Abstract
Although many drugs inhibit the replication of Epstein-Barr virus (EBV) in cell culture systems, there is still no drug that is effective and approved for use in primary EBV infection. More recently, maribavir (MBV), an l-ribofuranoside benzimidazole, has been shown to be a potent and nontoxic inhibitor of EBV replication and to have a mode of action quite distinct from that of acyclic nucleoside analogs such as acyclovir (ACV) that is based primarily on MBV's ability to block the phosphorylation of target proteins by EBV and human cytomegalovirus protein kinases. However, since the antiviral mechanisms of the drug are complex, we have carried out a comprehensive analysis of the effects of MBV on the RNA expression levels of all EBV genes with a quantitative real-time reverse transcription-PCR-based array. We show that in comparisons with ACV, the RNA expression profiles produced by the two drugs are entirely different, with MBV causing a pronounced inhibition of multiple viral mRNAs and with ACV causing virtually none. The results emphasize the different modes of action of the two drugs and suggest that the action of MBV may be linked to indirect effects on the transcription of EBV genes through the interaction of BGLF4 with multiple viral proteins.
After nearly three decades of study, there is still no drug approved for use in primary Epstein-Barr virus (EBV) infection despite the fact that many drugs inhibit the replication of this virus in cell culture, in part because of the complex pathogenesis of infectious mononucleosis (16). The first drug shown to inhibit EBV replication was phosphonoacetic acid (19, 35, 57), but the most intensively studied has been the acyclic nucleoside analog acyclovir (ACV) (8, 10, 39, 48). More recently, 1-H-β-l-ribofuranoside-2-isopropylamino-5,6-dichlorobenzimidazole(maribavir [MBV], also known as benzimidavir), which represents a new class of antiviral drugs, has been extensively examined. While other antiviral drugs inhibit multiple members of the herpesvirus family, MBV inhibits the replication of only human cytomegalovirus (HCMV), a member of the betaherpesvirus subfamily, and EBV, a member of the gammaherpesvirus subfamily (3, 64, 68). ACV is an effective inhibitor of several herpesviruses but not HCMV; ganciclovir, another acyclic nucleoside analog, is a potent inhibitor of HCMV and EBV replication, but its use is restricted because of significant toxicity (26). There is, therefore, a strong interest in MBV because of its favorable toxicity profile as well as efficacy (32, 63). Moreover, since EBV as well as HCMV are reactivated and produce disease in immunosuppressed patients such as bone marrow and organ transplant recipients (38, 55), MBV is a promising agent with a dual potential.
The potency of MBV against HCMV is equal to or better than that of ganciclovir or phosphonoformic acid (Foscarnet). It is much more potent than ACV, and more importantly, it is very active against HCMV strains that are resistant to either of these drugs (42, 64). Therefore, MBV is an excellent candidate to prevent and treat HCMV diseases in high-risk patients based on its efficacy and safety profile in phase 1 clinical trials (32, 63). A phase 2 clinical trial showed that MBV significantly decreased HCMV infection and prevented HCMV disease in allogeneic stem cell transplant recipients. It did so without causing myelosuppression or renal toxicity (65).
Unlike nucleoside analogs, MBV does not target the HCMV DNA polymerase, and unlike its parent compound, 1-H-β-d-ribofuranoside-2-bromo-5,6-dichlorobenzimida-zole (BDCRB), it does not block the cleavage of high-molecular-weight viral DNA concatemers in proviral genomes by the terminase complex, which is composed of UL56 and UL89 (3, 27, 29, 68). Instead, MBV inhibits the protein kinase (PK) activity of UL97 (measured by the autophosphorylation and phosphorylation of histone). Single point mutations in UL97 render HCMV resistant to MBV (2, 3, 6). MBV also blocks HCMV capsid egress from the nucleus and induces highly refractile bodies (aggregates of tegument proteins) in infected cells. As both phenomena are also observed in cells infected with HCMV-UL97 knockout mutant viruses (27, 51), it is likely that MBV interferes with some principal functions of UL97 PK. Many targets of UL97 are probably unknown, although recent data suggest that UL97 can interact with the HCMV pp65 tegument protein as well as cellular proteins such as EF-1delta, among others (21, 24). MBV may also exert its anti-HCMV effects through additional pathways since point mutations of UL27, an HCMV protein without a known function (and not conserved in EBV), also lead to low-level resistance to MBV (7).
Work with its homolog, the EBV PK BGLF4, showed that MBV could affect the essential replication gene, early antigen-diffuse (EA-D [BMRF1], the EBV DNA polymerase processivity factor), by inhibiting its hyperphosphorylation (15, 68). However, as indicated above, the mode of action of the drug is complex, with multiple potential targets, which also include EBV maturation proteins.
EA-D is an early gene and exists in at least three different molecular masses, ranging from 49 to 54 kDa, due to phosphorylation during EBV replication (5, 37, 50). When EBV replication is induced in Akata cells, EA-D is phosphorylated in a temporal fashion and is dominated by the hyperphosphorylated form (54 kDa) at 12 h postinduction (hpi). In addition, PK induces the phosphorylation of a smaller 52-kDa form. The smallest form (49 kDa) is thought of as being nonphosphorylated but is probably phosphorylated to some extent by cellular kinases (9). The two larger sizes were confirmed to be hyperphosphorylated forms based on results of phosphatase treatment (15, 68). In the presence of MBV, only 49-kDa EA-D is detected in induced Akata cells (14, 15). Since UL97 also phosphorylates UL44 (viral DNA polymerase processivity factor for HCMV) (28, 40), and MBV inhibits UL97 kinase activity (3), it is reasonable to hypothesize that the reduced levels of the hyperphosphorylated and hypophosphorylated forms of EA-D produced by MBV are due to the inhibition of BGLF4. However, since MBV was previously reported not to inhibit BGLF4 kinase function directly either in in vitro kinase assays or upon the transient coexpression of the two proteins (14, 15), its effects on EA-D during lytic replication are apparently indirect. Therefore, mechanisms of the anti-EBV activity of MBV and its inhibition of phosphorylated forms of EA-D remain elusive. However, it can be expected that BGLF4 plays diversified yet critical roles in EBV replication and pathogenesis.
