Abstract
The arenavirus small RING finger Z protein has been shown to be the main driving force of budding for several arenaviruses. This Z budding activity was found to be mediated by the late (L)-domain motifs P(T/S)AP and PPXY, located at the C terminus of Z. Here, we show that the Z protein of Tacaribe virus (TACV), a New World arenavirus, buds efficiently from cells despite lacking the canonical L-domain motifs P(T/S)AP and PPXY. Likewise, potential L-domain motifs ASAP and YLCL present in TACV Z did not exhibit any significant contribution to TACV Z budding activity. Budding of TACV Z was Tsg101 independent but required the activity of Vps4A/B. These results indicate that TACV Z utilizes a budding mechanism distinct from that reported for other arenaviruses.
Arenaviruses are enveloped viruses with a bisegmented, negative-strand (NS) RNA genome and a life cycle restricted to the cell cytoplasm (1). Each RNA segment uses an ambisense coding strategy to direct the expression of two genes that are in opposite orientations and separated by a noncoding intergenic region. The large segment (7.2 kb) encodes the late (L) protein, an RNA-dependent RNA polymerase, and the small RING finger protein Z, which is the counterpart of the matrix (M) protein found in many enveloped NS RNA viruses. The small segment (3.5 kb) encodes the viral nucleoprotein (NP) and the glycoprotein precursor (GPC). The GPC is processed by the cellular protease S1P into GP1 and GP2 (13). Trimers of GP1/GP2 form the spikes that decorate the virus surface and mediate cell entry via receptor-mediated endocytosis (12).
Arenaviruses merit significant interest both as tractable experimental model systems to study acute and persistent viral infections (18, 28) and as clinically important human pathogens. The Old World virus Lassa virus (LASV) and several New World (NW) arenaviruses cause hemorrhagic fever (HF) disease in humans, posing a serious public health problem (1). LASV is estimated to infect several hundred thousand individuals yearly in the regions of West Africa where it is endemic, resulting in a high number of Lassa fever (LF) cases associated with significant mortality and high morbidity. Notably, increased travel to and from regions of endemicity has led to the importation of LF into metropolitan areas, where the virus is not endemic, around the globe (11). Likewise, the NW arenavirus Junin virus causes Argentine HF, a severe illness with hemorrhagic and neurological manifestations and a fatality rate of 15 to 30% (7, 22, 27), while the NW Machupo and Guanarito arenaviruses have emerged as causative agents of HF in Bolivia and Venezuela, respectively (22). Public health concerns about HF arenavirus infections are exacerbated by the lack of licensed vaccines and by the fact that current antiarenavirus therapies are limited to the use of the nucleoside analogue ribavirin, which is only partially effective and is associated with significant side effects. Therefore, it is important to develop novel drugs to combat human pathogenic arenaviruses.
Similarly to many other enveloped NS RNA viruses, arenavirus infectious particles bud from the plasma membranes of infected cells. Evidence indicates that the M proteins of many enveloped NS RNA viruses play critical roles in virus budding (3). Accordingly, many of these M proteins are, in the absence of any other virus polypeptide, competent in budding and can form virus-like particles (VLPs). Budding of M proteins is often directed by L-domain motifs, which most frequently correspond to one of the following sequences: P(T/S)AP, PPXY, YXXL, or FPIV (3). Efficient M-mediated budding requires interactions between viral L domains and host factors, many of them involved in the cellular multivesicular body (MVB) sorting pathway (3). MVB formation requires the activity of a network of cytoplasmic protein complexes known as endosomal sorting complexes required for transport (ESCRT). Tsg101 is a component of the ESCRT-1 complex and plays a key role in the biogenesis of MVB. An AAA ATPase, Vps4, which is present in humans as two isoforms, Vps4A and Vps4B, binds to components of ESCRT-3 and mediates dissociation of ESCRT-3 complex from the endosomal membrane. We (20, 26) and others (24) have documented that Z is the driving force of arenavirus budding. As with many other bona fide viral budding proteins, all known arenavirus Z proteins contain P(T/S)AP, PPXY, or both, L-domain motifs that play a critical role in Z budding (20, 24). Tacaribe virus (TACV) Z protein, however, constitutes a unique exception, as it lacks both P(T/S)AP and PPXY L-domain motifs (Fig. 1A).
