Abstract
Understanding the mechanisms by which herpes simplex virus (HSV) evades host immune defenses is critical to defining new approaches for therapy and prevention. We performed transcriptional analyses and immunocytochemistry on sequential biopsy specimens of lesional tissue from the acute through the posthealing phases of recurrent mucocutaneous HSV-2 infection. Histological analysis of these biopsy specimens during the acute stage revealed a massive infiltration of T cells, as well as monocytes/macrophages, a large amount of myeloid, and a small number of plasmacytoid dendritic cells, in the dermis of these lesional biopsy specimens. Type I interferon (IFN-β and IFN-α) was poorly expressed and gamma IFN (IFN-γ) potently induced during time periods in which we detected abundant amounts of HSV-2 antigens and HSV-2 RNA. IFN-stimulated genes were also markedly upregulated, with expression patterns that more closely matched those in primary human fibroblasts treated by IFN-γ than those in fibroblasts treated by IFN-β. Transcriptional arrays of the same lesional biopsy sites during healing and at 2 and 4 weeks posthealing revealed no HSV nucleic acids or antigen; however, there was persistent expression of IFN-γ, with very low levels of IFN-β and IFN-α. The findings of extremely low levels of IFN-α and IFN-β, despite the presence of a large number of cells capable of synthesizing these substances, suggest a potent alteration in host defense during HSV-2 infection in vivo. HSV-2's blockade of the innate immune system's production of type I IFN may be a major factor in allowing the virus to break through host mucosal defenses.
The pathogenesis of reactivating herpes simplex virus type 2 (HSV-2) infection at mucosal sites in humans is not well understood. Recent studies have shown frequent and rapid reactivation and clearance of HSV-2 in genital tissue (27, 41, 47), suggesting that local host responses in the periphery are imperfect determinants of viral clearance. Most recently, HSV-2-specific T cells have been shown to persist in skin for weeks after the healing of a reactivation episode (15, 47). The mechanism by which HSV-2 evades such local host defense even for short time periods is unclear.
Nearly 3 decades ago, antiviral activities in vesicle fluid during recurrent acute HSV infection in 18 of 19 patients were demonstrated using a plaque reduction assay (32). Type I interferon (IFN) was shown using type I IFN neutralizing antibodies to be the source of this antiviral effect in <50% of such persons. Significant amounts of T lymphocytes and monocytes/macrophages were seen to infiltrate near the infected epidermis during HSV reactivation (4), and a recent study found plasmacytoid dendritic cells (pDCs) near the infected epidermis during HSV reactivation (5), suggesting that host innate immune cells play a role in host mucosal defense to recurrent HSV infection. However, the interaction of host mucosal innate immune systems with reactivating HSV-2 during acute infection is poorly understood.
IFNs are a family of related cytokines that are classified according to receptor specificity and sequence homology. Type I IFN consists of multiple IFN-α subtypes, IFN-β, and several less-understood species, including IFN-κ, IFN-ɛ, IFN-ο, IFN-τ, and IFN-δ. IFN-γ is the only type II IFN. The more recently described type III IFN includes three IFN-λ gene products (37). Type I IFNs are the first line of host innate defense against many types of viral infection (9, 10). Although most cell types can secrete type I IFNs in response to viral infection, certain cell types, such as pDCs, generate especially high levels of type I IFN (24). IFN-γ, a potent immunoregulatory cytokine, is mainly secreted by T lymphocytes and NK cells. There is evidence that other cell types, such as antigen-presenting cells, can also synthesize IFN-γ (38).
Genetic studies with humans and mice have shown that host innate immune responses, particularly the host type I IFN response, play important roles in controlling HSV infection. Humans with mutations in Stat1, TLR3, or UNC-93B, whose gene products are involved in the production of or response to type I IFN, are prone to HSV-induced encephalitis (3, 7, 45). Mouse models also suggest that type I IFN is important in controlling acute HSV infection, and several HSV-encoded gene products, such as ICP0 and ICP34.5, antagonize host type I IFN antiviral activities (19). In addition, several IFN-stimulated genes (ISGs), such as ISG15 and OAS1, have been shown to be important in controlling acute HSV infection in mice (1, 20).
In a previous study, we showed that IFN-β and IFN-γ induce overlapping yet distinct gene expression patterns in primary human fibroblasts (FB) (33). In this paper, we utilized a combination of transcriptional profiling and immunocytochemistry studies of skin biopsy specimens taken directly from genital HSV-2 lesions during the acute and posthealing phases of genital herpes to study the temporal expression patterns of IFNs (IFN-β, IFN-α, and IFN-γ) and ISGs. We found that despite the massive infiltrates into the infected epidermis of monocytes/macrophages, myeloid dendritic cells (mDCs), and a small number of pDCs, IFN-β and IFN-α are poorly expressed while IFN-γ and ISGs are strongly expressed in genital lesions, suggesting that HSV-2 reduces type I IFN production in vivo.
MATERIALS AND METHODS
Human subjects and specimens.
Study subjects were recruited into a study protocol approved by the University of Washington Institutional Review Board. Written consent was obtained from all subjects. Three-millimeter punch biopsy specimens were obtained during symptomatic recurrences from active lesion sites, from newly healed lesions, and from the same sites at 2 and 4 weeks posthealing, as previously described (47). For biopsy specimens taken from acute lesions, half of the biopsy specimen included the vesicle area and the other half covered the immediately adjacent erythematous skin area. In the posthealing time period, biopsy specimens were obtained from the predominant lesion area, usually contiguous to the prior biopsy specimens. Control skin biopsy specimens were taken from normal epithelialized genital skin obtained at the same time from the anatomic site opposite from that of HSV reactivation.
RNA extraction, amplification, and hybridization of cRNA to Illumina bead arrays.