In this study we used a different approach to gain a fuller perspective of the complex antiviral mechanisms of MBV by analyzing the expressions of all EBV-specific genes during the lytic program in Akata cells treated with MBV in comparison with ACV. For this purpose, we took advantage of our established quantitative real-time reverse transcription-PCR-based array that is able to measure the viral mRNA levels expressed by all the genes in the EBV genome (31). We show that while the majority of EBV genes are expressed at significant levels after the induction of lytic EBV replication, the EBV RNA expression profiles found with the two drugs are entirely different. The results emphasize the distinct modes of action of MBV and ACV and suggest that the mechanism of action of MBV may be linked to its indirect effects on the transcription of EBV genes through the interaction of BGLF4 with multiple viral proteins (69).
MATERIALS AND METHODS
Cells.
Akata is an EBV-positive cell line derived from Burkitt's lymphoma (gift of Lindsey Hutt-Fletcher). The cells were cultured in RPMI 1640 medium (Mediatech, Inc., Manassas, VA) with 10% fetal calf serum (HyClone, Logan, UT), 2 mM l-glutamine, and antibiotics (culture medium) at 37°C in 5% CO2. For the induction of lytic infection, the number of cells was adjusted to 2 × 106 cells/ml in culture medium, and goat F(ab′)2 fragment to human immunoglobulin G (IgG) (anti-human IgG, 100 μg/ml; MP Biomedicals, LLC, Solon, OH) was used to cross-link cell surface IgG (43, 66). Cells were kept at 37°C for 1 h with occasional mixing before readjusting to 1 × 106 cells/ml. Antiviral drugs were diluted in culture medium and added with anti-human IgG. Culture medium with corresponding concentrations of MBV or ACV was used to readjust cell concentrations. MBV was obtained from GlaxoSmithKline (Research Triangle Park, NC), and ACV was purchased from Sigma-Aldrich (St. Louis, MO).
RNA preparation.
Akata cells were collected, resuspended in 1 ml Tri reagent (Sigma-Aldrich, St. Louis, MO), and frozen at −80°C before extraction according to the manufacturer's recommendations. Briefly, 200 μl of chloroform was added to tubes containing harvested cells in Tri reagent, vortexed, and transferred into phase-lock tubes (Eppendorf, Brinkman Instruments, NY). After centrifugation, supernatant fluid containing RNA was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1 mixture, pH 5.2; Fisher Scientific, Inc., Pittsburgh, PA) and once with chloroform. RNA was precipitated with an equal volume of isopropanol (Sigma, St. Louis, MO) containing 1 μl of GlycoBlue tracer (Ambion, Inc., Austin, TX) and suspended in 1× Tris-EDTA buffer (10 mM Tris, 1 mM EDTA [pH 7.5]; Fisher Scientific). RNA quality was checked by monitoring of the A260/A280 ratio and by electrophoresis on agarose gels.
mRNA enrichment and reverse transcription.
The mRNA fraction was enriched with an Oligotex mRNA minikit (Qiagen, Inc., Valencia, CA) according to the manufacturer's protocol except that mRNA was eluted in 0.1× Tris-EDTA (pH 7.5). Reverse transcription was performed with a high-capacity cDNA archive kit (Applied Biosystems, Austin, TX) in a single cycle of a four-step reaction in a thermocycler (Eppendorf AG, Hamburg, Germany). The remaining RNA was digested with RNase H (Epicenter, Madison, WI).
Real-time QPCR array.
For real-time quantitative PCR (QPCR), 2.5 μl of primer mix was combined with 7.5 μl SYBR green 2× PCR mix (Applied Biosystems) and 5 μl cDNA and subjected to real-time QPCR by using an MJR Opticon2 cycler under standard cycling conditions. Data were normalized to cellular reference genes and analyzed by unsupervised clustering as described previously (49).
Unsupervised hierarchical clustering.
For each biological replicate series, the raw threshold cycle values were normalized to the median of four reference genes for each sample to yield the difference in cycle threshold (dCT). These values were clustered by gene with the use of a correlation metric and depicted as heat maps (12).
Statistical analysis.
Statistical analysis was conducted using R, version 2.8.0. First, we analyzed each biological replicate separately.
(i) 2005 dCT data set.
Based on density plots, we eliminated genes with mean dCT values of <−10 or >10. The resulting mean dCT values were approximately normally distributed (see Fig. S1 in the supplemental material), with a minimum dCT value of −3.5, a maximum dCT value of 12.3, a mean dCT value of 3.7, a median dCT value 3.6, a first-quadrant dCT value of 1.8, and a third-quadrant dCT value of 5.4.
(ii) 2008 dCT data set.
Based on density plots, we eliminated genes with mean dCT values of <−5 or >9.5. The resulting mean dCT values were approximately normally distributed, with a minimum dCT value of −9.7, a maximum dCT value of 15.9, a mean dCT value of 3.6, a median dCT value of 3.1, a first-quadrant dCT value of 1.3, and a third-quadrant dCT value of 5.5 (see Fig. 3).
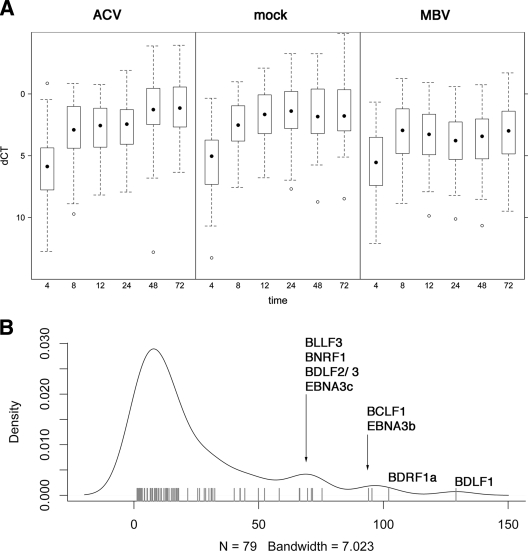
FIG. 3.