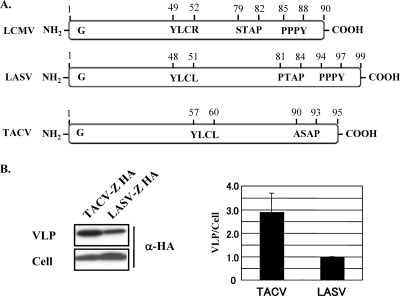
FIG. 1.
Arenavirus Z protein and its budding efficiency. (A) Schematic representation of Z proteins from LCMV, LASV, and TACV. G corresponds to the strictly conserved glycine residue found at position 2 of all known arenavirus Z proteins. The locations of bona fide PTAP (LASV), PPPY (LASV and LCMV), and YXXL (LASV and TACV) L-domain motifs are indicated, as well as that of the L-like domain motifs STAP (LCMV) and ASAP (TACV). (B) Budding activity of TACV Z protein. 293T cells were transfected with 0.25 μg of either pC-TACV-Z-HA or pC-LASV-Z-HA. At 36 h posttransfection, tissue culture supernatants were collected, and VLPs and total cell lysates were prepared as described previously (2). Levels of Z proteins in VLPs and cell lysates were determined by WB using an antibody to HA (sc-7392; Santa Cruz). The budding efficiency of LASV Z was set at 1.0. The data are averages and standard deviations from three independent experiments.
Budding efficiency of TACV Z protein.
To examine whether TACV Z could bud efficiently from cells, we used the plasmid pCAGGS (17) to express a C-terminally hemagglutinin (HA)-tagged TACV Z protein and compared its budding efficiency with that of LASV Z HA (20) using a previously described method (2). Briefly, we transfected 293T cells (4 × 105) with 0.25 μg of each plasmid, and 36 h later we collected VLP-containing tissue culture supernatants and cells. After clarification from cell debris (1,500 × g; 5 min), VLPs were collected by ultracentrifugation (100,000 × g; 30 min at 4°C) through a 20% sucrose cushion. Cells and VLPs were resuspended in lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 8.0], 62.5 mM EDTA, 0.4% sodium deoxycholate) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting (WB) using an anti-HA mouse monoclonal antibody (sc-7392; Santa Cruz). TACV Z exhibited about three-times-higher budding efficiency than LASV Z (Fig. 1B). This result indicated that like lymphocytic choriomeningitis virus (LCMV) and LASV, TACV Z is a bona fide budding protein.
Effect of G2A mutation and 2-OHM treatment on TACV Z budding.