To prepare RNA from acute lesion or healed lesion biopsy specimens, half of a biopsy specimen was homogenized with a Qiagen RNeasy microkit lysis buffer, using a Pro200 homogenizer (PRO Scientific, Oxford, CT). Extraction of total RNA followed the manufacturer's protocol. The quality of total RNA was analyzed by Agilent picochips, and RNA whose RNA quality index was above 6 was used. Total RNA (150 ng) was then used to prepare cRNA by using an Illumina TotalPrep RNA amplification kit (Ambion, Austin, TX). The size distribution of cRNA was analyzed by Agilent nanochips. In all the biopsy specimens that were used for arrays, the amplified cRNA had a Gaussian distribution with an average size of 1.2 kb. The cRNA (750 ng) was then used to hybridize to Illumina HumanRef8_v2 bead arrays in the Center for Array Technology (CAT) at the University of Washington, per the manufacturer's protocol.
Analysis of bead array data.
Raw data were imported to Beadstudio (version 2.3; Illumina). Control summaries were generated by Beadstudio to analyze the quality of the hybridization. Data passing this initial quality control step were exported and normalized by the lumi package in R, a statistical platform for data analysis (6). Normalized data were visualized and analyzed by a hierarchical clustering method (with complete linkage and Euclidean distance) using SpotFire DecisionSite for functional genomics (version 9.1.1). The GoMiner program (44) was used to identify the enriched functional categories (gene ontology [GO] terms) in the differentially expressed genes.
Quantitative reverse transcription-PCR (RT-PCR).
Total RNA was isolated from primary human FB or lesional biopsy specimens by using a Qiagen RNeasy minikit per the manufacturer's instructions. First-strand cDNA was then synthesized from total RNA (250 ng) by using a high-capacity cDNA synthesis kit from Applied Biosystems (Foster City, CA). Two microliters of the final 50 μl cDNA was used for each reaction in a 96-well TaqMan PCR plate. TaqMan probes for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), IFN-β, IFN-α1/IFN-α21, IFN-γ, DDX58, OAS1, INDO, and GBP5 were ordered from Applied Biosystems (the assay identification numbers are Hs02758991_g1, Hs01077958_s1, Hs00353738_s1, Hs00174143_m1, Hs01061433_m1, Hs00973640_m1, Hs00158032_m1, and Hs00369472_m1). HSV RNA and DNA were measured by TaqMan PCR as previously described (42, 47). To test the sensitivity of IFN TaqMan PCRs, cDNA clones (for INFB1, LIFESEQ90179925; for INFA-21/1, 7262162; and for INFG, LIFESEQ95113490) containing corresponding amplicon sequences were purchased from Open Biosystems (Open Biosystems, Huntsville, AL). After the plasmids were purified by using a QIAprep spin miniprep kit (Qiagen, Valencia, CA), quantified by optical density reading, and linearized by enzyme digestion, 10-fold serial dilutions were made to create standard curves and to test the sensitivity. Each 30-μl PCR mixture consists of 1.5 μl of 20× primer-probe mix, 15 μl 2× QuantiTect multiplex PCR master mix (Qiagen), and 2 μl of cDNA. All three PCRs used for this study were able to detect 10 copies of cDNA consistently. Standard curves consisting of dilution series of 105, 104, 103, 104, 102, and 10 were then used to quantify IFN (IFN-β, IFN-α1, and IFN-γ) expression in control and lesion biopsy specimens.
Immunofluorescent staining.
The staining of the biopsy specimens with the antibody for the HSV whole antigen has previously been described (47). Biopsy specimens were snap frozen on optimal cutting temperature compound (Sakura Finetek USA, Inc., Torrance, CA) and stored at −80°C until use. As previously described, we sectioned frozen skin biopsy specimens into 7-μm slices before they were fixed and permeabilized in acetone for 20 min at −20°C. Tissue was incubated in blocking buffer for 30 min and the primary antibody applied overnight at 4°C. After being washed in phosphate-buffered saline three times, the samples were incubated with fluorescence-labeled secondary antibody at room temperature for 1 h, washed, counterstained with DAPI (4′,6-diamidino-2-phenylindole; Fluka), and mounted in Mowiol 40-88 containing 2.5% 1,4-diazabicyclo[2,2,2]-octane (DABCO; Sigma). The primary antibodies used in this study were specific for HSV-2 (rabbit polyclonal, Dako), BDCA2 (goat polyclonal; R&D systems), DC-LAMP (mouse monoclonal; Beckman Coulter), DC-SIGN-AF647 (mouse monoclonal; e-Bioscience), CD14 (mouse monoclonal; BD Pharmingen), CD68 (mouse monoclonal; Dako), and CD163 (mouse monoclonal; ABCAM). The secondary antibodies were donkey anti-mouse Alexa Fluor 488, donkey anti-mouse Alexa Fluor 647, donkey anti-rabbit Alexa Fluor 594, and goat anti-mouse horseradish peroxidase coupled with Alexa Fluor-labeled tyramide.
Culturing and IFN treatment of primary human FB.
Primary human FB were propagated in Waymouth MB 752/1 medium (Invitrogen, Carlsbad, CA) containing 20% fetal bovine serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). FB were seeded into six-well dishes at a density of 1.5 × 105 cells per well so that cells were about 70% confluent the following day for IFN treatment. FB were either treated with 100 U/ml of human IFN-β1a (PBL Biomedical Laboratories, Piscataway, NJ), treated with 100 U/ml of human recombinant IFN-γ (Roche Diagnostics, Indianapolis, IN), or left untreated 48 h before the total RNA extraction using a Qiagen RNeasy minikit.
Microarray data accession number.
The array data have been deposited into the NCBI Gene Expression Omnibus (GEO) database under accession number GSE18527.
