Abstract
Hepatitis B virus (HBV) biosynthesis involves the transcription of the 3.5-kb viral pregenomic RNA, followed by its reverse transcription into viral DNA. Consequently, the modulation of viral transcription influences the level of virus production. Nuclear receptors are the only transcription factors known to support viral pregenomic RNA transcription and replication. The coactivator peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) and corepressor small heterodimer partner (SHP) have central roles in regulating energy homeostasis in the liver by modulating the transcriptional activities of nuclear receptors. Therefore, the effect of PGC1α and SHP on HBV transcription and replication mediated by nuclear receptors was examined in the context of individual nuclear receptors in nonhepatoma cells and in hepatoma cells. This analysis indicated that viral replication mediated by hepatocyte nuclear factor 4α, retinoid X receptor α (RXRα) plus peroxisome proliferator-activated receptor α (PPARα), and estrogen-related receptor (ERR) displayed differential sensitivity to PGC1α activation and SHP inhibition. The effects of PGC1α and SHP on viral biosynthesis in the human hepatoma cell line Huh7 were similar to those observed in the nonhepatoma cells expressing ERRα and ERRγ. This suggests that these nuclear receptors, potentially in combination with RXRα plus PPARα, may have a major role in governing HBV transcription and replication in this cell line. Additionally, this functional approach may help to distinguish the transcription factors in various liver cells governing viral biosynthesis under a variety of physiologically relevant conditions.
In natural infection, hepatitis B virus (HBV) transcription and replication is essentially restricted to the hepatocytes in the livers of humans and a limited number of primates (15, 19, 33, 36, 41). HBV tropism is probably restricted at the level of entry by the viral receptor, which likely has a limited tissue distribution (10, 33). In addition, transcription of the viral genome limits HBV biosynthesis to cells expressing the nuclear receptors required for viral pregenomic RNA synthesis and replication (13, 40). The nuclear receptors present in hepatocytes that regulate HBV transcription include both ligand-dependent and orphan nuclear receptors which lack known ligands (16, 22, 30, 40). Long-chain fatty acids are ligands for peroxisome proliferator-activated receptor α (PPARα), which links HBV biosynthesis to energy homeostasis (9). Bile acids are ligands for farnesoid X receptor α (FXRα), further linking HBV biosynthesis to lipid metabolism (29, 30). Hepatocyte nuclear factor 4α (HNF4α) and estrogen-related receptor (ERR) are orphan nuclear receptors, which like PPARα and FXRα can display alteration in transcriptional activities in response to the coactivator peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) and the corepressor small heterodimer partner (SHP) (1, 23). PGC1α is critical for the activation of liver gluconeogenesis and therefore couples HBV transcription and replication to liver carbohydrate metabolism and whole-body energy homeostasis (43). SHP expression is activated by bile acids via FXRα and tumor necrosis factor α through AP1, leading to the inhibition of the activities of multiple nuclear receptors (6, 11, 14, 24). Therefore, SHP may regulate HBV biosynthesis in response to changing lipid metabolism or inflammatory signals within the liver (28).
Several nuclear receptors expressed in the liver have been shown to support HBV biosynthesis in nonhepatoma cell lines (see Fig. 1 to 6) (27a, 40). However, it is unclear which of these nuclear receptors are critical to supporting viral transcription and replication in hepatocytes in vivo. Conditional deletion of HNF4α in the liver of neonatal HBV transgenic mice demonstrated that this nuclear receptor was essential for viral biosynthesis (21). However, the early developmental loss of HNF4α is associated with decreased expression of a variety of additional nuclear receptors capable of supporting viral biosynthesis. Therefore, it is unclear if the loss in HBV transcription and replication observed in the liver-specific HNF4α-null HBV transgenic mouse is due directly to the loss of HNF4α or to the indirect effects on other nuclear receptors (18). Similarly, it is apparent that hepatoma cells can support HBV biosynthesis, but it has not been established which transcription factors present in these cells, but not in nonhepatoma cells, are responsible for supporting viral pregenomic RNA synthesis (4, 39, 40).
FIG. 1.
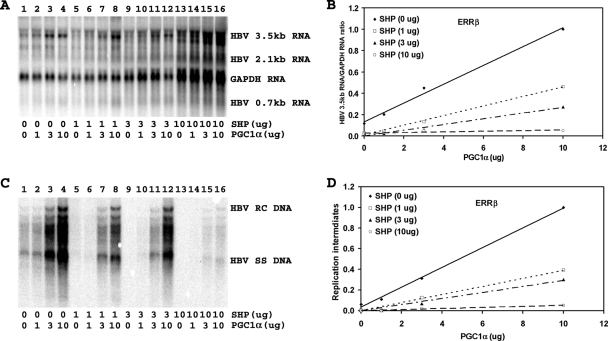
Effect of PGC1α and SHP expression on HBV biosynthesis in the human hepatoma cell line Huh7. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or with the HBV DNA (4.1-kbp) construct plus the PGC1α and SHP expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the 3.5-kb HBV RNA results from four independent experiments. Trend lines were calculated using linear regression analysis. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediate results from four independent experiments. Trend lines were calculated using linear regression analysis.
