Abstract
The oncogenic microRNA miR-155 is upregulated by several oncogenic viruses. The precursor of miR-155, termed bic, was first observed to cooperate with myc in chicken B-cell lymphomas induced by avian leukosis proviral integrations. We identified another oncogenic retrovirus, reticuloendotheliosis virus strain T (REV-T), that upregulates miR-155 in chicken embryo fibroblasts. We also observed very high levels of miR-155 in REV-T-induced B-cell lymphomas. To study the role of miR-155 in these tumors, we identified JARID2/Jumonji, a cell cycle regulator and part of a histone methyltransferase complex, as a target of miR-155. The overexpression of miR-155 decreased levels of endogenous JARID2 mRNA. We confirmed that miR-155 directly targets both human and chicken JARID2 by assaying the repression of reporters containing the JARID2 3′-untranslated regions. Further, the overexpression of a sponge complementary to miR-155 in a tumor cell line increased endogenous JARID2 mRNA levels. The overexpression of JARID2 in chicken fibroblasts led to decreased cell numbers and an increase in apoptotic cells. The overexpression of miR-155 rescued cells undergoing cytopathic effect caused by infection with subgroup B avian retroviruses. Therefore, we propose that miR-155 has a prosurvival function that is mediated through the downregulation of targets including JARID2.
The expression of microRNA-155 (miR-155), the first identified oncomiR, is mediated by a number of different oncogenic viruses. Retroviral integration into bic, the precursor of miR-155, first was observed in 1989 in avian leukosis virus (ALV)-induced bursal lymphomas, which also had integrations in the myc locus (6). bic also cooperates with myc in chicken lymphomas induced by retroviral vectors expressing both genes (53). The overexpression of miR-155 in B cells of transgenic mice also led to B-cell tumors (8). One of the hallmarks of oncogenes is that they often are incorporated into the genomes of oncogenic viruses and/or are deregulated by viruses. Recently, Epstein-Barr virus has been shown to upregulate miR-155 through the NF-κB pathway (38, 63). In addition, the Kaposi's sarcoma herpes virus (KSHV) and Marek's disease virus (MDV) have been shown to encode homologs of miR-155, called miR-K12-11 (KSHV) (19, 50) and MDV-miR-M4 (MDV) (37, 65), which share the important seed sequence with miR-155.
The expression of many oncogenes is deregulated as a result of insertional mutagenesis (6, 43, 46, 62). However, tumors can arise because of either the amplified expression of oncogenes or the decreased expression of tumor suppressors. For a tumor suppressor to be identified by insertional mutagenesis, both alleles need to be deregulated, which is unlikely. Therefore, it is likely that microRNAs upregulated in tumors target and repress the expression of tumor suppressors.
miR-155 is overexpressed in many human tumors, including many Hodgkin's lymphomas (12, 14, 18, 23, 27, 58), which also are characterized by the amplification of the c-rel locus (17). We investigated whether reticuloendothelosis virus strain T (REV-T), which expresses the viral homolog of Rel, v-rel (21), could induce miR-155 expression. It has been shown that v-Rel, which is a member of the NF-κB family (16), regulates downstream genes through the activation of the AP-1 transcription factor family (28). miR-155 expression also is regulated by the AP-1 (64) and NF-κB (56) transcription factor families. We observed that infecting chicken embryo fibroblasts (CEFs) with the REV-T virus led to the overexpression of both miR-155 and its precursor RNA. We also observed that miR-155 is overexpressed to high levels in v-Rel-induced B-cell lymphomas.
miR-155 is expressed in normal lymphoid and hematopoietic tissues in chickens and humans, especially in activated immune cells. Most of the validated targets of miR-155, such as c-Maf (45), AID (10, 54), Pu.1 (57), SOCS1 (34), interleukin-1 (4), and IKKɛ (33, 56), have been implicated in mediating functions of miR-155 in the immune system. It also has been shown that the induction of miR-155 negatively regulates normal myelopoiesis and erythropoiesis (15). Other targets of miR-155, like Ets-1, a protooncogene, and Meis1 mediate megakaryopoiesis (42). A few targets of miR-155, like SHIP1 and C/EBP, have been implicated in myeloproliferative disorders (7, 40), and tumor protein p53 inducible nuclear protein 1 (Tp53INP1) is involved in pancreatic cancer (18).
To further investigate the mechanism of miR-155 action in these virus-induced tumors, we identified and validated JARID2, a cell cycle regulator (25, 51) and part of a histone methyltransferase complex (49), as a direct target of miR-155. The two target sequences in the 3′-untranslated regions (3′UTR) of JARID2 are highly conserved evolutionarily. We used a retrovirus-driven miR-155 sponge to sequester miR-155 in a REV-T-induced B-cell tumor line and observed the upregulation of endogenous JARID2 mRNA. Furthermore, the overexpression of JARID2 led to apoptosis in CEFs. Surprisingly, we observed that miR-155 can rescue cells from cytotoxicity due to infection with subgroup B avian retroviruses, which activate the tumor necrosis factor death receptor (3, 9, 59). Thus, we propose that miR-155 has a prosurvival function mediated through the inactivation of targets including JARID2. This may be important in oncogenesis involving oncogenes that can induce apoptosis, such as Myc (13).
MATERIALS AND METHODS
Plasmids.
Exon 2a of the chicken bic gene (accession number AF182318) was cloned into the pcDNA3.1(+) (Invitrogen) expression vector (referred to as pBIC) and into the RCAS(A) retroviral vector (22) [referred to as RCAS(A)miR-155]. The entire 3′UTR of target genes were amplified by PCR from genomic DNA and cloned into pMIR-report (Ambion) downstream of the luciferase open reading frame (ORF). The 3′UTR of human JARID2 was cloned into psiCHECK2 (Promega) downstream of the Renilla Luciferase ORF. miR-155 target sites (nucleotides [nt] 3 to 5) were mutated by QuikChange mutagenesis in these constructs.
To inhibit miR-155 function in tumor cell lines, we constructed a miR-155 sponge. Eight copies of the sequence 5′CCC CTA TCA CCT AAA GCA TTA A GTCGAC A 3′, complementary to miR-155 and with a bulge from nt 9 to 12, was inserted into the 3′UTR of either enhanced green fluorescent protein (eGFP) or MS2 expression vectors. The eGFP-miR-155 sponge was also cloned into the RCAS(A) retroviral vector at the ClaI site. The directionality of the insert was verified by PCR and sequencing.
A full-length clone of JARID2, in the pCMV-Sport 6 vector, was obtained from the ATCC (catalog number 9121501).