RESULTS
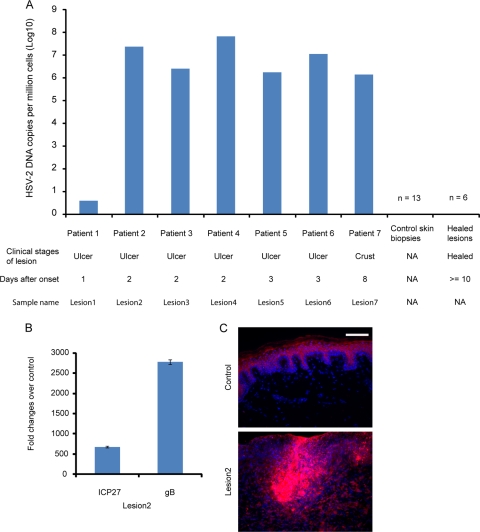
To understand the temporal gene expression patterns of host immune response during the progression of HSV-2 reactivation at mucocutaneous sites, particularly the expression of type I and type II IFNs and ISGs, we analyzed the whole-genome transcriptional profiles of four skin biopsy specimens at acute lesion stages and six skin biopsy specimens at posthealing stages from four patients. After our analyses of these biopsy specimens, we subsequently performed selected quantitative RT-PCR of HSV-2 gene expression of three different patients during their acute episodes of reactivation. We detected HSV DNA in tissues from all seven acute lesions but not in any of the posthealing or control biopsy specimens (Fig. 1A). We also found high levels of RNA from ICP27 and gB, two HSV-encoded gene products, in all seven lesional biopsy specimens (Fig. 1B) but not in biopsy specimens from healed lesions or in control biopsy specimens. Similarly, HSV antigen expression at the infected epidermis was found in all seven lesional biopsy specimens (Fig. 1C).
FIG. 1.
Significant presence of HSV-2 DNA and expression of HSV-2 RNA and antigens in lesion biopsy specimens from individuals with recurrent mucocutaneous HSV-2 infection. (A) Detection of HSV-2 DNA by quantitative PCR in skin biopsy specimens during the acute (lesional) and healed stages of HSV-2 reactivation and in control biopsy specimens that are derived from genital skin in non-HSV-2-affected areas. The seven biopsy specimens taken during acute lesion stages from seven different patients are referred to as lesions 1, 2, 3, 4, 5, 6, and 7 in subsequent experiments. NA, not applicable. (B) Expression of two HSV genes (ICP27 and gB) in lesion 2 detected by real-time RT-PCR. The expression of ICP27 and gB in lesional biopsy specimens was normalized to that of human rRNA. The error bars represent 1 standard deviation from the average values obtained from triplicate technical replicates. (C) Immunofluorescent staining for HSV antigen by using polyclonal antibody in lesion 2 (bottom panel) and noninfected genital skin (top panel). Blue, DAPI; red, HSV-2. Scale bar, 50 μM.
Hierarchical clustering analysis of genes that show differential expression in lesion biopsy specimens.
To examine changes in host gene expression during lesion progression, we used the contralateral normal genital skin biopsy specimen as the control reference to generate expression ratios of more than 22,000 genes on Illumina Human Ref8_v2 bead arrays for all the biopsy specimens. A gene was defined as differentially expressed if it had a more-than-twofold change over its control biopsy specimen; 2,625 genes met the criteria among the four lesion biopsy specimens in which HSV RNA was detected. A hierarchical clustering analysis shows that the lesion biopsy specimens from four different patients during the acute stage of reactivation have striking similarities in expression patterns and that the expression levels of these genes vary over the course of healing (Fig. 2A).
FIG. 2.
Transcriptional profiling of skin biopsy specimens at lesion and posthealing stages. (A) Hierarchical clustering of differentially expressed genes in four acute lesion samples relative to their matched controls. We performed transcriptional arrays that assayed the expression of more than 22,000 genes on four acute lesion biopsy specimens and six healed biopsy specimens and the uninfected control biopsy specimens from the same patients and identified 2,625 genes with twofold-or-more differences in expression between lesion and control biopsy specimens. Genes with decreased expression are shown in green, and those with increased expression in comparison to that of the control biopsy specimens are shown in red. Expression data from both acute lesion and healed biopsy specimens are shown on the heat map. (B) List of significantly enriched GO terms (false discovery rate [FDR], <0.05) as defined by GoMiner in the set of differentially expressed genes shown in panel A.
To identify enriched functional categories in the set of differentially expressed genes relative to all the genes on the Illumina HumanRef8_2 bead arrays, we annotated these genes to the GO terms and identified the enriched GO terms by using the GoMiner program (Fig. 2B). Out of more than 10,000 GO terms used to annotate the set of differentially expressed genes, 461 GO terms are highly enriched (false discovery rate, ≤0.05) (data not shown). Consistent with the significant lymphocyte infiltration at acute lesion stages (4, 47), we find genes associated with such GO terms as “lymphocyte activation” to be highly enriched in our data set. As expected, the transcription arrays also indicated major alterations in innate immune responses during the course of lesion progression. As both types of IFNs and ISGs play important roles in host innate immune response, we concentrated our initial analysis on evaluating these genes during the progression of HSV-2 reactivation.
Type I and type II IFNs show significantly different expression levels in acute lesion biopsy specimens.
