Abstract
Most poxviruses express multiple proteins containing ankyrin (ANK) repeats accounting for a large superfamily of related but unique determinants of poxviral tropism. Recently, select members of this novel family of poxvirus proteins have drawn considerable attention for their potential roles in modulating intracellular signaling networks during viral infection. The rabbit-specific poxvirus, myxoma virus (MYXV), encodes four unique ANK repeat proteins, termed M-T5, M148, M149, and M150, all of which include a carboxy-terminal PRANC domain which closely resembles a cellular protein motif called the F-box domain. Here, we show that each MYXV-encoded ANK repeat protein, including M-T5, interacts directly with the Skp1 component of the host SCF ubiquitin ligase complex, and that the binding of M-T5 to cullin 1 is indirect via binding to Skp1 in the host SCF complex. To understand the significance of these virus-host protein interactions, the various binding domains of M-T5 were mapped. The N-terminal ANK repeats I and II were identified as being important for interaction with Akt, whereas the C-terminal PRANC/F-box-like domain was essential for binding to Skp1. We also report that M-T5 can bind Akt and the host SCF complex (via Skp1) simultaneously in MYXV-infected cells. Finally, we report that M-T5 specifically mediates the relocalization of Akt from the nucleus to the cytoplasm during infection with the wild-type MYXV, but not the M-T5 knockout version of the virus. These results indicate that ANK/PRANC proteins play a critical role in reprogramming disparate cellular signaling cascades to establish a new cellular environment more favorable for virus replication.
Myxoma virus (MYXV) is a rabbit-specific poxvirus that has proven to be a useful model system to study the mechanism by which virus-encoded immunoregulatory proteins function to manipulate the various host immune responses during the course of viral infection (50). In its long-term evolutionary host (Sylvilagus sp.), MYXV causes a benign disease localized to the site of inoculation, but when the virus infects European rabbits (Oryctolagus cuniculus), it causes a rapid systemic and highly lethal infection called myxomatosis (13). The success of MYXV as a pathogen can be attributed to the ability of the virus to effectively avoid recognition and clearance by the immune systems of susceptible rabbit hosts. At the level of individual virus-infected cells, poxviruses, like MYXV, are particularly adept at binding and entering most mammalian cells, where they attempt to establish a favorable intracellular environment, which promotes viral replication. Thus, the ability of poxviruses to reconfigure or disable the various host antiviral responses of the infected cell directly dictates the outcome of a viral infection at the cellular level (28). To this end, poxviruses possess a large genomic capacity, and all encode a unique repertoire of immunoregulatory and host-interactive proteins that have evolved to specifically mediate a broad range of cellular processes critical for successful viral replication. To date, a large collection of poxvirus-encoded immunoregulatory proteins have been identified and characterized, including virokines, viroreceptors, signaling modulators, and inhibitors of various antiviral responses, such as apoptotic pathways and interferon signaling (43). More recently, a novel category of poxvirus ankyrin (ANK) repeat proteins have drawn considerable attention for their potential roles in modulating intracellular signaling networks during viral infection (48, 49, 53).
With the exception of poxviruses, the ANK motif is not commonly reported in viruses, although numerous examples have been identified in eukaryotic, bacterial, and archaeal proteins (6). The ANK motif, a tandemly repeated consensus module of approximately 33 amino acid residues, has been demonstrated to mediate diverse protein-protein interactions between cellular proteins having a broad spectrum of functional roles (32, 42). Solved crystal structures have revealed a conserved fold structure of the ANK repeat unit, by which each repeat forms a characteristic helix-loop-helix structure with a beta-hairpin/loop region projecting out from the helices at a 90° angle (3, 16, 19, 26). However, the ANK fold appears to be defined by its structure rather than any conserved biological function since there is no specific conserved substrate or binding partner structure that is universally recognized by members of the superfamily.
The majority of poxviral ANK repeat-containing proteins also include a conserved carboxy-terminal PRANC (pox protein repeats of ankyrin C terminus) motif, which closely resembles a cellular protein motif called the F-box domain (30). Characterized as substrate adaptors, F-box-containing host proteins function to recruit cellular substrate proteins to the SCF ubiquitin-ligase complex (named after their main components, Skp1, cullin 1 [CUL1], and an F-box protein), where the substrates selected by the complex are ubiquitinated and targeted for degradation by the proteasome (21, 45, 60). The process of selective ubiquitination is an essential regulatory step for many cellular processes, and the human genome encodes more than 70 different F-box proteins, which collectively are thought to specifically target a broad collection of cellular substrates for delivery to the SCF complex to initiate turnover (62).
Accounting for the largest family of poxviral proteins, almost all chordopoxviruses encode multiple ANK repeat-containing proteins, some of which have been defined as viral host range or virulence factors (30). For example, canarypox virus encodes 51 ANK repeat proteins, accounting for greater than 20% of the genome; however, most other poxviruses express less than a half dozen ANK repeat proteins (52). MYXV encodes four unique ANK repeat proteins, termed M-T5, M148, M149, and M150, all of which have been described as virulence factors for myxomatosis in rabbits (5, 8, 33). The MYXV host range factor M-T5 was first characterized for its ability to regulate viral tropism within rabbit lymphocytes and, later, some classes of human cancer cell lines (33, 51). In human cancer cells, the direct physical interaction between M-T5 and the host cell Akt was shown to be a key restriction determinant for MYXV tropism in a subset referred to as type II cancer cells (56). Furthermore, M-T5 was shown to be functionally interchangeable with a host ANK repeat protein called PIKE-A, and the activation of Akt by either the viral M-T5 or the host PIKE-A protein was critical for MYXV permissiveness in type II human cancer cells (57). M-T5 was also demonstrated to protect MYXV-infected cells from virus-induced cell cycle arrest, a property which was linked to its ability to interact with a member of the host cell SCF complex called CUL1 (20). Unlike M-T5, no specific host binding partners or target substrates have yet been identified for M148, M149, or M150. However, in tumor necrosis factor alpha (TNF-α)-stimulated cells, M150 was shown to colocalize in the nucleus with NF-κB p65, suggesting that this MYXV protein may modulate the NF-κB pathway (8).
In this study, we demonstrate that M-T5, M148, M149, and M150 all have functional carboxy-terminal PRANC/F-box-like domains and that each one can interact directly with the Skp1 component of the host SCF complex. We further examined the various binding domains of M-T5 and identified ANK repeats I and II as being important for interaction with Akt, whereas the PRANC/F-box-like domain was essential for binding to Skp1. We also show that the previously reported interaction of M-T5 with CUL1 was in fact, indirect linking of M-T5 to the host SCF complex via Skp1. More specifically, we investigated the ability of M-T5 to function as a molecular scaffold to link disparate cellular binding partners together within a single complex and report that the viral protein binds Akt and the SCF complex (via Skp1) simultaneously in MYXV-infected cells. Finally, we demonstrate that M-T5 specifically mediates the relocalization of Akt from the nucleus to the cytoplasm during MYXV infection. These results suggest that ANK/PRANC proteins, such as M-T5, play a critical role in reprogramming disparate cellular signaling cascades to establish a new cellular environment more favorable for viral replication.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney (HEK) 293, HeLa, HOS, and Caki cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (heat inactivated), 100 U penicillin/ml, and 100 μg/ml streptomycin at 37°C in 5% CO2. The recombinant MYXVs (vMyx; strain Lausanne) used in this study include vMyx-lac (36) and vMyx-gfp (20), control viruses expressing wild-type M-T5. Additionally, vMyx-T5KO (33), which contains LacZ between M11L and M12, and vMyx-T5KOgfp (57), in which the enhanced green fluorescent protein (gfp) cassette is inserted between M135 and M136 in the vMyx-T5KO background (and thus contains both LacZ and gfp), both of which fail to express M-T5 due to targeted disruption of both copies of the M-T5 open reading frame (ORF) (M005R/L), were used. All viruses were propagated and titrated by focus formation on baby green monkey kidney (BGMK) cells as described previously (36).
Plasmids.
Vectors encoding MYXV genes M-T5, M148, M149, and M150 were PCR amplified from viral genomic DNA and subcloned into pDONR221 (Invitrogen). By the use of Gateway recombination (Invitrogen), the viral ORFs were transferred to the expression plasmid pANT7_nHA, which was kindly provided by the Harvard Institute of Proteomics (HIP). The plasmids encoding human CUL1, Skp1, and Akt, all fused to C-terminal glutathione S-transferase (GST) (pANT7_cGST), were also received from HIP; CUL1 tagged with a hemagglutinin (HA-CUL1) or Flag (Flag-CUL1) epitope was a gift provided by Y. Xiong (University of North Carolina) and Z. Q. Pan (Mount Sinai School of Medicine, NY), respectively. M-T5 ORF fragment constructs were amplified from viral genomic DNA by PCR. N-terminal M-T5 deletions were generated using the forward primers listed in Table 1 and the reverse primer MT5rev (5′-AAACTCGAGCGCGTGTATCTTTAC-3′ [XhoI site underlined]). Alternatively, internal M-T5 deletions were generated by amplifying the right and left flanks of the deletion region by use of the primer sets listed in Table 1. Afterwards, a second round of PCR was performed using the forward primer MT5for (5′-AAAGGATCCATGGATCTATACGGG-3′ [BamHI site underlined]) and the reverse primer MT5rev. PCR products were digested with BamHI and XhoI, gel purified (Qiagen), and cloned directly into the vector pcDNA3.1(+)/myc-His A (Invitrogen). The identity of all clones was confirmed by sequence analysis, and expression of fusion proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
TABLE 1.
