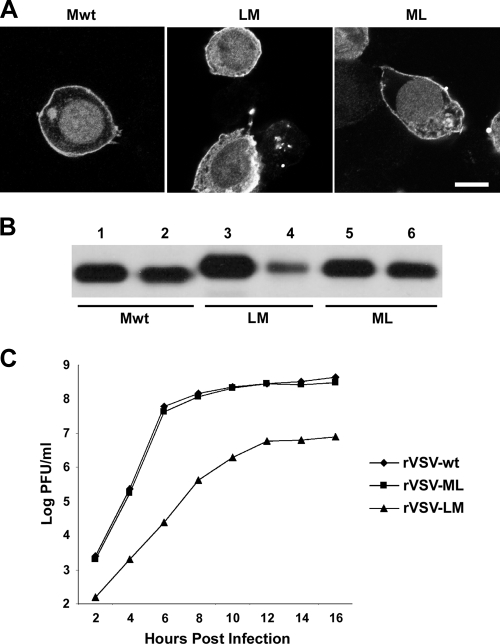
To investigate virus uncoating and the fate of released M protein, we constructed fluorescent M proteins utilizing Lumio technology in which a tetracysteine (CCRECC-) Lumio tag was fused to the N or C terminus of M (LM or ML protein, respectively). To assess the cellular distribution and assembly function of these tagged proteins, we compared them to M-wt protein by immunofluorescence staining (Fig. 1A) and in a budding assay (Fig. 1B). There was no discernible difference between the distributions of the tagged M proteins and M-wt (Fig. 1A). When examined for budding activity, the N-terminally tagged LM showed a modest reduction in the amount of protein released from cells (Fig. 1B, lane 4). The C-terminally tagged ML, on the other hand, had wt budding activity (Fig. 1B, lane 6).
FIG. 1.
Expression of LM and ML and rVSV characterization. (A) M proteins expressed in BHK-21 cells were stained with anti-M MAb 23H12 conjugated to rhodamine and imaged using LSCM (bar, 5 μm). A 1-μm optical section through the middle of each cell is shown. (B) M protein budding assay. Cell extracts (lanes 1, 3, and 5) and supernatants (lanes 2, 4, and 6) from cells expressing M-wt, LM, or ML were immunoprecipitated with M MAb after [35S]methionine labeling for 16 h and analyzed by SDS-PAGE and fluorography. (C) One-step growth curve of rVSV-wt, rVSV-LM, and rVSV-ML.
To determine if LM or ML could support virus assembly, we replaced the M-wt gene with the Lumio-tagged versions in the viral genome. Both viruses (rVSV-LM and rVSV-ML) were easily recovered, demonstrating that M proteins with small N- or C-terminal tags are functional for virus replication and assembly. However, there was a growth defect observed for rVSV-LM, whereas the growth of rVSV-ML was similar to that of rVSV-wt (Fig. 1C). Because rVSV-ML had growth properties indistinguishable from those of rVSV-wt, we used it to make fluorescent rVSV and for the uncoating studies described below.
Fluorescent labeling of rVSV-ML.
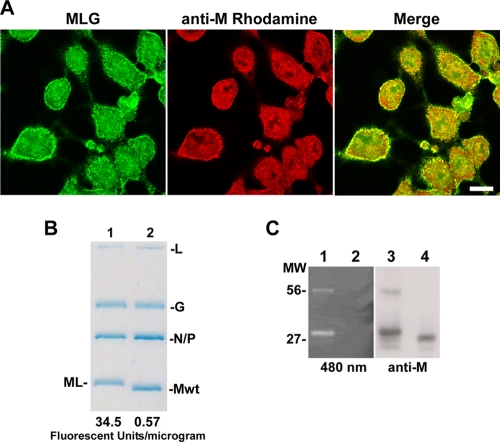
Lumio Green is a small, membrane-permeable fluorophore which fluoresces green when covalently bound to the thiol pairs in the Lumio tag (1, 14, 16). To fluorescently label rVSV-ML, we initially mixed purified virus with the Lumio reagent using protocols similar to those described by others to label human immunodeficiency virus virions encoding a Lumio-tagged integrase protein (2). However, we found that no labeling occurred, suggesting that the Lumio tag was sequestered or incapable of interacting with the fluorophore within virions. We reasoned that the Lumio tag may be more accessible for labeling within cells before assembly into rVSV-ML virions; therefore, we added Lumio reagent to cells infected with rVSV-ML at 5 hpi. The fluorophore-containing medium was removed 1 h later, and the cells were examined by confocal microscopy after immunofluorescence staining with M MAb 23H12 (Fig. 2A). We observed that the ML protein bound the fluorophore and that the Lumio Green signal colocalized with M protein primarily at the plasma membrane where virus assembly occurs (Fig. 2A, merge). Similar results were also observed when the red fluorophore Lumio Red reagent was used (data not shown).
FIG. 2.
Labeling of rVSV-ML with Lumio Green. (A) LSCM of BHK-21 cells infected with rVSV-ML, labeled with Lumio Green reagent for 1 h at 5 hpi, and stained with M MAb (bar, 10 μm). (B and C) SDS-PAGE analysis of 107 PFU of rVSV-MLG and rVSV-wt virions stained with Coomassie blue. The relative fluorescence signal/μg is shown below each lane in panel B. Similarly prepared samples were also visualized by excitation at 480 nm on a Storm 860 fluorescence imager (C, lanes 1 and 2) or analyzed by immunoblot analysis for M (anti-M, lanes 3 and 4). Lanes 1 and 3 contain rVSV-MLG, and lanes 2 and 4 contain rVSV-wt. The M MAb-reactive and fluorescent bands at approximately 56 kDa are likely artificially induced ML dimers formed as a result of the reduced concentration of β-mercaptoethanol used in the sample buffer (see Materials and Methods for details). MW, molecular weight (in thousands).