(A) Box plot of EBV mRNA levels at the indicated times after induction in the presence or absence of treatment. Times (hpi) are shown on the horizontal axis, and relative mRNA levels (dCT) are shown on the vertical axis. “dCT” indicates the mRNA abundance relative to the median of the housekeeping genes in the array. Lower dCT levels correspond to higher mRNA levels on a log2 scale. The dot indicates the median, boxes approximate the 25% and 75% percentiles, and whiskers indicate 1.5× the interquartile range. Outliers are indicated by open circles. (B) Density distribution of the cumulative differences between ACV and MBV for all EBV genes. The cumulative difference for each mRNA was calculated as the sum over all time points of the differences (ddCT) between both treatments. More differentially regulated genes exhibit a higher cumulative difference (located further to the right) (P ≤ 0.05). The top differentially regulated genes are indicated by name.
Two-way analysis of variance was conducted to determine the effect of time and treatment. The P value was adjusted for multiple comparisons using Bonferroni correction. We excluded the zero time points from statistical comparisons, since there was no difference in treatment. Next, we used a generalized linear model to identify the individual genes in the array independently and q value comparison (56) to adjust for multiple comparisons.
Immunofluorescence microscopy.
Cells were washed once with ice-cold phosphate-buffered saline and deposited onto 14-well 5-mm glass slides to create a cell monolayer (Erie Scientific Co., Portsmouth, NH). Cells were fixed with cold acetone and stained with monoclonal EA-D antibody (Capricorn Products, Inc., Portland, ME), monoclonal gB (gp125) antibody, or monoclonal gp350/250 antibody (Millipore Corp., Billerica, MA). Isothiocyanate-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA) was applied, and photography was performed with an Axioskop 2 microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY) at a 200× amplification.
Protein lysates and immunoblotting.
Cells were washed once with ice-cold phosphate-buffered saline and digested in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 5 mM dithiothreitol, 0.2 mM Na3VaO4, 100 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany) (15, 68). Protein lysates were separated by electrophoresis on sodium dodecyl sulfate-10% polyacrylamide gels and transferred onto a PVDF-Plus membrane (0.45 μm; GE Water & Process Technologies, Trevose, PA) with a Trans-Blot SD semidry transfer cell (Bio-Rad, Hercules, CA). The membrane was preincubated in blocking buffer of 5% fat-free milk in Tris-buffered saline solution with 0.1% Tween 20 (pH 7.6). The membrane was probed with monoclonal EA-D antibody or monoclonal antibody to β-actin (Sigma) overnight at 4°C and was then probed with horseradish peroxidase-conjugated anti-mouse secondary antibody. Protein levels were visualized with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL) and Blue Basic Autorad film (ISC BioExpress, Kaysville, UT).
RESULTS
MBV but not ACV inhibits phosphorylation of EA-D in lytic infection.
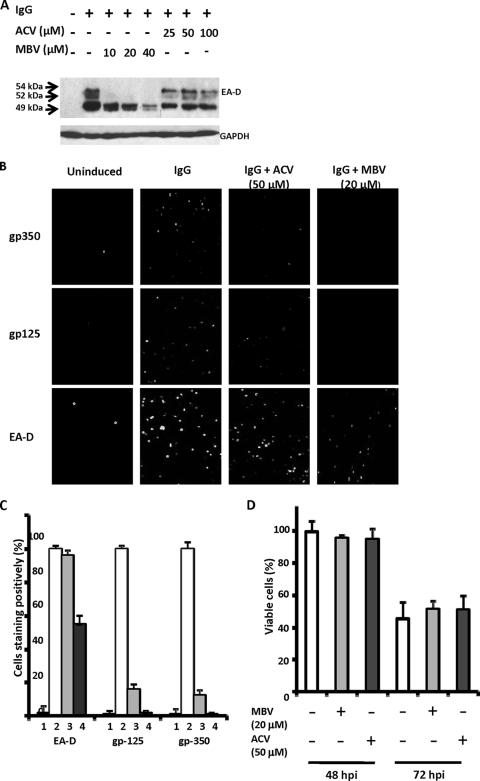
The cross-linking of B-cell receptors expressed on latently infected Akata cells by anti-human IgG treatment efficiently induces lytic EBV replication. This system has been used to analyze the effects of anti-EBV drugs including ACV, phosphonoacetic acid, and MBV (15, 66, 68). We selected concentrations of MBV and ACV that were sufficient to inhibit EBV replication without significant toxicity, as determined previously. Effects of the drugs on EA-D expression were analyzed by immunoblotting analyses of whole-cell extracts collected 24 hpi, when the three forms of EA-D are at their highest level. EA-D was not detected in uninduced Akata cells, whereas three bands corresponding to molecular masses of 54, 52, and 49 kDa could be detected in induced cells (Fig. 1A). The 54- and 52-kDa forms represent EA-D hyperphosphorylated and hypophosphorylated by EBV PK; the 49-kDa form is not phosphorylated by viral PK. MBV (20 μM) almost completely inhibited the appearance of the 54-kDa and 52-kDa EA-D products and also reduced the level of 49-kDa EA-D but to a lesser extent and in a dose-dependent manner; in contrast, ACV (50 μM) had no effect on EA-D protein, in agreement with previous results (15, 68). Throughout this study, the concentrations used are ∼20 times greater than the 50% inhibitory concentrations (0.3 to 10 μM for ACV and 0.15 to 1.1 μM for MBV) and about five times less than the 50% cytotoxicity concentrations (250 μM for ACV and 94 μM for MBV) (13, 14, 39, 47).
FIG. 1.
MBV inhibits the appearance of EA-D as well as EBV gp125 and gp350, but ACV inhibits only gp125 and gp350. (A) Akata cells in log phase were exposed to anti-IgG (IgG) in the presence or absence of MBV or ACV and harvested 24 hpi. Protein lysates equivalent to 1.5 × 105 cells were separated on sodium dodecyl sulfate-10% polyacrylamide gels, transferred onto a PVDF-Plus membrane, and immunoblotted with monoclonal EA-D antibody. (B) Akata cells were induced as described above (A); harvested at 48 hpi; washed; dropped onto 14-well glass slides; stained with monoclonal EA-D, gp125, and gp350 antibodies; and examined by immunofluorescence. A representative set of photographs is shown. Experiments were repeated three times. (C) Percentages of 500 to 1,000 cells that stained positively were calculated from the experiment depicted in B and are presented graphically; percentages of positive cells treated with IgG only were set as 100%. Columns labeled 1 represent uninduced cells; columns labeled 2 represent cells induced with IgG; columns labeled 3 represent cells treated with ACV; columns labeled 4 represent cells treated with MBV. (D) Cells from the experiment described above (B) were stained after 48 and 72 hpi with 0.2% trypan blue for 5 min at room temperature. Viable cells were determined by the exclusion of trypan blue stain, and percentages were calculated by counting 500 to 1,000 cells. Percentages of viable cells treated with IgG only at 48 hpi were set as 100%.