List of primers used to PCR amplify M-T5 constructs
| Primer | Sequence |
|---|---|
| MT5for | AAAGGATCCATGGATCTATACGGG |
| MT5rev | AAACTCGAGCGCGTGTATCTTTAC |
| ΔNfor | AAAGGATCCATGCGGGATACCCCCTTTCGC |
| ΔN-Ifor | AAAGGATCCATGGTTCGTGGTTCTCGTACG |
| ΔN-IIfor | AAAGGATCCATGGCGCCAGATGGACG |
| ΔN-IIIfor | AAAGGATCCATGCGGGGAACGGACGGATACG |
| ΔN-IVfor | AAAGGATCCATGCCAGACAAGACGTACGGG |
| ΔN-Vfor | AAAGGATCCATGGGATTACACGCGACC |
| ΔN-VIfor | AAAGGATCCATGGTGAACTTTCTGGGG |
| ΔN-VIIfor | AAAGGATCCATGTACACGAGGGCGTTGAACG |
| ΔILrev | AAACGAGAACCACGAACCGAACATAGTTCGTCCGG |
| ΔIRfor | AAACCGGACGAACTATGTTCGGTTCGTGGTTCTCG |
| ΔIILrev | AAACGTCCATCTGGCGCCATAGAACCACGAACTCCG |
| ΔIIRfor | AAACGGAGTTCGTGGTCCTATGGCGCCAGATGGACG |
| ΔI-IILrev | AAACGTCCATCTGGCGCCATCGAACATAGTTCGTCCGG |
| ΔI-IIRfor | AAACCGGACGAACTATGTTCGATGGCGCCAGATGGACG |
| ΔIII-Vlrev | AAAGGTCGCGTGTAATCCATCTGGCGCCATCGCG |
| ΔIII-Vrfor | AAACGCGATGGCGCCAGATGGATTACACGCGACC |
| ΔVI-VIILrev | AAACGTTCAACGCCCTCGTGTAGAGAGAGTTTTGTTCG |
| ΔVI-VIIRfor | AAACGAACAAAACTCTCTCTACACGAGGGCGTTGAACG |
| ΔPRANCrev | AAACTCGAGCCCAGGCGTGTCCAGCC |
Immunoblot analysis.
Samples were separated by SDS-PAGE. Separated proteins were transferred to nitrocellulose and blocked with 5% skim milk in phosphate-buffered saline (PBS) with 0.1% Tween 20 (PBST). Primary antibodies were diluted in 5% skim milk-PBST and incubated with membranes overnight at 4°C. Membranes were washed and incubated for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies diluted 1:5,000 in 5% milk-PBST. Immunoreactive proteins were detected by chemiluminescence (PerkinElmer). Antibodies used included monoclonal antibodies specific for HA (12CA5; Roche) and myc (9E10; Santa Cruz Biotechnology) epitopes. Polyclonal antibodies used included those for detecting human Skp1 and Akt (Cell Signaling) and GST (NeoMarkers). Horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies were obtained from Jackson ImmunoResearch Laboratories. Densitometric levels of Akt were detected by Molecular Imaging software (Kodak).
GST pull-down assay.
For GST fusion proteins, plasmids were expressed by in vitro transcription-translation according to the manufacturer's protocol (Promega) and were incubated with glutathione agarose beads for 2 h. Beads were pelleted by centrifugation and washed five times, and the immunocomplexes were resolved by SDS-PAGE. Immunoblot analyses were performed as described above, using the appropriate antibodies.
Y2H experiments.
The details of the human cDNA libraries and the yeast two-hybrid (Y2H) screening methods have been reported elsewhere (31).
Immunoprecipitation assay.
Samples were incubated with protein A/G beads (Pierce) at 4°C for 1 h in a preclear phase. After centrifugation, supernatants were removed and incubated with the specified antibody at 4°C overnight with shaking. The next day, protein A/G beads were added and further incubated at 4°C for 2 h. The beads were washed three times with lysis buffer, resuspended in 2× SDS loading buffer with β-mercaptoethanol, and boiled for 5 min to elute the proteins. The eluted supernatants were resolved by SDS-PAGE and analyzed by Western blotting.
AlphaScreen binding protocol.
Cells were seeded into six-well plates at a density of 5 × 105 cells per well in complete growth medium with 10% fetal bovine serum. Transfections were performed with Effectene (Qiagen) in accordance with the manufacturer's instructions. Cultured cells were collected, and cell lysis was prepared as previously described. All AlphaScreen assays described were performed in triplicates in 384-well white opaque plates (PerkinElmer), using PBS containing 0.1% bovine serum albumin as the buffer. For the detection of HA fusion proteins, the HA detection kit containing anti-HA-coated acceptor beads (PerkinElmer) was used. Equal concentrations of acceptor beads and streptavidin donor beads were used at a final concentration of 20 μg/ml in a final volume of 25 μl per well. First, 5 μl of cell lysate, followed by biotinylated anti-HIS (10 nM) or biotinylated anti-Flag antibody (25 nM) and acceptor beads in buffer, were added to each well and incubated for 2 h at room temperature. A total of 5 μl of a 1:50 dilution of the donor beads was then added to give a final volume of 25 μl, and the mixture was incubated at room temperature for 2 h. All additions and incubations were made under subdued lighting conditions due to the photosensitivity of the beads, and finally, the assay plates were read in an EnVision plate reader (PerkinElmer).
siRNA transfections.
Cellular monolayers were transfected with the indicated small interfering RNA (siRNA) per manufacturer's protocol (Dharmacon), and 48 h later, cells were infected with either vMyx-lac or vMyx-T5KO at a multiplicity of infection (MOI) of 0.1. Inoculum was allowed to adsorb for 1 h before being removed; cells were then washed and supplemented with Dulbecco's modified Eagle's medium. Cells were incubated at 37°C and 48 h postinfection (hpi) focus formation was examined under a Leica fluorescence microscope as a measure of viral replication.
Subcellular localization of Akt and CUL1.
HeLa cells were grown on glass coverslips to 70% confluence and were transiently transfected with HA-CUL1 using Effectene (Qiagen) according to the manufacturer's recommendations. Two days later, cells were mock infected or infected with vMyx-lac or vMyx-T5KO at an MOI of 5. At 4 hpi, cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 and blocked with 5% fetal bovine serum. HA-CUL1 expression was detected by indirect immunofluorescence by the use of monoclonal antibody specific for the HA epitope (1:100 dilution) and a Texas Red-conjugated goat anti-mouse secondary antibody (1:1,000 dilution; Jackson ImmunoResearch). Detection of endogenous Akt was determined by polyclonal Akt antibody (1:100 dilution) and the fluorescein isothiocyanate-conjugated goat anti-rabbit secondary antibody (1:1,000; Jackson ImmunoResearch). Cells were mounted with Vectashield (Vector Laboratories) containing the fluorescent marker DAPI (4′,6-diamidino-2-phenylindole) to assay nuclear localization and were examined under a spinning-disk confocal microscope (Olympus), using the appropriate filters. Nuclear and cytoplasmic extractions were performed according to manufacturer's recommendations (Thermo Scientific).
RESULTS
MYXV ANK repeat proteins all contain a functional carboxy-terminal PRANC/F-box-like domain and bind Skp1.
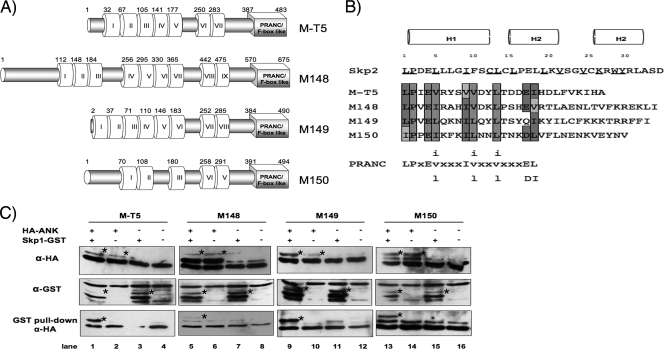
With the exception of Molluscipoxvirus, all sequenced members from each of the poxvirus genera encode multiple ANK repeat-containing proteins, the majority of which include a PRANC/F-box-like domain located at the carboxy terminus (30). MYXV, a member of the Leporipoxvirus genus, encodes four ANK/PRANC-containing proteins: M-T5, M148, M149, and M150 (7, 58). Similar to other poxviral ANK/PRANC proteins, the four MYXV proteins range from 400 to 700 amino acids in length, possess 5 to 10 copies of the ANK motif clustered toward the amino terminus of each protein, and include a single carboxy-terminal PRANC/F-box-like motif (Fig. 1A). However, amino acid sequence alignment of M-T5, M148, M149, and M150 demonstrates that the proteins otherwise exhibit little sequence similarity to one another (33.4 to 43.4% similarity; 18.8 to 23.3% identity) (Table 2). M148 was the least similar ORF among the four MYXV ORFs and appears to contain an additional 179 residues at the amino terminus, accounting for an extra two to four ANK repeats.
FIG. 1.
MYXV ANK repeat proteins include a functional C-terminal PRANC domain that binds Skp1. (A) Schematic representation of the four ANK repeat-containing proteins encoded by MYXV. Predicted ANK repeats and putative carboxyl-terminal PRANC/F-box-like domains are represented by white and gray boxes, respectively (24, 41). The first amino acid residue of each predicted ANK repeat and amino acid length of the corresponding proteins are indicated. (B) Alignment of the amino acid sequences of the carboxy-terminal region of each MYXV ANK repeat protein. A consensus PRANC/F-box-like sequence adapted from Mercer et al. (30) is shown with lowercase letters representing variant positions, and X indicates nonconserved positions. Dark-shaded letters represents residues identical to the poxviral consensus motif, while conserved substitutions are denoted with light shading. Nonconserved differences contain no shading. The F-box α-helical secondary structures are represented by H1, H2, and H3 (37, 63). Known residues of Skp2 that contact the linker protein Skp1 are underlined (40). (C) In vitro coupled transcription-translation was used to express the indicated (+) combination of plasmids, which include Skp1-GST and HA-tagged MYXV ANK repeat proteins M-T5, M148, M149, and M150. Samples were subjected to a GST pull-down assay. Precipitates and total lysates were resolved by SDS-PAGE and probed with anti-HA antibody to detect coprecipitated viral proteins (bottom row). Expression of MYXV ANK repeat proteins (top row) and Skp1 (middle row) was confirmed by immunoblotting with antibody against HA (anti-HA) and GST (anti-GST) epitopes, respectively. Bands of interest are indicated with stars. α, anti.
