Abstract
Infection of host cells with human cytomegalovirus (HCMV) induces cell cycle dysregulation. Two HCMV immediate-early (IE) proteins, IE1-72 and IE2-86, are promiscuous transactivators that have been implicated in the dysregulatory events. Cellular p53 protein is accumulated to high levels in HCMV-infected cells, but the indicative marker of p53 transcriptional activity, p21, is markedly decreased. Both IE1-72 and IE2-86 were able to transactivate the p53 promoter and interact with p53 protein in DNA-transfected or HCMV-infected cells. HCMV UL84, a multiregulatory protein expressed in early periods of HCMV infection, also interacted with p53. HCMV IE1-72 prevented or disrupted p53 binding to p53-specific DNA sequences, while IE2-86 and/or UL84 enhanced p53 binding and induced supershift of this DNA-protein complex. Both HCMV IE1-72 and IE2-86 were able to inhibit p53-dependent transcriptional activation in plasmid-transfected cells. IE1-72, rather than IE2-86, was found to be responsible for p21 downregulation in HCMV-infected HEL cells. DNA transfection analysis using IE1-72 mutants revealed that exon 2/3 and the zinc finger region of IE1-72 are essential for IE1-72's effect on the repression of p53-dependent transcriptional activation. These data suggest that HCMV IE1-72 and/or IE2-86 transactivates the p53 promoter and induces p53 accumulation, but HCMV IE1-72 represses the p53 transactivation activity by a unique binding hindrance mechanism different from that of IE2-86. Thus, various modes of viral IE proteins and p53 interactions might result in multiple outcomes, such as stimulation of cellular DNA synthesis, cell cycle progression and cell cycle arrest, and prevention of program cell death.
Human cytomegalovirus (HCMV), a member of the herpesvirus family, is the leading viral cause of birth defects and poses a serious threat in immunocompromised patients and in organ transplant recipients (35, 43). One of the distinctive features of HCMV is that it affects many aspects of host cell regulation and metabolism, including stimulation of the synthesis of cellular DNA, RNA, and proteins (16, 53). In productive infection, HCMV gene expression occurs in three temporally regulated phases, termed the immediate-early (IE), early, and late phases (53, 73). The IE proteins IE1-72 and IE2-86 are alternatively spliced gene products of the major IE gene that play important roles in the regulation of viral and cellular gene expression. Expression of HCMV IE1-72 and IE2-86 alters the cell cycle to generate an environment conducive to stimulating quiescent cells to enter G1/S phase (82), arresting cell cycle progression (45), inducing cellular macromolecule synthesis (7, 8), and resisting apoptotic stimuli (83).
IE1-72 has been shown to be a strong activator of the major IE promoter and can stimulate transcription of many viral and cellular promoters (12, 14, 27, 68, 77). Expression of IE1-72 also induces p53 accumulation in infected fibroblasts, resulting in the alteration of cell cycle control (8).
IE2-86 protein has been identified as an essential promiscuous transcriptional transactivator and repressor molecule that autoregulates IE gene expression by binding to cis repression signal sequences of the major IE promoter (11, 58, 59). Studies have demonstrated an interaction between HCMV IE2-86 and cellular p53 that correlates with HCMV infection and coronary restenosis (66, 67). Expression of IE2-86 also induces p53 accumulation in fibroblasts (8, 54, 55), resulting in cellular proliferation (8). IE2-86 has been shown to interact with p53 in vitro and in vivo (3, 66, 70), and this interaction results in the downregulation of p53's transactivation function (66, 70). IE2-86, but not IE1-72, increases the half-life of p53 and reduces that of mdm2 (80). There has been no report of direct binding of IE1-72 to p53. The exact nature of the interaction between IE proteins and p53 remains to be elucidated.
Cellular protein p53 has been known as a tumor suppressor gene product (17, 31, 42) and cell cycle inhibitor (48). It is known to regulate transcription of a variety of genes, including genes involved in cell cycle regulation, DNA repair, and programmed cell death (28, 52, 62). Elevated levels of p53 were observed in cell lines transformed by a variety of agents, including DNA and RNA tumor viruses, and chemical carcinogens (for reviews, see reference 61). Although several studies have shown that p53 levels are rapidly active and stabilized after HCMV infection in several cell types (36, 44, 55, 67), there is no significant activation of p53 downstream targets (4, 36). On the other hand, p53 appears to play some functional roles in the life cycle of HCMV. It was shown that p53 was involved in the regulation of viral UL94 protein expression (75). Fortunato and her colleagues demonstrated that cells infected with HCMV in the absence of p53 (p53−/− HF cells) produced fewer infectious viral particles (5). Viral protein production, trafficking, and viral encapsidation delays were found in cells lacking p53. By microarray analyses, Fortunato's group further revealed that expression of 22 viral genes was affected by the absence of p53, and the presence of p53 is important for both IE and early viral gene expression mostly at IE and early times postinfection (28). These influences are probably due to the combination of direct and indirect interaction of p53 with the virus proteins and genome (28).
Our study here was originally designed to assess the hypothesis that p53 controls the initiation of viral DNA replication by binding to a p53-specific DNA binding site located within the origin of HCMV DNA replication (oriLyt), and the expression of HCMV IE1-72 protein will allow the removal of p53 from oriLyt or from a IE1-72/p53 complex to facilitate the initiation of HCMV DNA replication. Therefore, this study was performed first to investigate the effect of IE1-72 and IE2-86 on p53 promoter activity and on the sequence-specific binding activity of p53, as well as to define the regions of IE1-72 that mediate its effect on p53. Although IE1-72 and IE2-86 exerted similar transcriptional repression effects on a p53-responsive promoter, their mechanisms of repression were different. IE1-72 disrupted p53 binding to its DNA-specific binding sequences while IE2-86 enhanced this binding. Whether the binding of IE1-72 disrupts p53 binding to its DNA-specific binding sequences will facilitate the initiation of HCMV DNA replication has to be further investigated.
MATERIALS AND METHODS
Cell culture and virus.
Human embryonic lung (HEL-299; ATCC CCL137) cells and a human osteosarcoma cell line, Saos-2 (ATCC HTB-85), were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL, Gaithersburg, MD) and penicillin-streptomycin at 37°C in a 6% CO2 incubator. Infection with the HCMV Towne strain (P-34; gift of Stanley Plotkin) was performed as described previously (33). A multiplicity of infection (MOI) of 2 to 5 was used for all experiments. Cells and virus were incubated for 90 min at 37°C in a 6% CO2 incubator. Unabsorbed free virus was removed, and DMEM with 4% heat-inactivated FBS was added to the infected culture.
Plasmids.
The IE1-72, IE2-86, and UL84 eukaryotic expression vectors (pcDNA3IE1-72, pcDNA3IE2-86, and pcDNA3UL84) and prokaryotic expression vectors (pGEX-IE1-72, pGEX2T-IE2-86, and pGEX2T-UL84) were described previously (21, 30, 60, 78). The IE1-72 full-length and mutant expression vectors (pSG5IE1, pSG5IE1exon2/3, pSG5IE1exon3, pSG5IE1LZ, pSG5IE1acidic, pSG5IE1ZF, and pSG5IE1350-420) were gifts from J. Sinclair and have been described elsewhere (29, 60, 78). The p53-responsive reporter constructs, pG13CAT (where CAT is chloramphenicol acetyltransferase) for the wild type and MG15CAT for the mutant, were gifts from B. Vogelstein and were described previously (38, 79). The p53 expression plasmids, pC53-SN3 for wild-type p53 and pC53-SCX3 for mutant p53, were also gifts from B. Vogelstein and were as described previously (2). Recombinant His-tagged p53 protein was expressed in baculovirus as described previously (2). The p53 promoter-CAT construct (wild type) was a gift from Shannon Kenney. This promoter has a 600-bp insert obtained by PCR from the human endothelial cell p53 promoter region using the sequences identified by Tuck and Crawford (71).