Cells and transfection.
Secondary CEFs were cultured in medium 199 (Invitrogen) with 2% tryptose phosphate broth (Sigma), 1% chick serum, 1% calf serum, and 1% antibiotics. KBMC cells (31) and CM758 cells (47), which are B-cell tumor lines that were derived from v-rel induced tumors, and BK3A cells, S13 cells, H1 cells, and S234S4 cells (32, 48), which are B-cell tumor lines that were derived from ALV integrations, were cultured in RPMI 1640 (GIBCO) with 10% tryptose phosphate, 5% calf serum, 2% chick serum, and 1% antibiotics.
CEFs were transfected by electroporation or DEAE-dextran transfection. Briefly, 1.5 × 106 cells were resuspended in OPTI-MEM (GIBCO) and placed in a 4-mm cuvette. Cells were electroporated using a BTX electroporator set to 240 V, 1,500 μF, and 129 Ω. CEFs and KBMC were transfected with dextran as described elsewhere (30).
Viruses.
RCAS(B) and RCAS(B)BIC [referred to as RCAS(B)miR-155] are described elsewhere (53). Viral stocks were frozen at −80°C and briefly thawed before infection. Infections were verified by a reverse transcriptase (RT) assay, which has been described elsewhere (26).
Plasmids containing RCAS(A), RCAS(A)miR-155, RCAS(A)eGFP, and RCAS(A)eGFP-miR-155-sponge were transfected separately into CEFs by electroporation. Cells were cultured for 1 week before virus production was confirmed by RT assay. Two weeks after transfection, medium was collected and centrifuged to remove cell debris. Half of the medium was frozen at −80°C. The remaining half was ultracentrifuged at 23,000 rpm in a Beckman SW27 rotor for 2 h to pellet virus. Viral RNA was isolated from virus particles using RNA Bee (Tel-Test) per the manufacturer's protocol. The presence of eGFP and miR-155-sponge in viral particles was verified by RT-PCR.
REV-T virus was harvested from conditioned medium of CM758 cells.
Luciferase assay.
Luciferase activity was quantified using the Luciferase assay system (Promega) according to the manufacturer's protocols. Briefly, 2.5 × 105 CEFs were seeded in a 12-well plate the day before transfection. Eighty nanograms of Luciferase and 40 ng of Renilla plasmids were used in all transfections. miR-155 was expressed by transfecting pBIC at various amounts from 80 to 4,000 ng. Two days after transfection, cells were lysed in 300 μl passive lysis buffer.
Splinted ligation.
miR-155 levels were assayed by splinted ligation, a DNA-mediated RNA ligation, as described previously (35). The sequence of the bridge oligonucleotide is 5′ GAA TGT CAT AAC CGT ACC CTA TCA CTA TTA GCA TTA A 3′, and the sequence of the ligation oligonucleotide is 5′ ACG GTT ATG ACA TTC 3′.
RNase protection assay.
In vitro transcription of the bic probe was carried out as described elsewhere (30). RNA isolation and RNase protection assays were done as described previously (30). In brief, total cellular RNA was harvested using RNA-Bee (Tel-Test) per the manufacturer's instructions. RNA was resuspended in RNase digestion buffer and hybridized with probe overnight at 37°C. RNA then was digested with 10 U/ml RNase T1 (Calbiochem) and 5 μg/ml RNase A (Calbiochem) at 30°C. After 45 min, the digestion was stopped by the addition of sodium dodecyl sulfate and Proteinase K (Roche) and incubated at 37°C for 15 min. RNA was extracted with phenol-chloroform-isoamyl alcohol, precipitated with ethanol, resuspended in 95% formamide dye, and denatured for 5 min at 95°C. Samples were loaded onto a 15% acrylamide, 8 M urea gel and electrophoresed at 15 W for 2 to 4 h. RNA levels were quantified using a Typhoon 9410 PhosphorImager (Amersham).
Real-time qRT-PCR.
Total RNA was extracted from cells using RNA Bee per the manufacturer's protocol. For quantitative reverse transcription-PCR (qRT-PCR), 1 μg of total RNA was reverse transcribed using Random Hexamers (Integrated DNA Technologies) and Moloney murine leukemia virus RT (Promega) in the presence of RNase inhibitor (Promega). Ten percent of each reverse transcription reaction mixture was used for PCR amplification on a Bio-Rad iCycler using iTaq SYBR green mix (Bio-Rad). Primer pairs were designed in exons flanking an intron of at least 5 kb in size to eliminate genomic DNA amplification. The efficiency of each primer pair was optimized and validated relative to results for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control primers.
Cell cycle analysis by fluorescence-activated cell sorting.
Transfected cells were trypsinized, pelleted, and washed with 1× phosphate-buffered saline (PBS). Cells then were fixed in 70% ethanol and stored at −20°C. On the day of analysis, fixed cells were pelleted and washed with cold 1× PBS. Cells were resuspended in 1 μg/ml propidium iodide (PI; Sigma) and 2 μg/ml RNase A (Calbiochem). Cells were incubated in the dark at room temperature for 15 min and then analyzed on a FACS Calibur (Becton Dickinson). Cells were sorted on forward scatter and side scatter. All live cells, as determined by scatter plots, were gated for GFP expression. GFP-positive cells were gated for PI fluorescence to analyze the cell cycle profile.
RESULTS
miR-155 was upregulated in v-rel-derived B-cell lymphomas and by REV-T.
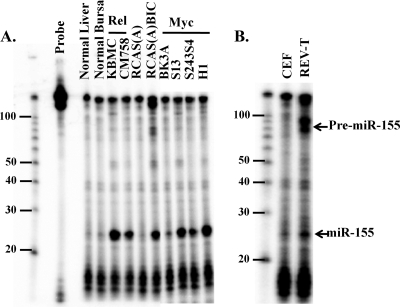
Retroviruses induce tumors either by overexpressing an oncogene encoded in the retroviral genome or by integrating into cellular genomic loci and deregulating the expression of a host protooncogene. It has been shown previously that long-latency tumors resulting from ALV infection have clonal integrations into the c-myc and bic loci (6). The retrovirus REV-T encodes the oncogene v-rel and also induces B-cell lymphomas (60). Since ALV-induced tumors have upregulated miR-155, we investigated whether REV-T-derived lymphomas also have deregulated miR-155 expression. As shown in Fig. 1A, miR-155 was upregulated in KBMC and CM758 cell lines derived from REV-T-induced tumors (31, 47) compared to levels for the normal bursa control. miR-155 levels were fourfold higher in KBMC cells than in Hodgkin's lymphoma cell lines (12) overexpressing miR-155 (data not shown). miR-155 also was upregulated in three out of four cell lines from ALV-induced tumors that have been shown previously to have integrations into the c-myc locus (Fig. 1A) (32, 48). For comparison, we show levels of miR-155 expression in cells infected with RCAS(A)miR-155 and the control RCAS(A) (Fig. 1A).