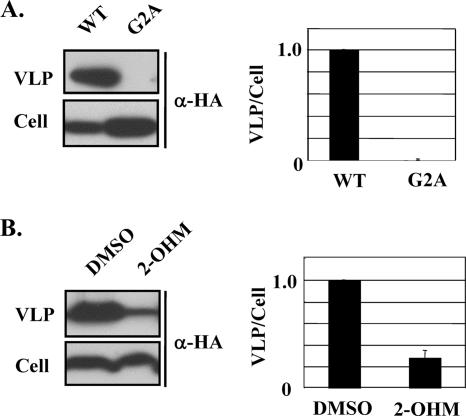
A wide range of viral proteins are myristoylated (16), a posttranslational modification that can significantly influence protein function. Data from studies on both the sequences of known myristoylated proteins and the sequence requirements for N-myristoyltransferase activity indicate that the N-terminal acceptor amino acid for protein myristoylation is a glycine residue at position 2 (G2) (15). Accordingly, the mutation G2A prevents protein myristoylation. Previously, we showed that myristoylation of the LCMV and LASV Z proteins was essential for their budding activity (21). Notably, comparison of all known arenavirus Z-protein sequences, including that of TACV Z, revealed the presence of a strictly conserved G2 in the context of a consensus myristoylation signal (5). We therefore sought to define the role of the G2 residue in the budding activity of TACV Z. For this, we first compared the budding activity between wild-type (WT) TACV Z and TACV Z(G2A) (Fig. 2A). Consistent with our previous findings with LASV and LCMV Z proteins (21), we observed slightly higher cellular expression levels of TACV Z(G2A) than WT TACV Z. Likewise, the G2A mutation also resulted in a TACV Z protein with budding activity below the detection levels of our assay (Fig. 2A). This finding indicated that G2 plays a critical role in TACV Z budding activity, which is likely related to its role as myristoylation acceptor site. To further evaluate the role of myristoylation in TACV Z budding, we examined the effect of dl-2-hydroxymyristic acid (2-OHM) on TACV Z budding. 2-OHM is a myristic acid analog that becomes metabolically activated in cells to form 2-hydroxymyristoyl-coenzyme A, a potent inhibitor of N-myristoyltransferase (19). Budding of TACV Z was inhibited in cells treated with 2-OHM (Fig. 2B). At the drug concentration used (100 μM), no effect on cell viability or cellular expression levels of Z protein was observed.
FIG. 2.
Effect of G2A mutation and 2-OHM treatment on TACV Z budding. (A) 293T cells were transfected with 0.25 μg of either pC-TACV-Z-HA or pC-TACV-Z(G2A)-HA. At 36 h posttransfection, levels of Z proteins in VLPs and total cell lysates were determined by WB as for Fig. 1. The budding efficiency of WT TACV Z was set at 1.0. (B) 293T cells were transfected with pC-TACV-Z-HA and treated with 2-OHM (100 μM) or vehicle alone (dimethyl sulfoxide) for 36 h before levels of Z budding were determined. The budding efficiency of TACV Z in vehicle-treated cells was set at 1.0. The data are averages and standard deviations from three independent experiments.
Role of the L-like domain ASAP motif on TACV Z budding.
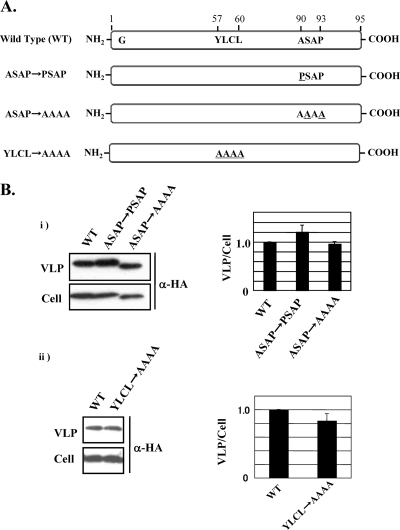
The budding activities of LCMV and LASV Z proteins were reported to depend on the presence of the canonical L-domain motifs P(T/S)AP and PPPY (Fig. 1A) (20, 24). TACV Z has the motif ASAP instead of P(T/S)AP (Fig. 1A); hence, we examined whether ASAP could functionally replace P(T/S)AP, thus providing TACV Z protein with budding activity. For this, we generated two TACV Z mutants, one with a change of ASAP to PSAP (pC-TACV-Z ASAPPSAP) and one with a change of ASAP to AAAA (pC-TACV-Z ASAPAAAA), and determined their budding activities compared to that of WT TACV Z (Fig. 3A). The mutant and WT TACV Z proteins exhibited similar budding activities, indicating that ASAP does not make a significant contribution to TACV Z budding activity (Fig. 3B, panel i).
FIG. 3.
Role of the L-like domain on TACV Z budding. (A) Schematic representation of all TACV Z mutants used in this study. (B) Budding activity of TACV Z with mutations in ASAP (panel i) and YLCL (panel ii) motifs. 293T cells were transfected with 0.25 μg of pC-TACV-Z, pC-TACV-Z ASAPPSAP, pC-TACV-Z ASAPAAAA, or pC-TACV-Z YLCLAAAA; all Z-expressing constructs were HA tagged at their C termini. At 36 h posttransfection, VLPs and total cell lysates were prepared and Z expression levels were determined by WB as for Fig. 1. The budding efficiency of WT TACV Z was set at 1.0. The data are averages and standard deviations from three independent experiments.