Antibodies.
Monoclonal antibodies to p53, DO-1 (Santa Cruz Biotechnology, Santa Cruz, CA) and Ab-4 (CalBiochem, La Jolla, CA), were used. Antibodies to IE1-72, IE2-86, and UL84 were generated in our laboratory as described previously (30, 39).
Protein expression.
Recombinant p53 protein was produced in SF21 insect cells and purified as described previously (20). The IE1-72, IE2-86, and UL84 glutathione S-transferase (GST) fusion proteins, GST-IE1-72, GST-IE2-86, and GST-UL84, were propagated and purified as described previously (21). Briefly, GST fusion proteins were purified from corresponding plasmid-transfected Escherichia coli BL21 bacterial lysates by loading to glutathione-Sepharose 4B columns (Pharmacia Biotech, Piscataway, NJ) after clarification by centrifugation. The fusion protein-loaded columns were washed five times with 5 volumes of an ice-cold phosphate-buffered saline (PBS) with 1% Triton X-100 (Sigma, St. Louis, MO). The GST fusion proteins were then eluted with 20 mM reduced glutathione (Boehringer Mannheim Corporation, Indianapolis, IN) in 100 mM Tris-HCl (pH 8.0) solution, dialyzed, and stored at −70°C until used.
Immunoprecipitation.
Approximately 200 μg of HCMV-infected cell lysate was precleared for 1 h with Pansorbin cells (CalBiochem, La Jolla, CA). Monoclonal antibody and 30 μl of protein G beads (Pharmacia, Uppsala, Sweden) were added, and the volume was increased to 0.5 ml with ELB+ buffer (0.25 M NaCl, 0.1% Nonidet P-40, 0.05 M HEPES [pH 7.0], 1 mM phenylmethylsulfonyl fluoride, 5 mM EDTA, 0.5 mM dithiothreitol). The incubation was performed at 4°C for 4 h with constant mixing. Beads were spun down and washed six times with ELB+ buffer. The beads were mixed with Laemmli sodium dodecyl sulfate (SDS) sample buffer and boiled. Supernatant was run on SDS-polyacrylamide gel electrophoresis gels. Western blot analysis was used to detect proteins in the immunoprecipitated complexes by the appropriate antibody.
p53 EMSA.
An electrophoretic mobility shift assay (EMSA) was performed as described previously (78). In brief, baculovirus-expressed p53 protein alone or together with various amounts of GST-IE1-72, GST-IE2-86, or GST-UL84 were incubated in a binding buffer (10 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, 10% glycerol, 1 mM dithiothreitol) plus 7.5 mM MgCl2, 0.1 mg of poly(dI-dC) (Pharmacia Biotech), and a 32P-labeled, wild-type p53 (5′-CCTGCCTGGACTTGCCTGG-3′; two copies) or mutant p53 (5′-CCTTAATGGACTTTAATGG-3′; two copies) double-stranded oligonucleotide probe for 15 min. The samples were electrophoresed on a 5% polyacrylamide gel, vacuum dried, and subjected to autoradiography using X-ray films with intensifier screens at −70°C. For EMSAs in which the GST-IE1-72, GST-IE2-86, or GST-UL84 fusion protein was added, equal protein amounts of the different fusion proteins and GST alone were incubated in the above-mentioned reaction mixture. Titrations of the fusion products and different incubation conditions were used to determine the optimal conditions for these experiments.
Microarray.
HEL cells were infected at an MOI of 4 per cell of HCMV. The infected cells were harvested at 0, 6, 12, 24, 48, 72, and 96 h postinfection (hpi). Total RNA was extracted from infected cells using Trizol (Invitrogen, Carlsbad, CA) at each time point. Total RNA (2 μg) was reverse transcribed with a Chemiluminescent RT-IVT Labeling kit (Applied Biosystems, Foster City, CA) and hybridized to the Human Genome Survey Microarray (Applied Biosystems). For each time point, data were initially normalized, and differential gene expression at each time point was calculated by comparing the differences between expression levels at each time point and the level in mock-infected HEL cells at 0 hpi.
IE1-72 and IE2-86 knockdown by siRNA.
HCMV IE1-72 and IE2-86 were knocked down in HEL cells after HCMV infection by small interfering RNAs (siRNAs) as described previously (41, 80). In brief, the IE1-72 siRNA 5′GGUUAUCAGUGUAAUGAAGdTdT-3′ and complement 5′CUUCAUUACACUGAUAACCdTdT-3′ targeting HCMV IE1-72 as well as the IE2-86 siRNA oligonucleotide 5′-AAACGCAUCUCCGAGUUGGACdTdT-3′ and complement 5′-GUCCAACUUGGAGAUGCGUUUdTdT-3′ targeting HCMV IE2-86 were synthesized by Dharmacon (Lafayette, CO) as described previously (74). HEL cells were grown to confluence in 100-mm dishes at 37°C in an atmosphere of 6% CO2 and serum starved for 24 h. Dishes of cells were transfected with 10 μl of Lipofectamine 2000 and 5 μl of 20 μM siRNA, according to the manufacture's instructions (Invitrogen). Four hours later transfected cells were washed twice with DMEM, and the medium was replaced with DMEM. At 24 h posttransfection cells were infected with 2 to 5 PFU/cell of HCMV. The plates were incubated for an additional 16 h prior to harvesting for Western blot analysis.
UV treatment of HEL cells.
Nearly confluent HEL cell cultures (95 to 99% confluence) were washed once with PBS. Cells were UV irradiated (30 mJ/cm2) with UV cross-linker after PBS was removed, and DMEM was then added to plates. UV-irradiated HEL cells were harvested 8 h post-UV treatment for Western blot analysis.
Western blot analysis.
Equal protein amounts of lysates per time point were mixed in Laemmli sample buffer and were loaded onto 10% SDS-polyacrylamide gels. Proteins were separated by electrophoresis and transferred overnight at 18 V to Immobilon-P Transfer Membrane (Millipore, Bedford, MA) blots. The blots were blocked for 1 h in 5% (wt/vol) nonfat dry milk dissolved in PBS. The blots were then probed with primary antibody for 1 h in PBS and were washed three times with PBS-0.1% Tween 20. After the washing step, the blots were probed with secondary antibody(horseradish peroxidase-conjugated anti-mouse immunoglobulin G; Sigma, St. Louis, MO). The blots were again washed extensively in PBS-0.1% Tween 20 and then in PBS alone and subsequently were developed by enhanced chemiluminescence (Amersham, Buckinghamshire, England) according to the manufacturer's protocol.
CAT assay.
For transfection and infection experiments, HEL cells were seeded onto 100-mm-diameter plates at 1 × 106 cells. The following day, cells were transfected with plasmid DNA via calcium phosphate-mediated transfection. For each 100-mm-diameter plate, 1 μg of p53 promoter-CAT plasmid along with 1 μg of pCMV-Gal was mixed in a final volume of 500 μl of HEPES-buffered saline (50 mM HEPES, 280 mM NaCl, 10 mM KCl, 12 mM dextrose, 1.5 mM Na2HPO4, pH 7.4) and 500 μl of 0.25 M calcium chloride solution. The DNA-calcium phosphate complexes were added dropwise to the cultured cells. All transfections were done in triplicate and were allowed to proceed overnight. The next day, the transfectant was removed, and cells were washed once with 10 ml of DMEM. The cells were subsequently infected with HCMV at the MOIs shown in Fig. 1B. Following a 90-min adsorption period, 10 ml of DMEM supplemented with 4% heat-inactivated FBS was added to each 100-mm-diameter plate. Cells were harvested at the appropriate time points postinfection in 250 μl of 1× cell lysis buffer (Promega, Madison, WI). For CAT assays, cell extract was mixed with acetylenzyme coenzyme A (Boehringer Mannheim) and [14C]chloramphenicol (New England Nuclear), and CAT assays were performed as described previously (24). Samples were standardized by using a Promega galactosidase enzyme assay system as suggested by the manufacturer. Absorbance at 420 nm for each sample was determined with a Beckman DU-70 spectrophotometer. In the experiments using the cotransfection assay to study p53 promoter or p53-responsive promoter activations, 1 μg of the p53 promoter-CAT construct or p53-responsive CAT construct (pG13CAT or pMG15CAT) and various IE1-72 wild-type or mutant plasmids were transfected into Saos-2 cells.