FIG. 1.
miR-155 is upregulated in v-rel- and c-myc-induced B-cell lymphomas and by REV-T. (A) Total RNA from cells and tissues was harvested, and miR-155 levels were assayed by RNase protection. KBMC and CM758 are v-rel-derived tumor cell lines. CEFs were infected with RCAS(A) and RCAS(A)miR-155. BK3A, S13, S234S4, and H1 are c-myc-induced cell lines. miR-155 levels were compared to those for RCAS(A) and normal liver and bursa controls. (B) CEFs were infected with REV-T virus harvested from CM758 cells. Total RNA was harvested after confirming positive infection by RT assay, and miR-155 levels were assayed by RNase protection.
To further investigate if increased levels of miR-155 in v-rel-derived cell lines were REV-T mediated, REV-T virus produced from cell line CM758 was used to infect CEFs in culture. Ten days after infection, positive infection was confirmed by RT assay, total RNA was isolated, and miR-155 levels were assayed by RNase protection. miR-155 levels were upregulated 2.4-fold compared to levels for uninfected controls; however, the levels of pre-miR-155 were upregulated more than 20-fold in the REV-T infections (Fig. 1B). This indicates that REV-T induces the transcription of the bic gene and perhaps the microRNA-processing system is overwhelmed in CEFs, which could explain the greatly increased amount of pre-miR-155.
Jumonji/JARID2 3′UTR has miR-155 target sites that are highly conserved evolutionarily.
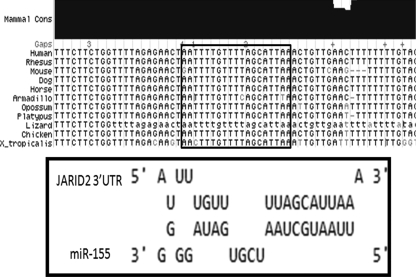
Since miR-155 is upregulated in retrovirus-induced B-cell lymphomas along with many other human cancers, it is important to identify mRNA targets of miR-155 that might play a role in tumorigenesis. Using miRBASE (20) and RNA hybrid (44) target prediction programs, JARID2 was identified as a putative miR-155 target. There is remarkable conservation of the JARID2 3′UTR sequence from humans to chickens within and around the first predicted miR-155 target site (Fig. 2). This site also is conserved across multiple species, from humans to Xenopus, as shown by the multiple sequence alignment (Fig. 2). Interestingly, the chicken 3′UTR of JARID2 has only one predicted miR-155 target, whereas there are two in human, mouse, rhesus monkey, and dog. All of these species share the site that is 104 nt from the stop codon (Fig. 2), but the chicken 3′UTR is smaller than the other 3′UTRs by 900 bp, as annotated in PubMed. However, a BLAT analysis of the second target sequence in the human JARID2 3′UTR revealed that this sequence also is present downstream of the chicken JARID2 annotated 3′ end. Thus, both miR-155 predicted target regions are very well conserved across species, including chicken.
FIG. 2.
JARID2 3′UTR has conserved (Cons) miR-155 target sites. The miR-155 target site was predicted using RNAhybrid (44) and miRBase (20). The miR-155 target site was mapped using the UCSC BLAT genome browser, and the corresponding Vista plot showing conservation across species is displayed. X_tropicalis, Xenopus tropicalis.
Overexpression of miR-155 downregulated endogenous JARID2 mRNA in infected CEFs.
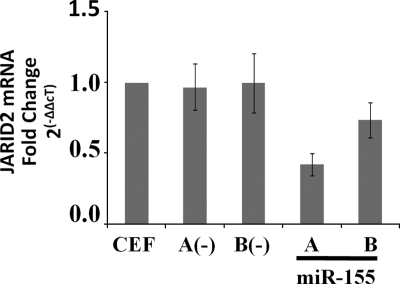
Since JARID2 has conserved target sequences for miR-155, we first investigated whether endogenous levels of JARID2 were affected by the overexpression of miR-155. CEFs were infected with control viruses [RCAS(A) and RCAS(B)] and miR-155 overexpression viruses [RCAS(A)miR-155 and RCAS(B)miR-155]. Infection was verified by RT assay, and total cellular RNA was harvested at least 10 days after infection. miR-155 levels were assayed by RNase protection (data not shown), and JARID2 mRNA levels were assayed by real-time RT-PCR. Interestingly, the overexpression of miR-155 with RCAS(A)miR-155 downregulated JARID2 mRNA to 40% of the level in uninfected and control infected cells (Fig. 3). A lower decrease in JARID2 mRNA levels was seen with RCAS(B)miR-155. These results are similar to those of previous reports of microRNA-dependent mRNA degradation in other systems (1). However, since vertebrate microRNA-mediated silencing is not thought to involve endonucleolytic cleavage, miR-155-mediated JARID2 mRNA degradation probably is carried out by mRNA degradation machinery (41).
FIG. 3.
JARID2 mRNA is downregulated by miR-155 in CEFs. CEFs were infected with RCAS(A), RCAS(B), RCAS(A)miR-155, and RCAS(B)miR-155 and passaged for 10 days. Total RNA was harvested, and JARID2 mRNA levels were determined by qRT-PCR and normalized to results for GAPDH mRNA. Relative changes (n-fold) were calculated by the ΔΔCT method; data represent means ± standard deviations from five experiments.
Reporters with JARID2 3′UTR were repressed by miR-155.
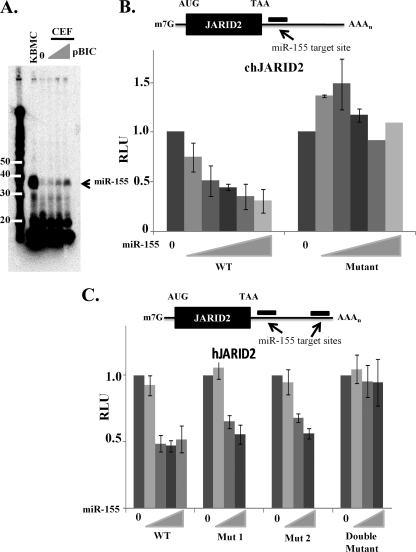
To test whether JARID2 is directly targeted by miR-155, the 3′UTR of both chicken and human JARID2 mRNAs were cloned into firefly luciferase expression vectors. Luciferase levels were assayed in the presence of various concentrations of miR-155 expressed from plasmid pBIC. Levels of miR-155 were assayed using a technique called splinted ligation, which involves the ligation of a radioactive DNA oligonucleotide (14 nt) to the microRNA. This caused miR-155 to migrate as 35 nt on polyacrylamide gel electrophoresis (Fig. 4A). Luciferase-3′UTR, Renilla luciferase, pBIC, and vector control plasmids were transfected into CEFs with DEAE-dextran. Transfections were always controlled for the total amount of DNA. Firefly luciferase levels were assayed 3 days posttransfection and normalized to cotransfected control Renilla luciferase.