Role of the YLCL motif in TACV Z budding.
The YXXL motif has been shown to operate as an L domain in a variety of viruses (4, 8-10, 23). Notably, Z proteins from the majority of known arenaviruses, including TACV, have a YLCL motif located within the RING domain of Z. To examine whether this YLCL motif could functionally provide TACV Z protein with budding activity, we generated a TACV Z mutant with the change YLCL to AAAA (pC-TACV-Z YLCLAAAA) and determined its budding activity compared to that of WT TACV Z (Fig. 3B, panel ii). The mutant and WT TACV Z proteins exhibited similar budding activities, indicating that YLCL does not make a significant contribution to TACV Z budding activity.
Roles of Tsg101 and Vps4A/B in TACV Z budding.
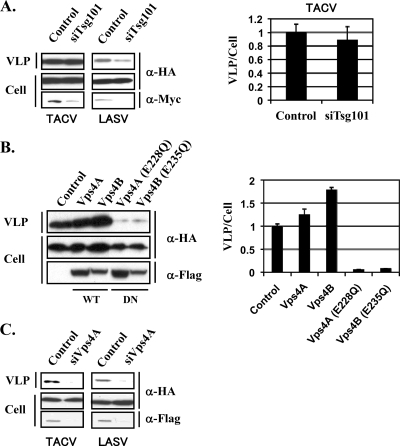
Next, we examined whether, as with LCMV and LASV Z proteins, TACV Z budding activity also required the interaction of Z with members of the MVB sorting pathway. Tsg101 is a component of the ESCRT-1 complex known to participate in the cellular MVB sorting pathway (6). The L-domain motif P(T/S)AP can bind to a ubiquitin E2 variant domain present within Tsg101, and this interaction has been shown to allow the association of a variety of viral budding proteins, including HIV-1 Gag, Ebola virus VP40, and LASV Z, with the MVB sorting pathway (6, 14, 20, 26). To examine the contribution of Tsg101 to TACV Z budding, we used a small interfering RNA (siRNA)-based approach. For this we used pS-Tsg101 to direct intracellular expression of a siRNA that specifically and effectively targeted Tsg101 expression (20). Expression of Tsg101 siRNA, but not a control siRNA, caused a severe reduction in Tsg101 expression levels in cells transfected with a plasmid (pTsg101) expressing a Myc-tagged Tsg101 (Fig. 4A). However, TACV Z budding activity was not affected by reduced expression levels of Tsg101 (Fig. 4A), whereas, consistent with previous findings (20, 26), LASV Z budding in cells expressing the Tsg101 siRNA was decreased dramatically (Fig. 4A). Vps4A/B is an AAA ATPase that has a key function in the endocytic pathway for degradation of membrane proteins. Vps4 function is closely linked to ESCRTs, which are multiprotein complexes required for the sorting of endocytosed transmembrane proteins into intraluminal vesicles. Mutant forms of Vps4 defective in ATPase activity have been shown to be potent inhibitors of budding in a variety of viruses (3). To assess the contribution of Vps4A/B to TACV Z budding, we examined the effect of WT and dominant negative forms of Vps4A and Vsp4B on TACV Z budding activity. Overexpression of WT forms of Vps4A/B slightly increased the budding activity of TACV Z (Fig. 4B), whereas overexpression of dominant negative forms [Vps4A(E228Q) and Vps4B(E235Q)] caused a dramatic reduction of TACV Z budding activity (Fig. 4B). Consistent with these results, siRNA-mediated knockdown of Vps4A using previously described conditions (26) resulted in reduced budding of TACV Z and also, in agreement with previous findings (26), of LASV Z (Fig. 4C).
FIG. 4.