To produce fluorescent virus, Lumio Green was added to infected cells at 4 hpi, and virus released into the medium was harvested at 18 hpi. The labeled virus, rVSV-MLG, was purified from supernatants, titers were determined, and fluorescence was quantified using a fluorometer. To determine if the labeled rVSV-MLG virus had a level of infectivity similar to that of identically treated rVSV-wt, we examined the protein profiles from 1 × 107 PFU of the two viruses after electrophoresis on SDS-polyacrylamide gels (Fig. 2B). The migration of ML was slightly slower than that of M-wt because of the additional 17 amino acids of the Lumio tag. Coomassie blue staining indicated that the two virus preparations had similar amounts of protein, but the rVSV-MLG had a 60-fold higher fluorescence signal than rVSV-wt virus. To determine whether the fluorescent signal from the rVSV-MLG virions was due to specific binding of the Lumio reagent to the ML protein, we visualized fluorescent proteins from purified virus after separation by SDS-PAGE. Fluorescent protein bands were observed only with the rVSV-MLG virus, and the bands that corresponded to the MLG protein were also detected in an immunoblot using the M-specific MAb 23H12 (Fig. 2C, lanes 1 and 3, respectively). A higher-molecular-weight band migrating at ∼56 kDa was also seen in the MLG lanes by fluorescence and immunoblot analysis. This band likely corresponds to an ML dimer that may result from the formation of spurious disulfide bonds between SDS-denatured ML monomers due to the reduced amount of β-mercaptoethanol in the protein sample buffer, which was required to retain the fluorophore on the MLG protein. Overall, these data demonstrated that rVSV-MLG was specifically labeled on the M protein and therefore should be useful to examine virus uncoating.
Transient loss of MLG fluorescence during rVSV entry.
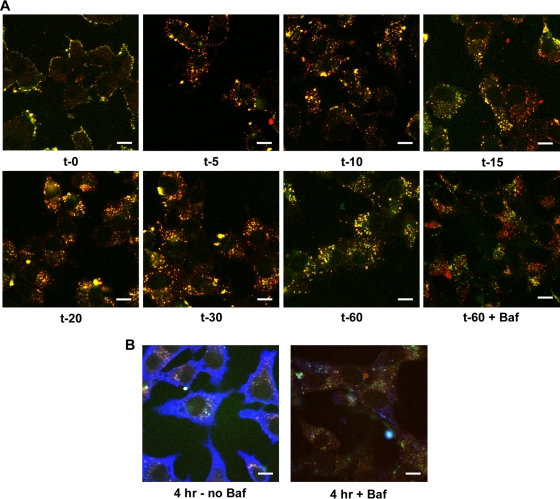
To examine virus entry into live cells, rVSV-MLG and Tfn-TR were adsorbed onto cells at 4°C to prevent endocytosis. A set of plates that had medium containing bafilomycin A1, which prevents endosome acidification and therefore fusion and uncoating of rVSV-MLG, was examined in parallel. After adsorption, virus and Tfn entry were synchronized by removing the inoculum and adding medium prewarmed to 37°C (with or without inhibitor). The cells were then examined without fixation using confocal microscopy at various times after initiation of entry up to 60 min (Fig. 3A). At t-0, prior to the addition of prewarmed medium, no endocytosis had occurred, as confirmed by the sensitivity of Tfn-TR to release by acid washing (data not shown). After a 5-min incubation, the virus and Tfn-TR had been endocytosed and were clearly seen in small endocytic vesicles (Fig. 3A). By 20 min the Tfn-positive endosomes had become perinuclear (Fig. 3A). Of note, MLG continued to remain associated with Tfn-positive endocytic structures until the end of the experiment. This was not due to a lack of uncoating, as shown by new N protein synthesis in cells that were not treated with bafilomycin A1 (Fig. 3B). Importantly, bafilomycin A1 prevented N protein synthesis (Fig. 3B) but did not prevent rVSV-MLG endocytosis, as shown by the perinuclear localization of MLG fluorescence in Tfn-positive endosomes (Fig. 3A). A detailed analysis of MLG trafficking during entry and uncoating of rVSV-MLG will be described elsewhere (Mire et al., unpublished data).
FIG. 3.
Live-cell confocal microscopy of rVSV-MLG entry. (A) Cells were inoculated with rVSV-MLG and Tfn-TR in ice-cold medium (with or without bafilomycin A1) for 90 min. The cells were washed once with cold medium, and then medium prewarmed to 37°C (with or without inhibitor) was added to initiate virus entry, or the cells were examined directly on an ambient temperature stage for the t-0 time point. For t-5 through t-60, the cells were examined by LSCM on a heated stage at the indicated times using separate 35-mm glass-bottomed dishes for each time point to reduce signal loss by photobleaching (bar, 10 μm). (B) Two plates from panel A were incubated for 4 h in medium with (+Baf) or without (no Baf) bafilomycin A1. The cells were fixed, permeabilized, and stained for VSV N protein using MAb 10G4 and a secondary anti-mouse antibody conjugated to Alexa Fluor 633. The extensive blue in the absence of bafilomycin A1 indicates new N protein synthesis.