FIG. 4.
JARID2 3′UTR is sensitive to miR-155 in a dose-dependent manner. (A) Splinted ligation to assay levels of miR-155 expressed from pBIC. Based on target prediction programs, the chicken JARID2 3′UTR contains one target sequence (B) and the human JARID2 3′UTR contains two target sequences (C). The entire 3′UTR of JARID2 mRNA was cloned downstream of firefly luciferase (WT). Cells (5 × 105) were cotransfected with 80 ng of firefly luciferase plasmid, 40 ng of renilla luciferase plasmids, and increasing amounts of pBIC (80 to 4,000 ng). Ratios on the x axis indicate the amount of reporter vector to microRNA vector. pcDNA3.1(+) was used as a filler to keep the total amount of DNA per transfection constant. Relative luciferase units (RLU) is a ratio of firefly luciferase values normalized to renilla luciferase values; data represent means ± standard deviations from six experiments.
The chicken 3′UTR was sensitive to miR-155; luciferase activity was downregulated in a miR-155 dose-dependent manner, from 20% at a low concentration to about 80% at higher concentrations (Fig. 4B). To be certain that this repression was dependent on miR-155, we mutated three nucleotides (nt 3 to 5) in the 3′UTR target site that are complementary to the miR-155 seed sequence. As expected, this completely abrogated miR-155-mediated repression (Fig. 4B).
Similar experiments also were performed with the 3′UTR of human JARID2 mRNA. The human JARID2 3′UTR has two predicted miR-155 binding sites. Similarly to the chicken 3′UTR, the human 3′UTR was sensitive to miR-155 in a dose-dependent manner (Fig. 4C). To demonstrate that this was a direct effect, both putative binding sites for miR-155 were mutated (nt 3 to 5), generating both single and double mutants. The single mutants both still were responsive to miR-155 but at slightly higher concentrations than those of the wild-type 3′UTR (Fig. 4C). The double mutant was completely unresponsive to miR-155, indicating that both sites in the JARID2 3′ UTR are direct targets of miR-155 (Fig. 4C).
Endogenous JARID2 was repressed by miR-155 in KBMC cells.
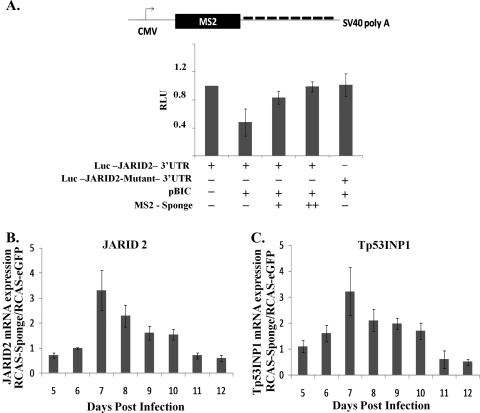
In order for a target to be regulated by miR-155, the transcription of this target must take place in the same cell in which miR-155 is expressed. Therefore, it was necessary to investigate the effects of endogenous miR-155 on endogenous mRNA levels of targets like JARID2. Hence, we wanted to look at levels of JARID2 in cell types endogenously overexpressing miR-155, like KBMC. It was difficult to transfect these B-cell lines with antagomirs (chemically modified anti-miRs) with reasonable efficiency to knock down miR-155 function. Hence, we decided to construct a miR-155 sponge, with the idea that sequestering endogenous miR-155 could alleviate the repression of endogenous targets and prevent their degradation (11).
Eight miR-155 target sites with a bulge from nt 9 to 12 were cloned into the 3′UTR of the MS2 viral coat protein. We first investigated the functionality of the miR-155 sponge by the luciferase assay. In the presence of miR-155, luciferase levels of JARID2 3′UTR were reduced. With increasing amounts of miR-155 sponge, miR-155 function was inhibited and the repression of luciferase JARID2 3′UTR was abrogated (Fig. 5A).
FIG. 5.
Inhibition of JARID2 and Tp53INP1 mRNA was abrogated in the presence of miR-155 sponge. (A) Eight bulged targets sites of miR-155 were cloned into the 3′UTR of MS2 coat protein. CEFs were transfected with 80 ng of firefly luciferase, 40 ng of renilla luciferase, and 500 ng of pBIC. Two different concentrations of MS2-sponge plasmid were transfected (400 and 800 ng). pcDNA3.1(+) was used as a filler to keep the total amount of DNA per transfection constant. Relative luciferase units (RLU) is a ratio of firefly luciferase values normalized to renilla luciferase values; data represent means ± standard deviations from three experiments. CMV, cytomegalovirus; SV40, simian virus 40. (B and C) KBMC cells (1 × 105) were infected with either RCAS(A)eGFP or RCAS(A)eGFP-sponge virus. Cells then were passaged continuously and total RNA harvested on the indicated days. (B) JARID2 mRNA and (C) Tp53INP1 mRNA were assayed by qRT-PCR and normalized to GAPDH mRNA results. Differences (n-fold) were calculated as a ratio of mRNA levels in RCAS(A)eGFP-sponge-infected cells to RCAS(A)eGFP-infected control cells; data represent means ± standard deviations from five experiments.
Since KBMC cells were difficult to transfect, RCAS(A) retroviral vectors overexpressing eGFP-sponge were generated. KBMCs were infected with either RCAS(A)eGFP or RCAS(A)eGFP-miR-155-sponge on day 0. Total RNA was collected every day starting 5 days postinfection. Both JARID2 and Tp53INP1 (another validated miR-155 target [18]) mRNA levels were assayed by qRT-PCR, using GAPDH as a control. All experimental samples were performed in duplicate and normalized to GAPDH, and changes (n-fold) were calculated by the ΔΔCT method (61). The ratios of RCAS(A)eGFP-sponge to RCAS(A)eGFP were calculated. Endogenous levels of JARID2 and Tp53INP1 mRNA increased in the presence of the miR-155 sponge, indicating that they are physiologically relevant targets of miR-155 (Fig. 5B and C). However, a decreased effect was seen after day 7 of infection. We think endogenous miR-155 is targeting the miR-155 sponge and downregulating retroviral production.