MBV inhibits the intracellular appearance of EA-D as well as EBV gp125 and gp350, but ACV inhibits only gp125 and gp350.
We next examined the effects of MBV and ACV on the expression of the early (EA-D) and the late (gp125 and gp350) viral proteins by immunofluorescence staining. EA-D was detected in the nucleus, while both gp125 and gp350 were detected in the cytoplasm. However, some EA-D could be detected in cytoplasm. On average, 39% ± 5% of cells entered the lytic cycle, as indicated by the detection of EA-D, compared with 2% ± 1% of cells without induction, and 27% ± 3% and 26% + 3% of cells expressed the gp125 and gp350 proteins, respectively (<1% without induction). MBV (20 μM) reduced the number of cells expressing EA-D by about 50% and almost eliminated the expression of gp125 and gp350 (Fig. 1B and C). In contrast, ACV (50 μM) had virtually no effect on the percentage of cells expressing EA-D and inhibited gp125 and gp350 to a lesser extent, as shown by immunostaining and summarized graphically (Fig. 1B and C). There was little effect on cell viability in the presence of either drug at 48 hpi. By 72 hpi viable cells were reduced by about 50%, as expected, after the induction of lytic replication (Fig. 1D). These findings clearly show the potent inhibitory effect of MBV on the phosphorylation of EA-D during lytic infection and the lack of such an effect produced by ACV. Both drugs inhibited the expression of the two late viral glycoproteins, but MBV had a considerably greater effect, probably due to its greater antiviral potency.
MBV inhibits EBV transcription genome wide, most distinctly a subset of gamma transcripts.
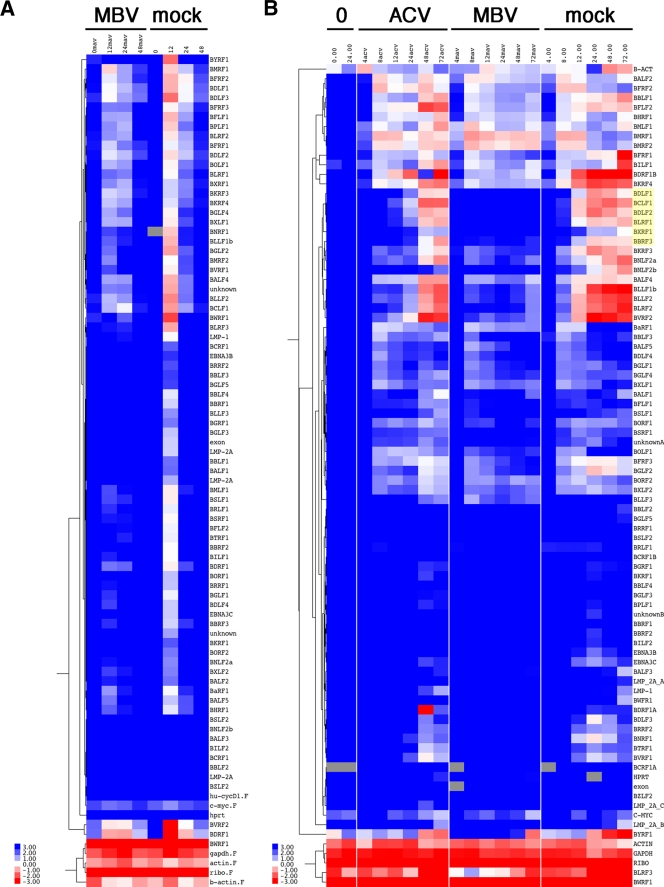
To assess the effect of MBV on EBV transcription, we used EBV-specific real-time QPCR arrays as described previously (31). We conducted two biological-replicate experiments and analyzed the resulting mRNA levels separately by unsupervised hierarchical clustering (Fig. 2). Note that no data were removed due to quality control issues. Results were surprising: MBV treatment resulted in the reduction of EBV mRNA levels, whereas the effect of ACV treatment was negligible (Fig. 2B). The MBV effect was evident at early time points but most pronounced for certain gamma transcripts: BDLF1, BCLF1, BDLF2, BLRF1, BXRF1, and BBRF3. The reason why the inhibition of these genes stands out is that their RNAs are undetectable in uninduced cells and then accumulate to very high levels, as indicated in Fig. 2B.
FIG. 2.
(A) Heat map diagram of unsupervised hierarchical clustering of EBV mRNA levels at different times after induction in the presence or absence of MBV (2005 data set). (B) Heat map diagram of unsupervised hierarchical clustering of EBV mRNA levels at different times after induction in the presence of MBV, ACV, or no drug (2008 data set). Red indicates higher and blue indicates lower levels than means of dCT values. Note that no data were removed due to quality control.
The raw data were further processed by statistical analysis. First, we analyzed each biological replicate separately. A subset of good dCT data for only the EBV genes and not the reference genes was derived by removing extremely abundant (e.g., BWRF1 repeat) transcripts or those below the limit of detection. As expected, the overall data revealed a striking time dependence of EBV transcription (P < 10−14, adjusted for multiple comparisons) (Fig. 3A), providing evidence that F(ab)2-IgG treatment of Akata cells induced a coordinated cascade of EBV gene transcription. Figure 3A shows low and identical levels at 4 h for mock- and MBV-treated cells. Upon induction in the absence of MBV, all EBV mRNAs were induced up to 48 h. By 72 h, the induction of the mRNA was stopped by the effects of viral replication in the cells. In the presence of MBV, overall EBV mRNA levels increased initially (8 h) but not thereafter. The difference between MBV and ACV treatment was significant, with a P value of ≤4.02 × 10−7. There was no statistical difference in overall EBV gene expression levels between ACV and mock treatments, which served as a negative control. These results demonstrate that MBV inhibits EBV transcription.