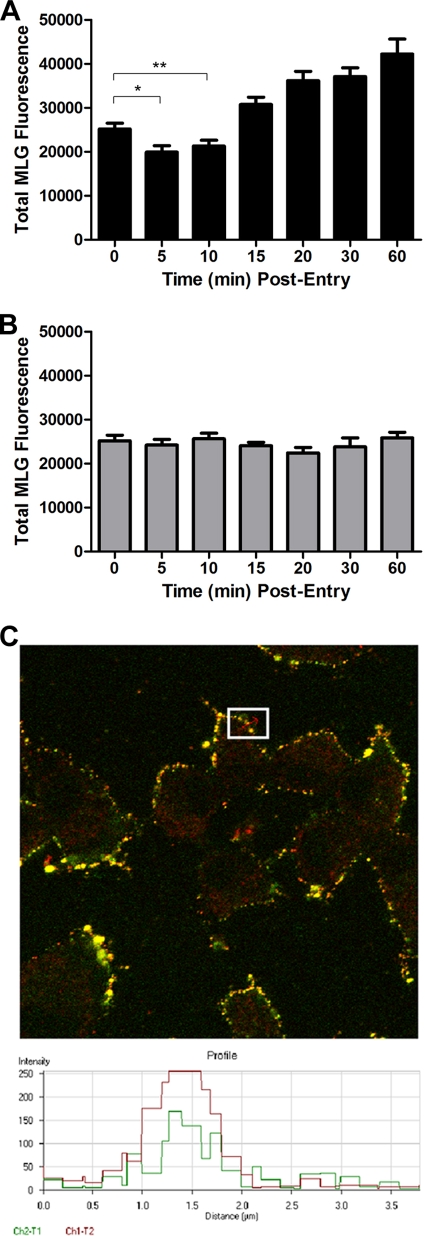
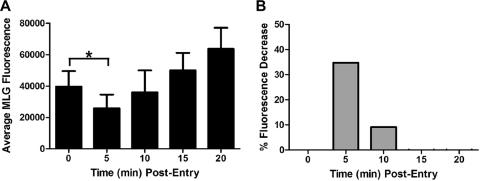
During the course of these studies we observed that at early times after initiation of virus entry, there appeared to be a loss of Lumio Green fluorescence in the cells that were incubated in medium lacking bafilomycin A1. The fluorescence signal then appeared to increase at later times (e.g., 20 to 60 min postentry). To determine if there were, indeed, changes in fluorescence of MLG during entry, we quantified the MLG fluorescence signal in 20 individual Tfn-positive endosomes that contained rVSV-MLG at each time point. For quantification, the images for each time point were collected using identical detector, gain, and threshold settings, which allowed us to compare fluorescence signals between time points. As shown in Fig. 4A, there was a small but significant drop in fluorescence at 5 to 10 min postentry. This was followed by a steady increase in fluorescence from 15 to 60 min. Interestingly, there was no loss of MLG fluorescence in the cells treated with bafilomycin A1 (Fig. 4B) even though the virus was endocytosed (Fig. 3A). Figure 4C shows an example of a profile from which MLG fluorescence quantification was performed.
FIG. 4.
Average fluorescence of rVSV-MLG during entry. (A) The average Lumio Green fluorescence from approximately 20 Tfn-TR-positive endosomes was determined using the Profile function of the Zeiss LSM510 software. Significance was determined using an unpaired, two-tailed t test (*, P = 0.0108; **, P = 0.0457). (B) The average Lumio Green fluorescence from 20 Tfn-TR-positive endo-somes in cells treated with bafilomycin A1. (C) An example of fluorescence quantification from one region of the plasma membrane that contained colocalized Tfn-TR and rVSV-MLG fluorescence from the t-0 time point. The inset shows a region from which one quantification data set was obtained. The graph at the bottom shows pixel intensity as a function of distance (length of the red arrow within the inset region). The portion of the profile that had a continuous stretch of pixel intensity over 100 units for at least 180 nm was used to quantify total MLG fluorescence. The threshold setting (e.g., detector gain) for all images was identical and was set such that no pixel saturation occurred in the green channel, allowing accurate MLG fluorescence signal quantification.
To provide additional confidence that the reduction in fluorescence at early times postentry was authentic, we quantified the MLG fluorescence from three independent experiments performed on three separate days using 30 individual Tfn-TR-positive endosomes per experiment; therefore, 90 endosomes were examined, providing high statistical power for the analysis. Only MLG fluorescence signals that colocalized with Tfn-TR were used for quantification, which ensured that the fluorescence measured was due to virus in an endosome. As shown in Fig. 5A, the largest reduction in fluorescence occurred at 5 min postentry, where there was a 35% reduction in fluorescence compared to that at t-0 (Fig. 5B). This difference was statistically significant, with a P value less than 0.0001 using an unpaired, two-tailed t test. By 10 min the fluorescence intensity began to increase, and by 15 min postentry the MLG fluorescence was higher than the initial value at t-0. We hypothesize that the loss of fluorescence at 5 min may be due to exposure of MLG within virions to reduced pH since there was no reduction seen in cells treated with bafilomycin A1 (Fig. 4B). These data suggested that during endocytosis the interior of VSV virions may become acidified, similar to the process in influenza virus (20). The recovery of fluorescence to the initial input levels may correspond to exposure of MLG to neutral pH in the cytoplasm. The subsequent increase observed after 10 min may correspond to the dissociation of RNPs from MLG protein (e.g., virus uncoating). This aspect will be described in detail in another article (Mire et al., unpublished data).
FIG. 5.
Quantification of rVSV-MLG fluorescence during entry. (A) Average fluorescence of MLG virus within Tfn-TR-positive endosomes was determined every 5 min for 20 min from 90 endosomes. The data are from three separate experiments performed on three separate days (∼30 endosomes per experiment); therefore, the averages from 90 different endosomes are shown. Significance was determined using an unpaired, two-tailed t test (*, P < 0.0001). (B) Percent loss of fluorescence relative to t-0. The change in fluorescence from panel A was converted to percent loss using the following formula: [I(t-0) − I(t-x)/I(t-0)] × 100, where I is fluorescence intensity and t-x is time at 5, 10, 15, or 20 min postentry.
Low pH-induced reduction in fluorescence in vitro.