Roles of Tsg101 and Vps4A/B in TACV Z budding. (A) 293T cells were cotransfected with pC-TACV-Z-HA or pC-LASV-Z-HA (0.1 μg) and pC-Tsg101-myc (0.2 μg) together with pS-Tsg101 (0.2 μg) or control plasmid pSUPER (0.2 μg). Myc-tagged Tsg101 was detected by WB using a mouse monoclonal antibody to Myc. Levels of Z protein in VLPs and total cell lysates were determined by WB as for Fig. 1. (B) 293T cells were cotransfected with pC-TACV-Z-HA and the indicated Flag-tagged Vps4-expressing constructs or control plasmid pcDNA3.1. Flag-tagged Vps4 proteins were detected by WB using a rabbit serum to Flag (no. 162150; Cayman). Levels of Z protein in VLPs and cell lysates were determined by WB as for Fig. 1. The data are averages and standard deviations from three independent experiments. (C) 293T cells were transfected with a specific Vps4A (siVps4A) or control siRNA (100 pmol/ml), and 24 h later cells were cotransfected with pC-TACV-Z-HA or pC-LASV-Z-HA (0.1 μg) and pVps4A-Flag (0.03 μg), together with a specific Vps4A or control siRNA (50 pmol/ml). Flag-tagged Vps4A was detected as described above. Levels of Z protein in VLPs and cell lysates were determined by WB as for Fig. 1.
Previous reports have shown that Z plays a key role in arenavirus budding and that this activity was mediated by the presence of canonical L-domain motifs [P(T/S)AP and PPXY] within Z (20, 24, 26). Here we have shown that although TACV Z does not possess these canonical L domains, it exhibits strong budding activity. G2 of TACV Z plays a critical role in budding, suggesting that myristoylation of TACV Z is necessary for its budding activity, as previously reported for LCMV and LASV Z proteins (21, 25). We have also shown that ASAP and YLCL motifs do not exhibit L-domain activities related to budding in the context of TACV Z protein. In addition, our findings revealed that TACV Z does not utilize Tsg101 but does depend on Vps4A/B activity for its budding activity. These findings suggest that, as with LCMV and LASV, budding of TACV requires the participation of the ESCRT machinery of the cell but involves different specific ESCRT components that remain to be determined.
Progress in understanding the mechanisms underlying cell egress of enveloped viruses has shown that different viruses can exhibit a significant degree of commonality regarding the virus-cell protein interactions that direct viral budding. This knowledge has led to the consideration of this critical step of the virus life cycle as a potential novel target for the development of antiviral drugs. Budding of both the prototypic arenavirus LCMV and the HF virus LASV was shown to be driven by the virus Z protein in a process mediated by the interaction between canonical L-domain motifs [P(T/S)AP and PPXY] within Z and members of the cellular MVB pathway (20, 24, 26). Based on sequence comparison among known Z proteins, it is predicted that budding of pathogenic NW arenaviruses would be driven by an interaction between the L-domain motif P(T/S)AP of Z and components of the cellular MVB pathway. However, our finding that the Z protein of the NW TACV exhibits a strong budding activity that is not mediated by the canonical L-domain motif P(T/S)AP, PPXY, or YXXL raises the possibility that in response to antiviral drugs targeting L-domain-MVB protein interactions, human-pathogenic arenavirus variants resistant to these drugs could arise by acquiring the ability to use a different budding pathway. It is also quite plausible that in the absence of functional known canonical L-domain motifs, other sequence motifs could exhibit bona fide L-domain activity. A detailed understanding of mechanisms underlying TACV Z budding should facilitate the development of targeting strategies to counteract the potential ability of arenaviruses to use more than one budding pathway.
Acknowledgments
This work was supported by NIH grant R01 AI047140-06 to J.C.T. S.U. was supported by the Japan Society for the Promotion of Science (JSPS) Research Fellowship for Young Scientists and by NIH grant T32-AI0735419.