TABLE 2.
Identity/similarity matrix of MYXV ANK repeat-containing proteins
| Protein | Similaritya score (%) for: |
|||
|---|---|---|---|---|
| M-T5 | M148 | M149 | M150 | |
| M-T5 | 33.6 | 43.3 | 44.2 | |
| M148 | 18.8 | 38.0 | 33.4 | |
| M149 | 23.2 | 20.1 | 36.3 | |
| M150 | 23.3 | 19.1 | 20.8 | |
Values above and below the diagonal gap are similarity and identity scores, respectively.
Notably, the PRANC/F-box-like domains for each of the MYXV proteins share a high degree of sequence similarity to one another (Fig. 1B). The PRANC motif is comparably shorter than the established F-box consensus sequence common to eukaryotes, but still contains a few invariant positions; these include positions 1 (leucine), 2 (proline), 9 (isoleucine or valine), and 13 (leucine or valine) (30). Based on cocrystal structures, the cellular F-box is usually composed of three α-helices; however, the poxviral version of the motif is frequently truncated and lacks the third helix (47). Many of the invariant residues within helices H1 and H2 are well conserved in the MYXV PRANC/F-box-like domains, but little sequence similarity was maintained within α-helix H3 (Fig. 1B). Despite the lack of H3, the 2-α-helix PRANC/F-box-like domain found in poxviral proteins can still be capable of interacting faithfully with the cellular SCF complex (47).
The presence of a carboxy-terminal PRANC/F-box-like motif suggests that the four ANK repeat proteins encoded by MYXV likely interact with the SCF complex by means of binding to an adaptor protein, such as Skp1. To determine if these viral proteins have a functional PRANC/F-box-like domain, we tested their ability to bind host Skp1 protein, using a GST pull-down assay. A coupled in vitro transcription-translation (TNT) protocol was used to coexpress Skp1-GST, which is tagged at the C terminus with GST, plus one of the MYXV ANK/PRANC proteins fused to a common N-terminal HA tag (HA-MT5, HA-M148, HA-M149, or HA-M150). Samples were then incubated with GST-coated beads to pull down Skp1-GST fusions, and complexes were resolved by SDS-PAGE and immunoblotted with anti-HA antibody to analyze binding of HA-tagged MYXV proteins. As predicted, all four of the MYXV-encoded ANK/PRANC proteins were detectable by the GST pull-down assay only when coexpressed in the presence of Skp1-GST (Fig. 1C, lanes 1, 5, 9, and 13). However, no MYXV-encoded ANK/PRANC proteins were detected following GST pull-down in the absence of Skp1-GST expression (Fig. 1C, lanes 2, 6, 10, and 14). It should be noted that the binding of HA-M148 to Skp1-GST is difficult to appreciate; however, when this interaction was further examined by the AlphaScreen assay, the signal produced by the interaction between M148 and Skp1 was comparable to M-T5, M149, and M150 (data not shown). Our findings suggest that M-T5, M148, M149, and M150 each contain a functional PRANC/F-box-like domain, which can interact with the host Skp1 as previously reported with other poxviral ANK repeat proteins (48, 49, 53).
Y2H screen for potential cellular binding partners of M-T5.
Although PRANC proteins are predicted to manipulate the cellular ubiquitination machinery, surprisingly little is known about the molecular mechanism by which this largest family of poxvirus proteins functions during viral infection. Aside from Skp1, identification of additional binding partners, either cellular or viral, has resulted in little success and coincidently has hindered the study of this superfamily of proteins. The Y2H system has become an increasingly important tool to identify and map novel protein-protein interactions. For example, systematic Y2H analysis of the unique variola virus genes with human cDNA libraries revealed the presence of a novel viral inhibitor of NFκB1/p105 (31). A similar Y2H screen was performed using various human cDNA libraries to identify potential human cellular binding partners of M-T5, the best characterized of the four MXYV ANK repeat proteins. Multiple independent human cDNA libraries were analyzed for interactions, and the results from this screen identified a total of 13 potential human binding partners for M-T5 (Table 3). Notably, neither of the two previously identified binding partners for M-T5, namely, CUL1 or Akt, were identified in this Y2H screen, but the Skp1 component of the host SCF complex (that includes CUL1) was picked up in this screen.
TABLE 3.
Potential cellular binding partners of MYXV-encoded M-T5 identified by a Y2H screen of human cDNA libraries
| Designation | Name | NCBI RefSeq no. |
|---|---|---|
| BUB1B | Budding uninhibited by benzimidazoles 1 homolog β | NM_001211 |
| CBX3(183) | Chromobox homolog 3, isoform 1 | NM_007276 |
| ESPN | Epsin | NM_031475 |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | NM_001032456 |
| KIF4A | Kinesin family member 4A | NM_012310 |
| KIN13A(1749) | Kinesin-13A2 | NM_022113 |
| NEB | Nebulin | NM_004543 |
| PGD | 6-Phosphogluconate dehydrogenase | NM_002631 |
| PKM2(531) | Pyruvate kinase, muscle isoform 1 | NM_002654 |
| RAD50 | DNA repair and recombination protein Rad50 | NM_005732 |
| SCYL1BP1 | SCY1-like 1 binding protein 1 | NM_152281 |
| SKP1A(164) | S-phase kinase-associated protein 1A, isoform b | NM_006930 |
| SNX4 | Sorting nexin 4 | NM_003794 |
Creation of M-T5 deletion constructs for protein partner binding studies.
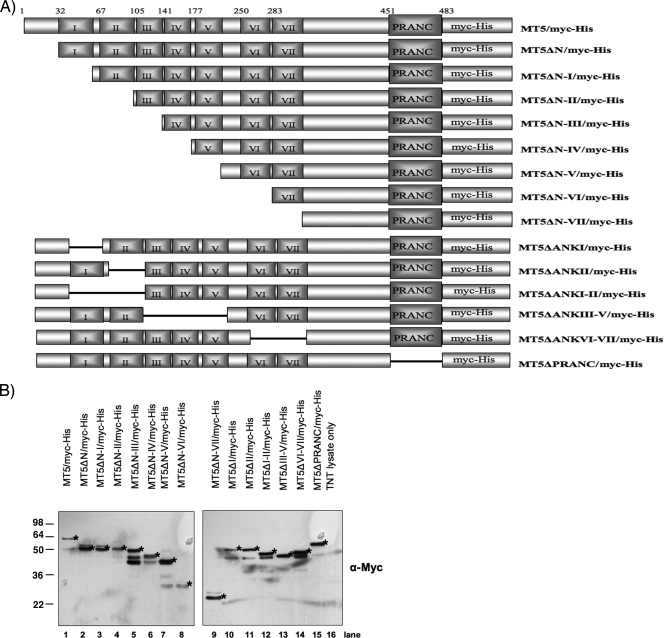
The intricate relationship between virus-encoded proteins and components of the host cell signaling networks can have a profound impact on poxvirus tropism. To investigate the significance of these virus-host protein interactions in controlling virus tropism, a collection of myc-His (C-terminus)-tagged M-T5 fragment constructs were created and used to map the binding domain(s) critical for interaction with the cellular proteins (Fig. 2A). Briefly, ANK repeats of M-T5 were systematically deleted from the amino termini of the M-T5 ORF and fused to a carboxy-terminal myc-His tag. To complement, an additional set of plasmids was constructed, which includes internal ANK repeat deletions, M-T5 with the PRANC/F-box-like domain only (MT5ΔN-VII), and M-T5 without the PRANC/F-box-like domain (MT5ΔPRANC) (Fig. 2A). Upon completion, each plasmid was sequence verified, and protein expression was confirmed by Western blot analysis with anti-myc antibody (Fig. 2B). It is interesting to note that some plasmids produced multiple bands, which we predict are shorter M-T5 protein fragments that were translated from an alternative start codon by the polymerase during the in vitro coupled transcription-translation reaction. Furthermore, no prominent bands were observed when an empty vector was used (Fig. 2B, lane 16), suggesting that these particular bands are not the result of nonspecific binding from the anti-myc antibody.
FIG. 2.
Summary of constructs employed in this study. (A) Schematic representation of the M-T5 constructs used during this study. The first residue for each of the seven predicted ANK repeats and the carboxy-terminal PRANC/F-box-like domain is indicated. All plasmids contain a carboxy-terminal myc-His tag. (B) Immunoblot analysis of M-T5 constructs expressed by in vitro coupled transcription-translation, detected by a myc-specific antibody. Bands of interest are indicated with stars. α, anti.
M-T5 domains are needed to bind the host cell SCF complex.
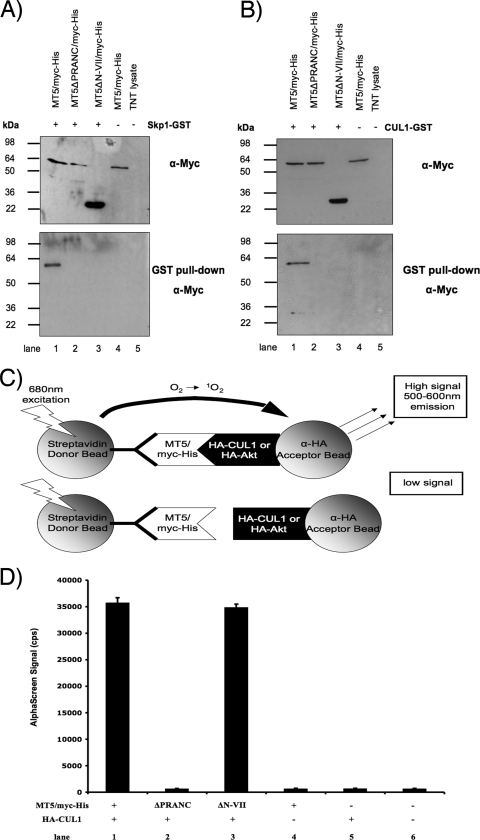
To determine whether the PRANC/F-box-like domain of M-T5 was responsible for the observed binding between Skp1 and M-T5 (Fig. 1C), GST pull-downs were performed. Skp1 fused to GST (Skp1-GST) and one of the various M-T5 myc-His-tagged constructs (MT5, MT5ΔPRANC, or MT5ΔN-VII) were coexpressed by in vitro coupled transcription-translation and protein complexes isolated using GST-coated beads. After washing the beads, samples were resolved by SDS-PAGE and probed with an anti-myc antibody. Coprecipitation of Skp1-GST and M-T5 was observed only when full-length M-T5 (MT5/myc-His) was expressed (Fig. 3A, lane 1). The truncated M-T5 constructs (MT5ΔPRANC/myc-His or MT5ΔN-VII/myc-His) failed to interact with Skp1 (Fig. 3A, lanes 2 and 3). Another essential component of the SCF complex, CUL1, has been previously reported to form a complex with M-T5 (20). When the interaction between CUL1 and M-T5 was examined by GST pull-down, the results were equivalent to Skp1 binding. In other words, CUL1-GST coprecipitated with full-length M-T5 (MT5/myc-His) but not with the truncated M-T5 constructs (Fig. 3B).