The effects mirror the first data set (see the supplemental material). The time dependence was significant, with a P value of ≤10−14 (adjusted for multiple comparisons). The time zero sample for mock-treated cells had unusually high levels of housekeeping mRNAs; hence, the mean dCT numbers are lower. This becomes evident in Fig. S1 in the supplemental material but was corrected for in the heat map. Because this first replicate had fewer data points, we did not use it for statistical analysis. In summary, MBV causes an extensive reduction in levels of transcription of EBV mRNAs.
Our statistical analysis derives power from taking into account all time points rather than doing a pairwise comparison, which would in fact omit most of the data. There were no statistically significant differences at any time or for any mRNA between ACV- and mock-treated cells. There were statistically significant differences across all mRNAs and time points in aggregate (Fig. 3A) between ACV and MBV at a P value of ≤10−7 based on two-way analysis of variance (time and drug).
We then used a linear model to determine for each mRNA individually whether it was differentially regulated between ACV and MBV. We added these differences for each time point and plotted the cumulative differences in Fig. 3B. As can be seen, these differences have a long-tailed distribution (one large peak and three small peaks). The largest peak comprises those mRNAs that are not significantly changed (P ≥ 0.05 after adjustment for multiple comparisons); the smaller peaks on the right in Fig. 3B comprise mRNAs that differ significantly with the different drug treatments. We identified the most differentially changed mRNAs by name in Fig. 3B, based on the shape of the density distribution rather than a single fixed cutoff.
The genes that exhibited the largest cumulative differences under MBV compared to ACV treatments across all time points were BDLF1, BDRF1a, BCLF1, EBNA3B, BDLF2, BLLF3, BDLF3, EBNA3C, and BNRF1 (Fig. 3B). The identities (name and temporal class) and functions, if known, of these genes are shown in Table 1, which shows the changes in mRNA levels relative to levels of uninduced cells (time zero) for each of these viral genes. The changes are the basis for the statistical analyses. For each of the nine genes, mRNA levels increased over time after induction in the absence of drug (labeled mock). Their mRNA levels still increased in the presence of ACV. This increase appears to be less dramatic and delayed upon ACV treatment. However, the differences between ACV- and mock-treated cells were often within twofold and did not rise to the strict level of significance in our statistical analysis. In contrast, MBV inhibited any increase in mRNA levels upon the induction of viral lytic replication, with the exception being BLLF3, where both ACV and MBV seemed to delay mRNA downregulation at late times. This outcome would be expected if the BLLF3 downregulation were mediated by a late viral gene, the accumulation of which is inhibited by either drug. At this time, we do not know the identity of this factor.
TABLE 1.
Changes in transcription of genes produced by MBV and ACVa
| Gene | Function | Treatment | Fold change at hpib |
% Change from GAPDHc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 8 | 12 | 24 | 48 | 72 | ||||
| EBNA 3B | Transcription factor (latent) | Mock | 1 | 1 | 6 | 68 | 131 | 80 | 37 | 7 |
| ACV | 1 | 1 | 4 | 7 | 22 | 22 | 15 | 2 | ||
| MBV | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | ||
| EBNA 3C | Transcription factor (latent) | Mock | 1 | 2 | 10 | 84 | 119 | 83 | 53 | 14 |
| ACV | 1 | 1 | 7 | 12 | 32 | 64 | 51 | 6 | ||
| MBV | 1 | 3 | 7 | 4 | 2 | 3 | 4 | 2 | ||
| BDLF1 | Minor capsid protein (late) | Mock | 1 | 1 | 7 | 45 | 130 | 111 | 65 | 40 |
| ACV | 1 | 1 | 2 | 8 | 23 | 80 | 75 | 18 | ||
| MBV | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | ||
| BDRF1a | Packaging (late) | Mock | 1 | 3 | 20 | 52 | 73 | 67 | 66 | 10 |
| ACV | 1 | 2 | 18 | 20 | 29 | 2,972 | 96 | 14 | ||
| MBV | 1 | 4 | 17 | 12 | 10 | 12 | 17 | 4 | ||
| BCLF1 | Major capsid protein (late) | Mock | 1 | 1 | 11 | 62 | 100 | 127 | 93 | 72 |
| ACV | 1 | 1 | 7 | 13 | 31 | 130 | 129 | 42 | ||
| MBV | 1 | 1 | 4 | 3 | 2 | 4 | 4 | 6 | ||
| BDLF2 | Structural protein (late) | Mock | 1 | 1 | 15 | 91 | 182 | 135 | 87 | 58 |
| ACV | 1 | 1 | 6 | 14 | 38 | 113 | 99 | 27 | ||
| MBV | 1 | 1 | 4 | 5 | 3 | 4 | 5 | 5 | ||
| BLLF3 | dUTPase (early) | Mock | 1 | 86 | 323 | 234 | 25 | 13 | 16 | 2 |
| ACV | 1 | 6 | 257 | 137 | 111 | 830 | 1,017 | 7 | ||
| MBV | 1 | 375 | 1,330 | 722 | 554 | 719 | 874 | 31 | ||
| BDLF3 | gp150 (late) | Mock | 1 | 2 | 87 | 950 | 4,859 | 2,157 | 914 | 8 |
| ACV | 1 | 2 | 14 | 36 | 187 | 1,798 | 865 | 2 | ||
| MBV | 1 | 4 | 34 | 95 | 49 | 54 | 61 | 1 | ||
| BNRF1 | Tegument protein (late) | Mock | 1 | 1 | 17 | 120 | 199 | 121 | 49 | 23 |
| ACV | 1 | 1 | 9 | 17 | 41 | 79 | 55 | 11 | ||
| MBV | 1 | 1 | 6 | 6 | 4 | 4 | 4 | 2 | ||
Largest cumulative difference across all time points.
Compared to uninduced (fold = 1.8ddCT). ddCT, difference in dCT.
Average over the time course for each drug compared to GAPDH (percent = 1.8dCT/100).
In Table 1, we also provide a relative measure of the mean abundance of each of these viral mRNAs vis-à-vis GAPDH (glyceraldehyde-3-phosphate dehydrogenase) levels. This shows that the mRNAs for each of these genes are relatively abundant (7 to 72% for mock-treated cells) and thus within the linear range of our assay. This summary measure also confirms the more detailed comparison based on changes at multiple time points. Overall, ACV does not inhibit mRNA levels by more than twofold, whereas MBV inhibits viral mRNA levels 5- to 40-fold.