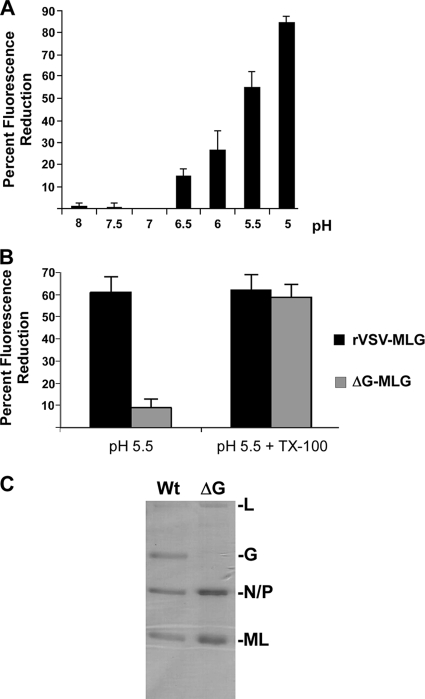
The loss of fluorescence intensity of rVSV-MLG during entry was interesting when one considers that the pKa of Lumio Green when bound by the tetracysteine tag is 5.4 (16). We reasoned that the low pH within endosomes (41) could be responsible for the reduction in fluorescence of MLG if the viral envelope becomes porous to protons during entry. To determine whether pH has an effect on the fluorescence of rVSV-MLG virions, we mimicked the drop in pH that occurs during endocytosis by acidifying a suspension of intact MLG virus in vitro and measured fluorescence as a function of pH. When excited at a wavelength of 508 nm in HMN buffer at pH 7, 1 μg of rVSV-MLG virions gave an average fluorescence reading of 34.5 ± 4.7. The fluorescence signals were similar at pHs of 7.5 and 8.0; however, we found that below pH 7 the fluorescence of rVSV-MLG progressively decreased as the buffer became more acidic (Fig. 6A). These data support the hypothesis that the loss of Lumio Green fluorescence quantified in Fig. 4 and 5 was pH dependent. Interestingly, at a pH of 6, which is approximately the pH threshold where G protein-induced fusion occurs, we observed a similar loss of fluorescence intensity in vitro as we observed by microscopy in cells at t-5 (29% loss versus 35% loss, respectively). We also observed that the pH-dependent loss of fluorescence was similar to the pH-dependent curve for VSV G protein-induced membrane fusion (53). This similarity further suggested that G protein contributed to the loss in fluorescence and indicated that both in vitro and in vivo MLG was being exposed to acidic pH within intact virus particles.
FIG. 6.
pH and G protein dependence of fluorescence loss in vitro. (A) rVSV-MLG virions were suspended in HMN buffered to the indicated pH; the fluorescence was read in triplicate, and the percent change from the fluorescence measured at pH 7 was calculated as described in the legend of Fig. 5. Standard deviations are shown. (B) Percent fluorescence loss for rVSV-MLG and ΔG-MLG virions at pH 5.5, plus or minus Triton X-100 (TX-100), relative to fluorescence at pH 7. Samples were measured in triplicate, and standard deviations are shown. (C) Coomassie-blue stained SDS-PAGE of rVSV-MLG and ΔG-MLG virions. The amount of virus in each lane corresponded to 35 fluorescence units. Note the absence of G protein in the ΔG virion sample.
Loss of MLG fluorescence in vitro is G protein dependent.
To determine if G protein was required for the loss of MLG fluorescence, we compared the responses of rVSV-MLG and bald ΔG-MLG virions to incubation in buffer at neutral and low pHs. ΔG-MLG was treated as described above, and fluorescence was measured. In this experiment the fluorescence signal of the G-containing virus was reduced 57% at pH 5.5 while bald ΔG virus lost only 8% (Fig. 6B). This suggested that ΔG virus is not permeable to protons and indicated that the loss of MLG fluorescence at low pH requires G protein. To ensure that the MLG in ΔG particles was pH responsive, we treated both ΔG and G-containing MLG virus with Triton X-100, which completely solubilizes the viral envelope and would expose the MLG in ΔG virions to the acidic environment. As shown in Fig. 6B, when virus was solubilized with Triton X-100, the pH-dependent loss of fluorescence of ΔG-MLG was similar to that for rVSV-MLG virions. Notably, there was no additional reduction in fluorescence for rVSV-MLG virus after the addition of Triton X-100, indicating that acidification of the virion interior was complete at the pHs used in intact virions when G protein was present. These results strongly indicated that in the absence of G protein, the viral envelope did not allow penetration of protons and did not result in the pH-dependent reduction in MLG fluorescence, but once the membrane was solubilized, the loss of fluorescence was similar to that of virions containing G protein. Thus, the viral membrane is sufficiently intact in our virus preparations to prevent exposure of MLG to acidic pH when G protein is not in the viral envelope.
To ensure that the observed difference in response of G versus ΔG viruses was not due to a difference in the incorporation of Lumio Green into the virions, we examined the protein composition of the two virus preparations using Coomassie blue-stained SDS-polyacrylamide gels. In the virus preparation used for these experiments, there was slightly more N, P, and MLG in the ΔG sample than in the rVSV preparation when values were normalized to the fluorescence signal (Fig. 6C), indicating that the difference seen in Fig. 7B was not due to insufficient amounts of MLG protein in the sample.
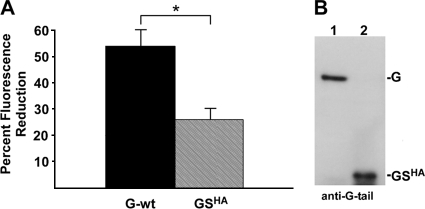
FIG. 7.
Effect of GSHA on fluorescence loss. (A) Percent fluorescence loss for G- and GSHA-complemented ΔG-MLG virions at pH 5.5 relative to fluorescence at pH 7. *, P < 0.0001. (B) Immunoblot using an antipeptide antibody to the cytoplasmic tail of G protein to assess the relative levels of G and GSHA incorporated into complemented ΔG-MLG virions. The amount of virus loaded for each sample corresponded to 35 fluorescence units measured at pH 7.0.
The G ectodomain is important for loss of MLG fluorescence.