FIG. 3.
Skp1 and CUL1 members of the SCF complex bind M-T5 via the PRANC/F-box-like domain. The specified (+) combination of plasmids that include HA-Skp1 (A) or HA-CUL1 (B) and a variety of myc-His-tagged fragments of M-T5 was transiently transfected into HEK 293 cells or expressed by in vitro coupled transcription-translation. Samples were subjected to a GST pull-down assay, resolved by SDS-PAGE, and probed with anti-myc antibody to detect coprecipitated proteins. Protein expression of HA-CUL1, HA-Skp1, and M-T5 fragments was detected using anti-HA and anti-myc antibodies, respectively. (C) Principle of AlphaScreen detection for the interaction between an HA fusion host protein (CUL1 or Akt) and the myc-His-tagged M-T5 fragment constructs. Excitation of a donor bead causes the conversion of ambient oxygen to the singlet state. If a biomolecular interaction brings the acceptor bead within close proximity (>200 nm) to the donor bead, a cascade of chemiluminescence occurs, resulting in the emission of a characteristic fluorescent signal between 520 and 620 nm. Conversely, if binding between bead-bound proteins does not occur, the distance between donor and acceptor beads is too great for the oxygen singlet to migrate, and no emission signal is produced. Biotin-conjugated anti-myc antibody is used to indirectly link streptavidin-coated acceptor beads to myc-His-tagged protein. (D) Cell lysates were harvested after 48 h and incubated with biotin-conjugated anti-myc antibody and donor/acceptor beads before an AlphaScreen assay was performed. Each sample was performed in triplicate, and the standard deviation is represented by the error bars. α, anti.
To further verify the GST pull-down experiments by an independent protein-protein interaction method, the bead-based assay AlphaScreen was used to examine binding interactions. As shown schematically in Fig. 3C, activation of the donor beads at 680 nm results in the generation of excited singlet oxygen radicals, which migrate to react with chemiluminescers on the acceptor bead, subsequently triggering a cascade of chemical reactions that induce the release of light (520 to 620 nm) from activated fluorophores. For this reaction to occur, the two beads must be within close proximity (<200 nm) via specific interactions of the protein-protein complexes coupled to them (Fig. 3C, upper panel). However, if the bound test proteins do not interact, no light is emitted by the unstimulated chemiluminescers on the acceptor bead (Fig. 3C, lower panel).
To investigate the interaction between M-T5 and the SCF complex, cells were cotransfected with HA-CUL1 and one of the various M-T5 constructs (MT5, MT5ΔPRANC, or MT5ΔN-VII). The day following transfection, cell lysates were collected and prepared for protein-protein interaction analysis. AlphaScreen acceptor beads, coupled to an HA-specific antibody, were added to the cell lysate along with an anti-myc antibody coupled to biotin. Two hours later, AlphaScreen donor beads coated with streptavidin were added, and again, the sample was allowed to incubate for a couple of hours before the luminescent signal was measured by an EnVision plate reader. The presence of a distinct signal was detectable only when the PRANC/F-box-like domain of M-T5 was present for binding CUL1 (Fig. 3D, lanes 1 and 3). In contrast, the signal was dramatically reduced when the PRANC/F-box-like domain of M-T5 was deleted (Fig. 3D, lane 2). Interestingly, interaction between the M-T5 construct containing only the PRANC/F-box-like domain (MT5ΔN-VII/myc-His) and CUL1-GST was observed by AlphaScreen; however, when this same construct was previously examined by GST pull-down, this M-T5 PRANC-only construct was unable to bind either Skp1 or CUL1 (Fig. 3A and B, lane 3). Furthermore, MT5ΔN-VII/myc-His dissociated more rapidly from Skp1-GST than did MT5/myc-His when binding was examined by Biacore (data not shown). Thus, we propose that the half-life of the PRANC-only M-T5 complex with CUL1 might be shorter than that of the full-length M-T5 protein with CUL1 and, thus, be detectable only by AlphaScreen and not by GST pull-down. Together, these results demonstrate that Skp1 and CUL1 are both potential cellular binding partners of M-T5 and that deletion of the carboxy-terminal PRANC/F-box-like domain from M-T5 clearly abolishes protein-protein interactions with both partners. However, since reticulocyte lysates possess endogenous untagged SCF complex proteins, we wanted to determine whether the CUL1 binding to M-T5 might be indirect, via interactions to the Skp1 component of the SCF complex.
Skp1 serves as the direct adaptor protein that binds M-T5 to the SCF complex.
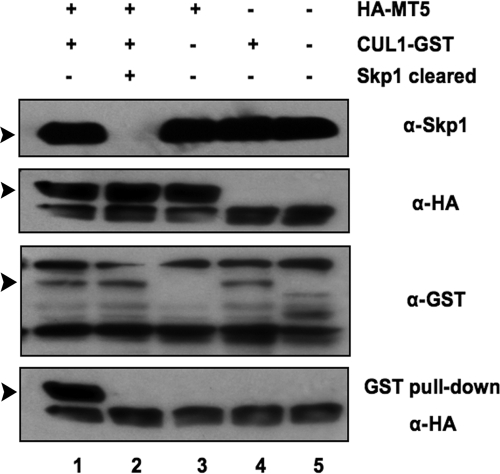
In the SCF complexes, Skp1 generally serves as a common adaptor that directly links the various F-box proteins to CUL1. M-T5 has been shown by immunoprecipitation and pull-down assays to coprecipitate with both Skp1 and CUL1, suggesting that M-T5 might bind indirectly to CUL1 through the direct interaction with Skp1 within the SCF complex. Unfortunately, binding assays that utilize tagged proteins expressed in transfected cells in vivo or in reticulocyte translation assays in vitro are unable to discriminate between direct and indirect interactions of the cellular proteins and M-T5, because of the presence of endogenous pools of these proteins. Thus, HA-MT5 coprecipitated with CUL1-GST following GST pull-down when coexpressed by in vitro coupled transcription-translation (Fig. 4, lane 1). However, when endogenous Skp1 was cleared first by immunoprecipitation with anti-Skp1 antibody from the TNT lysate prior to the coexpression of HA-MT5 and CUL1-GST, the binding of M-T5 to CUL1 was not detected following GST pull-down (Fig. 4, lane 2). This result clearly indicates that CUL1 binds to M-T5 only when Skp1 (either tagged or untagged) is present.
FIG. 4.
Binding of M-T5 to the SCF complex is dependent upon Skp1. The plasmids HA-MT5 and CUL1-GST were expressed by in vitro coupled transcription-translation and subjected to a GST pull-down assay. Precipitates and total lysate were immunoblotted and probed with anti-HA antibody to detect coprecipitated proteins. Prior to the addition of plasmids, Skp1 was precleared by immunoprecipitation from the TNT lysate using anti-Skp1 antibody (lane 2). Expression levels of the endogenous Skp1, HA-MT5, and CUL1-GST were confirmed using the designated antibodies. Bands of interest are indicated with arrowheads. α, anti.
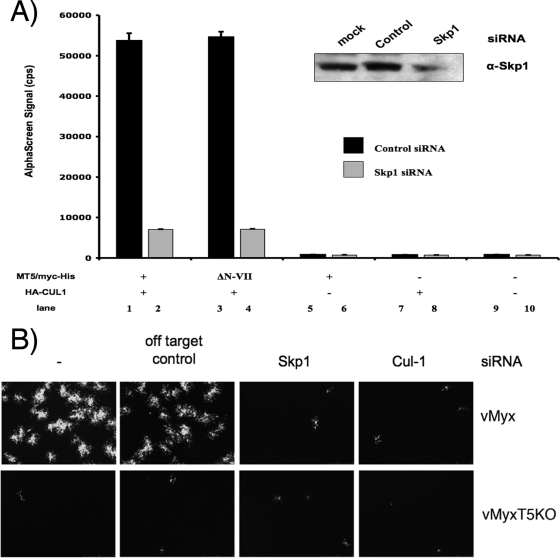
To further confirm the role of Skp1 as the direct adaptor and its ability to coordinate the binding of M-T5 and CUL1/SCF, siRNAs were employed to downregulate the expression of Skp1 in HEK 293 cells. Cells were cotransfected with Skp1-specific siRNA, plus plasmids that express HA-CUL1 and one of the various M-T5 constructs (MT5, MT5ΔPRANC, or MT5ΔN-VII), and cells were harvested 2 days later and assayed by AlphaScreen for protein-protein binding. Knockdown of Skp1 expression (Fig. 5A, insert) significantly decreased the binding interaction between the PRANC/F-box-like motif of M-T5 and CUL1, as demonstrated by a reduction in AlphaScreen signal (Fig. 5A, lanes 2 and 4). In contrast, transfection of control siRNA did not interrupt the binding interaction between the viral and cellular proteins (Fig. 5A, lanes 1 and 3).
FIG. 5.