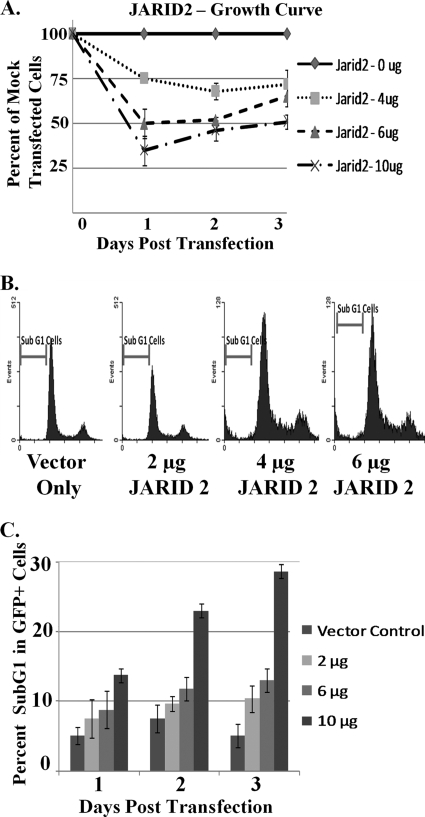
JARID2 decreased cell numbers when overexpressed in CEFs.
JARID2 has been shown to be a transcriptional repressor that potentiates the effect of the tumor suppressor retinoblastoma-mediated repression of E2F responsive genes (25, 51). We propose that miR-155 mediates its oncogenic effects in part by the downregulation of JARID2. This would partly alleviate the repression of E2F genes and increase their cycling capability. To test this hypothesis, we overexpressed JARID2 in CEFs and counted the total cell numbers. The overexpression of JARID2 protein was confirmed by Western blotting with an anti-hJARID2 antibody (data not shown). As expected, the total cell numbers decreased with increasing amounts of JARID2 (Fig. 6A), indicating a block in the cell cycle or an increase in cell death.
FIG. 6.
Overexpression of JARID2 decreased cell number through apoptosis. CEFs were transfected with various amounts of JARID2 as indicated. The total amount of DNA was kept constant by using pCMV-SPORT as a control vector. (A) Upon transfection, cells were split equally into three 12-mm dishes. Cells were counted each day using a hemacytometer; data represent means ± standard deviations from six experiments. (B) Along with JARID2, cells were cotransfected with an eGFP expression vector. On the day of analysis, cells were fixed and treated with RNase A and PI. GFP and cells were analyzed for cell cycle profile by PI staining. The number of cells in sub-G1, G1, S, and G2/M phase were counted. (C) Representation of GFP+ sub-G1 cells with increasing amounts of JARID2. Data are represented as the percentage of GFP+ cells (transfected cells); data represent means ± standard deviations from four experiments.
To investigate the reason for decreased cell numbers, cell cycle analysis by PI staining followed by flow-cytometric analysis was performed. Briefly, CEFs were transfected with increasing amounts of JARID2 together with 1 μg of GFP and control vector, keeping the total amount of DNA constant throughout the experiment. Cells were fixed, treated with RNase A, and stained with PI every day posttransfection for 3 days. Cells were sorted on a BD FACS Calibur, and 20,000 events were collected. All dead cells were identified by gating out small cells on a side scatter versus forward scatter plot. Single (nonclumped) cells were identified and gated for on an FL2-A versus FL2-W plot, with FL2 measuring PI fluorescence. To select for transfected cells, all cells expressing GFP at a level 10-fold higher than that of mock-transfected cells were gated. The PI fluorescence of these cells was plotted and analyzed by gating specific peaks (Fig. 6B). Interestingly, there was no change in the G1 (57% ± 1.2%), S (9.1% ± 2.2%), or G2/M (14.2% ± 4.2%) phases of the cell cycle upon the overexpression of JARID2, suggesting that the growth rate was not changed, contrary to what had been reported previously for other systems (25, 51). However, we observed an increase in apoptotic cells upon the overexpression of JARID2, as identified by the sub-G1 cell population (Fig. 6C). Therefore, JARID2 overexpression promotes cell apoptosis, and miR-155 likely plays an oncogenic role by downregulating JARID2, along with other possible targets.
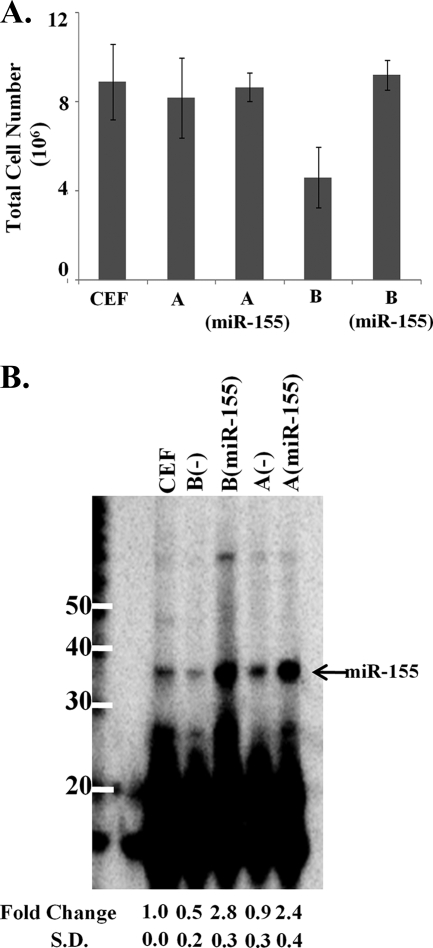
miR-155 abrogated cytopathic effects of infection with subgroup B retroviruses.
RCAS(B) is a subgroup B retroviral vector derived from the Rous sarcoma virus Schmidt-Ruppin strain that is used to overexpress genes in avian cells (22). We used this expression vector to express miR-155 from the bic exon 2a in CEFs, as described in earlier studies (53). Seven days after infection, the control plates infected with RCAS(B) had about half the number of cells as either the uninfected plates or those infected with a subgroup A vector, RCAS(A) (Fig. 7A). It has been reported previously that infection with subgroup B retroviruses results in cytopathic effects correlated with high levels of unintegrated proviral DNA (59). The binding of the envelope protein of subgroup B viruses to its receptor (tumor necrosis factor alpha) has been shown to trigger a cascade that decreases the cell number of infected cells as well as bystander cells (3, 9) and results in apoptosis with condensed nuclei in the absence of NF-κB synthesis (5).