Note that this selection reflects both the level of the mRNA and the magnitude of the decrease. These are biomarkers that distinguish MBV from ACV. However, profiling experiments, in general, cannot exclude the possibility that different genes, which change less dramatically in response to treatment, are rate limiting for the replicative cycle and mediate the MBV effect.
DISCUSSION
Currently available antiviral drugs that might be used to treat EBV infection can be divided into two main classes based on whether they target herpesvirus DNA polymerases or not (14, 38, 47, 55). Acyclic nucleoside and phosphonated nucleotide analogs are incorporated into viral DNA and cause DNA chain termination during viral DNA synthesis. These analogs have much greater affinity for viral than cellular DNA polymerase (10, 47). Pyrophosphate analogs compete with nucleotides in binding to pyrophosphate-binding sites of DNA polymerase. ACV is an acyclic guanosine analog that has consistently been shown to have high anti-EBV activity even though unlike herpes simplex virus (HSV), it cannot be efficiently phosphorylated by the EBV thymidine kinase (BXLF1) (11, 20, 39, 44, 68). Our early work showed that the anti-EBV effect is possibly due to the exquisite sensitivity of EBV DNA polymerase so that even a small amount of triphosphorylated ACV is effective (48).
The second group of drugs includes quite different compounds with distinct structures, foremost among them being MBV and its parent, d-ribonucleoside (BDCRB), each of which has a completely different mode of action (3, 14, 64). MBV was developed in place of BDCRB, the development of which was abandoned since BDCRB is rapidly inactivated metabolically (61). MBV inhibits the replication of EBV and HCMV; the effects on HCMV involve the inhibition of enzymatic activity of the HCMV PK UL97, discovered initially through drug resistance caused by a mutation of the gene. While MBV inhibits the hyperphosphorylation of EA-D, the essential EBV DNA processivity factor, by the EBV PK BGLF4, in cell culture (Fig. 1A) (4, 15), the drug does not inhibit the enzymatic activity of BGLF4 in vitro (14). Therefore, the inhibitory mechanism of MBV for EBV remains far from being understood.
BGLF4 belongs to the family of conserved herpesvirus PKs, which includes HCMV UL97, HSV UL13, and HSV US3 (17). UL97 and BGLF4 might be expected to have many functions in common, including some carried out by HSV's two kinases. HSV US3 phosphorylates a host of viral and cellular proteins involved in blocking the apoptosis of infected cells (36), and it may be needed for the optimal growth of HSV (52, 53). HSV UL13 is able to phosphorylate several viral and cellular genes, and its biological roles are still under investigation. HSV UL13 phosphorylates US3 (22) and is also required for the optimal growth of HSV (58). Both HCMV UL97 and EBV BGLF4 are essential genes (18, 27, 51). BGLF4 is functionally more similar to HCMV UL97 than to HSV UL13 despite a low degree of amino acid identity (54). It is likely that BGLF4 and UL97 share common functions in viral replication since BGLF4-specific small interfering RNAs inhibit the nuclear egress of EBV capsid (18), similar to the phenotype of cells infected with UL97-deleted HCMV (27).
These two MBV-sensitive viral kinases have more than one target and can phosphorylate cellular in addition to viral proteins. HCMV UL97 phosphorylates cellular elongation factor 1δ (25); histone H2B (2); p32 and nuclear lamins A, B, and C (41); and HCMV pUL44 (28, 40) and pUL69 (60). EBV BGLF4 is able to autophosphorylate and to phosphorylate many exogenous viral substrates such as EBNA-LP (23), EBNA2 (67), and BZLF1 (1) in addition to EA-D (4, 15) and also ganciclovir (Q. Meng, S. Hagemeier, J. S. Pagano, and S. Kenney, unpublished data) as well as cellular targets including elongation factor 1δ (24), condensin, topoisomerase II (33), replication origin-binding protein MCM4 (30), lamin A (34), histone protein H2AX (59), and interferon regulatory factor 3 (62). At present, the significance of the interaction between BGLF4 and these targets for EBV replication is undefined. Moreover, it is very likely that the list of BGLF4 targets will continue to grow, and more functions of BGLF4 will be defined, since another 19 EBV proteins have recently been reported to interact with BGLF4 in protein chip arrays (69), and most if not all of these proteins are phosphorylated by BGLF4. However, whether MBV blocks the functions of these substrates is largely unknown.
Our results indicate that MBV inhibits the phosphorylation of EA-D; reduces the protein expression of EA-D, gp125, and gp350; and inhibits EBV transcription in induced Akata cells. It is unclear why some EBV transcripts are downregulated to a greater extent than others. Many of the most highly downregulated transcripts are adjacent to each other in the EBV genome and may be under the control of the same promoter. No known EBV transcription factors were found to be downregulated. However, cellular transcription factors are involved in the regulation of transcription of many EBV genes and may be a target for MBV. MBV treatment also results in reduced EA-D phosphorylation, a known target for the EBV PK, suggesting that BGLF4 may be inhibited by the drug and is involved in decreased EBV transcription. Similar to the effects of MBV treatment, EBV mutant virus in which EA-D is knocked out does not express the late viral gp125 and gp350 genes (46). In contrast, the knockout of BGLF4 has almost no effect on the expression of EBV early and late genes including gp125 (45). Future work is designed to address the interaction of MBV and BGLF4 and to evaluate the mechanisms through which MBV downregulates viral transcripts.
Supplementary Material
Acknowledgments
We thank Lindsey Hutt-Fletcher for Akata cell lines and technical advice and Anuradha Gullapalli for technical assistance with immunofluorescence staining.
This work was supported by funding from the Leukemia and Lymphoma Foundation (grant 6021) and NIH grants DE018304 to D.P.D. and HL64851-06 to J.S.P.