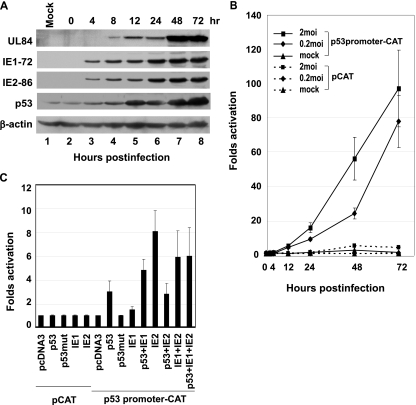
FIG. 1.
Transactivation of p53 promoter by HCMV IE1-72 and IE2-86. (A) Western blots demonstrating expression of IE1-72, IE2-86, UL84, and p53 in HCMV-infected HEL cells harvested and lysed at the times (hours postinfection) indicated. M, mock infected. (B) Results of CAT assays demonstrating activation of p53 promoter in lysates from HEL cells harvested at the indicated times after HCMV infection. Bars indicate standard deviations. (C) CAT assays from lysates of Saos-2 cells transfected with combinations of target promoter construct, p53 promoter-CAT, and plasmids encoding wild-type p53, mutant p53 (p53mut), IE1-72, and IE2-86, as indicated. Values shown are means plus standard deviations for three experiments, each run with triplicate samples. pcDNA3, transfection with empty plasmid.
RESULTS
Kinetics of HCMV IE1-72, IE2-84, and UL84 protein expressions and the induction of the accumulation of p53 in HCMV-infected HEL cells.
The expression of HCMV IE1-72, IE2-86, UL84, and p53 in HCMV-infected HEL cells was determined at various time points postinfection by Western blot analysis. IE1-72 and IE2-86 were detected as early as 4 hpi (Fig. 1A, lanes 3 to 8), while UL84 was detected starting at 8 hpi (Fig. 1A, lanes 4 to 8). IE1-72 and IE2-86 levels steadily increased throughout the infection time course. The amount of UL84 increased at the early stage of viral DNA replication and reached a plateau at 48 and 72 hpi. Increases in p53 were first observed at 4 hpi; the protein levels were then increased in parallel with both IE1-72 and IE2-86, reaching a plateau at 48 hpi.
HCMV IE proteins transcriptionally transactivate the p53 promoter.
Using CAT assays on lysates from HCMV-infected HEL cells, we determined that p53 promoter activity was enhanced by IE protein expression in a dose-dependent manner (Fig. 1B). When HEL cells were first transfected with the p53 promoter-CAT reporter plasmid and then infected with HCMV 24 h later, p53 promoter activity was activated throughout the entire HCMV infection course. There were nearly 80- and 100-fold enhancements in p53 promoter-CAT activity at 72 hpi with MOIs of 0.2 and 2, respectively. There was only basal p53 promoter activity in mock-infected cells and less than fivefold activation with pCAT (without p53 promoter sequence) control transfection in HCMV-infected cells.
The role of IE1-72 and IE2-86 in p53 promoter activation was analyzed by cotransfection of p53-deficient Saos-2 cells with IE plasmids and wild-type (pC53-SN3) or mutant p53 (pC53-SCX3) expression vectors. CAT assays were then used to determine p53 promoter activity (Fig. 1C) because the Saos-2 cell line was p53−/− (deletion of the p53 coding sequence in Saos-2 cell line was demonstrated previously by Masuda et al. [49] and Scheffner et al. [64]), and the DNA transfection efficiency is greater in Saos-2 than in HEL cells. In Saos-2 cells, p53 promoter activity was activated approximately threefold by the cotransfection with the wild-type human p53 expression vector but not by cotransfection with mutant p53, which suggested that wild-type p53 activates its own promoter. p53 promoter activity was activated only slightly by IE1-72 but approximately eightfold by IE2-86. Cotransfection of cells with IE1-72 and p53 together activated p53 promoter activity four- to sixfold. While IE1-72 transactivated p53 promoter additively with p53, a negative effect was found when IE2-86 and p53 were transfected together, with up to a 40% decrease in CAT activity compared to IE2-86 alone. There was only basal CAT activity in cells cotransfected with the pCAT control vector and the various IE or p53 expression plasmids.
HCMV infection downregulates p53 downstream target gene expression.
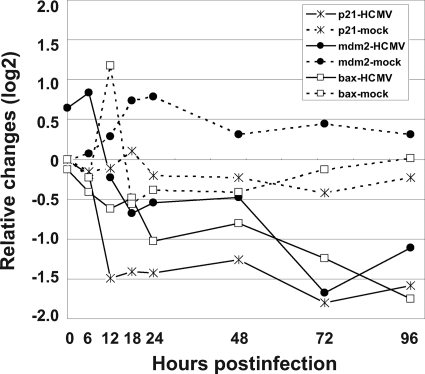
mRNA expression levels of genes containing p53-responsive elements p21, mdm2, and Bax were measured in HCMV-infected HEL cells by human genomic microarray assay, and relative changes in expression are reported in Fig. 2. The expression levels of all three of these p53-responsive genes were decreased at least twofold compared with those in mock-infected HEL cells between 6 to 96 hpi.
FIG. 2.
mRNA expression pattern of p53-responsive genes, p21, mdm2, and Bax, in HEL cells infected with the Towne strain of HCMV. mRNA levels of each gene were detected by microarray assay as described in Materials and Methods. Values shown are the differences between those in HCMV- or mock-infected HEL cells at the indicated time and in mock-infected HEL cells at 0 hpi.
p53 interacts with IE1-72 and IE2-86 in HEL cells infected with the Towne strain of HCMV.
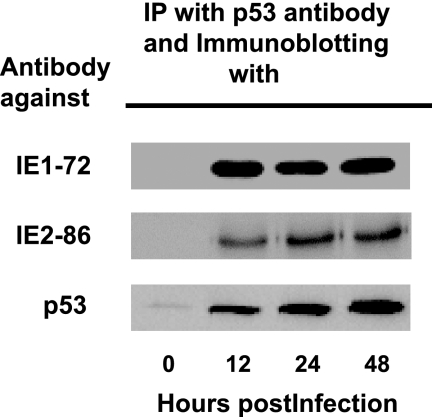
To examine the interaction between HCMV IE proteins and p53, we performed immunoprecipitation of lysates of HCMV-infected cells with anti-p53 antibody and immunoblotting with anti-IE1-72 or anti-IE2-86 antibody. IE1-72 and IE2-86 proteins coprecipitated with p53 in immunoprecipitation preparations from HCMV-infected lysates collected at 12, 24, and 48 hpi (Fig. 3). p53-containing complexes from mock-infected lysates did not contain the IE1-72 and IE2-86 proteins (Fig. 3, 0 hpi).
FIG. 3.
p53 interacts with HCMV IE1-72 and IE2-86 in HEL cells infected with the Towne strain of HCMV. Western blots demonstrating immunoreactivity to IE1-72, IE2-86, or p53 in complexes immunoprecipitated (IP) with anti-p53 antibody from lysates of HEL cells infected with HCMV and harvested at the indicated times postinfection.
Both HCMV IE1-72 and IE2-86 interact with HCMV UL84.