Previously, we reported the recovery of a virus encoding a truncated version of G in which approximately 90% of the ectodomain was deleted. We called this protein GS (for G-stem), and it is composed of the 42 membrane-proximal amino acids of the G protein ectodomain together with the transmembrane and cytoplasmic tail domains (47). To determine if the entire ectodomain of G was required for the loss of fluorescence observed at low pH, we produced virus containing GS and MLG by complementation of ΔG-MLG virions. For these experiments we used GSHA, which has an N-terminal HA tag (23), to monitor GSHA surface expression, and we used ΔG virus complemented with G-wt as a control. Equivalent amounts of the two viruses were suspended in HMN buffer, the pH was adjusted to 5.5, and then the reduction in fluorescence relative to that at pH 7 was determined. We observed a smaller reduction of fluorescence with the GSHA-complemented virus than with G-wt virus (Fig. 7A); however, the reduction was approximately twice that seen with ΔG-MLG virions. This difference was not due to differences in the amount of GS incorporated compared to full-length G-wt, as determined by immunoblot analysis using a peptide-specific antibody to the cytoplasmic tail of G (Fig. 7B). These data indicated that the entire ectodomain of G was required for maximal loss of MLG fluorescence, suggesting that conformational changes induced in the ectodomain contribute to the acidification of the virion interior rather than acid-induced changes in the viral envelope containing only G membrane anchors.
Low pH enhances release of M protein from VSV virions.
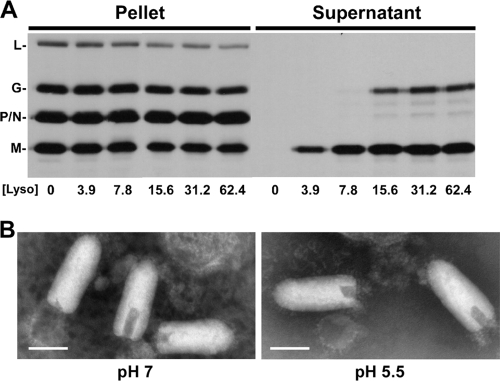
We hypothesized that acidification of the virus interior may be important for virus uncoating, as had been shown for influenza virus (20, 45, 49, 57). Therefore, to determine if the loss in fluorescence observed when rVSV-MLG is exposed to low pH could represent a biologically relevant acidification of the virion interior, we asked whether internal viral proteins are affected when the virus is exposed to low pH. Before we could examine this question, we first determined conditions that would partially permeabilize the viral envelope without completely solubilizing the membrane, which is defined by the release of G protein into the supernatant. Lysolecithin (lysophosphatidylcholine) inserts into membranes and causes transient pores at low concentrations (33, 42). To establish the concentration of lysolecithin that could permeabilize virions without solubilizing the viral envelope, we used 35S-labeled rVSV-wt virions suspended in HMN buffered to pH 5.5 and treated the virus with different concentrations of lysolecithin. After incubation for 10 min on ice, the virions were pelleted through a sucrose cushion, and proteins that were released into the supernatant were TCA precipitated and examined by SDS-PAGE (Fig. 8A). At concentrations above ∼15 μg/ml, we observed some release of both G and M proteins, but at concentrations below 7.8 μg/ml lysolecithin only some M protein was released (Fig. 8A). Based on these results, we used 3.9 μg/ml lysolecithin for our analysis. To determine if lysolecithin-treated virions remained intact, we examined the pelleted material by negative-stain TEM and found that the release of M from the virions was not due to disruption of virions (Fig. 8B).
FIG. 8.
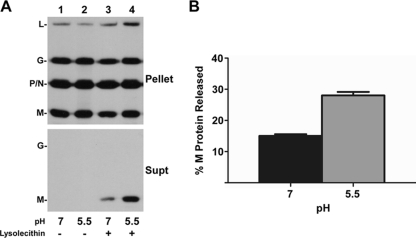
Lysolecithin treatment of rVSV-wt. (A) Autoradiograph of [35S]methionine-labeled rVSV-wt treated with different concentrations of lysolecithin in HMN buffered to pH 5.5. The mixtures were ultracentrifuged over a sucrose cushion, the supernatant was TCA precipitated, and both the pellet and supernatant fractions were analyzed by SDS-PAGE. (B) Negative-stain TEM of rVSV-wt after incubation in HMN buffered to pH 7 or pH 5.5 and treatment with lysolecithin at a concentration of 3.9 μg/ml. Magnification, ×100,000. Bar, 100 nm.
To determine if there was a difference in the amount of M released into the supernatant at pH 7 versus pH 5.5, we treated 1 μg of virions with or without 3.9 μg/ml lysolecithin in HMN buffered to pH 7 or pH 5.5 for 10 min on ice and separated proteins released into the supernatant from virion-associated proteins by ultracentrifugation. There was no protein released from virus in the absence of lysolecithin at either pH (Fig. 9A, lanes 1 and 2). Notably, there was an increase in the amount of M released from lysolecithin-permeabilized virus at pH 5.5 compared to that at pH 7.0 (Fig. 9A, lanes 1 and 2). Quantification of the amount of M released, as determined by the ratio of M protein in the supernatant to N protein in the pellet, indicated 15% (± 1 standard deviation) at pH 7 and 28% (± 2 standard deviations) at pH 5.5 (Fig. 9B). Therefore, low pH enhanced but was not absolutely required for release of M protein from lysolecithin-permeabilized virions.
FIG. 9.
Lysolecithin treatment of rVSV-wt at pH 7 and pH 5.5. (A) Representative autoradiographs of [35S]methionine-labeled rVSV-wt treated at a concentration of 0 or 3.9 μg/ml lysolecithin in HMN buffered to pH 7 or pH 5.5 and processed as described in the legend of Fig. 8. (B) Quantification of M protein released relative to the amount of N protein found in the pellet. Data are the means of three independent experiments, and standard deviations are shown. Statistical analysis of the raw data indicated the difference was significant (P < 0.0005).
DISCUSSION
The mechanism by which RNPs and M protein associate during assembly and dissociate during uncoating is not fully understood for VSV and many other enveloped viruses that have matrix proteins. A well-studied model for this group of viruses is the uncoating of influenza virus, which requires acidification of the virion interior to dissociate the M1 matrix protein from the RNP core prior to establishing a productive infection (27, 38). Influenza virion acidification is accomplished by the M2 protein, which is a proton-selective ion channel (20, 45, 49). To understand the uncoating mechanism of VSV, we utilized a recombinant virus encoding a C-terminally tagged M protein that could be labeled with a pH-sensitive fluorophore. This gave us the ability to follow the distribution of fluorescent M protein during virus entry and uncoating.