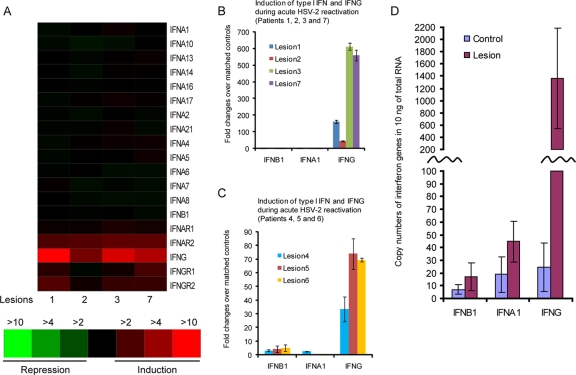
We found marked differences between the inductions of type I and type II IFNs in the lesional biopsy specimens. As shown in Fig. 3A, we found no evidence for induction of either IFN-α or IFN-β in lesional tissue. In contrast, there were 3- to 15-fold increases of IFN-γ in all four lesion biopsy specimens. The receptors for both type I and type II IFNs had modest increases in expression in these biopsy specimens. Since the gene arrays are less sensitive and have less dynamic ranges than real-time RT-PCR, the same RNA was converted to cDNA and the expression of type I and type II IFNs was assayed by real-time PCR, using the synthesized cDNA as a template. The RT-PCR analyses confirmed the microarray data, as IFN-α1/IFN-α21 and IFN-β expression levels were within twofold relative to those of the control and healed biopsy specimens. In contrast, IFN-γ expression was upregulated from 45- to 500-fold relative to that of controls (Fig. 3B) and persisted over time (see below and Fig. 6). We then assayed the expression of type I and II IFNs by using three additional acute lesion biopsy specimens from three different patients. HSV-2 DNA and RNA were present in these three biopsy specimens in concentrations similar to those for patient 2 (Fig. 1A and data not shown). These three early biopsy specimens also exhibited weak induction of IFN-β (3- to 5-fold) and IFN-α1 (less than 2-fold) yet significantly increased expression of IFN-γ (30- to 70-fold) in comparison to that of the matched contralateral normal genital skin biopsy specimens (Fig. 3C). To estimate the copy numbers of IFN-β, IFN-α1, and IFN-γ in the four lesion biopsy specimens with matched controls shown in Fig. 3B, the standard curves were generated using IFN-β-, IFN-α1-, and IFN-γ-expressing plasmids (see Materials and Methods for details), and the copy numbers of IFNs in the cDNA used in TaqMan PCR shown in Fig. 3B were calculated according to standard curves. The cDNA was synthesized from 250 ng total RNA, and 1/25 of the cDNA was used to estimate the copy numbers of IFNs. There were fewer than 50 copies of IFN-β, IFN-α1, and IFN-γ in control biopsy specimens, and similar numbers of IFN-β and IFN-α1 were found in the four lesion biopsy specimens; however, more than 1,300 copies of IFN-γ on average were detected in the lesion biopsy specimens (Fig. 3D). In summary, IFN-γ expression is strongly upregulated from early lesions to crust stages during the development of mucocutaneous lesions induced by HSV-2 reactivation, but type I IFNs show little change over the same time course in comparison to matched contralateral normal genital skin biopsy specimens.
FIG. 3.
Human skin biopsy specimens taken from acute HSV-2 lesions show little type I IFN expression but strong IFN-γ expression. (A) Expression of type I IFN (indicated as IFN-α or IFN-β) and type II IFN (IFN-γ) and their receptors (IFN-αR and IFN-γR) in four acute lesion biopsy specimens relative to that in their matched control biopsy specimens in arrays. (B) Real-time RT-PCR for assaying the expression of IFN-β, IFN-α1, and IFN-γ in the same set of acute lesion biopsy specimens as shown in panel A. (C) Real-time RT-PCR for assaying the expression of type I IFN (IFN-β and IFN-α1) and IFN-γ in three additional patients from whom lesion biopsy specimens were sampled 2, 3, and 3 days, respectively, after the onset of lesion. The three patients from whom these three acute lesion biopsy specimens were taken are different from the four patients represented in panels A and B. The error bars represent 1 standard deviation from the average values obtained from triplicate technical replicates. (D) Detection of copy numbers of IFN-β, IFN-α1, and IFN-γ in control and lesion biopsy specimens. To calculate the copy numbers of IFNs in the four lesion biopsy specimens with matched controls that were used for microarrays shown in panel A and TaqMan RT-PCR shown in panel B, standard curves were generated using IFN-β, IFN-α1, and IFN-γ expressing plasmid DNA and the same primer-probe sets used in panels B and C (see Materials and Methods for details). The error bars represent 1 standard deviation from the average values obtained from the four control or lesion biopsy specimens.
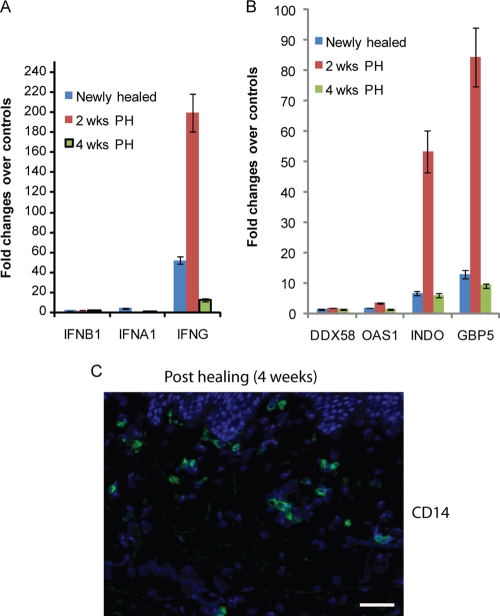
FIG. 6.
Healed lesion biopsy specimens from patients with recurrent HSV-2 infection have little type I IFN, but high levels of IFN-γ and IFN-γ-associated ISGs persist in specimens newly healed and at 2 and 4 weeks posthealing. (A) Real-time RT-PCR detected little type I IFN (IFN-α1 and IFN-β) yet significant upregulation of IFN-γ in three healed biopsy specimens (newly healed and at 2 and 4 weeks posthealing [wks PH]). (B) Expression patterns of four ISGs, DDX58, OAS1, INDO, and GBP5, in the same three healed biopsy specimens as described for panel A. The error bars represent 1 standard deviation from the average values obtained from triplicate technical replicates. (C) Detection of CD14 (green)-positive cells in the dermis area of a healed biopsy specimen (4 weeks posthealing). Blue, DAPI staining. Scale bar, 50 μM.