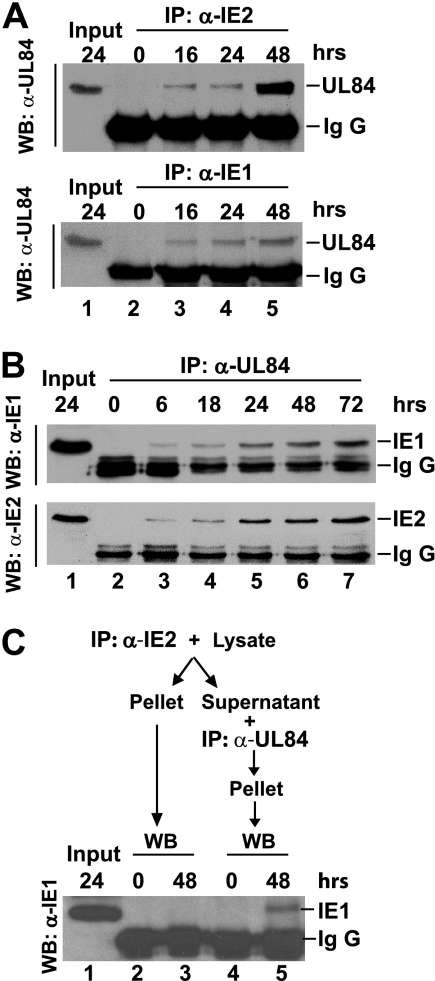
UL84 is an HCMV-specific phosphor-protein which is not only associated with DNA polymerase but also carries UTPase activity. It is required for oriLyt-dependent DNA replication and virus growth (13, 22). When HCMV-infected HEL cell lysates were immunoprecipitated with either anti-HCMV IE1-72 or IE2-86 antibody and then immunoblotted with anti-UL84 antibody, UL84 was found to coprecipitate with either IE1-72 or IE2-86 in lysates collected at 16 hpi and thereafter (Fig. 4A). When lysates were immunoprecipitated with anti-HCMV UL84 antibody and then immunoblotted with anti-IE1-72 or anti-IE2-86 antibody, both IE1-72 and IE2-86 were detected in immunoprecipitation products from lysates collected at 6 hpi and thereafter (Fig. 4B). These results suggested that both IE1-72 and IE2-86 interacted with UL84. To test if a complex comprised of all three proteins, IE1-72, IE2-86 and UL84, is formed in HCMV-infected cells, the lysates of HCMV-infected HEL cells were immunoprecipitated first with anti-IE2-86, and the supernatant from the first immunoprecipitation was subsequently immunoprecipitated with anti-UL84 antibody. The IE1-72 protein was detected in the anti-UL84 immunoprecipitation complex but not in the first anti-IE2-86 immunoprecipitation complex (Fig. 4C). These data indicate that the bindings of IE1-72 and IE2-86 to UL84 are mutually exclusive.
FIG. 4.
Both HCMV IE1-72 and IE2-86 interacted with HCMV pUL84. (A) Western blots demonstrating the presence of UL86 in complexes immunoprecipitated with anti-IE2-86 (upper panel) and anti-IE1-72 (lower panel) antibodies from lysates of HCMV-infected cells harvested at the indicated times postinfection. (B) Western blots (WB) demonstrating IE1-72 and IE2-86 immunoreactivity in complexes immunoprecipitated (IP) with anti-HCMV UL84 antibody from lysates of HCMV-infected HEL cells harvested at the indicated times postinfection. (C) Lysates of HCMV-infected cells harvested at the time indicated were immunoprecipitated first with anti-IE2 antibody, and the supernatant was subsequently immunoprecipitated with anti-UL84 antibody. Both immunoprecipitated complexes were subjected to Western blotting to detect the presence of IE1-72 with anti-IE1 antibody. α, anti; IgG, immunoglobulin G.
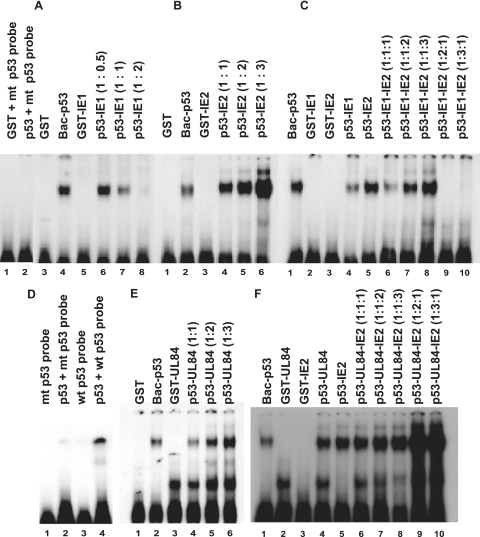
HCMV IE1-72 prevents p53 binding to p53-specific DNA binding sequences while IE2-86 and/or UL84 is capable of enhancing the ability of p53 to bind to p53-specific DNA sequences and inducing supershift.
To assess the effect of IE1-72 on the binding of p53 to p53-specific binding sequences, an EMSA was performed with recombinant p53 derived from baculovirus (Bac-p53), prokaryotically expressed IEs and UL84 proteins (GST fusion proteins), and a radioactively labeled p53-specific oligonucleotide probe. EMSA results showed that Bac-p53 bound specifically to p53 consensus sequences (Fig. 5) but not to a mutant p53 probe (Fig. 5D). GST, GST-IE1, GST-IE2, or GST-UL84 alone could not bind to the wild-type p53 DNA probe. When Bac-p53 was mixed with GST-IE1-72, the degree of p53 binding to DNA probe was decreased with increases in the amount of GST-IE1-72 (Fig. 5A, lanes 6 to 8). p53 binding to p53 wild-type probe was almost completely disrupted in EMSAs when two times the amount of GST-IE1 (0.35 μg/reaction mixture) was mixed with Bac-p53 (0.17 μg/reaction mixture) (Fig. 5, lane 8). When Bac-p53 was mixed with GST-IE2-86 (Fig. 5B, lanes 4 to 6) or GST-UL84 (Fig. 5E, lanes 4 to 6), p53 DNA binding was increased in a dose-dependent manner with the addition of increasing amounts of GST-IE2-86 or GST-UL84. In view of the fact that IE1-72, IE2-86, and UL84 are expressed during the IE and early period in HCMV-infected cells (demonstrated in Fig. 1A) and that IE1-72 and IE2-86 and UL84 exerted different or opposite effects on p53 DNA binding activity, we then performed an experiment to determine the effects of various combinations of IE1-72 and of IE2-86 and UL84 simultaneously on p53 DNA binding activity, and the results are shown in Fig. 5C and F. When fixed amounts of IE1-72 and p53 were used for EMSAs, increasing amounts of IE2-86 enhanced p53 binding in a dose-dependent manner (Fig. 5C, lanes 6 to 8). On the other hand, when the amounts of input IE2-86 and p53 were fixed, p53-DNA binding decreased or was disrupted drastically with increases in the amount of input IE1-72 (Fig. 5C, lanes 9 and 10). IE2-86 and UL84, independently and in combination, enhanced p53 binding in a dose-dependent fashion (Fig. 5F, lanes 4 to 10), and increased enhancement was observed when the input UL84 level was increased by two- or threefold (Fig. 5F, lanes 9 and 10).
FIG. 5.
HCMV IE1-72 prevented p53 binding to p53-specific DNA binding sequences while IE2-86 and/or UL84 enhanced binding. EMSA was performed after purified recombinant proteins were mixed, as indicated, in binding buffer containing 32P-labeled double-stranded oligonucleotide probe, as described in Materials and Methods. The effect of increasing the amounts of these proteins alone or in various combinations on p53 binding to p53-specific DNA binding sequences is shown: IE1-72 (A), IE2-86 (B), IE1-72 plus IE2-86 (C), HCMV UL84 (E), and UL84 plus IE2-86 (F). Numbers in parentheses indicate ratios (wt/wt or wt/wt/wt) of proteins in the indicated reaction. Panel D shows the specific binding of p53 to 32P-labeled double-stranded wild-type (wt) p53 oligonucleotide probe and the inability of p53 to bind mutant probe (mt).