Using a synchronized entry assay, we observed a loss in the fluorescence intensity of rVSV-MLG virions soon after the initiation of virus entry. The reduction in fluorescence peaked at approximately 5 min postentry (Fig. 4 and 5) and was followed by a recovery of fluorescence levels similar to that seen prior to entry after 10 to 15 min postentry, with a steady increase in fluorescence above input levels between 15 through 60 min postentry. When we considered the absence of fluorescence loss in cells treated with bafilomycin A1 (Fig. 4B) and the pKa of Lumio Green when bound to the tetracysteine motif of the Lumio tag (16), we hypothesized that the acidic environment of endosomes may be responsible for the loss of fluorescence. Indeed, we observed a similar loss of fluorescence in vitro when the pH was reduced from 7 to 5.5. The observation that the percentage of fluorescence loss seen within cells at t-5 min postentry was similar to the loss of fluorescence observed at pH 6 in vitro, which is the approximate pH of early endosomes (17) where the bulk of VSV fusion is thought to occur (24, 50), was striking. We also observed that the curve for the decrease in fluorescence intensity was similar to pH curves seen for VSV G protein pH-dependent cell-cell (53) and virus-cell (3) fusion, further suggesting a correlation between pH-dependent conformational changes in G and the loss of MLG fluorescence at acidic pH. We then compared the loss of fluorescence intensity for bald ΔG-MLG virions with that of ΔG-MLG complemented with either full-length G protein or a large ectodomain deletion mutant (GSHA) at acidic pH and found that the full G protein ectodomain was needed for maximal pH-dependent loss of fluorescence in vitro.
The requirement for full-length G protein was interesting and somewhat paralleled the M2 ion channel-dependent acidification of influenza virions (28, 45). During influenza virus entry, the M2 proton channel results in acidification of the virion interior. The low pH results in a decrease in the interaction between the M1 (matrix) protein of influenza virus and nucleocapsids, causing dissociation of the M1 protein from RNPs, which is required for import of RNPs into the nucleus (6, 27, 38). Our observations that low pH can change the fluorescence of MLG within VSV particles in a G protein- and pH-dependent manner suggests that exposure to low pH may affect the association of the M protein within VSV RNPs in a similar manner.
Previous studies have found that the permeability of cells expressing the spike proteins of several different enveloped viruses including VSV, Sindbis virus, Semliki Forest virus (SFV), and Ebola virus is increased when the cells are exposed to acidic pH (18, 26, 29). The involvement of the SFV spike protein in membrane disruption at low pH has been extensively studied using cell permeabilization assays, whole-cell patch-clamp recordings, and dual voltage-clamp intracellular current flow monitoring (29, 31), and it was proposed that these changes were due to pore formation by the E1-E2-E3 protein trimers that make up the SFV spike (30). However, the mechanism by which other viral envelope proteins induce membrane permeabilization is not well understood. Based on these and other studies describing the viroporin activity of VSV G protein (26), our data support the idea that G protein may induce transient pores in the viral membrane while undergoing pH-dependent conformational changes or changes in the higher-order oligomeric state induced by low pH or from the movement of G protein trimers to the virion poles, as observed when purified virus is treated with low pH and examined by rotary-shadow and negative-stain electron microscopy (5). These pH-dependent changes may cause enough disruption of the envelope to allow protons to acidify the interior of the virion, thereby resulting in the loss of MLG fluorescence reported here.
It was previously shown that M protein exists in a dynamic equilibrium with RNPs when virions are disrupted with Triton X-100 and that its association with nucleocapsids was not affected by low pH (36). Consistent with these observations, we found that skeletons treated with pH 5.5 to 6.0 did not release any of the tightly bound M protein from RNPs (data not shown). Even though these results suggested that pH has no effect on the population of M protein that is tightly bound to RNPs, we reasoned that low pH may affect a different population of M inside the virion, in particular, the M associated with the viral envelope that acts as a bridge between the viral envelope and the RNP core (8, 35, 56). Indeed, virions treated with lysolecithin (which permeabilizes but does not solubilize membranes) in the presence of low pH showed an enhanced release of M protein compared to virions treated similarly at neutral pH. It is important to note that low pH is not absolutely necessary for uncoating to occur since VSV pseudotypes having envelope proteins that induce membrane fusion at neutral pH and that fuse at the cell surface are infectious (4, 25, 40, 52); however, the relative infectivity of VSV pseudotypes entering via pH-independent fusion proteins compared to entry by pH-dependent fusion proteins has not been determined.
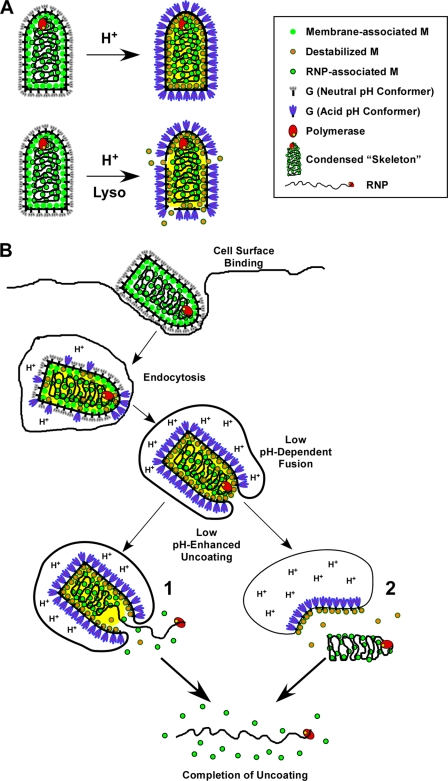
From this study we propose the following model for VSV uncoating (Fig. 10): as virions are exposed to the low-pH environment within endosomes during entry, G protein undergoes conformational changes (Fig. 10A and B), and these changes induce pore formation, thereby allowing protons to penetrate the viral envelope. Virion acidification then reduces the interaction between M protein that bridges the viral envelope and the RNP skeletons (Fig. 10). Acidification may trigger a conformational change in membrane-associated M protein that reduces the interaction between adjacent M protein monomers and/or between envelope-associated M proteins and the RNP within the virion just before (as shown) or concomitant with fusion. This reduced affinity of membrane-bound M protein would enhance release of the skeleton into the cytosol. The small amount of M protein remaining on the released skeleton could then dissociate from the RNP in the cytoplasm of newly infected cells, which would be devoid of M protein, as previously suggested (36), thereby allowing transcription to occur and effectively initiating the replication program.