FIG. 7.
RCAS(B) viral infection leads to decreased cell growth and a decrease in miR-155. (A) CEFs were infected with RCAS(A) and RCAS(B) retroviral vectors, plus retroviral vectors expressing miR-155, and passaged for 7 days. After confirming positive infection by an RT assay, cells were counted using a hemacytometer. Data represent means ± standard deviations from six experiments. (B) miR-155 levels in CEFs infected with corresponding viruses were assayed by splinted ligation and relative intensity determined by PhosphorImager analysis. Data represent means ± standard deviations from six experiments.
Interestingly, the overexpression of miR-155 using the RCAS(B) retroviral vector resulted in the complete abrogation of this virus-induced cytopathic effect, restoring the total cell number to the levels of uninfected cells and those infected with the noncytopathic subgroup A retrovirus (Fig. 7A). To further assess cell proliferation, 1 × 106 infected cells and controls were plated and counted each day for 3 days. Cells infected with RCAS(B) appeared to grow more slowly than uninfected controls (data not shown). However, overexpressing miR-155 restored the proliferative capacity, and the cells grew at the same rate as the controls. When miR-155 was expressed from either RCAS(A) or RCAS(B) vector, its expression was threefold higher than the endogenous level in uninfected CEFs (Fig. 7B).
We also found that endogenous miR-155 levels were decreased twofold in cells infected with the RCAS(B) vector alone compared to that of uninfected cells (Fig. 7B). It is possible that miR-155 plays a protective role in the survival of uninfected cells. This may be consistent with previously published results that the overexpression of miR-155 is oncogenic (8, 53). We propose that miR-155 mediates its effects by downregulating an apoptotic pathway or otherwise promoting cell survival.
DISCUSSION
miR-155 is upregulated in retrovirus-induced B-cell lymphomas.
miR-155 was first identified as a second locus, in addition to c-myc, of ALV integrations in bursal lymphomas (6). These integrations were clonal, suggesting that they were important for the development of the tumor. We identified two different B-cell lines derived from REV-T induced B-cell lymphomas overexpressing miR-155 (Fig. 1A) and also observed that REV-T induced miR-155 expression in infected CEFs (Fig. 1B). REV-T encodes the oncogene v-rel (31), which is the cellular homolog of c-rel that is amplified in 50% of Hodgkin's lymphomas (17). miR-155 also is upregulated in Hodgkin's lymphoma (12), and therefore it is probable that miR-155 is regulated by Rel in Hodgkin's lymphoma. It has been shown previously that v-rel exerts downstream effects through the transcription factor AP-1 (28). B-cell receptor activation also leads to an increase in bic/miR-155 transcript levels through AP-1 (64). Therefore, in both v-rel-derived chicken B-cell lymphomas and human Hodgkin's lymphoma, miR-155 likely is upregulated due to the overexpression of the oncogene Rel through downstream transcription factors such as AP-1.
miR-155 downregulated endogenous JARID2 mRNA in tumors.
The overexpression of miR-155 led to decreased endogenous JARID2 mRNA levels, as well as a decrease in reporter protein levels. Both the chicken and human JARID2 3′UTRs were sensitive to miR-155, indicating that the regulation of JARID2 by miR-155 was conserved. The 3′UTR region around the miR-155 target sites also was very well conserved across species (Fig. 2), indicating that these regions probably are important in JARID2 regulation. The inhibition of miR-155 using a retroviral sponge increased JARID2 mRNA levels in v-rel-derived chicken B-cell lymphomas, indicating a potential role for JARID2 in suppressing tumorigenesis, along with other known targets of miR-155, like Tp53INP1 (18), SHIP (7, 40), and C/EBP (7). Since JARID2 has been shown to be part of a histone methyltransferase complex (49), inhibiting JARID2 could lead to an epistatic change in the tumor progenitor cells.
miR-155 suppresses apoptosis and cooperates with Myc to induce cell proliferation.
The infection of CEFs with the subgroup B retrovirus has been shown to cause cytopathic effects in cells during acute infection (59). We and others have observed that chronically infected RCAS(B) cells grow much slower than uninfected cells and/or die in culture (59). The binding of the ALV-B envelope protein to its receptor activates cell death in the absence of NF-κB synthesis (5). However, when miR-155 was overexpressed using the same RCAS(B) virus, this induction of cell death and decrease in cell number was abrogated. Interestingly, miR-155 is downregulated in RCAS(B) cells compared to levels in uninfected cells. Therefore, the repression of JARID2 along with other proapoptotic targets, like BACH-1, Bim, LDOC1 (50), and IKKɛ (33, 56) by miR-155 may protect against RCAS(B)-induced apoptosis.
It has been shown previously that miR-155 cooperates with Myc in oncogenesis (6, 53). However, the reason for this cooperative effect has not been elucidated. It has been shown that JARID2 potentiates the retinoblastoma repression of E2F-responsive genes, such as Cyclin D1, D2, and D3 (25), and that it acts in a histone methyltransferase complex (49). Interestingly, c-Myc exerts control over the cell cycle by regulating these same genes either transcriptionally or by enhancing the functional complex Cyclin D-CDK4 (36). However, the deregulated expression of myc not only promotes cell proliferation but also apoptosis (13), which is thought to act as a safeguard protecting the organism from tumors that might otherwise arise as a consequence of a mutation at a myc gene locus (2). Since JARID2 and Myc both regulate Cyclin D as well as many other genes, it is possible that JARID2 is antagonizing Myc and preventing Myc-induced oncogenesis. In this study, we see predominantly the proapoptotic effects of JARID2 when overexpressed in CEFs. Since c-Myc also can induce apoptosis (13), miR-155 may cooperate with c-myc by suppressing JARID2-mediated apoptosis. This would increase the capacity of c-myc to increase cell proliferation, which ultimately could lead to tumor formation.
miR-155 may inhibit apoptosis in germinal centers resulting in Hodgkin's lymphoma.
B cells mature into plasma/effector B cells in the dark zone of germinal centers. Here, upon activation, B-cell receptors (BCR) are subject to hypermutation and class switching from immunoglobulin M to other immunoglobulins. Upon hypermutation, cells undergo selection for the presence of a functional BCR. Functional BCR expressing cells class switch and divide and enter the circulation. B cells with unfavorable mutations resulting in nonfunctional BCR undergo apoptosis (55). Hodgkin's lymphoma cells are characterized by Hodgkin and Reed-Sternberg cells (29). These cells are defective B cells that have escaped the negative selection because they acquired other mutations to suppress apoptosis, like amplifications of the c-rel locus (24) and FAS gene mutations (39). It is possible that some of these mutations lead to an upregulation of miR-155 and, therefore, the downregulation of JARID2 and other proapoptotic targets. This could lead to an inhibition of the apoptotic pathway and an increase in the tumorigeneic potential of these B cells.