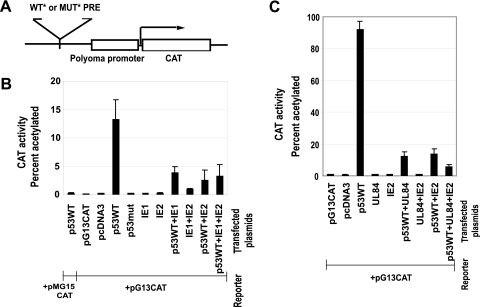
HCMV IE1-72, IE2-86, and UL84 proteins downregulate p53-dependent transcriptional activation in Saos-2 cells.
To determine the effects of IE1-72, IE2-86, and UL84 on p53 transcriptional activation in eukaryotic cells, the CAT activities were measured in p53-deficient Saos-2 cells following cotransfection with wild-type or mutant p53-responsive CAT reporters (pG13CAT or pMG15CAT) and various combinations of p53-, UL84- and IE-expressing plasmids (Fig. 6). In control experiments, wild-type p53 could not transactivate the mutant p53 reporter (Fig. 6B). Transfection with wild-type pG13CAT or parental vector (pcDNA3) alone had no CAT activity. The presence of mutant p53, IE1-72, or IE2-86 alone could not transactivate pG13CAT. Additionally, the combination IE2-86 and IE1-72 did not significantly transactivate pG13CAT. On the other hand, wild-type p53 alone activated the wild-type pG13CAT reporter. When wild-type p53 plasmid was cotransfected with IE1-72, IE2-86, or both IE1-72 and IE2-86 expression plasmids together, transcriptional activation activity of p53 was suppressed by 70% (IE1-72), 81% (IE2-86), and 76% (IE1-72 plus IE2-86), compared with transcriptional activation activity of p53 alone (Fig. 6B).
FIG. 6.
HCMV IE1-72, IE2-86, and UL84 downregulated p53-dependent gene transactivation in Saos-2 cells. (A) Diagram of target constructs showing wild-type (pG13CAT) and mutant (pMG15CAT) CAT reporter plasmids. WT* PRE, wild-type p53 responsive element with 13 copies of 5′-CCTGCCTGGACTTGCCTGG-3′ in plasmid pG13CAT (also called pSK-45-13-2-PyCAT); MUT* PRE, mutant PRE with 15 copies of 5′-CCTTAATGGACTTTAATGG-3′ in plasmid pMG15CAT (also called pSK89-15-1-PyCAT). (B and C) CAT assays of lysates from p53-negative Saos-2 cells cotransfected with combinations of expression vectors for IE1-72 and IE2-86 (B) and for IE2-86 and UL84 (C), along with wild-type p53 (p53WT) or mutant p53 (p53mut), as indicated, and with expression vectors for the target reporter constructs described in panel A, as indicated.
The effects of UL84 in similar CAT experiments are summarized in Fig. 6C. In cotransfection of the wild-type p53 plasmid with UL84 or IE2-86 alone or with UL84 and IE2-86 in combination, the transcriptional activation activity of p53 was suppressed by 87% (UL84 alone), 84% (IE2-86), and 95% (UL84 plus IE2-86) compared to the transcriptional activation activity of p53 alone.
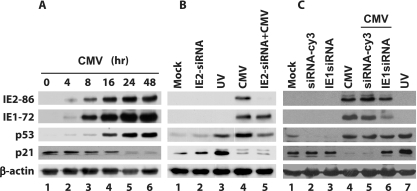
HCMV IE1-72, rather than IE2-86, was responsible for p21 downregulation in IE1-72 and IE2-86 siRNA-treated HEL cells infected with HCMV.
Since p21 is a downstream target of p53, we assessed the effect of HCMV IE1-72 and IE2-86 on p21 expression, as determined by levels of p21 measured by Western blot analysis. p21 was detected in confluent HEL cells (0 hpi) as well as in early stages of HCMV-infected HEL cells (before 16 hpi) (Fig. 7A). However, the p21 protein level was remarkably decreased and hardly detected at 24 hpi and thereafter. On the other hand, p53 expression was detected in HCMV-infected HEL cells as early as 4 hpi and continuously increased.
FIG. 7.
IE1-72 rather than IE2-86 was responsible for p21 downregulation in IE2-86 siRNA-treated HEL cells infected with HCMV. (A) Western blot demonstrating the levels of IE1-72, IE2-86, p53, and p21 in HCMV-infected HEL cells harvested at the indicated times postinfection. (B) Western blots demonstrating levels of IE1-72, IE2-86, p53, and p21 at 16 hpi in mock-infected HEL cells (lane 1), uninfected HEL cells pretreated with IE2-86 siRNA (lane 2), UV-treated HEL cells (lane 3), HCMV-infected HEL cells (lane 4), and HCMV-infected HEL cells that were pretreated with siRNA (lane 5). (C) Western blots demonstrating levels of IE1-72, IE2-86, p53, and p21 at 16 hpi in mock-infected HEL cells (lane 1), uninfected HEL cells pretreated with siRNA-Cy3 (lane 2) and IE1-72 siRNA (lane 3), HCMV-infected HEL cells (lane 4), HCMV-infected HEL cells that were pretreated with siRNA-Cy3 (negative control) and IE1-72 siRNA (lanes 5 and 6), and UV-treated HEL cells (lane 7).
We then utilized an IE1-72 and IE2-86 siRNA knockdown to study the role of HCMV IE proteins in the inhibition of p21 expression in HCMV-infected cells. Pretreatment of HEL cells with IE2-86 and IE1-72 siRNA did not appear to alter baseline p21 levels (Fig. 7B, lane 2, and C, lane 3, respectively). p21 expression was decreased at 16 hpi in HCMV-infected HEL cells that were pretreated with IE2-86 siRNA (Fig. 7B, lane 5). On the other hand, p21 expression was increased in HCMV-infected HEL cells that were pretreated with IE1 siRNA (Fig. 7C, compare lane 6 to lanes 4 and 5). Lane 3 of Fig. 7B and lane 7 of Fig. 7C are positive controls in which HEL cells were treated with UV. The resulting data suggest that IE1-72, but not 1E2-86, plays an important role in the decrease of p21 levels in HCMV-infected HEL cells.
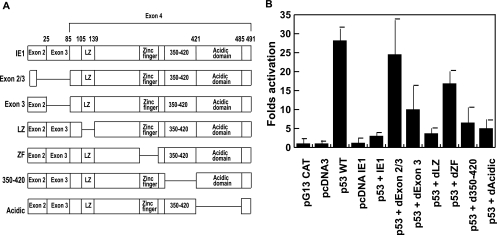
Exon 2/3 and zinc finger domains of the HCMV IE1-72 protein are required for IE1-72 repression of p53-dependent transcriptional activation.
In order to determine the domains of IE1-72 necessary for the suppression of p53 transcriptional activity, we cotransfected Saos-2 cells with pG13CAT, wild-type p53, and full-length IE1-72 or various deletion mutant plasmids of IE1-72. Lysates of transfected cells were prepared, and CAT activity was measured. Transfection of cells with pG13CAT alone or with empty vector (pcDNA3) plus pG13CAT revealed no CAT activity. pcDNA3IE1-72 with pG13CAT had a basal level of CAT activity. Wild-type p53 increased activation of pG13CAT an average of 28-fold. Addition of full-length of IE1-72 to wild-type p53 in cotransfection resulted in a dramatic decrease in CAT activity, from 28-fold activation down to an average of 1.2-fold activation. Cotransfection of wild-type p53 with mutant IE1-72s with deletions in exon 2/3, exon 3, the leucine zipper domain, the zinc finger domain, residues 350 to 420, or the acidic domain showed an average of 24.5-, 10.0-, 3.7-, 16.8-, 6.5-, and 5.0-fold activation, respectively (Fig. 8). These data indicate that only a mutation in exon 2/3 could lead to complete activation of p53, in comparison with the suppressive effect of full-length IE1-72 on the transactivation of p53. Deletion mutants in exon 3 and the zinc finger domain of IE1-72 led to partial restoration of p53 activity. These results indicate that IE1-72 exon 2, exon 3, and zinc finger domains play important roles in the suppression of p53 transcriptional activation.