FIG. 10.
Models for low-pH-enhanced VSV uncoating. (A) Depiction of how low pH induces the release of M protein in vitro from lysolecithin-treated virus. When virions are exposed to low pH, G protein undergoes conformational changes (shown by gray G trimers becoming blue trimers). We propose that during this transition, proton-permeable pores allow acidification of the virion interior (indicated by the yellow background), causing changes in M protein (represented by green circles changing to circles with orange centers). This pH-dependent change in M protein reduces affinity of M for the RNP skeleton. If the solution also contains low concentrations of lysolecithin (e.g., 3.9 μg/ml), then the viral envelope is partially permeabilized, allowing the release of the low-affinity form of M into the supernatant. (B) Model for VSV uncoating in vivo. Virus binds to the cell surface and is endocytosed. As the endosome becomes acidified, G begins to undergo conformational changes that induce pores to form in the viral envelope allowing acidification of the virion interior. This acidification causes changes in M protein that affect the contacts between the condensed RNP and membrane-bound M proteins (depicted by orange circles). When the pH is low enough to convert G trimers to the acid conformation, membrane fusion is induced. We propose that acidification of the virion interior allows the release of RNPs from membrane-bound M protein. The release of RNPs could occur either concomitant with (1) or after (2) completion of membrane fusion. Once RNP skeletons are released from the membrane-bound M, the more tightly bound RNP-associated M (green circles with black rims) is released into the cytoplasm. In both cases the bulk of the envelope-associated M protein remains bound to the fused endosomal membrane (Mire et al., unpublished data).
In summary, we report that low pH causes a reduction in the fluorescence of MLG within VSV particles as they transit through cellular endosomes and that low pH enhances the release of M from lysolecithin-treated virions. Together, these data suggest that pH is one of the regulatory elements involved in VSV uncoating. Although this effect is accomplished by a different mechanism than that of influenza virus, our findings suggest that low pH may be a trigger for, or may enhance, the uncoating of other enveloped viruses that enter the host cell through the endocytic pathway.
Acknowledgments
We thank Carolyn Matthews for technical assistance and for management of the LSCM facility and Elizabeth Matheny and Erika Dillard for comments during manuscript preparation. We also thank Sharon Frase for TEM assistance, which was performed at the University of Memphis Integrated Microscopy Center.
This work was supported in part by NIH grant AI22470 to J.M.W. The LSCM facility was made available through NCRR grant RR13725 to M.A.W.
Footnotes
Published ahead of print on 23 September 2009.
REFERENCES
- 1.Adams, S. R., R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis, and R. Y. Tsien. 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124:6063-6076. [DOI] [PubMed] [Google Scholar]
- 2.Arhel, N., A. Genovesio, K. A. Kim, S. Miko, E. Perret, J. C. Olivo-Marin, S. Shorte, and P. Charneau. 2006. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat. Methods 3:817-824. [DOI] [PubMed] [Google Scholar]
- 3.Blumenthal, R., A. Bali-Puri, A. Walter, D. Covell, and O. Eidelman. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. J. Biol. Chem. 262:13614-13619. [PubMed] [Google Scholar]
- 4.Boritz, E., J. Gerlach, J. E. Johnson, and J. K. Rose. 1999. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J. Virol. 73:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. C., W. W. Newcomb, and S. Lawrenz-Smith. 1988. pH-dependent accumulation of the vesicular stomatitis virus glycoprotein at the ends of intact virions. Virology 167:625-629. [PubMed] [Google Scholar]
- 6.Bui, M., G. Whittaker, and A. Helenius. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasco, L. 1981. Modification of membrane permeability induced by animal viruses early in infection. Virology 113:623-629. [DOI] [PubMed] [Google Scholar]
- 8.Chong, L. D., and J. K. Rose. 1993. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J. Virol. 67:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinton, G. M., S. P. Little, F. S. Hagen, and A. S. Huang. 1978. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell 15:1455-1462. [DOI] [PubMed] [Google Scholar]
- 10.Das, S. C., D. Panda, D. Nayak, and A. K. Pattnaik. 2009. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged M protein allows dynamic imaging of M protein and virus uncoating in infected cells. J. Virol. 83:2611-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De, B. P., G. B. Thornton, D. Luk, and A. K. Banerjee. 1982. Purified matrix protein of vesicular stomatitis virus blocks viral transcription in vitro. Proc. Natl. Acad. Sci. USA 79:7137-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flood, E. A., and D. S. Lyles. 1999. Assembly of nucleocapsids with cytosolic and membrane-derived matrix proteins of vesicular stomatitis virus. Virology 261:295-308. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaietta, G., T. J. Deerinck, S. R. Adams, J. Bouwer, O. Tour, D. W. Laird, G. E. Sosinsky, R. Y. Tsien, and M. H. Ellisman. 2002. Multicolor and electron microscopic imaging of connexin trafficking. Science 296:503-507. [DOI] [PubMed] [Google Scholar]
- 15.Green, T. J., X. Zhang, G. W. Wertz, and M. Luo. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357-360. [DOI] [PubMed] [Google Scholar]
- 16.Griffin, B. A., S. R. Adams, and R. Y. Tsien. 1998. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269-272. [DOI] [PubMed] [Google Scholar]
- 17.Gruenberg, J., and F. R. Maxfield. 1995. Membrane transport in the endocytic pathway. Current Opinion in Cell Biology 7:552-563. [DOI] [PubMed] [Google Scholar]
- 18.Han, Z., J. M. Licata, J. Paragas, and R. N. Harty. 2007. Permeabilization of the plasma membrane by Ebola virus GP2. Virus Genes 34:273-281. [DOI] [PubMed] [Google Scholar]