Knockdown of Skp1 with siRNA disrupts binding of M-T5 to the SCF complex. (A) HEK 293 cells were transfected with either control (dark bars) or Skp1-specific (light bars) siRNA and the indicated (+) combination of plasmids. Harvested after 48 h, cell lysates were assayed for binding by AlphaScreen as described in the legend for Fig. 3. Knockdown of Skp1 protein expression by the siRNA treatment in HEK 293 cells was determined by Western blot analysis (inset). (B) The type II human cancer cell line 786-0 (56) was mock transfected or transfected with either control or Skp1- or CUL1-specific siRNA and, 24 h later, was infected with vMyx-gfp or vMyx-T5KOgfp at an MOI of 0.1. Viral focus formation was measured at 48 hpi by fluorescence microscopy. α, anti.
Collectively, these results demonstrate that the interaction between CUL1 and M-T5 is dependent upon the adaptor protein Skp1, which functionally bridges M-T5 with CUL1 and the SCF complex. Interestingly, siRNA-mediated knockdown of either Skp1 or CUL1 expression in the human type II cancer cell line 786-0 considerably blocked replication of vMyx-gfp at 48 hpi compared to cells transfected with control siRNA (Fig. 5B). No replication of vMyx-T5KOgfp was observed (Fig. 5B) since these particular type II cancer cells express low levels of endogenous phosphorylated Akt and do not normally support MYXV replication in the absence of M-T5 (56). These findings demonstrate how the interaction between M-T5 and the SCF complex significantly contributes to the permissiveness of MYXV replication in these cells. Furthermore, these results provide further insights into the molecular mechanism by which M-T5 functions via the host cell SCF complex during viral infection.
Identification of the M-T5 ANK repeats essential for binding to cellular Akt.
Almost all poxviruses express multiple proteins containing ANK repeats, thus accounting for a large superfamily of related but unique poxviral gene products (30). Detailed analyses of the M-T5 sequence predicted the presence of approximately seven ANK repeat domains within the amino terminus and central regions of the protein (Fig. 1A). The functional significance for any of the specific ANK domains in M-T5 has yet to be identified; however, it is reasonable to predict that they facilitate protein-protein interactions with host targets critical for viral replication. Since the ANK repeat is one of the most common protein-protein interaction motifs found in nature (32), it can be hypothesized that one or more of these ANK domains function as the docking site for binding viral or cellular substrate proteins.
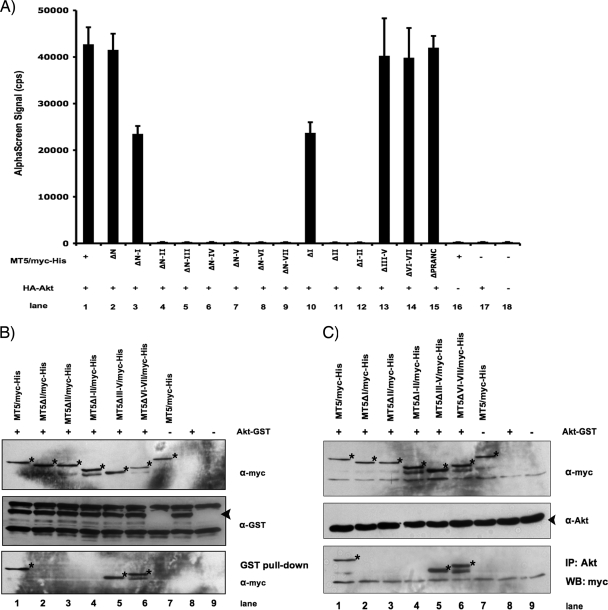
The host protein, Akt, was previously demonstrated to bind M-T5, by coimmunoprecipitation assays; however, the details of this interaction were not fully investigated at the molecular level (56). To investigate this protein-protein interaction in greater detail, HEK 293 cells were cotransfected with one of the various M-T5 fragment constructs fused to myc/His (Fig. 2A) together with HA-Akt, and cell lysates were assayed for binding by AlphaScreen. The presence of a distinct emission signal was detectable when full-length M-T5 (MT5/myc-His) was available for binding HA-Akt (Fig. 6A, lane 1); likewise, deletion of the first 31 amino acids at the amino terminus of M-T5 (MT5ΔN/myc-His) did not affect binding (Fig. 6A, lane 2). However, deletion of the first ANK repeat (MT5ΔN-I/myc-His and MT5ΔI/myc-His) reduced the binding affinity between the two proteins by approximately half (Fig. 6A, lanes 3 and 10). Furthermore, deletion of both ANK I plus II completely abolished the interaction between M-T5 and Akt, as demonstrated by the significant decrease in the emission signal for all the constructs tested (Fig. 6A, lanes 4 to 9 and 12). Interestingly, internal deletion of ANK II only (MT5ΔII/myc-His) was also able to ablate binding (Fig. 6A, lane 11); however, whenever ANK I and II were both present, the interaction between M-T5 and Akt appeared to be maintained (Fig. 6A, lanes 1, 2, and 13 to 15).
FIG. 6.
ANK repeats I and II of M-T5 are critical for binding Akt. The plasmids HA-Akt and a variety of myc-His-tagged M-T5 fragments (Fig. 2A) were transiently transfected into HEK 293 cells or expressed by in vitro coupled transcription-translation in the indicated (+) combinations. (A) Cell lysates were collected 48 h following plasmid transfection and assayed for binding by AlphaScreen as described in the legend for Fig. 3. (B) GST recombinant proteins were pulled down with glutathione agarose beads, and the associated proteins were resolved by SDS-PAGE and analyzed by immunoblotting with anti-myc antibody. (C) Endogenous untagged Akt protein was immunoprecipitated with anti-Akt antibody, and samples were analyzed by SDS-PAGE and probed with anti-myc antibody to detect coprecipitated proteins. The specified antibodies were used to detect the protein expression of HA-Akt, Akt-GST, endogenous Akt, and myc-His-tagged M-T5 fragments. Bands of interest are indicated with stars and arrowheads. α, anti.
Next, GST pull-downs were performed to confirm the interaction between Akt and the ANK I and II domains of M-T5. Individual M-T5-myc/His fragment constructs were coexpressed with Akt-GST by in vitro coupled transcription-translation and incubated with GST-coated beads overnight. Precipitated complexes were resolved by SDS-PAGE and only those M-T5 constructs which contained both ANK I and II were detected when immunoblotted with an anti-myc antibody (Fig. 6B, lanes 1, 5, and 6). Similarly, coimmunoprecipitation of M-T5 fragment constructs containing both ANK I and II was observed when endogenous untagged Akt was immunoprecipitated using an Akt-specific antibody (Fig. 6C, lanes 1, 5, and 6). In both experiments, interaction between Akt and M-T5 fragments lacking ANK I and/or II was not detected (Fig. 6B and C, lanes 2, 3, and 4). Taken in aggregate, these data clearly demonstrate that the composite ANK I-II domain of M-T5 is critical for binding to cellular Akt.
M-T5 functions as a molecular scaffold that binds Akt and the SCF complex simultaneously.
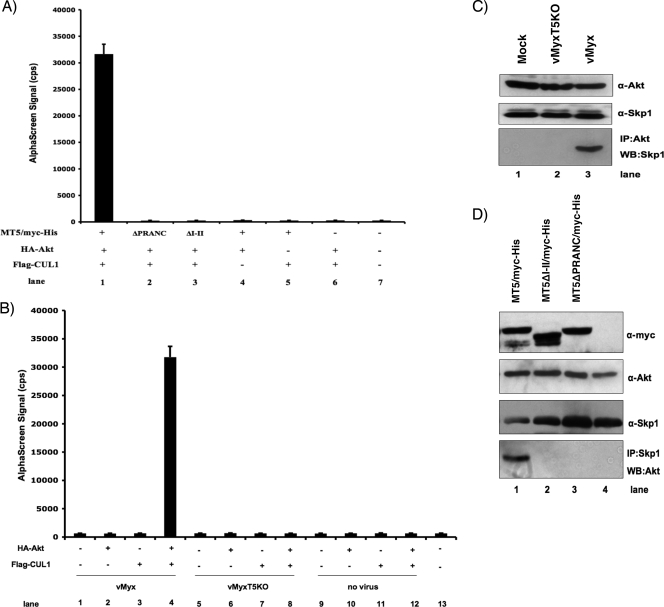
Functional diversity among ANK repeat proteins suggests that the motif is far more important structurally as scaffolding modules than in an enzymatic role (42). Thus, the potential of viral ANK repeat proteins to operate as a molecular scaffold may provide poxviruses a mechanism to generate novel protein-protein interactions fundamental to successful viral replication. Results from this study and previous reports have shown that M-T5 can independently bind CUL1/Skp1/SCF (Fig. 3) (20) and Akt (Fig. 5) (56); however, the potential ability of M-T5 to bind both cellular partner complexes simultaneously had not been examined. To determine if M-T5 functions as a molecular scaffold that can link Akt directly with the SCF complex, HEK 293 cells were cotransfected with HA-Akt, Flag-CUL1, and one of the various myc-His-tagged M-T5 constructs (MT5, MT5ΔPRANC, or MT5ΔN-VII). The following day, cell lysates were collected and assayed for binding by AlphaScreen. In Fig. 7A, we show that HA-Akt and Flag-CUL1 do not directly interact (Fig. 7A, lane 6); however, the addition of M-T5 (MT5/myc-His) elevated the emission signal dramatically (Fig. 7A, lane 1). In contrast, deletion of either the PRANC/F-box-like domain (Fig. 7A, lane 2) or the ANK I and II domain (Fig. 7A, lane 3) from M-T5 significantly reduced the capacity of the viral M-T5 protein to bridge HA-Akt with Flag-CUL1. As a control, cells cotransfected with MT5/myc-His and either HA-Akt or Flag-CUL1 alone did not produce a significant emission signal when binding was analyzed (Fig. 7A, lanes 5 and 6). Although Flag-CUL1 will indirectly bind to the acceptor bead via the biotin-conjugated anti-Flag antibody, the donor bead which is coated with an HA-specific antibody will not recognize the myc-His epitope expressed by M-T5. As a result, the interaction between M-T5 and CUL1 was not detected based on the design of this experiment. The same principle applies to HA-Akt, which binds directly to the donor beads.