This is The Scripps Research Institute paper 20139 from the Department of Immunology and Microbial Science.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Buchmeier, M. J., C. J. Peters, and J. C. de la Torre. 2007. Arenaviridae: the virus and their replication, p. 1792-1827. In D. L. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 2.Capul, A. A., and J. C. de la Torre. 2008. A cell-based luciferase assay amenable to high-throughput screening of inhibitors of arenavirus budding. Virology 382:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, B. J., and R. A. Lamb. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciancanelli, M. J., and C. F. Basler. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J. Virol. 80:12070-12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farazi, T. A., G. Waksman, and J. I. Gordon. 2001. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276:39501-39504. [DOI] [PubMed] [Google Scholar]
- 6.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 7.Harrison, L. H., N. A. Halsey, K. T. McKee, Jr., C. J. Peters, J. G. Barrera Oro, A. M. Briggiler, M. R. Feuillade, and J. I. Maiztegui. 1999. Clinical case definitions for Argentine hemorrhagic fever. Clin. Infect. Dis. 28:1091-1094. [DOI] [PubMed] [Google Scholar]
- 8.Honeychurch, K. M., G. Yang, R. Jordan, and D. E. Hruby. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J. Virol. 81:7310-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui, E. K., S. Barman, D. H. Tang, B. France, and D. P. Nayak. 2006. YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42. J. Virol. 80:2291-2308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Irie, T., Y. Shimazu, T. Yoshida, and T. Sakaguchi. 2007. The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J. Virol. 81:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isaacson, M. 2001. Viral hemorrhagic fever hazards for travelers in Africa. Clin. Infect. Dis. 33:1707-1712. [DOI] [PubMed] [Google Scholar]
- 12.Kunz, S. 2009. Receptor binding and cell entry of Old World arenaviruses reveal novel aspects of virus-host interaction. Virology 387:245-249. [DOI] [PubMed] [Google Scholar]
- 13.Lenz, O., J. ter Meulen, H. D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 15.Maurer-Stroh, S., B. Eisenhaber, and F. Eisenhaber. 2002. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 317:523-540. [DOI] [PubMed] [Google Scholar]
- 16.Maurer-Stroh, S., and F. Eisenhaber. 2004. Myristoylation of viral and bacterial proteins. Trends Microbiol. 12:178-185. [DOI] [PubMed] [Google Scholar]
- 17.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 18.Oldstone, M. B. 2002. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Curr. Top. Microbiol. Immunol. 263:83-117. [DOI] [PubMed] [Google Scholar]
- 19.Paige, L. A., G. Q. Zheng, S. A. DeFrees, J. M. Cassady, and R. L. Geahlen. 1990. Metabolic activation of 2-substituted derivatives of myristic acid to form potent inhibitors of myristoyl CoA:protein N-myristoyltransferase. Biochemistry 29:10566-10573. [DOI] [PubMed] [Google Scholar]
- 20.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez, M., D. L. Greenwald, and J. C. de la Torre. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443-11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters, C. J. 2002. Human infection with arenaviruses in the Americas. Curr. Top. Microbiol. Immunol. 262:65-74. [DOI] [PubMed] [Google Scholar]
- 23.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strecker, T., R. Eichler, J. Meulen, W. Weissenhorn, H. Dieter Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 77:10700-10705. (Erratum, 77:12927.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strecker, T., A. Maisa, S. Daffis, R. Eichler, O. Lenz, and W. Garten. 2006. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol. J. 3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urata, S., T. Noda, Y. Kawaoka, H. Yokosawa, and J. Yasuda. 2006. Cellular factors required for Lassa virus budding. J. Virol. 80:4191-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissenbacher, M. C., R. P. Laguens, and C. E. Coto. 1987. Argentine hemorrhagic fever. Curr. Top. Microbiol. Immunol. 134:79-116. [DOI] [PubMed] [Google Scholar]
- 28.Zinkernagel, R. M. 2002. Lymphocytic choriomeningitis virus and immunology. Curr. Top. Microbiol. Immunol. 263:1-5. [DOI] [PubMed] [Google Scholar]