IFN-γ has a dominant role in inducing the expression of ISGs in acute lesion biopsy specimens.
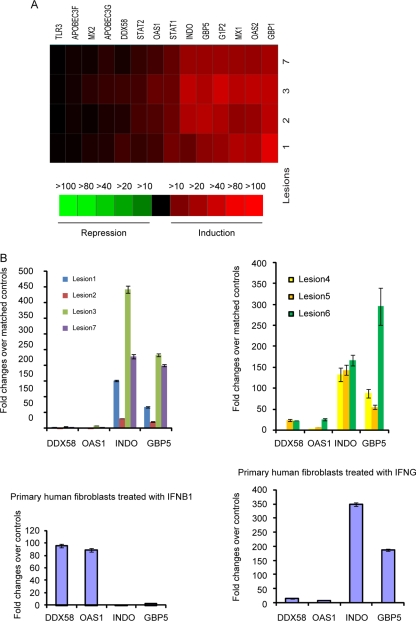
Our previous studies have shown that IFN-β and IFN-γ induce overlapping yet distinct expression patterns of ISGs (33). As such, we analyzed in detail the expression patterns of ISGs in acute lesional biopsy specimens. Several ISGs known to be induced by IFN-γ, such as INDO, GBP1, and GBP5, were much more strongly expressed in lesional tissue than other ISGs, such as DDX58 and OAS1, known to be induced by type I IFN (Fig. 4A). To evaluate these changes more closely, we performed quantitative RT-PCR to assay the expression of four ISGs, DDX58, OAS1, INDO, and GBP5, in all seven acute lesion biopsy specimens; the results are consistent with those seen in transcriptional arrays (Fig. 4B, top panel). To more directly examine the effects of type I and type II IFNs on the expression of these ISGs, we compared the expression data in lesion biopsy specimens to our previously published data for these four genes for which we performed RT-PCR with primary human FB treated with IFN-β or IFN-γ (33). Consistent with our results showing that type I IFNs are barely expressed in lesional tissues, we find that the expression patterns of these four ISGs in the seven lesional biopsy specimens more closely match those in human primary FB treated with IFN-γ than those in cells treated with IFN-β (Fig. 4B, bottom panel). The data suggest that IFN-γ, rather than type I IFNs, has a dominant role in inducing the expression of ISGs during the development of recurrent HSV-2 mucocutaneous lesion, which is consistent with our results showing that IFN-γ but not type I IFNs is upregulated in lesional biopsy specimens.
FIG. 4.
Expression patterns of ISGs in acute lesion biopsy specimens. (A) Expression patterns of ISGs that showed twofold or greater increases in expression in any of the four acute lesion biopsy specimens relative to that in their matched controls in transcriptional arrays. (B) Confirmation of array data for select ISGs by RT-PCR. Expression patterns are shown for two ISGs with large changes in expression (INDO and GBP5) and two ISGs with moderate to low increases in expression (OAS1 and DDX58) in seven lesion biopsy specimens as detected by real-time RT-PCR (top panel) and for the four ISGs in primary human FB treated with IFN-β (100 U/ml) or IFN-γ (100 U/ml) for 48 h in comparison to the level for untreated cells (bottom panel) (29). The error bars represent 1 standard deviation from the average values obtained from triplicate technical replicates.
Microarray and immunocytochemical evidence of monocytes/macrophages, mDCs, and pDCs in acute lesion biopsy specimens.
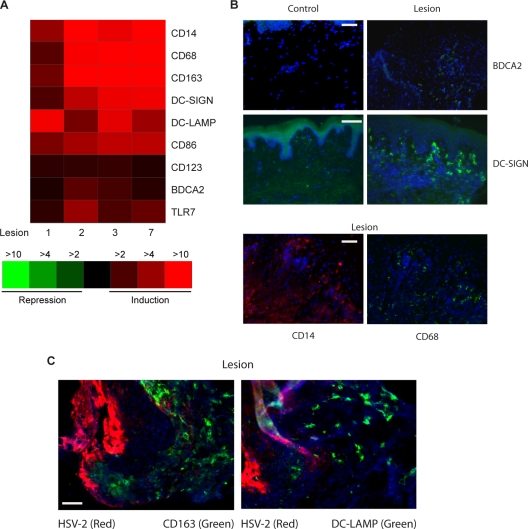
To evaluate the potential sources of IFN production in lesional tissue, we analyzed the expression patterns of surface markers known to be associated with inflammatory cells. As shown in Fig. 5A, CD14, CD68, and CD163, transcripts known to be highly expressed in monocytes/macrophages, were upregulated more than 10-fold in three of the four lesional biopsy specimens; DC-SIGN, DC-LAMP, and CD86, which are abundant in mature DCs, were upregulated more than fivefold on average, and BDCA2, a specific marker for pDCs, was upregulated about twofold.
FIG. 5.
Significant presence of monocytes/macrophages, mDCs, and pDCs in acute lesion biopsy specimens from patients with recurrent HSV-2 infection. (A) Expression patterns of markers for monocytes/macrophages, mDCs, and pDCs in lesional biopsy specimens from transcriptional arrays. The changes for each lesion biopsy specimen are calculated in comparison to its matched control biopsy specimen. (B) Detection of BDCA2 (green, top two panels)-, DC-SIGN (green, middle two panels)-, CD14 (red, bottom left panel)-, and CD68 (green, bottom right panel)-positive cells in the dermis area close to the infected (lesional biopsy specimens) or uninfected (control biopsy specimens) epidermis by ICC. Blue, DAPI staining. (C) Significant infiltration of CD163- and DC-LAMP-positive cells to the HSV-2-infected epidermis during acute HSV-2 ulcerations. Blue, DAPI staining. Scale bar, 50 μM.