- 19.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helenius, A. 1992. Unpacking the incoming influenza virus. Cell 69:577-578. [DOI] [PubMed] [Google Scholar]
- 21.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeetendra, E., K. Ghosh, D. Odell, J. Li, H. P. Ghosh, and M. A. Whitt. 2003. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 77:12807-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeetendra, E., C. S. Robison, L. M. Albritton, and M. A. Whitt. 2002. The membrane-proximal domain of vesicular stomatitis virus G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 76:12300-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johannsdottir, H. K., R. Mancini, J. Kartenbeck, L. Amato, and A. Helenius. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83:440-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn, J. S., M. J. Schnell, L. Buonocore, and J. K. Rose. 1999. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 254:81-91. [DOI] [PubMed] [Google Scholar]
- 26.Kasermann, F., and C. Kempf. 1996. Low pH-induced pore formation by spike proteins of enveloped viruses. J. Gen. Virol. 77:3025-3032. [DOI] [PubMed] [Google Scholar]
- 27.Kemler, I., G. Whittaker, and A. Helenius. 1994. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology 202:1028-1033. [DOI] [PubMed] [Google Scholar]
- 28.Lamb, R. A., S. L. Zebedee, and C. D. Richardson. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627-633. [DOI] [PubMed] [Google Scholar]
- 29.Lanzrein, M., N. Kasermann, and C. Kempf. 1992. Changes in membrane permeability during Semliki Forest virus induced cell fusion. Biosci. Rep. 12:221-236. [DOI] [PubMed] [Google Scholar]
- 30.Lanzrein, M., A. Schlegel, and C. Kempf. 1994. Entry and uncoating of enveloped viruses. Biochem. J. 302:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanzrein, M., R. Weingart, and C. Kempf. 1993. pH-dependent pore formation in Semliki forest virus-infected Aedes albopictus cells. Virology 193:296-302. [DOI] [PubMed] [Google Scholar]
- 32.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, Y., and S. I. Chan. 1977. Effect of lysolecithin on the structure and permeability of lecithin bilayer vesicles. Biochemistry 16:1303-1309. [DOI] [PubMed] [Google Scholar]
- 34.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157-167. [PubMed] [Google Scholar]
- 35.Lenard, J., and R. Vanderoef. 1990. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J. Virol. 64:3486-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyles, D. S., and M. O. McKenzie. 1998. Reversible and irreversible steps in assembly and disassembly and vesicular stomatitis virus: equilibria and kinetics of dissociation of nucleocapsid-M protein complexes assembled in vivo. Biochemistry 37:439-450. [DOI] [PubMed] [Google Scholar]
- 37.Lyles, D. S., M. O. McKenzie, P. E. Daptur, K. W. Grant, and W. G. Jerome. 1996. Complementation of M gene mutants of vesicular stomatitis virus by plasmid-derived M protein converts spherical extracellular particles into native bullet shapes. Virology 217:76-87. [DOI] [PubMed] [Google Scholar]
- 38.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 39.Matlin, K., H. Reggio, A. Helenius, and K. Simons. 1982. The pathway of vesicular stomatitis entry leading to infection. J. Mol. Biol. 156:609-631. [DOI] [PubMed] [Google Scholar]
- 40.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767-773. [DOI] [PubMed] [Google Scholar]
- 41.Mellman, I., R. Fuchs, and A. Helenius. 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55:663-700. [DOI] [PubMed] [Google Scholar]
- 42.Miller, M. R., J. J. Castellot, and A. B. Pardee. 1978. A permeable animal cell preparation for studying macromolecular synthesis. DNA synthesis and the role of deoxyribonucleotides in S phase initiation. Biochemistry 17:1073-1080. [DOI] [PubMed] [Google Scholar]
- 43.Newcomb, W. W., and J. C. Brown. 1981. Role of the vesicular stomatitis virus matrix protein in maintaining the viral nucleocapsid in the condensed form found in native virions. J. Virol. 39:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newcomb, W. W., G. J. Tobin, J. J. McGowan, and J. C. Brown. 1982. In vitro reassembly of vesicular stomatitis virus skeletons. J. Virol. 41:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto, L. H., L. J. Holsinger, and R. A. Lamb. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517-528. [DOI] [PubMed] [Google Scholar]
- 46.Rigaut, K. D., D. E. Birk, and J. Lenard. 1991. Intracellular distribution of input vesicular stomatitis virus proteins after uncoating. J. Virol. 65:2622-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 49.Shimbo, K., D. L. Brassard, R. A. Lamb, and L. H. Pinto. 1996. Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys. J. 70:1335-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sieczkarski, S. B., and G. R. Whittaker. 2003. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333-343. [DOI] [PubMed] [Google Scholar]
- 51.Sun, X., V. K. Yau, B. J. Briggs, and G. R. Whittaker. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338:53-60. [DOI] [PubMed] [Google Scholar]
- 52.Tatsuo, H., K. Okuma, K. Tanaka, N. Ono, H. Minagawa, A. Takade, Y. Matsuura, and Y. Yanagi. 2000. Virus entry is a major determinant of cell tropism of Edmonston and wild-type strains of measles virus as revealed by vesicular stomatitis virus pseudotypes bearing their envelope proteins. J. Virol. 74:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza and vesicular stomatitis virus. J. Cell Biol. 89:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, T., and J. Lenard. 1981. Interaction of wild-type and mutant M protein vesicular stomatitis virus with nucleocapsids in vitro. Biochemistry 20:1349-1354. [DOI] [PubMed] [Google Scholar]
- 56.Zakowski, J. J., and R. R. Wagner. 1980. Localization of membrane-associated proteins in vesicular stomatitis virus by use of hydrophobic membrane probes and cross-linking reagents. J. Virol. 36:93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhirnov, O. P. 1990. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology 176:274-279. [DOI] [PubMed] [Google Scholar]