FIG. 8.
Exon 2/3 and the zinc finger of HCMV IE1-72 protein are required for repression of p53-dependent transactivation. (A) Schematic map of IE1-72 full-length and deletion mutants. (B) CAT assays of lysates from p53-negative Saos-2 cells cotransfected with the expression vector for the wild-type p53 response element pG13CAT and combinations of expression vectors for p53, IE1-72, and IE1-72 mutants, as indicated. d350-420, mutant with a deletion of residues 350 to 420; dAcidic, mutant with a deletion of the acidic domain; WT, wild type; LZ, leucine zipper; ZF, zinc finger.
DISCUSSION
Molecular and cytogenetic studies have provided convincing evidence that accumulation of wild-type p53 is able not only to inhibit proliferation but also to induce cellular apoptosis in many types of cells (2, 34, 51). HCMV, like other DNA tumor viruses, is able to morphologically transform cells in vitro and stimulate host cell macromolecular synthesis in virus-infected cells (16). Previous studies have demonstrated an increase in p53 protein levels during HCMV infection, suggesting that p53 is targeted for regulation during HCMV infection (19, 55, 67). Because HCMV infection activates host cell enzymes involved in DNA replication and cellular proliferation pathways, it was hypothesized that p53 might be specifically targeted for binding by viral proteins to activate or inactivate its apoptotic-inducing properties. Our results demonstrate that HCMV infection activates the p53 promoter as well as p53 protein synthesis beginning at the IE stage of viral infection (Fig. 1), supporting the previous observation that the p53 promoter could be activated by p53 protein itself (15).
It has been demonstrated that IE2-86 can bind to and transcriptionally inactivate the p53 protein (67, 70). On the other hand, the positive effect of IE2-86 on p53 promoter activity has been published (54). Similarly, our data presented here show that the IE1-72 protein not only binds p53 but also inactivates p53-dependent transcriptional activity. These results and those from other studies (8, 54, 72) indicate that HCMV IE proteins play important roles in p53 induction as well as transcriptional regulation of p53 function during the IE period of HCMV infection. The inhibitory effect of IE2-86 on p53 transcriptional activity may be partly explained by the direct binding of IE2-86 to p53 (70). In addition, it was demonstrated that IE2-86 also binds to mdm2 and facilitates mdm2 degradation. Thus, it can lead to the accumulation of p53 in HCMV-infected HEL cells (80).
Coimmunoprecipitation with anti-p53 antibody revealed that p53 complexes with both IE1-72 and IE2-86. In the case of IE2-86, it can bind p53 directly (3, 70) and interact with a variety of transcription factors including TATA-binding protein (9), TFIIB (26), TAF130 (46, 47), Sp1 (46, 47), CREB (40), and Egr-1 (76). IE2-86 binds p53 but does not abrogate G1 checkpoint function (3). On the other hand, there has been no previous report of direct binding between p53 and IE1-72. In our current study, we have demonstrated that IE1-72 can be coprecipitated with p53 (Fig. 3) and that the IE1-72 and p53 interaction might lead to the dissociation of p53 from its cognitive DNA binding site, thus leading to the downregulation of p21 expression, as shown in Fig. 7. The IE1-72 protein can interact with the p107 protein and alleviates the transcriptional repression of the E2F-responsive promoter (60).
The demonstration of HCMV IE1-72 inhibition of p53 binding to p53-specific DNA binding sequences with dose dependency is interesting (Fig. 5A, lanes 6 to 8). The possibility of the competition for specific DNA binding between IE1-72 and p53 molecules can be ruled out because GST-IE1-72 alone had no binding activity to p53-specific DNA binding sequences (Fig. 5A, lane 5). There is no direct evidence that IE1-72, IE2-86, and UL84 bind to the p53-responsive element. Binding inhibition is more likely due to conformational changes of p53 resulting from the IE1-72 and p53 interaction. In contrast, under similar experimental conditions, IE2-86 and UL84 enhanced p53 binding to p53-specific DNA sequences with dose dependency (Fig. 5B and E, respectively). Although supershift was observed in EMSAs as the result of the interaction of IE2-86 and UL84 together with p53, the amount of supershifted product was relatively less than the nonsupershift portion. The specific binding was increased comparably with increases in IE2-86 and UL84 (Fig. 5B and E, respectively). The binding enhancement by IE2-86 and UL84 may be due to a conformational change or a modification of p53 by IE2-86 and UL84 that enhances the p53 binding to its specific DNA sequences. UL84 may act as an initiator protein for viral DNA synthesis at the origin of HCMV DNA replication (oriLyt) (57) and interacts with IE2-86 and regulates the transactivation activity of IE2-86 by interfering with binding of IE2-86 to its target (13). Similar to IE2-86, Epstein-Barr virus BZLF1 directly binds p53 and inhibits its ability to activate a reporter containing p53 binding motifs (79) but does not inhibit p53-specific DNA binding (50).
As expected, in DNA transfection experiments IE1-72 and IE2-86 inhibited wild-type p53 from transactivation of the p53-responsive promoter (Fig. 6). These inhibitory effects could again be explained by (i) IE1-72 inhibition of p53 binding to p53-specific DNA binding sequences, as described above, and (ii) IE2-86 enhancement of p53 binding to p53-specific DNA binding sequences but prevention of formation of transcriptionally active complexes, as suggested by previous reports (67, 70). Molecular interaction with p53 could not explain the suppressive effect of p53 on the transcriptional activity because some molecules such as apoptotic specific protein of p53 enhance the DNA binding and transactivator function of p53 (63). IE1-72 appears to be dominant over IE2-86 in regulating p53 DNA binding activity (Fig. 5C), but the mechanism remains to be further elucidated. As shown in Fig. 4, UL84 complexes with not only IE1 but also IE2. However, a complex containing all three components (IE1, IE2, and UL84) was not found (Fig. 4C). This result implied that the binding site of IE1 or IE2 to UL84 is within the common region of exon 2 and exon 3 shared by both IE proteins. Therefore, it is likely that UL84 only has one region for binding to exon 2 and exon 3 of either IE1 or IE2; thus, UL84 could not bind to both IE1 and IE2 simultaneously. To date, the functional role of each UL84-IE1 and UL84-IE2 complex is still unclear.
Castillo et al. (8) reported that p53 protein levels increased in fibroblasts following the expression of IE1-72 and that the observed accumulation of p53 protein in IE1-72-expressing cells may account for the inability of IE1-72 to induce S phase and delay cell cycle exit. Steady-state levels of p21, which is the indicative marker of p53 transcriptional activity, are decreased in HCMV-infected fibroblasts, as indicated in this study (Fig. 7A) and as reported by others (10, 56). Our observation that IE1-72 disrupts p53 binding and that IE2-86 enhances p53 binding might result in the in vivo experimental data shown in Fig. 7 indicating that IE1-72, but not IE2-86, interacts with p53 to prevent or disrupt p53 binding to its cognitive site to downregulate p21 expression in the HCMV-infected cell. In cases of either IE1-72 or IE2-86 transduction, the relative amounts of p21 are increased (6, 65). Tanaka et al. (69) showed that p21 was increased in smooth muscle cells by IE1-72 and p53 expression. However, in the presence of high levels of p53 and HCMV IE proteins, the HCMV-infected cells are mostly in an active metabolic and proliferative state, depending on the cell cycle status and cell types. Furthermore, both IE1-72 and IE2-86 proteins can bind to different members of the retinoblastoma protein (pRb) family to activate E2F-responsive promoters and cyclin-dependent kinases. IE1-72 interacts with the p107 protein while IE2-86 binds to pRb (8, 25, 37). IE1-72 and IE2-86 expression can alleviate the repression of E2F transcriptional activity mediated by p107 and pRb, respectively (18, 37, 60). The binding of IE1-72 to p107 displaces cyclin E/Cdk2 from p107; released cyclin E/Cdk2 is able to function as an active kinase, but IE2-86 cannot activate the kinase activity (81).