Footnotes
Published ahead of print on 16 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 80:5125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek, M. C., P. M. Krosky, and D. M. Coen. 2002. Relationship between autophosphorylation and phosphorylation of exogenous substrates by the human cytomegalovirus UL97 protein kinase. J. Virol. 76:11943-11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, M. R., S. J. Chang, H. Huang, and J. Y. Chen. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 74:3093-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, M. S., G. Milman, and S. D. Hayward. 1985. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J. Virol. 56:860-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, S., and G. I. Marousek. 2008. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J. Virol. 82:246-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, S., G. I. Marousek, A. E. Senters, M. G. Davis, and K. K. Biron. 2004. Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir. J. Virol. 78:7124-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby, B. M., J. E. Shaw, G. B. Elion, and J. S. Pagano. 1980. Effect of acyclovir [9-(2-hydroxyethoxymethyl)guanine] on Epstein-Barr virus DNA replication. J. Virol. 34:560-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daibata, M., and T. Sairenji. 1993. Epstein-Barr virus (EBV) replication and expressions of EA-D (BMRF1 gene product), virus-specific deoxyribonuclease, and DNA polymerase in EBV-activated Akata cells. Virology 196:900-904. [DOI] [PubMed] [Google Scholar]
- 10.Datta, A. K., B. M. Colby, J. E. Shaw, and J. S. Pagano. 1980. Acyclovir inhibition of Epstein-Barr virus replication. Proc. Natl. Acad. Sci. USA 77:5163-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, A. K., and J. S. Pagano. 1983. Phosphorylation of acyclovir in vitro in activated Burkitt somatic cell hybrids. Antimicrob. Agents Chemother. 24:10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrichs, C., J. Neyts, G. Gaspar, E. De Clercq, and P. Wutzler. 2004. Evaluation of antiviral activity against human herpesvirus 8 (HHV-8) and Epstein-Barr virus (EBV) by a quantitative real-time PCR assay. Antivir. Res. 62:121-123. [DOI] [PubMed] [Google Scholar]
- 14.Gershburg, E., K. Hong, and J. S. Pagano. 2004. Effects of maribavir and selected indolocarbazoles on Epstein-Barr virus protein kinase BGLF4 and on viral lytic replication. Antimicrob. Agents Chemother. 48:1900-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gershburg, E., and J. S. Pagano. 2002. Phosphorylation of the Epstein-Barr virus (EBV) DNA polymerase processivity factor EA-D by the EBV-encoded protein kinase and effects of the l-riboside benzimidazole 1263W94. J. Virol. 76:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershburg, E., and J. S. Pagano. 2005. Epstein-Barr virus infections: prospects for treatment. J. Antimicrob. Chemother. 56:277-281. [DOI] [PubMed] [Google Scholar]
- 17.Gershburg, E., and J. S. Pagano. 2008. Conserved herpesvirus protein kinases. Biochim. Biophys. Acta 1784:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershburg, E., S. Raffa, M. R. Torrisi, and J. S. Pagano. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 81:5407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granlund, D. J., and G. R. Pearson. 1977. Membrane antigen expression in Epstein-Barr virus-infected Raji cells in the presence of phosphonoacetic acid. Virology 83:217-220. [DOI] [PubMed] [Google Scholar]
- 20.Gustafson, E. A., A. C. Chillemi, D. R. Sage, and J. D. Fingeroth. 1998. The Epstein-Barr virus thymidine kinase does not phosphorylate ganciclovir or acyclovir and demonstrates a narrow substrate specificity compared to the herpes simplex virus type 1 thymidine kinase. Antimicrob. Agents Chemother. 42:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamil, J. P., and D. M. Coen. 2007. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 81:10659-10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, K., A. Yokoyama, Y. Tohya, H. Akashi, Y. Nishiyama, and Y. Kawaguchi. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 84:3381-3392. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1δ. J. Virol. 77:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi, Y., T. Matsumura, B. Roizman, and K. Hirai. 1999. Cellular elongation factor 1δ is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J. Virol. 73:4456-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimberlin, D., and R. Whitley. 2007. Antiviral therapy of HSV-1 and -2, p. 1153-1174. In A. Arvin, A. G. Campadelli-Fiumeeta, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom.
- 27.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krosky, P. M., M. C. Baek, W. J. Jahng, I. Barrera, R. J. Harvey, K. K. Biron, D. M. Coen, and P. B. Sethna. 2003. The human cytomegalovirus UL44 protein is a substrate for the UL97 protein kinase. J. Virol. 77:7720-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudoh, A., T. Daikoku, Y. Ishimi, Y. Kawaguchi, N. Shirata, S. Iwahori, H. Isomura, and T. Tsurumi. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 80:10064-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurokawa, M., S. K. Ghosh, J. C. Ramos, A. M. Mian, N. L. Toomey, L. Cabral, D. Whitby, G. N. Barber, D. P. Dittmer, and W. J. Harrington, Jr. 2005. Azidothymidine inhibits NF-kappaB and induces Epstein-Barr virus gene expression in Burkitt lymphoma. Blood 106:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalezari, J. P., J. A. Aberg, L. H. Wang, M. B. Wire, R. Miner, W. Snowden, C. L. Talarico, S. Shaw, M. A. Jacobson, and W. L. Drew. 2002. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob. Agents Chemother. 46:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, C. P., J. Y. Chen, J. T. Wang, K. Kimura, A. Takemoto, C. C. Lu, and M. R. Chen. 2007. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J. Virol. 81:5166-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, C. P., Y. H. Huang, S. F. Lin, Y. Chang, Y. H. Chang, K. Takada, and M. R. Chen. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 82:11913-11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemon, S. M., L. M. Hutt, and J. S. Pagano. 1978. Cytofluorometry of lymphocytes infected with Epstein-Barr virus: effect of phosphonoacetic acid on nucleic acid. J. Virol. 25:138-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, J. C., E. I. Choi, and J. S. Pagano. 1985. Qualitative and quantitative analyses of Epstein-Barr virus early antigen diffuse component by Western blotting enzyme-linked immunosorbent assay with a monoclonal antibody. J. Virol. 53:793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ljungman, P. 2002. Prevention and treatment of viral infections in stem cell transplant recipients. Br. J. Haematol. 118:44-57. [DOI] [PubMed] [Google Scholar]
- 39.Long, M. C., D. J. Bidanset, S. L. Williams, N. L. Kushner, and E. R. Kern. 2003. Determination of antiviral efficacy against lymphotropic herpesviruses utilizing flow cytometry. Antivir. Res. 58:149-157. [DOI] [PubMed] [Google Scholar]
- 40.Marschall, M., M. Freitag, P. Suchy, D. Romaker, R. Kupfer, M. Hanke, and T. Stamminger. 2003. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 311:60-71. [DOI] [PubMed] [Google Scholar]
- 41.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357-33367. [DOI] [PubMed] [Google Scholar]
- 42.McSharry, J. J., A. McDonough, B. Olson, C. Talarico, M. Davis, and K. K. Biron. 2001. Inhibition of ganciclovir-susceptible and -resistant human cytomegalovirus clinical isolates by the benzimidazole l-riboside 1263W94. Clin. Diagn. Lab. Immunol. 8:1279-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore, S. M., J. S. Cannon, Y. C. Tanhehco, F. M. Hamzeh, and R. F. Ambinder. 2001. Induction of Epstein-Barr virus kinases to sensitize tumor cells to nucleoside analogues. Antimicrob. Agents Chemother. 45:2082-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murata, T., H. Isomura, Y. Yamashita, S. Toyama, Y. Sato, S. Nakayama, A. Kudoh, S. Iwahori, T. Kanda, and T. Tsurumi. 2009. Efficient production of infectious viruses requires enzymatic activity of Epstein-Barr virus protein kinase. Virology 389:75-81. [DOI] [PubMed] [Google Scholar]
- 46.Neuhierl, B., and H.-J. Delecluse. 2006. The Epstein-Barr virus BMRF1 gene is essential for lytic virus replication. J. Virol. 80:5078-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagano, J. S. 1995. Epstein-Barr virus: therapy of active and latent infection, p. 155-195. In D. J. Jefferies and E. De Clercq (ed.), Antiviral chemotherapy. John Wiley & Sons Ltd., New York, NY.