FIG. 7.
M-T5 binds Akt and SCF components simultaneously. The indicated (+) combinations of plasmids were used to transiently transfect HEK 293 cells or were expressed by in vitro coupled transcription-translation. (A) After being harvested 48 h after transfection, cell lysates were subjected to an AlphaScreen binding assay as described in the legend for Fig. 3. (B) Cells were infected with vMyx-lac (lanes 1 to 4) or vMyx-T5KO (lanes 4 to 8) or were mock infected (lanes 9 to 12) the day following transfection. At 48 hpi, cell lysates were collected and assayed for protein-protein binding by AlphaScreen. (C) Antibody specific for Akt (anti-Akt) was used to immunoprecipitate Akt bound protein complexes in mock-infected cells or cells infected with vMyx-T5KO or vMyx-lac. Immunoblot analysis by SDS-PAGE detected the resulting protein complexes with anti-Skp1 antibody. (D) Endogenous Skp1 protein was immunoprecipitated (IP) with anti-Skp1 antibody in samples that expressed HA-MT5 by in vitro coupled transcription-translation. Detection of Akt coimmunoprecipitation was determined by Western blot analysis by the use of Akt-specific antibody (anti-Akt). Protein expression of HA-CUL1, HA-Skp1, and M-T5 fragments was detected using the designated antibodies. α, anti.
To confirm the ability of M-T5 to simultaneously bind the cellular proteins in virus-infected cells, HEK 293 cells were first cotransfected with HA-Akt plus Flag-CUL1 and then infected with either vMyx-lac or vMyx-T5KO the following day. Specific binding interaction between Akt and CUL1, as indicated by the elevated emission signal, was observed only when cells were infected with wild-type MYXV (Fig. 7B, lane 4). In contrast, cells that were mock infected (Fig. 7B, lane 12) or infected with MYXV that does not express the M-T5 gene product (Fig. 7B, lane 8) produced only a low emission signal, indicating that the M-T5 protein bridges Akt with components of the SCF complex in MYXV-infected cells. These results are congruent with the M-T5 binding data (Fig. 3 and 5) and suggest that the viral M-T5 protein has the capacity to bind Akt and CUL1-containing SCF simultaneously during viral infection.
Next, to verify that Skp1 is also within the SCF complex that binds Akt in the presence of M-T5, HEK 293 cells were either mock infected or infected with vMyx-lac (that expresses M-T5) or vMyx-T5KO, and at 48 hpi, untagged cellular Akt was immunoprecipitated from the cellular lysate with an anti-Akt antibody. Coimmunoprecipitation of Skp1 was observed only in cells infected with the wild-type virus that expresses M-T5, and not in mock- or vMyx-T5KO-infected cells (Fig. 7C). Similarly, coimmunoprecipitation of endogenous Akt was detected only when full-length M-T5 (MT5/myc-His) was coexpressed by in vitro coupled transcription-translation (Fig. 7D, lane 1). In contrast, Akt coimmunoprecipitation with Skp1 was not identified in TNT lysates that expressed either MT5ΔI-II or MT5ΔPRANC (Fig. 7D, lanes 2 and 3). Collectively, the results clearly demonstrate that M-T5 operates as a scaffolding protein to bridge two distinct cellular binding partners—Akt and the SCF complex—that contain both CUL1 and Skp1, during viral infection.
Cellular localization of Akt is influenced by M-T5 during virus infection.
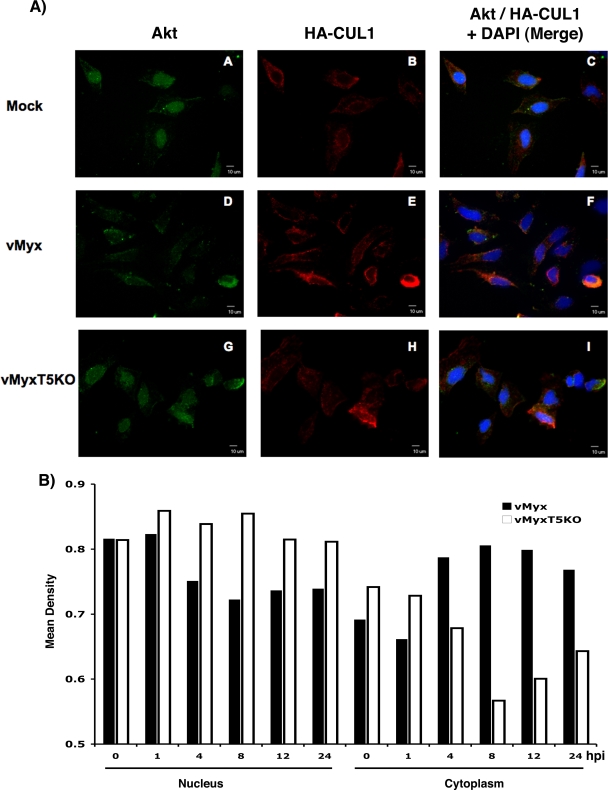
The potential capacity of poxviral ANK repeat proteins to operate as molecular scaffolds to link disparate cellular partners together may result in novel cross communication between distinct host signaling pathways that are normally independent of one another. Such modifications of the host cellular signaling networks, induced by poxvirus-encoded host range genes, would thus be critical in establishing an environment within the host crucial for successful virus replication. Confocal immunofluorescence microscopy was used to examine the ability of M-T5 to alter the cellular localization of Akt and/or members of the SCF complex in the context of a viral infection (Fig. 8A). Briefly, HeLa cells were transfected with HA-CUL1 and, 2 days later, were mock infected or infected with vMyx-lac or vMyx-T5KO for 4 h (M-T5 is expressed early during infection). Cells were fixed with 4% paraformaldehyde and permeabilized with 1% Triton X-100. Then, rabbit polyclonal anti-Akt antibody (to monitor untagged cellular Akt) and a mouse monoclonal antibody against HA (to monitor the HA-tagged CUL1) were applied to the samples, and the protein localizations were assessed by counterstaining with fluorescently labeled secondary antibodies. In both the mock control and vMyx-T5KO-infected cells, Akt was abundantly detected in the nucleus, colocalizing with the DNA stain DAPI. Smaller amounts of Akt also appeared to be located at the plasma membrane, where the protein is thought to reside when phosphorylated and activated. In contrast, within cells infected with wild-type MYXV that expresses M-T5, significant relocalization of Akt, predominantly from the nucleus to the cytoplasm, was observed. In contrast, we did not observe any obvious cellular relocalization of CUL1 in either the presence or absence of virus. Thus, in MYXV-infected cells, there was no obvious relocalization of CUL1-containing SCF complex (which remained in the cytoplasm), but a fraction of the nuclear pool of Akt was specifically relocalized from the nucleus to the cytoplasm only when M-T5 was present. Thus, the results support our previous data and demonstrate the ability of M-T5 to simultaneously interact with both Akt and the SCF complex. To further examine the relocalization of Akt following vMyx-lac infection, the cellular distribution of Akt was examined. HeLa cells were infected with either vMyx-lac or vMyx-T5KO, and cells were collected at various time points after infection. Nuclear and cytoplasmic extracts were isolated and separated by SDS-PAGE, and blots were probed with an Akt-specific antibody. Cellular distribution of Akt was determined by quantifying the level of Akt present in both the nucleus and cytoplasm by densitometry (Fig. 8B). Initially, Akt resided predominantly within the nucleus; however, 4 hours following vMyx-lac infection, the cytoplasmic levels of Akt increased and remained elevated for up to 24 h, while during the same time period, the level of Akt present in the nucleus decreased. In stark contrast, relatively no change in the level of Akt localized to the nucleus was observed in cells infected with vMyx-T5KO. Furthermore, lower Akt levels were observed in the cytoplasmic samples after vMyx-T5KO infection. The data suggest that expression of M-T5 by MYXV appears to induce at least some of the nuclear Akt to migrate to the cytoplasm with the CUL1/SCF complexes.
FIG. 8.
Relocalization of Akt in MYXV-infected cells requires M-T5. (A) The cellular localization of endogenous Akt (fluorescein isothiocyanate [green]) and transiently transfected HA-CUL1 (Texas Red [red]) was assayed in HeLa cells mock infected (A to C) or infected with either vMyx-lac (D to F) or vMyx-T5KO (G to I). Nuclei (blue) were detected using DAPI reagent, as shown in merged panels C, F, and I. (B) HeLa cells were infected with either vMyx-lac (dark bars) or vMyx-T5KO (white bars) and collected at 0, 1, 4, 8, 12, and 24 hpi. Nuclear and cytoplasmic extracts were isolated and separated by SDS-PAGE, and blots were probed with an Akt-specific antibody. Cellular distribution of Akt was determined by quantifying the level of Akt present in both the nucleus and cytoplasm by densitometry (Molecular Imaging software; Kodak).
DISCUSSION
Generally speaking, most poxviruses exhibit a narrow or restricted host range in which the distinct tropism is linked with the unique repertoire of host range genes expressed by each individual virus (28). During poxviral infection, these host range genes contribute significantly to modulating the intracellular environment by specifically targeting a diverse array of cellular factors and pathways to establish optimal conditions for permissive viral replication (29, 43). Many of the known poxviral host range genes are not only functionally diverse but also demonstrate a wide spectrum of biochemical and structural characteristics (59). The ANK repeat has been noted within many poxvirus host range factors from different genera, such as K1L from vaccinia virus and CHOhr from cowpox virus, both of which are orthopoxviruses, and M-T5 of MYXV, which is a Leporipoxvirus. Generally, this unique family of poxvirus-encoded proteins are 400 to 600 amino acids in size and contain 5 to 10 ANK repeats and a carboxy-terminal PRANC/F-box-like domain (30). The cellular versions of the F-box domain commonly exist in concert with additional protein-protein interaction motifs, such as WD repeats or leucine-rich repeats (LRR), which are thought to mediate substrate specificity (22, 46). The presence of the ANK repeat motifs within F-box proteins is exclusive to poxviral proteins, suggesting that the collaborative role of these distinct binding domains likely provides poxviruses novel strategies to regulate key cellular pathways that mediate pathogenesis and tropism.