The IE2-86-p53 interaction did not impair epitope recognition for anti-p53 antibody with the specific site positioned in the DNA binding domain of p53 (3). Although IE2-86 strengthens the binding of p53 to the p53-responsive element (Fig. 5), transcriptional activity of p53 was inhibited by IE2-86 (Fig. 6). IE2-86 binds to the histone acetyltransferase domain of the p53 coactivators, p300 and CBP (CREB-binding protein), and blocks their acetyltransferase activity on both histones and p53. IE2-86 thus downregulates p53-dependent gene activation by inhibiting p300/CBP-mediated local histone acetylation (32). Therefore, the experiment on the interactions between the oligonucleotides containing the p53-responsive element and the purified p53 protein (shown in Fig. 5) might not be able to explain adequately the mechanism of HCMV-induced metabolic activation in HCMV-infected cells.
In mapping the domains of IE1-72 that are involved in suppression of transcriptional activation of p53 activity, we demonstrated that exons 2/3 and zinc finger domains within exon 4 of IE1-72 were responsible. In view of the fact that exons 2 and 3 are shared between IE1-72 and IE2-86 (7), it could be argued that IE2-86 should also have a suppressive effect on p53 activity. Nevertheless, we have recently found that the leucine zipper domain in exon 4 of IE1-72 is needed for IE1-72 and p107 interaction (81), while in this communication we have demonstrated that, in addition to exons 2 and 3, the zinc finger domain is needed for IE1-72's effect on the ability of p107 to affect transcriptional activation of E2F (37). Thus, additional domains other than exon 2 and exon 3 likely exist within exon 4 and exon 5 of major IE genes and may be needed for specific interactions between IE proteins and their counterparts for specific functions.
Exon 4 and exon 5 of HCMV IE genes are unique regions of IE1-72 and IE2-86, respectively. Both exon 4 and exon 5 contain a sequence motif called the zinc finger, with two Cys and two His residues. The orientations of the zinc finger of IE1-72 (HX2HXFX3LX2C4C) and IE2-86 (CX4CXL5FX3HX4H) are opposite to each other (23). The putative zinc finger of IE2-86 (from C428 to H452) is dispensable for DNA binding and autorepression (1), and the deletion mutant of IE2-86 (deletion of residues 325 to 448) containing zinc finger/LIM domain damage has no effect on the binding to p53 (80). Such distinct patterns of zinc finger arrangements may result in different consequences from IE1-72 and IE2-86 interactions with p53 protein with regard to p53's ability to bind to its specific DNA sequences.
In conclusion, we have shown that HCMV IE1-72 and IE2-86 can transactivate p53 promoter and are involved in the inhibition of p53-dependent transactivation by a possible conformational change in p53-DNA binding. Our data also suggest that IE1-72 represses the p53 transactivation activity by a unique binding hindrance mechanism different from that of IE2-86. We have also demonstrated that IE1-72, rather than IE2-86, is responsible for p21 downregulation in HCMV-infected HEL cells.
Acknowledgments
We thank John Sinclair for kindly providing the IE1-72 deletion mutant plasmid constructs, Shannon Kenney for p53 promoter construct, and B. Vogelstein for p53 expression plasmids pC53-SN3 and pC53-SCX3.
This work was supported by grants AI47678 and CA10914 from the National Institutes of Health (to E.-S. Huang), grant 97-I-01-03-A-053 from the Ministry of Science and Technology, and grant 21-2004-035-0 from Seoul National University Hospital, Republic of Korea (to E.-S. Hwang).
Footnotes
Published ahead of print on 23 September 2009.
REFERENCES
- 1.Asmar, J., L. Wiebusch, M. Truss, and C. Hagemeier. 2004. The putative zinc finger of the human cytomegalovirus IE2 86-kilodalton protein is dispensable for DNA binding and autorepression, thereby demarcating a concise core domain in the C terminus of the protein. J. Virol. 78:11853-11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. [DOI] [PubMed] [Google Scholar]
- 3.Bonin, L. R., and J. K. McDougall. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J. Virol. 71:5861-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 5.Casavant, N. C., M. H. Luo, K. Rosenke, T. Winegardner, A. Zurawska, and E. A. Fortunato. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castillo, J. P., F. M. Frame, H. A. Rogoff, M. T. Pickering, A. D. Yurochko, and T. F. Kowalik. 2005. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J. Virol. 79:11467-11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo, J. P., and T. F. Kowalik. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19-34. [DOI] [PubMed] [Google Scholar]
- 8.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caswell, R., C. Hagemeier, C. J. Chiou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., E. Knutson, A. Kurosky, and T. Albrecht. 2001. Degradation of p21cip1 in cells productively infected with human cytomegalovirus. J. Virol. 75:3613-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colletti, K. S., Y. Xu, I. Yamboliev, and G. S. Pari. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 280:11955-11960. [DOI] [PubMed] [Google Scholar]
- 14.Davis, M. G., S. C. Kenney, J. Kamine, J. S. Pagano, and E. S. Huang. 1987. Immediate-early gene region of human cytomegalovirus trans-activates the promoter of human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 84:8642-8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deffie, A., H. Wu, V. Reinke, and G. Lozano. 1993. The tumor suppressor p53 regulates its own transcription. Mol. Cell. Biol. 13:3415-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMarchi, J. M. 1983. Correlation between stimulation of host cell DNA synthesis by human cytomegalovirus and lack of expression of a subset of early virus genes. Virology 129:274-286. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, C. A., P. W. Hinds, and A. J. Levine. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57:1083-1093. [DOI] [PubMed] [Google Scholar]
- 18.Fortunato, E. A., M. H. Sommer, K. Yoder, and D. H. Spector. 1997. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J. Virol. 71:8176-8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman, P. N., S. E. Kern, B. Vogelstein, and C. Prives. 1990. Wild-type, but not mutant, human p53 proteins inhibit the replication activities of simian virus 40 large tumor antigen. Proc. Natl. Acad. Sci. USA 87:9275-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furnari, B. A., E. Poma, T. F. Kowalik, S. M. Huong, and E. S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, Y., K. Colletti, and G. S. Pari. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82:96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghazal, P., and J. A. Nelson. 1993. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis. Springer-Verlag, Berlin, Germany.