The increased expression of inflammatory cell-specific genes could be due to upregulation in individual cells or to increased numbers of inflammatory cells in the lesional tissue. To distinguish between these possibilities, we sought to visualize the different types of innate cells in lesional biopsy specimens by using immunofluorescent staining. As shown in Fig. 5B, we found larger amounts of inflammatory cells in lesional biopsy specimens than in control tissue. The biopsy specimens contain cells expressing BDCA2, indicating the presence of pDCs near the infected epidermis. In addition, large quantities of CD14- and CD68-positive cells and cells expressing DC-SIGN were seen in lesional biopsy specimens in the dermal area. Costainings of lesional biopsy specimens with HSV and CD163 or DC-LAMP antibodies show that host monocytes/macrophages (CD163 positive) and DCs (DC-LAMP positive) infiltrate the epidermis during the acute reactivation (Fig. 5C). Taken together, the transcription and ICC data indicate a marked infiltration of transcriptionally active monocytes/macrophages, myeloid, and pDCs in genital lesions.
Type I IFN is expressed at low levels, and IFN-γ is persistently induced in posthealing biopsy specimens.
To examine host immune response after virus clearance, we generated transcriptional profiles of biopsy specimens at posthealing stages (newly healed and at 2 weeks and 4 weeks posthealing). The array analysis revealed little expression of type I IFN in any of the posthealing stages and continual presence of IFN-γ at high levels in genital skin biopsy specimens from the sites of prior HSV-2 infection in comparison to the levels in genital skin not involved with HSV-2 reactivation (data not shown). We confirmed the low levels of type I IFN and high levels of IFN-γ by RT-PCR (Fig. 6A). The expression patterns of different ISGs are similar to those seen in acute ulcerations (Fig. 6B versus 4B), suggesting that IFN-γ still has a dominant role in inducing the expression of ISGs in healed lesion biopsy specimens. Notably, although still upregulated compared to those in uninvolved skin, the expression levels of IFN-γ and ISGs are significantly lower than those seen in acute ulcerations (Fig. 6A versus 3B and Fig. 6B versus 4B, respectively). Histological analyses by ICC showed persistent CD8/CD4 (data not shown) and monocyte/macrophage infiltration in posthealing biopsy specimens (Fig. 6C). Thus, as in acute ulcerations, the histological and transcriptional analyses of posthealing biopsy specimens were consistent.
DISCUSSION
Using the combination of in vivo transcriptional profiling with in vivo immunocytochemistry studies of lesional biopsy specimens, we show that HSV-2 appears capable of effectively evading the host innate immune system by suppressing type I IFN production. Whole HSV-2 lesional tissues contain large amounts of IFN-γ but surprisingly low levels of type I IFN, despite evidence, shown by ICC and transcriptional analysis, of a marked infiltration of the lesional biopsy specimens by monocytes/macrophages and mDCs and the significant presence of pDCs, all cell types known to produce type I IFN in response to HSV infection in vitro (28, 34, 39) and to systematic HSV infection in mouse models (36). Analysis of the array data showed that several type I IFN-induced ISGs, such as OAS1 and DDX58, were expressed at low levels, further strengthening the view that there is little type I IFN activity in lesional tissues.
Our study suggests that more-detailed explanations of the mechanism of this apparent blockade of type I IFN induction in vivo are warranted. pDC cells secrete large amounts of type I IFN in response to viral infection. However, at a local infection, such as a respiratory infection, other cell types may also play prominent roles in the synthesis of type I IFN. In a mouse model using nasal infection with Newcastle disease virus, it has been shown that alveolar macrophages and conventional dendritic cells, rather than pDCs, are the major producers of IFN-α (16). In a mouse model with systematic infection of HSV-1 and HSV-2, pDCs and TLR9 are responsible for early production of type I IFN, yet other types of innate cells, including mDCs and macrophages, produce IFN late (36). In a mouse model with vaginal HSV-2 infection, pDCs are recruited to the infection site and produce a significant amount of type I IFN in a TLR9-dependent manner (25). In the lesion biopsy specimens that we studied, CD14-positive monocytes/macrophages significantly infiltrate the infected epidermis, along with large numbers of mDCs and a small number of pDCs. When compared to these infection studies, the low expression level of type I IFN during the progression of HSV reactivation is surprising.
The expression patterns of IFNs (type I and type II) and ISGs during recurrent mucocutaneous HSV-2 infection differ from those under other inflammatory conditions. For example, transcriptional profiling of psoriasis lesional skin versus nonlesional skin biopsy specimens reveals significant upregulation of type I IFN (IFN-β and IFN-α) and type I IFN ISGs, such as OAS1 and MX1 (43). Thus, the host gene expression patterns that we described to occur in the acute lesions as well as over the course of healing are likely to be dictated by the interplay between the host and HSV-2 and are not the nonspecific effects of inflammation per se. The persistent expression of IFN-γ is consistent with other findings, achieved using biological techniques, that IFN-γ is the predominant species made in herpetic lesions. Our data are also consistent with our previous work showing that CD8 T cells persist from lesion to healed stages during the progression of HSV reactivation (47) and our recent data showing that HSV-specific CD4 T cells that persist for weeks after healing express IFN-γ when stimulated with inactivated HSV-2 antigens (46). We cannot yet define the major source of IFN-γ, whether it is from CD4 T cells, CD8 T cells and/or macrophages, or likely all three. With mouse studies, it has been shown that IFN-γ and CD8 T cells persist in latently infected ganglia and that IFN-γ plays important roles in the maintenance of HSV latency (14, 23). IFN-γ is also known to induce the robust expression of some ISGs, such as INDO and GBP5, therefore perhaps stimulating an antiviral state in the infected area. During HSV reactivation, IFN-γ seems to have a dominant role in inducing the expression of ISGs from lesion to posthealing stages. This finding is consistent with mouse studies which suggest that IFN-γ plays important roles in host defense during HSV infection (2, 12).