Across six of the eight genera of poxviruses that possess members which encode these ANK repeat proteins, the PRANC domain is relatively well conserved, which would suggest that this is a common mechanism used by most poxviruses to mediate regulation of the ubiquitin-proteasome pathway (48). Like many previously characterized cellular F-box proteins, all four MYXV-encoded ANK/PRANC repeat proteins were demonstrated to interact directly with the common core SCF adaptor component, Skp1 (Fig. 1C). These four MYXV proteins add to a growing list of ANK repeat-containing poxviral proteins, which bind to the Skp1 component of the SCF complex via a carboxy-terminal PRANC/F-box-like domain (48, 49, 53). However, the identification of other cellular substrates for the poxviral ANK/PRANC protein family members, either cellular or viral, has proven to be a major obstacle. For example, M150 of MYXV has been reported to colocalize with NF-κB in the nucleus following stimulation of TNF-α; however, direct binding studies have not yet been performed to verify this interaction (8). In another study, a Y2H screen was performed to identify cellular binding partners for the 68-kDa ANK repeat protein encoded by modified vaccinia virus Ankara; interestingly, 99% of the analyzed positive interactions were with cellular Skp1 (49). When the unique variola virus ORFs were screened by Y2H, one of the identified partners of an ANK repeat protein, called G1R, was identified as Skp1 (31). In this study, multiple independent human cDNA libraries were screened by Y2H for potential binding partners of M-T5, and a total of 13 unique potential host protein interactions were identified (Table 3). Aside from Skp1, all proteins were novel potential M-T5 binding partners and have not been previously reported, but their verification as true binding partners in virus-infected cells remains to be confirmed.
Significantly, Akt was not identified as an M-T5 binding partner by the Y2H screen; however, the protein-protein interaction between Akt and M-T5 may be strictly dependent upon the phosphorylation status of Akt (our unpublished data). Phosphorylation of Akt at residues Thr308 and Ser473 regulates the kinase activity and, consequently, may also affect the binding of M-T5. If this is the case, then proper phosphorylation of Akt would likely not occur during the Y2H screen, which may help to explain the failure of the Y2H screen with M-T5 to identify Akt as a potential binding partner. Furthermore, Skp1, but not CUL1, was identified as a direct M-T5 binding partner, reaffirming our contention that the direct binding interaction is between M-T5 and Skp1, while binding to CUL1 is indirect, by virtue of being a component of the larger SCF complex that is ubiquitously found in cells and cellular lysates. The Y2H system has become an increasingly important tool for identifying and mapping novel protein-protein interactions; however, like any high-throughput assay, all hits must be verified by other methods to eliminate false positives that are not biologically relevant. Future studies will focus on validating these potential binding partners of M-T5 identified by the Y2H screen, using complementary methods, such as AlphaScreen, coimmunoprecipitation, and GST pull-down experiments. Deconstructing the intricate interactions between virus host range proteins and their host interacting proteins will be particularly useful to decipher the targets for this class of viral ANK repeat proteins during viral infection.
Using a combination of experimental approaches, we successfully demonstrate that M-T5 binds to the SCF complex via the adaptor protein Skp1 by means of the carboxy-terminal PRANC/F-box-like domain (Fig. 3 and 4). Previous studies reported that cellular F-box proteins without WD or LRR motifs can also bind to SCF components in vivo (9). Likewise, the ORF008 protein encoded by ORF virus was able to directly interact with Skp1, even following the deletion of all the ANK repeats (48). Conversely, binding between components of the SCF complex and the ectromelia virus ANK repeat protein ECT005 was abrogated when the ANK repeat domains of the viral protein were removed (53). In the case of M-T5, CUL1 interacted with both M-T5 and a mutant of M-T5 lacking all the ANK repeats, when analyzed by AlphaScreen (Fig. 3D). However, the GST pull-down protocol could detect M-T5 binding with CUL1 only when the ANK repeats of M-T5 were present (Fig. 3A and B), suggesting that the ANK repeat domains may contribute to the stability of the interaction between M-T5 and components of the SCF complex. We propose that stability of the complex formed between the solitary PRANC/F-box-like domain of M-T5 and SCF is compromised in the absence of the ANK domains, making the more-transient interactions difficult to identify when using methods that rely upon the complex having a longer half-life, such as GST pull-downs or coimmunoprecipitations, to assay the complexes. AlphaScreen provides greater sensitivity and is much more rapid than the above-mentioned assays and, therefore, may be a more sensitive method to measure more-transient interactions between potential binding partners that have shorter half-lives.
The cellular SCF complex plays a critical role in the selective degradation of regulatory proteins that mediate the cell cycle, particularly the cyclin-dependent kinase inhibitor p27/Kip1 (12). Generally, substrates are recognized by the SCF complex after they have been phosphorylated on specific epitopes (11). Phosphorylation of p27/Kip1 at threonine 187 by active cyclin E- or cyclin A-CDK2 provides a recognition site for the binding of Skp2 and subsequent degradation via the ubiquitin-proteasome pathway (34, 44, 55). Consequently, in the absence of p27/Kip1, the cell cycle is permitted to progress beyond G1 arrest and enter S phase (55). During MYXV infection, when M-T5 is expressed, enhanced phosphorylation, ubiquitination, and degradation of p27/Kip1 were observed, whereas in the absence of M-T5, levels of p27/Kip1 were selectively increased, consistent with its reduced phosphorylation and ubiquitination. Moreover, cells infected with vMyx-T5KO specifically entered cell cycle arrest and accumulated at G0/G1, whereas in the presence of virus that expresses M-T5, the viral protein was shown to promote cell cycle progression beyond the G0/G1 checkpoint during virus infection (20). Note that p27/Kip1 is known to be a substrate of activated Akt and that the phosphorylation of p27/Kip1 at site threonine 157 by Akt causes retention of p27/Kip1 in the cytoplasm and failure to induce G1 arrest (14, 54). Recent studies have shown that Akt-dependent phosphorylation leads to the cytoplasmic translocation of Skp2 and assembly of the SCF complex (15, 25). Interestingly, the interaction between M-T5 and Akt has been demonstrated to induce the activation of Akt in certain human cancer cells called type II, which is a key determinant for permissive MYXV replication in these particular cells (56). The current results suggest that bridging of the SCF complex to Akt by M-T5, as demonstrated in Fig. 7, may directly enhance the activation of Akt that becomes localized to the SCF complex and contribute to the concurrent phosphorylation and proteolytic degradation of p27/Kip1. Interestingly, there is no evidence that Akt itself is subject to any excessive degradation as a consequence of its relocalization to the SCF complex, suggesting that the Akt/M-T5/SCF complex functions to increase the range of Akt targets, and not to ferry Akt into the proteasome.
The ability of M-T5 to function as a molecular bifunctional adaptor defines a novel mechanism by which a viral host range protein can influence two distinct signaling pathways, consequently altering the intracellular environment to promote viral replication. However, more details are necessary to further characterize the functional role of M-T5 and its coordinated interaction with the cellular SCF complex and various substrates, such as Akt, during viral infection. Recently, another poxvirus host range protein, CP77, has been demonstrated to bind both the SCF complex and the NF-κB subunit p65, which blocks NF-κB activation by TNF-α (10). The capacity of poxviral ANK/PRANC proteins to function as molecular scaffolds may coordinate the cross communication between host signaling pathways that normally function independently. At the molecular level, such viral proteins have the potential to reconfigure intracellular signaling networks into downstream effects which may have previously not existed due to spatial or temporal constraints but may be crucial for viral replication.
Poxviruses are known to specifically target and manipulate a variety of signaling networks that regulate critical cellular processes. In addition to the ANK/PRANC subfamily of proteins, poxviruses have evolved additional strategies to subvert the host ubiquitination pathway (64). For example, poxviral proteins which contain Broad-complex, Tramtrack, Bric-a-Brac (BTB), and kelch domains have been shown to interact with cullin 3 ubiquitin ligases (23, 61). Furthermore, the p28 protein encoded by ectromelia virus contains a single-subunit RING (really interesting new gene) finger which functions as a viral E3 ubiquitin ligase (18, 35). This protein is highly conserved among orthopoxviruses; however, in the vaccinia virus strains Copenhagen and Western Reserve and modified vaccinia virus Ankara, the p28 gene products are either truncated or interrupted. Another E3 ubiquitin ligase, M153R of MYXV, is a membrane-associated RING-CH (MARCH) protein that downregulates host cell surface expression of major histocompatibility complex class I, the proapoptotic factor CD95 (Fas), the activated leukocyte cell adhesion molecule, and CD4 (2, 17, 27). Many other viruses, including several small DNA tumor viruses (adenoviruses, papillomaviruses, and polyomaviruses), have also been demonstrated to manipulate the ubiquitin pathway by the use of various strategies (4). In fact, degradation of p53 by human papillomavirus type 16 and 18 E6 proteins was the first documented virally encoded protein to specifically target this signaling network (1, 38, 39).
In summary, viruses such as poxviruses encode a myriad of proteins that have the capacity to hijack the ubiquitin-proteasomal pathway, for example, to target and eliminate unwanted cellular proteins, such as inhibitors of the cell cycle or various antiviral factors that would otherwise function to block viral replication. Understanding the mechanisms by which viral host range proteins interact with host factors and signaling networks should continue to provide invaluable insights into how vital cellular networks can be reprogrammed by viral factors to increase virus survival at the cellular level.
Acknowledgments
This study was supported by a start-up grant awarded to G.M. from the University of Florida College of Medicine, NIH R01 AI080607 and NIH U54 AI057157 to the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (SERCEB). The Y2H screen was supported by contract DHHSN26620040057C to Myriad Genetics, Inc. S.J.W. received funding from the University of Florida Medical Guild Research Incentive Award.
We thank Sherin Smallwood for providing plasmids and John Barrett and Steven Nazarian for technical assistance.
Many of the authors are current employees of Myriad Genetics. Publication of this article may have a positive effect on the company's stock price and therefore financially benefit those authors.