Acknowledgments
This work was supported by NIH grant R01 CA124596 to K.L.B.
We thank Michael Edidin for discussions and suggestions, Maxine Linial for BK3A, H1, S13, and S243S4 cell lines, Karel Schat for the CM758 cell line, and Stephen Hughes for the RCASBP(A) plasmid. We also thank Johanna Withers for the review of the manuscript and Yingying Li for technical assistance.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553-563. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard, C., P. Staller, and M. Eilers. 1998. Control of cell proliferation by Myc. Trends Cell Biol. 8:202-206. [DOI] [PubMed] [Google Scholar]
- 3.Brojatsch, J., J. Naughton, H. B. Adkins, and J. A. T. Young. 2000. TVB receptors for cytopathic and noncytopathic subgroups of avian leukosis viruses are functional death receptors. J. Virol. 74:11490-11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceppi, M., P. M. Pereira, I. Dunand-Sauthier, E. Barras, W. Reith, M. A. Santos, and P. Pierre. 2009. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 106:2735-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi, Y., F. Diaz-Griffero, C. Wang, J. A. T. Young, and J. Brojatsch. 2002. An NF-κB-dependent survival pathway protects against cell death induced by TVB receptors for avian leukosis viruses. J. Virol. 76:5581-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clurman, B. E., and W. S. Hayward. 1989. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 9:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costinean, S., S. K. Sandhu, I. M. Pedersen, E. Tili, R. Trotta, D. Perrotti, D. Ciarlariello, P. Neviani, J. Harb, L. R. Kauffman, A. Shidham, and C. M. Croce. 2009. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein β are targeted by miR-155 in B cells of Eμ-miR-155 transgenic mice. Blood 114:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costinean, S., N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, and C. M. Croce. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 103:7024-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz-Griffero, F., S. A. Hoschander, and J. Brojatsch. 2003. Bystander killing during avian leukosis virus subgroup B infection requires TVBS3 signaling. J. Virol. 77:12552-12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsett, Y., K. M. McBride, M. Jankovic, A. Gazumyan, T.-H. Thai, D. F. Robbiani, M. Di Virgilio, B. R. San-Martin, G. Heidkamp, T. A. Schwickert, T. Eisenreich, K. Rajewsky, and M. C. Nussenzweig. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-IgH translocation. Immunity 28:630-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert, S. M., R. J. Neilson, and P. A. Sharp. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4:721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eis, P. S., W. Tam, L. Sun, A. Chadburn, Z. Li, M. F. Gomez, E. Lund, and J. E. Dahlberg. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 102:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 14.Fulci, V., S. Chiaretti, M. Goldoni, G. Azzalin, N. Carucci, S. Tavolaro, L. Castellano, A. Magrelli, F. Citarella, M. Messina, R. Maggio, N. Peragine, S. Santangelo, F. R. Mauro, P. Landgraf, T. Tuschl, D. B. Weir, M. Chien, J. J. Russo, J. Ju, R. Sheridan, C. Sander, M. Zavolan, A. Guarini, R. Foa, and G. Macino. 2007. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 109:4944-4951. [DOI] [PubMed] [Google Scholar]
- 15.Georgantas, R. W., R. Hildreth, S. Morisot, J. Alder, C.-g. Liu, S. Heimfeld, G. A. Calin, C. M. Croce, and C. I. Civin. 2007. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA 104:2750-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore, T. D. 1999. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene 18:6925-6937. [DOI] [PubMed] [Google Scholar]
- 17.Gilmore, T. D., D. Kalaitzidis, M.-C. Liang, and D. T. Starczynowski. 2004. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene 23:2275-2286. [DOI] [PubMed] [Google Scholar]
- 18.Gironella, M., M. Seux, M. J. Xie, C. Cano, R. Tomasini, J. Gommeaux, S. Garcia, J. Nowak, M. L. Yeung, K. T. Jeang, A. Chaix, L. Fazli, Y. Motoo, Q. Wang, P. Rocchi, A. Russo, M. Gleave, J. C. Dagorn, J. L. Iovanna, A. Carrier, M. J. Pebusque, and N. J. Dusetti. 2007. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA 104:16170-16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J.-T. A. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34:D140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoelzer, J. D., R. B. Franklin, and R. H. Bose, Jr. 1979. Transformation by reticuloendotheliosis virus: development of a focus assay and isolation of a nontransforming virus. J. Virol. 93:20-30. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio, M. V., M. Ferracin, C.-G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini, and C. M. Croce. 2005. MicroRNA gene expression deregulation in humans. Breast Cancer Res. 65:7065-7070. [DOI] [PubMed] [Google Scholar]
- 24.Joos, S., C. K. Menz, G. Wrobel, R. Siebert, S. Gesk, S. Ohl, G. Mechtersheimer, L. Trumper, P. Moller, P. Lichter, and T. F. E. Barth. 2002. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 99:1381-1387. [DOI] [PubMed] [Google Scholar]
- 25.Jung, J., T.-G. Kim, G. E. Lyons, H.-R. C. Kim, and Y. Lee. 2005. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J. Biol. Chem. 280:30916-30923. [DOI] [PubMed] [Google Scholar]
- 26.Kane, S. E., and K. Beemon. 1987. Inhibition of methylation at two internal N6-methyladenosine sites caused by GAC to GAU mutations. J. Biol. Chem. 262:3422-3427. [PubMed] [Google Scholar]
- 27.Kluiver, J., S. Poppema, D. de Jong, T. Blokzijl, G. Harms, S. Jacobs, B. Kroesen, and A. van den Berg. 2005. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 207:243-249. [DOI] [PubMed] [Google Scholar]
- 28.Kralova, J., A. S. Liss, W. Bargmann, and H. R. Bose, Jr. 1998. AP-1 factors play an important role in transformation induced by the v-rel oncogene. Mol. Cell. Biol. 18:2997-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Küppers, R., K. Rajewsky, M. Zhao, G. Simons, R. Laumann, R. Fischer, and M. L. Hansmann. 1994. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA 91:10962-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBlanc, J. J., and K. L. Beemon. 2004. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 78:5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, R. B., J. McClure, B. Rup, D. W. Niesel, R. F. Garry, J. D. Hoelzer, K. Nazerian, and H. R. Bose. 1981. Avian reticuloendotheliosis virus: Identification of the hematopoietic target cell for transformation. Cell 25:421-431. [DOI] [PubMed] [Google Scholar]