Whether the lack of type I IFN production in acute lesion biopsy specimens is directly related to a specific HSV-2 gene product is unclear but worthy of investigation. HSV is a large DNA virus known to encode several gene products that enable viral evasion of host innate immune response (18). HSV infection in cell cultures induces host type I IFN response, which is subsequently shut down by HSV-encoded gene products (30, 31). Several studies have shown that HSV-1 encodes ICP0, an immediate-early gene product that can inhibit the transcription regulatory functions of IRF3 (8, 22, 29). ICP27, another immediate-early gene product, has been shown to antagonize type I IFN signaling (13). HSV-encoded late gene products γ-34.5 and Us11 inhibit the antiviral functions of PKR (11, 35). Us3, an HSV-encoded kinase, is shown to block the expression of IFN-γ-dependent genes (21). Lastly, vhs, a tegument protein and HSV-encoded late gene product, degrades both host and viral mRNA (17) and in mouse models it seems to play important roles in HSV pathogenesis (40). Elucidating HSV immune evasion mechanisms of the innate immune system at mucosal surfaces during recurrent infection may provide new strategies for antiviral drug and HSV vaccine development. For example, a recent human clinical trial demonstrated that topical resiquimod, a TLR7 and TLR8 agonist, decreases HSV-2 genital shedding, suggesting that type I IFN may play a critical role in controlling HSV reactivation (26). The ability to obtain genital lesion tissue in concert with techniques for isolating and characterizing lesion-infiltrating cells may allow the effective dissection of HSV-2 strategies for evading host innate immune systems.
Acknowledgments
This work was supported by National Institutes of Health grants AI-30731 and AI-42528 and by the Tietze Foundation. We declare that we have no competing financial interests.
We thank Michael Remington and Christine Johnson for clinical assistance and Rachel Tompa for editing the writing.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Austin, B. A., C. James, R. H. Silverman, and D. J. Carr. 2005. Critical role for the oligoadenylate synthetase/RNase L pathway in response to IFN-beta during acute ocular herpes simplex virus type 1 infection. J. Immunol. 175:1100-1106. [DOI] [PubMed] [Google Scholar]
- 2.Cantin, E., B. Tanamachi, H. Openshaw, J. Mann, and K. Clarke. 1999. Gamma interferon (IFN-γ) receptor null-mutant mice are more susceptible to herpes simplex virus type 1 infection than IFN-γ ligand null-mutant mice. J. Virol. 73:5196-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casrouge, A., S. Y. Zhang, C. Eidenschenk, E. Jouanguy, A. Puel, K. Yang, A. Alcais, C. Picard, N. Mahfoufi, N. Nicolas, L. Lorenzo, S. Plancoulaine, B. Senechal, F. Geissmann, K. Tabeta, K. Hoebe, X. Du, R. L. Miller, B. Heron, C. Mignot, T. B. de Villemeur, P. Lebon, O. Dulac, F. Rozenberg, B. Beutler, M. Tardieu, L. Abel, and J. L. Casanova. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314:308-312. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham, A. L., R. R. Turner, A. C. Miller, M. F. Para, and T. C. Merigan. 1985. Evolution of recurrent herpes simplex lesions. An immunohistologic study. J. Clin. Investig. 75:226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaghy, H., L. Bosnjak, A. N. Harman, V. Marsden, S. K. Tyring, T. C. Meng, and A. L. Cunningham. 2009. Role for plasmacytoid dendritic cells in the immune control of recurrent human herpes simplex virus infection. J. Virol. 83:1952-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du, P., W. A. Kibbe, and S. M. Lin. 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547-1548. [DOI] [PubMed] [Google Scholar]
- 7.Dupuis, S., E. Jouanguy, S. Al-Hajjar, C. Fieschi, I. Z. Al-Mohsen, S. Al-Jumaah, K. Yang, A. Chapgier, C. Eidenschenk, P. Eid, A. Al Ghonaium, H. Tufenkeji, H. Frayha, S. Al-Gazlan, H. Al-Rayes, R. D. Schreiber, I. Gresser, and J. L. Casanova. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388-391. [DOI] [PubMed] [Google Scholar]
- 8.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 10.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, B., M. Gross, and B. Roizman. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, J., H. Ichimura, T. Iida, M. Minami, K. Kobayashi, M. Kita, C. Sotozono, Y. I. Tagawa, Y. Iwakura, and J. Imanishi. 1999. Kinetics of cytokine production in the cornea and trigeminal ganglion of C57BL/6 mice after corneal HSV-1 infection. J. Interferon Cytokine Res. 19:609-615. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, K. E., B. Song, and D. M. Knipe. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khanna, K. M., R. H. Bonneau, P. R. Kinchington, and R. L. Hendricks. 2003. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity 18:593-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koelle, D. M., M. Schomogyi, and L. Corey. 2000. Antigen-specific T cells localize to the uterine cervix in women with genital herpes simplex virus type 2 infection. J. Infect. Dis. 182:662-670. [DOI] [PubMed] [Google Scholar]
- 16.Kumagai, Y., O. Takeuchi, H. Kato, H. Kumar, K. Matsui, E. Morii, K. Aozasa, T. Kawai, and S. Akira. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27:240-252. [DOI] [PubMed] [Google Scholar]
- 17.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leib, D. A. 2002. Counteraction of interferon-induced antiviral responses by herpes simplex viruses. Curr. Top. Microbiol. Immunol. 269:171-185. [DOI] [PubMed] [Google Scholar]