Footnotes
Published ahead of print on 23 September 2009.
REFERENCES
- 1.Band, V., J. A. De Caprio, L. Delmolino, V. Kulesa, and R. Sager. 1991. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J. Virol. 65:6671-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartee, E., A. McCormack, and K. Fruh. 2006. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor, A. H., D. E. Piper, F. C. de la Brousse, S. L. McKnight, and C. Wolberger. 1998. The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279:1037-1041. [DOI] [PubMed] [Google Scholar]
- 4.Blanchette, P., and P. E. Branton. 2009. Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology 384:317-323. [DOI] [PubMed] [Google Scholar]
- 5.Blanié, S., J. Mortier, M. Delverdier, S. Bertagnoli, and C. Camus-Bouclainville. 2009. M148R and M149R are two virulence factors for myxoma virus pathogenesis in the European rabbit. Vet. Res. 40:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork, P. 1993. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins 17:363-374. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 8.Camus-Bouclainville, C., L. Fiette, S. Bouchiha, B. Pignolet, D. Counor, C. Filipe, J. Gelfi, and F. Messud-Petit. 2004. A virulence factor of myxoma virus colocalizes with NF-kappaB in the nucleus and interferes with inflammation. J. Virol. 78:2510-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cenciarelli, C., D. S. Chiaur, D. Guardavaccaro, W. Parks, M. Vidal, and M. Pagano. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9:1177-1179. [DOI] [PubMed] [Google Scholar]
- 10.Chang, S. J., J. C. Hsiao, S. Sonnberg, C. T. Chiang, M. H. Yang, D. L. Tzou, A. A. Mercer, and W. Chang. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J. Virol. 83:4140-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig, K. L., and M. Tyers. 1999. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72:299-328. [DOI] [PubMed] [Google Scholar]
- 12.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 13.Fenner, F., and F. Ratcliffe. 1965. Myxomatosis. Cambridge University Press, Cambridge, England.
- 14.Fujita, N., S. Sato, K. Katayama, and T. Tsuruo. 2002. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J. Biol. Chem. 277:28706-28713. [DOI] [PubMed] [Google Scholar]
- 15.Gao, D., H. Inuzuka, A. Tseng, R. Y. Chin, A. Toker, and W. Wei. 2009. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell Biol. 11:397-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorina, S., and N. P. Pavletich. 1996. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274:1001-1005. [DOI] [PubMed] [Google Scholar]
- 17.Guerin, J. L., J. Gelfi, S. Boullier, M. Delverdier, F. A. Bellanger, S. Bertagnoli, I. Drexler, G. Sutter, and F. Messud-Petit. 2002. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J. Virol. 76:2912-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, J., Q. Huang, X. Zhou, M. M. Shen, A. Yen, S. X. Yu, G. Dong, K. Qu, P. Huang, E. M. Anderson, S. Daniel-Issakani, R. M. Buller, D. G. Payan, and H. H. Lu. 2004. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J. Biol. Chem. 279:54110-54116. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, M. D., and S. C. Harrison. 1998. Structure of an IkappaBalpha/NF-kappaB complex. Cell 95:749-758. [DOI] [PubMed] [Google Scholar]
- 20.Johnston, J. B., G. Wang, J. W. Barrett, S. H. Nazarian, K. Colwill, M. Moran, and G. McFadden. 2005. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J. Virol. 79:10750-10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kipreos, E. T., and M. Pagano. 2000. The F-box protein family. Genome Biol. 1:REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 23.Kochneva, G., I. Kolosova, T. Maksyutova, E. Ryabchikova, and S. Shchelkunov. 2005. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch. Virol. 150:1857-1870. [DOI] [PubMed] [Google Scholar]
- 24.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, H. K., G. Wang, Z. Chen, J. Teruya-Feldstein, Y. Liu, C. H. Chan, W. L. Yang, H. Erdjument-Bromage, K. I. Nakayama, S. Nimer, P. Tempst, and P. P. Pandolfi. 2009. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat. Cell Biol. 11:420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luh, F. Y., S. J. Archer, P. J. Domaille, B. O. Smith, D. Owen, D. H. Brotherton, A. R. Raine, X. Xu, L. Brizuela, S. L. Brenner, and E. D. Laue. 1997. Structure of the cyclin-dependent kinase inhibitor p19Ink4d. Nature 389:999-1003. [DOI] [PubMed] [Google Scholar]
- 27.Mansouri, M., E. Bartee, K. Gouveia, B. T. Hovey Nerenberg, J. Barrett, L. Thomas, G. Thomas, G. McFadden, and K. Fruh. 2003. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 77:1427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFadden, G., and P. M. Murphy. 2000. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr. Opin. Microbiol. 3:371-378. [DOI] [PubMed] [Google Scholar]
- 30.Mercer, A. A., S. B. Fleming, and N. Ueda. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31:127-133. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed, M. R., M. M. Rahman, A. Rice, R. W. Moyer, S. J. Werden, and G. McFadden. 2009. Cowpox virus expresses a novel ankyrin repeat NF-κB inhibitor that controls inflammatory cell influx into virus-infected tissues and is critical for virus pathogenesis. J. Virol. 83:9223-9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Y. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mossman, K., S. F. Lee, M. Barry, L. Boshkov, and G. McFadden. 1996. Disruption of M-T5, a novel myxoma virus gene member of poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J. Virol. 70:4394-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller, D., C. Bouchard, B. Rudolph, P. Steiner, I. Stuckmann, R. Saffrich, W. Ansorge, W. Huttner, and M. Eilers. 1997. Cdk2-dependent phosphorylation of p27 facilitates its Myc-induced release from cyclin E/cdk2 complexes. Oncogene 15:2561-2576. [DOI] [PubMed] [Google Scholar]
- 35.Nerenberg, B. T., J. Taylor, E. Bartee, K. Gouveia, M. Barry, and K. Fruh. 2005. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J. Virol. 79:597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opgenorth, A., K. Graham, N. Nation, D. Strayer, and G. McFadden. 1992. Deletion analysis of two tandemly arranged virulence genes in myxoma virus, M11L and myxoma growth factor. J. Virol. 66:4720-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112:243-256. [DOI] [PubMed] [Google Scholar]
- 38.Scheffner, M., T. Takahashi, J. M. Huibregtse, J. D. Minna, and P. M. Howley. 1992. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J. Virol. 66:5100-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 40.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381-386. [DOI] [PubMed] [Google Scholar]
- 41.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 43.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 44.Sheaff, R. J., M. Groudine, M. Gordon, J. M. Roberts, and B. E. Clurman. 1997. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 11:1464-1478. [DOI] [PubMed] [Google Scholar]
- 45.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 46.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 47.Sonnberg, S., S. B. Fleming, and A. A. Mercer. 2009. A truncated 2-α-helix F-box present in poxvirus ankyrin repeat proteins is sufficient for binding the SCF1 ubiquitin ligase complex. J. Gen. Virol. 90(Pt 5):1224-1228. [DOI] [PubMed] [Google Scholar]
- 48.Sonnberg, S., B. T. Seet, T. Pawson, S. B. Fleming, and A. A. Mercer. 2008. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc. Natl. Acad. Sci. USA 105:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sperling, K. M., A. Schwantes, B. S. Schnierle, and G. Sutter. 2008. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology 374:234-239. [DOI] [PubMed] [Google Scholar]
- 50.Stanford, M. M., S. J. Werden, and G. McFadden. 2007. Myxoma virus in the European rabbit: interactions between the virus and its susceptible host. Vet. Res. 38:299-318. [DOI] [PubMed] [Google Scholar]
- 51.Sypula, J., F. Wang, Y. Ma, J. Bell, and G. McFadden. 2004. Myxoma virus tropism in human tumour cells. Gene Ther. Mol. Biol. 8:103-114. [Google Scholar]
- 52.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Buuren, N., B. Couturier, Y. Xiong, and M. Barry. 2008. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J. Virol. 82:9917-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viglietto, G., M. L. Motti, P. Bruni, R. M. Melillo, A. D'Alessio, D. Califano, F. Vinci, G. Chiappetta, P. Tsichlis, A. Bellacosa, A. Fusco, and M. Santoro. 2002. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 8:1136-1144. [DOI] [PubMed] [Google Scholar]
- 55.Vlach, J., S. Hennecke, and B. Amati. 1997. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 16:5334-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, G., J. W. Barrett, M. Stanford, S. J. Werden, J. B. Johnston, X. Gao, M. Sun, J. Q. Cheng, and G. McFadden. 2006. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc. Natl. Acad. Sci. USA 103:4640-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Werden, S. J., J. W. Barrett, G. Wang, M. M. Stanford, and G. McFadden. 2007. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J. Virol. 81:2340-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Werden, S. J., and G. McFadden. 2008. The role of cell signaling in poxvirus tropism: the case of the M-T5 host range protein of myxoma virus. Biochim. Biophys. Acta 1784:228-237. [DOI] [PubMed] [Google Scholar]
- 59.Werden, S. J., M. M. Rahman, and G. McFadden. 2008. Poxvirus host range genes. Adv. Virus Res. 71:135-171. [DOI] [PubMed] [Google Scholar]
- 60.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695:133-170. [DOI] [PubMed] [Google Scholar]
- 61.Wilton, B. A., S. Campbell, N. Van Buuren, R. Garneau, M. Furukawa, Y. Xiong, and M. Barry. 2008. Ectromelia virus BTB/kelch proteins, EVM150 and EVM167, interact with cullin-3-based ubiquitin ligases. Virology 374:82-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winston, J. T., D. M. Koepp, C. Zhu, S. J. Elledge, and J. W. Harper. 1999. A family of mammalian F-box proteins. Curr. Biol. 9:1180-1182. [DOI] [PubMed] [Google Scholar]
- 63.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, L., N. Y. Villa, and G. McFadden. 2009. Interplay between poxviruses and the cellular ubiquitin/ubiquitin-like pathways. FEBS Lett. 583:607-614. [DOI] [PubMed] [Google Scholar]