- 24.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagemeier, C., R. Caswell, G. Hayhurst, J. Sinclair, and T. Kouzarides. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagemeier, C., S. M. Walker, P. J. Sissons, and J. H. Sinclair. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385-2393. [DOI] [PubMed] [Google Scholar]
- 28.Hannemann, H., K. Rosenke, J. M. O'Dowd, and E. A. Fortunato. 2009. The presence of p53 influences the expression of multiple human cytomegalovirus genes at early times postinfection. J. Virol. 83:4316-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase α promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, Y. S., L. Xu, and E. S. Huang. 1992. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J. Virol. 66:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 32.Hsu, C. H., M. D. Chang, K. Y. Tai, Y. T. Yang, P. S. Wang, C. J. Chen, Y. H. Wang, S. C. Lee, C. W. Wu, and L. J. Juan. 2004. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 23:2269-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, E. S., S. T. Chen, and J. S. Pagano. 1973. Human cytomegalovirus. I. Purification and characterization of viral DNA. J. Virol. 12:1473-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isaacs, W. B., B. S. Carter, and C. M. Ewing. 1991. Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer Res. 51:4716-4720. [PubMed] [Google Scholar]
- 35.Jacobson, M. A., and J. Mills. 1988. Serious cytomegalovirus disease in the acquired immunodeficiency syndrome (AIDS). Clinical findings, diagnosis, and treatment. Ann. Intern. Med. 108:585-594. [DOI] [PubMed] [Google Scholar]
- 36.Jault, F. M., J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, R. A., A. D. Yurochko, E. E. Poma, L. Zhu, and E. S. Huang. 1999. Domain mapping of the human cytomegalovirus IE1-72 and cellular p107 protein-protein interaction and the possible functional consequences. J. Gen. Virol. 80:1293-1303. [DOI] [PubMed] [Google Scholar]
- 38.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B. Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827-830. [DOI] [PubMed] [Google Scholar]
- 39.Kowalik, T. F., A. D. Yurochko, C. A. Rinehart, C. Y. Lee, and E. S. Huang. 1994. Productive infection of human endometrial stromal cells by human cytomegalovirus. Virology 202:247-257. [DOI] [PubMed] [Google Scholar]
- 40.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, K., K. Jeon, J. M. Kim, V. N. Kim, D. H. Choi, S. U. Kim, and S. Kim. 2005. Downregulation of GFAP, TSP-1, and p53 in human glioblastoma cell line, U373MG, by IE1 protein from human cytomegalovirus. Glia 51:1-12. [DOI] [PubMed] [Google Scholar]
- 42.Levine, A. J., J. Momand, and C. A. Finlay. 1991. The p53 tumour suppressor gene. Nature 351:453-456. [DOI] [PubMed] [Google Scholar]
- 43.Light, J. A., and D. S. Burke. 1979. Association of cytomegalovirus (CMV) infections with increased recipient mortality following transplantation. Transplant Proc. 11:79-82. [PubMed] [Google Scholar]
- 44.Lokensgard, J. R., M. C. Cheeran, G. Gekker, S. Hu, C. C. Chao, and P. K. Peterson. 1999. Human cytomegalovirus replication and modulation of apoptosis in astrocytes. J. Hum Virol. 2:91-101. [PubMed] [Google Scholar]
- 45.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez, J., I. Georgoff, and A. J. Levine. 1991. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 5:151-159. [DOI] [PubMed] [Google Scholar]
- 49.Masuda, H., C. Miller, H. P. Koeffler, H. Battifora, and M. J. Cline. 1987. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc. Natl. Acad. Sci. USA 84:7716-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mauser, A., S. Saito, E. Appella, C. W. Anderson, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503-12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercer, W. E., M. T. Shields, M. Amin, G. J. Sauve, E. Appella, J. W. Romano, and S. J. Ullrich. 1990. Negative growth regulation in a glioblastoma tumor cell line that conditionally expresses human wild-type p53. Proc. Natl. Acad. Sci. USA 87:6166-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meulmeester, E., and A. G. Jochemsen. 2008. p53: a guide to apoptosis. Curr. Cancer Drug Targets 8:87-97. [DOI] [PubMed] [Google Scholar]
- 53.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegalovirus and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 54.Muganda, P., R. Carrasco, and Q. Qian. 1998. The human cytomegalovirus IE2 86 kDa protein elevates p53 levels and transactivates the p53 promoter in human fibroblasts. Cell. Mol. Biol. (Noisy-le-grand). 44:321-331. [PubMed] [Google Scholar]
- 55.Muganda, P., O. Mendoza, J. Hernandez, and Q. Qian. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noris, E., C. Zannetti, A. Demurtas, J. Sinclair, M. De Andrea, M. Gariglio, and S. Landolfo. 2002. Cell cycle arrest by human cytomegalovirus 86-kDa IE2 protein resembles premature senescence. J. Virol. 76:12135-12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pizzorno, M. C., P. O'Hare, L. Sha, R. L. LaFemina, and G. S. Hayward. 1988. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E. S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prives, C., and J. J. Manfredi. 1993. The p53 tumor suppressor protein: meeting review. Genes Dev. 7:529-534. [DOI] [PubMed] [Google Scholar]
- 62.Riley, T., E. Sontag, P. Chen, and A. Levine. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9:402-412. [DOI] [PubMed] [Google Scholar]
- 63.Samuels-Lev, Y., D. J. O'Connor, D. Bergamaschi, G. Trigiante, J. K. Hsieh, S. Zhong, I. Campargue, L. Naumovski, T. Crook, and X. Lu. 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 8:781-794. [DOI] [PubMed] [Google Scholar]
- 64.Scheffner, M., K. Munger, J. C. Byrne, and P. M. Howley. 1991. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA 88:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song, Y. J., and M. F. Stinski. 2005. Inhibition of cell division by the human cytomegalovirus IE86 protein: role of the p53 pathway or cyclin-dependent kinase 1/cyclin B1. J. Virol. 79:2597-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Speir, E., E. S. Huang, R. Modali, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1995. Interaction of human cytomegalovirus with p53: possible role in coronary restenosis. Scand. J. Infect. Dis. Suppl. 99:78-81. [PubMed] [Google Scholar]
- 67.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 68.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka, K., J. P. Zou, K. Takeda, V. J. Ferrans, G. R. Sandford, T. M. Johnson, T. Finkel, and S. E. Epstein. 1999. Effects of human cytomegalovirus immediate-early proteins on p53-mediated apoptosis in coronary artery smooth muscle cells. Circulation 99:1656-1659. [DOI] [PubMed] [Google Scholar]
- 70.Tsai, H. L., G. H. Kou, S. C. Chen, C. W. Wu, and Y. S. Lin. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J. Biol. Chem. 271:3534-3540. [PubMed] [Google Scholar]
- 71.Tuck, S. P., and L. Crawford. 1989. Characterization of the human p53 gene promoter. Mol. Cell. Biol. 9:2163-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, J., P. H. Marker, J. D. Belcher, D. E. Wilcken, L. J. Burns, G. M. Vercellotti, and X. L. Wang. 2000. Human cytomegalovirus immediate early proteins upregulate endothelial p53 function. FEBS Lett. 474:213-216. [DOI] [PubMed] [Google Scholar]
- 73.Wathen, M. W., and M. F. Stinski. 1982. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J. Virol. 41:462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiebusch, L., M. Truss, and C. Hagemeier. 2004. Inhibition of human cytomegalovirus replication by small interfering RNAs. J. Gen. Virol. 85:179-184. [DOI] [PubMed] [Google Scholar]
- 75.Wing, B. A., R. A. Johnson, and E. S. Huang. 1998. Identification of positive and negative regulatory regions involved in regulating expression of the human cytomegalovirus UL94 late promoter: role of IE2-86 and cellular p53 in mediating negative regulatory function. J. Virol. 72:1814-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoo, Y. D., C. J. Chiou, K. S. Choi, Y. Yi, S. Michelson, S. Kim, G. S. Hayward, and S. J. Kim. 1996. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J. Virol. 70:7062-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yurochko, A. D., S. M. Huong, and E. S. Huang. 1999. Identification of human cytomegalovirus target sequences in the human immunodeficiency virus long terminal repeat. Potential role of IE2-86 binding to sequences between −120 and −20 in promoter transactivation. J. Hum. Virol. 2:81-90. [PubMed] [Google Scholar]
- 78.Yurochko, A. D., T. F. Kowalik, S. M. Huong, and E. S. Huang. 1995. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J. Virol. 69:5391-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, Z., D. L. Evers, J. F. McCarville, J. C. Dantonel, S. M. Huong, and E. S. Huang. 2006. Evidence that the human cytomegalovirus IE2-86 protein binds mdm2 and facilitates mdm2 degradation. J. Virol. 80:3833-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang, Z., S. M. Huong, X. Wang, D. Y. Huang, and E. S. Huang. 2003. Interactions between human cytomegalovirus IE1-72 and cellular p107: functional domains and mechanisms of up-regulation of cyclin E/cdk2 kinase activity. J. Virol. 77:12660-12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]