- 32.Linial, M., N. Gunderson, and M. Groudine. 1985. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science 230:1126-1132. [DOI] [PubMed] [Google Scholar]
- 33.Lu, F., A. Weidmer, C.-G. Liu, S. Volinia, C. M. Croce, and P. M. Lieberman. 2008. Epstein-Barr virus-induced miR-155 attenuates NF-κB signaling and stabilizes latent virus persistence. J. Virol. 82:10436-10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, L.-F., T.-H. Thai, D. P. Calado, A. Chaudhry, M. Kubo, K. Tanaka, G. B. Loeb, H. Lee, A. Yoshimura, K. Rajewsky, and A. Y. Rudensky. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maroney, P. A., S. Chamnongpol, F. D. R. Souret, and T. W. Nilsen. 2007. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA 13:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateyak, M. K., A. J. Obaya, and J. M. Sedivy. 1999. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol. Cell. Biol. 19:4672-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan, R., A. Anderson, E. Bernberg, S. Kamboj, E. Huang, G. Lagasse, G. Isaacs, M. Parcells, B. C. Meyers, P. J. Green, and J. Burnside. 2008. Sequence conservation and differential expression of Marek's disease virus microRNAs. J. Virol. 82:12213-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Motsch, N., T. Pfuhl, J. Mrazek, S. Barth, and F. A. Grässer. 2007. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 4:131-137. [DOI] [PubMed] [Google Scholar]
- 39.Müschen, M., D. Re, A. Brauninger, J. Wolf, M.-L. Hansmann, V. Diehl, R. Kuppers, and K. Rajewsky. 2000. Somatic mutations of the CD95 gene in Hodgkin and Reed-Sternberg cells. Cancer Res. 60:5640-5643. [PubMed] [Google Scholar]
- 40.O'Connell, R. M., A. A. Chaudhuri, D. S. Rao, and D. Baltimore. 2009. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 106:7113-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orban, T. I., and E. Izaurralde. 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paolo, R., L. Valentina, P. Elvira, B. Mauro, P. Cesare, and M. Giovanna. 2008. MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. Br. J. Haematol. 143:570-580. [DOI] [PubMed] [Google Scholar]
- 43.Polony, T. S., S. J. Bowers, P. E. Neiman, and K. L. Beemon. 2003. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J. Virol. 77:9378-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehmsmeier, M., P. Steffen, M. Ha-Chsmann, and R. Giegerich. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez, A., E. Vigorito, S. Clare, M. V. Warren, P. Couttet, D. R. Soond, S. van Dongen, R. J. Grocock, P. P. Das, E. A. Miska, D. Vetrie, K. Okkenhaug, A. J. Enright, G. Dougan, M. Turner, and A. Bradley. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-585. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 47.Schat, K. A., W. D. Pratt, R. Morgan, D. Weinstock, and B. W. Calnek. 1992. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 36:432-439. [PubMed] [Google Scholar]
- 48.Schubach, W., and M. Groudine. 1984. Alteration of c-myc chromatin structure by avian leukosis virus integration. Nature 307:702-708. [DOI] [PubMed] [Google Scholar]
- 49.Shirato, H., S. Ogawa, K. Nakajima, M. Inagawa, M. Kojima, M. Tachibana, Y. Shinkai, and T. Takeuchi. 2009. A Jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem. 284:733-739. [DOI] [PubMed] [Google Scholar]
- 50.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's Sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, M., M. Kojima, K. Nakajima, R. Suzuki-Migishima, and T. Takeuchi. 2007. Functions of a jumonji-cyclin D1 pathway in the coordination of cell cycle exit and migration during neurogenesis in the mouse hindbrain. Dev. Biol. 303:549-560. [DOI] [PubMed] [Google Scholar]
- 52.Tam, W., S. Ben-Yehuda, and W. S. Hayward. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell. Biol. 17:1490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam, W., S. H. Hughes, W. S. Hayward, and P. Besmer. 2002. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J. Virol. 76:4275-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teng, G., P. Hakimpour, P. Landgraf, A. Rice, T. Tuschl, R. Casellas, and F. N. Papavasiliou. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, R. K., D. Re, J. Wolf, and V. Diehl. 2004. Part I: Hodgkin's lymphoma—molecular biology of Hodgkin and Reed-Sternberg cells. Lancet Oncol. 5:11-18. [DOI] [PubMed] [Google Scholar]
- 56.Tili, E., J.-J. Michaille, A. Cimino, S. Costinean, C. D. Dumitru, B. Adair, M. Fabbri, H. Alder, C. G. Liu, G. A. Calin, and C. M. Croce. 2007. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179:5082-5089. [DOI] [PubMed] [Google Scholar]
- 57.Vigorito, E., K. L. Perks, C. Abreu-Goodger, S. Bunting, Z. Xiang, S. Kohlhaas, P. P. Das, E. A. Miska, A. Rodriguez, A. Bradley, K. G. C. Smith, C. Rada, A. J. Enright, K.-M. Toellner, I. C. M. MacLennan, and M. Turner. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27:847-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. P. Cimmino, F. R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weller, S. K., and H. M. Temin. 1981. Cell killing by avian leukosis viruses. J. Virol. 39:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilhelmsen, K. C., K. Eggleton, and H. M. Temin. 1984. Nucleic acid sequences of the oncogene v-rel in reticuloendotheliosis virus strain T and its cellular homolog, the proto-oncogene c-rel. J. Virol. 52:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winer, J., C. K. S. Jung, I. Shackel, and P. M. Williams. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytesn vitro. Anal. Biochem. 270:41-49. [DOI] [PubMed] [Google Scholar]
- 62.Yang, F., R. R. Xian, Y. Li, T. S. Polony, and K. L. Beemon. 2007. Telomerase reverse transcriptase expression elevated by avian leukosis virus integration in B cell lymphomas. Proc. Natl. Acad. Sci. USA 104:18952-18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin, Q., J. McBride, C. Fewell, M. Lacey, X. Wang, Z. Lin, J. Cameron, and E. K. Flemington. 2008. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 82:5295-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin, Q., X. Wang, J. McBride, C. Fewell, and E. Flemington. 2008. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 283:2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, Y., Y. Yao, H. Xu, L. Lambeth, L. P. Smith, L. Kgosana, X. Wang, and V. Nair. 2009. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J. Virol. 83:489-492. [DOI] [PMC free article] [PubMed] [Google Scholar]