- 48.Pagano, J. S., and A. K. Datta. 1982. Perspectives on interactions of acyclovir with Epstein-Barr and other herpes viruses. Am. J. Med. 73:18-26. [DOI] [PubMed] [Google Scholar]
- 49.Papin, J., W. Vahrson, R. Hines-Boykin, and D. P. Dittmer. 2005. Real-time quantitative PCR analysis of viral transcription. Methods Mol. Biol. 292:449-480. [DOI] [PubMed] [Google Scholar]
- 50.Pearson, G. R., B. Vroman, B. Chase, T. Sculley, M. Hummel, and E. Kieff. 1983. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J. Virol. 47:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prichard, M. N., W. J. Britt, S. L. Daily, C. B. Hartline, and E. R. Kern. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 79:15494-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roizman, B., E. A. Davies, and R. J. Whitley. 2007. Herpes simplex virus, p. 2501-2602. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 54.Romaker, D., V. Schregel, K. Maurer, S. Auerochs, A. Marzi, H. Sticht, and M. Marschall. 2006. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J. Med. Chem. 49:7044-7053. [DOI] [PubMed] [Google Scholar]
- 55.Slifkin, M., S. Doron, and D. R. Snydman. 2004. Viral prophylaxis in organ transplant patients. Drugs 64:2763-2792. [DOI] [PubMed] [Google Scholar]
- 56.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Summers, W. C., and G. Klein. 1976. Inhibition of Epstein-Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J. Virol. 18:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka, M., Y. Nishiyama, T. Sata, and Y. Kawaguchi. 2005. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology 341:301-312. [DOI] [PubMed] [Google Scholar]
- 59.Tarakanova, V. L., V. Leung-Pineda, S. Hwang, C. W. Yang, K. Matatall, M. Basson, R. Sun, H. Piwnica-Worms, B. P. Sleckman, and H. W. Virgin. 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas, M., S. Rechter, J. Milbradt, S. Auerochs, R. Muller, T. Stamminger, and M. Marschall. 2009. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 90:567-578. [DOI] [PubMed] [Google Scholar]
- 61.Trofe, J., L. Pote, E. Wade, E. Blumberg, and R. D. Bloom. 2008. Maribavir: a novel antiviral agent with activity against cytomegalovirus. Ann. Pharmacother. 42:1447-1457. [DOI] [PubMed] [Google Scholar]
- 62.Wang, J. T., S. L. Doong, S. C. Teng, C. P. Lee, C. H. Tsai, and M. R. Chen. 2009. Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. J. Virol. 83:1856-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, L. H., R. W. Peck, Y. Yin, J. Allanson, R. Wiggs, and M. B. Wire. 2003. Phase I safety and pharmacokinetic trials of 1263W94, a novel oral anti-human cytomegalovirus agent, in healthy and human immunodeficiency virus-infected subjects. Antimicrob. Agents Chemother. 47:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, S. L., C. B. Hartline, N. L. Kushner, E. A. Harden, D. J. Bidanset, J. C. Drach, L. B. Townsend, M. R. Underwood, K. K. Biron, and E. R. Kern. 2003. In vitro activities of benzimidazole d- and l-ribonucleosides against herpesviruses. Antimicrob. Agents Chemother. 47:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winston, D. J., J. A. Young, V. Pullarkat, G. A. Papanicolaou, R. Vij, E. Vance, G. J. Alangaden, R. F. Chemaly, F. Petersen, N. Chao, J. Klein, K. Sprague, S. A. Villano, and M. Boeckh. 2008. Maribavir prophylaxis for prevention of cytomegalovirus infection in allogeneic stem cell transplant recipients: a multicenter, randomized, double-blind, placebo-controlled, dose-ranging study. Blood 111:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan, J., E. Cahir-McFarland, B. Zhao, and E. Kieff. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yue, W., E. Gershburg, and J. S. Pagano. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 79:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zacny, V. L., E. Gershburg, M. G. Davis, K. K. Biron, and J. S. Pagano. 1999. Inhibition of Epstein-Barr virus replication by a benzimidazole l-riboside: novel antiviral mechanism of 5,6-dichloro-2-(isopropylamino)-1-β-l-ribofuranosyl-1H-benzimidazole. J. Virol. 73:7271-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, J., G. Liao, L. Shan, J. Zhang, M. R. Chen, G. S. Hayward, S. D. Hayward, P. Desai, and H. Zhu. 2009. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J. Virol. 83:5219-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.