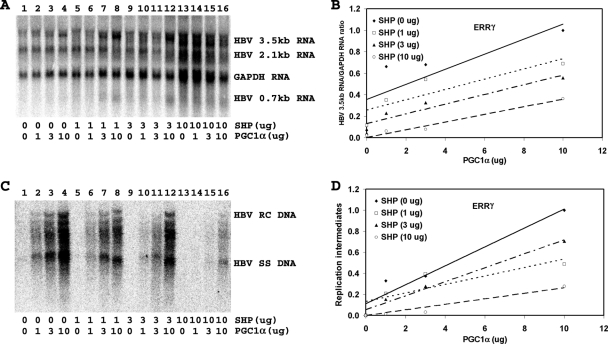
FIG. 6.
Effect of PGC1α and SHP expression on HBV biosynthesis in the human embryonic kidney cell line 293T expressing ERRγ. Cells were transfected with the HBV DNA (4.1-kbp) construct plus the ERRγ expression vector (lane 1) or the HBV DNA (4.1-kbp) construct plus the ERRγ, PGC1α, and SHP expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the 3.5-kb HBV RNA results from three independent experiments. Trend lines were calculated using linear regression analysis. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediate results from three independent experiments. Trend lines were calculated using linear regression analysis.
Given the importance of nuclear receptors and their associated coactivators and corepressors to liver energy homeostasis, it was of interest to investigate their relative importance in regulating HBV biosynthesis in hepatoma and nonhepatoma cells. Utilizing nonhepatoma cells, it was possible to investigate the sensitivity of HBV biosynthesis governed by the individual nuclear receptors to PGC1α and SHP modulation. This analysis indicated that the various nuclear receptors examined displayed distinct responses to various levels of coactivators and corepressors. By comparison, examination of the effect of PGC1α and SHP on HBV biosynthesis in a human hepatoma cell line, Huh7, suggested that ERRα or ERRγ alone or in combination with retinoid X receptor α (RXRα) plus PPARα activated the majority of the viral transcription and replication in this cell line. Therefore, this approach represents a means to examine the relative activities of nuclear receptors in liver cells contributing to HBV transcription and replication.
MATERIALS AND METHODS
Plasmid constructions.
The steps in the cloning of the plasmid constructs used in the transfection experiments were performed by standard techniques (35). HBV DNA sequences in these constructions were derived from the plasmid pCP10, which contains two copies of the HBV genome (subtype ayw) cloned into the EcoRI site of pBR322 (8). The HBV DNA (4.1-kbp) construct that contains 1.3 copies of the HBV genome includes the viral sequence from nucleotide coordinates 1072 to 3182 plus coordinates 1 to 1990 (40). This plasmid was constructed by cloning the NsiI/BglII HBV DNA fragment (nucleotide coordinates 1072 to 1990) into pUC13, generating pHBV(1072-1990). Subsequently, a complete copy of the 3.2-kbp viral genome linearized at the NcoI site (nucleotide coordinates 1375 to 3182 plus coordinates 1 to 1374) was cloned into the unique NcoI site (HBV nucleotide coordinate 1374) of pHBV(1072-1990), generating the HBV DNA (4.1-kbp) construct.
The pCMVHNF4α, pRS-hRXRα, pCMVPPARα-G, pSG5-mERRα, pcDNA3-2xFLAG-mERRβ, pSV5-mERRγ, pcDNA3-HA-hPGC1α, and pCMXSHP vectors express HNF4α, RXRα, PPARα-G, ERRα, ERRβ, ERRγ, PGC1α, and SHP polypeptides, respectively, from rat HNF4α, human RXRα, mouse PPARα-G, rat FXRα, mouse liver receptor homolog 1 (LRH1), mouse ERRα, mouse ERRβ, mouse ERRγ, human PGC1α, and mouse SHP cDNAs using the cytomegalovirus immediate-early promoter (pCMV, pCMX, and pcDNA3), the Rous sarcoma virus long terminal repeat (pRS), or the simian virus 40 early promoter (pSG5 and pSV5) (2, 5, 17, 24, 25, 27). The PPARα-G polypeptide contains a mutation in the PPARα cDNA, changing Glu282 to Gly, which may decrease the affinity of the receptor for the endogenous ligand. Consequently, this mutation increases the peroxisome proliferator-dependent (i.e., clofibric acid-dependent) activation of transcription from a peroxisome proliferator response element-containing promoter (27).
Cells and transfections.