- 19.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. Virgin IV. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 104:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, L., and B. Roizman. 2008. Expression of gamma interferon-dependent genes is blocked independently by virion host shutoff RNase and by US3 protein kinase. J. Virol. 82:4688-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, T., K. M. Khanna, B. N. Carriere, and R. L. Hendricks. 2001. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. J. Virol. 75:11178-11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Y. J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275-306. [DOI] [PubMed] [Google Scholar]
- 25.Lund, J. M., M. M. Linehan, N. Iijima, and A. Iwasaki. 2006. Cutting edge: plasmacytoid dendritic cells provide innate immune protection against mucosal viral infection in situ. J. Immunol. 177:7510-7514. [DOI] [PubMed] [Google Scholar]
- 26.Mark, K. E., L. Corey, T. C. Meng, A. S. Magaret, M. L. Huang, S. Selke, H. B. Slade, S. K. Tyring, T. Warren, S. L. Sacks, P. Leone, V. A. Bergland, and A. Wald. 2007. Topical resiquimod 0.01% gel decreases herpes simplex virus type 2 genital shedding: a randomized, controlled trial. J. Infect. Dis. 195:1324-1331. [DOI] [PubMed] [Google Scholar]
- 27.Mark, K. E., A. Wald, A. S. Magaret, S. Selke, L. Olin, M. L. Huang, and L. Corey. 2008. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J. Infect. Dis. 198:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melchjorsen, J., J. Siren, I. Julkunen, S. R. Paludan, and S. Matikainen. 2006. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-kappaB and IRF-3. J. Gen. Virol. 87:1099-1108. [DOI] [PubMed] [Google Scholar]
- 29.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215-2218. [DOI] [PubMed] [Google Scholar]
- 32.Overall, J. C., Jr., S. L. Spruance, and J. A. Green. 1981. Viral-induced leukocyte interferon in vesicle fluid from lesions of recurrent herpes labialis. J. Infect. Dis. 143:543-547. [DOI] [PubMed] [Google Scholar]
- 33.Peng, T., J. Zhu, Y. Hwangbo, L. Corey, and R. E. Bumgarner. 2008. Independent and cooperative antiviral actions of beta interferon and gamma interferon against herpes simplex virus replication in primary human fibroblasts. J. Virol. 82:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollara, G., M. Jones, M. E. Handley, M. Rajpopat, A. Kwan, R. S. Coffin, G. Foster, B. Chain, and D. R. Katz. 2004. Herpes simplex virus type-1-induced activation of myeloid dendritic cells: the roles of virus cell interaction and paracrine type I IFN secretion. J. Immunol. 173:4108-4119. [DOI] [PubMed] [Google Scholar]
- 35.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen, S. B., L. N. Sorensen, L. Malmgaard, N. Ank, J. D. Baines, Z. J. Chen, and S. R. Paludan. 2007. Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J. Virol. 81:13315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder, K., P. J. Hertzog, T. Ravasi, and D. A. Hume. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75:163-189. [DOI] [PubMed] [Google Scholar]
- 39.Siegal, F. P., N. Kadowaki, M. Shodell, P. A. Fitzgerald-Bocarsly, K. Shah, S. Ho, S. Antonenko, and Y. J. Liu. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 284:1835-1837. [DOI] [PubMed] [Google Scholar]
- 40.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wald, A., L. Corey, R. Cone, A. Hobson, G. Davis, and J. Zeh. 1997. Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J. Clin. Investig. 99:1092-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wald, A., M. L. Huang, D. Carrell, S. Selke, and L. Corey. 2003. Polymerase chain reaction for detection of herpes simplex virus (HSV) DNA on mucosal surfaces: comparison with HSV isolation in cell culture. J. Infect. Dis. 188:1345-1351. [DOI] [PubMed] [Google Scholar]
- 43.Yao, Y., L. Richman, C. Morehouse, M. de los Reyes, B. W. Higgs, A. Boutrin, B. White, A. Coyle, J. Krueger, P. A. Kiener, and B. Jallal. 2008. Type I interferon: potential therapeutic target for psoriasis? PLoS ONE 3:e2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeeberg, B. R., W. Feng, G. Wang, M. D. Wang, A. T. Fojo, M. Sunshine, S. Narasimhan, D. W. Kane, W. C. Reinhold, S. Lababidi, K. J. Bussey, J. Riss, J. C. Barrett, and J. N. Weinstein. 2003. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, S. Y., E. Jouanguy, S. Ugolini, A. Smahi, G. Elain, P. Romero, D. Segal, V. Sancho-Shimizu, L. Lorenzo, A. Puel, C. Picard, A. Chapgier, S. Plancoulaine, M. Titeux, C. Cognet, H. von Bernuth, C. L. Ku, A. Casrouge, X. X. Zhang, L. Barreiro, J. Leonard, C. Hamilton, P. Lebon, B. Heron, L. Vallee, L. Quintana-Murci, A. Hovnanian, F. Rozenberg, E. Vivier, F. Geissmann, M. Tardieu, L. Abel, and J. L. Casanova. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science 317:1522-1527. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, J., F. Hladik, A. Woodward, A. Klock, T. Peng, C. Johnston, M. Remington, A. S. Magaret, D. M. Koelle, A. Wald, and L. Corey. 2009. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat. Med. 15:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, J., D. M. Koelle, J. Cao, J. Vazquez, M. L. Huang, F. Hladik, A. Wald, and L. Corey. 2007. Virus-specific CD8+ T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J. Exp. Med. 204:595-603. [DOI] [PMC free article] [PubMed] [Google Scholar]