The human hepatoma Huh7 cell line and human embryonic kidney 293T cell line were grown in RPMI 1640 medium and 10% fetal bovine serum at 37°C in 5% CO2/air. Transfections for viral RNA and DNA analyses were performed as previously described (26) using 10-cm plates, containing approximately 1 × 106 cells. DNA and RNA isolation was performed 3 days posttransfection. In 293T cells, the transfected DNA mixture was composed of 5 μg of HBV DNA (4.1 kbp) plus 1.5 μg of the nuclear receptor expression vectors pCMVHNF4α, pRS-hRXRα, pCMVPPARα-G, pSG5-ERRα, pcDNA3-2xFLAG-mERRβ, and pSV5-mERRγ and various amounts of the pcDNA3-HA-hPGC1α and pCMXSHP expression vectors (2, 5, 17, 24, 25, 27). In Huh7 cells, 1.5 μg of nuclear receptor expression vector was omitted. Controls were derived from cells transfected with HBV DNA and the expression vectors lacking a nuclear receptor cDNA insert (31). All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear receptors RXRα and PPARα (40).
Characterization of HBV transcripts and viral replication intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (38), with minor modifications. For RNA isolation (7), the cells were lysed in 1.8 ml of 25 mM sodium citrate at pH 7.0, 4 M guanidinium isothiocyanate, 0.5% (vol/vol) sarcosyl, and 0.1 M 2-mercaptoethanol. After addition of 0.18 ml of 2 M sodium acetate, pH 4.0, the lysate was extracted with 1.8 ml of water-saturated phenol plus 0.36 ml of chloroform-isoamyl alcohol (dilution of 49:1). After centrifugation for 30 min at 3,000 rpm in a Sorvall RT6000, the aqueous layer was precipitated with 1.8 ml of isopropanol. The precipitate was resuspended in 0.3 ml of 25 mM sodium citrate at pH 7.0, 4 M guanidinium isothiocyanate, 0.5% (vol/vol) sarcosyl, and 0.1 M 2-mercaptoethanol and precipitated with 0.6 ml of ethanol. After centrifugation for 20 min at 14,000 rpm in an Eppendorf 5417C microcentrifuge, the precipitate was resuspended in 0.3 ml of 10 mM Tris hydrochloride at pH 8.0, 5 mM EDTA, and 0.1% (wt/vol) sodium lauryl sulfate and precipitated with 45 μl of 2 M sodium acetate plus 0.7 ml of ethanol.
For the isolation of viral DNA replication intermediates, the cells were lysed in 0.4 ml of 100 mM Tris hydrochloride, pH 8.0, and 0.2% (vol/vol) NP-40. The lysate was centrifuged for 1 min at 14,000 rpm in an Eppendorf 5417C microcentrifuge to pellet the nuclei. The supernatant was adjusted to 6.75 mM magnesium acetate plus 200 μg/ml DNase I and incubated for 1 h at 37°C to remove the transfected plasmid DNA. The supernatant was readjusted to 100 mM NaCl, 10 mM EDTA, 0.8% (wt/vol) sodium lauryl sulfate, and 1.6 mg/ml pronase and incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol, precipitated with 2 volumes of ethanol, and resuspended in 100 μl of 10 mM Tris hydrochloride, pH 8.0, and 1 mM EDTA. RNA (Northern) and DNA (Southern) filter hybridization analyses were performed using 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as described previously (35).
Statistical analysis.
Trend lines were calculated using linear regression analysis using the method of least squares (Microsoft Excel chart of a linear regression trend line). Correlation coefficients were optimized using an iterative process that identified the maximum possible r value for each combination of nuclear receptors at all expression levels of PGC1α and SHP (Microsoft Excel CORREL). Initially, theoretical levels of replication intermediates predicted for all pair-wise combinations of nuclear receptors were compared with the observed level of replication in Huh7 cells and were optimized at a 1% level of resolution for each pair of nuclear receptors. The optimal percentages for any two nuclear receptors yielding r values greater than the maximum r value for a single nuclear receptor (r = 0.95) were reported. Similarly, the levels of replication for all three-way combinations of nuclear receptors were compared with the observed level of replication in Huh7 cells and were optimized at a 1% level of resolution for each of the three nuclear receptors. The optimal percentages for any three nuclear receptors yielding an r value greater than the maximum r value for any pair of these nuclear receptors were reported. This process was repeated with the inclusion of additional nuclear receptors until no further increase in r value was observed.
RESULTS
PGC1α and SHP modulate HBV biosynthesis in human hepatoma Huh7 cells.
Nuclear receptors can support HBV biosynthesis in nonhepatoma cells (see Fig. 1 to 6) (27a, 40). The activity of these nuclear receptors has also been shown to be modulated by the coactivator PGC1α and the corepressor SHP under a variety of conditions (1, 23). In addition, PGC1α and SHP are critical regulatory control components for metabolic homeostasis in the liver (1, 23). Therefore, it is important to establish the potential role that PGC1α and SHP might have in regulating HBV biosynthesis under normal and altered physiological conditions. As an initial step toward understanding the possible importance of PGC1α and SHP in the regulation of HBV biosynthesis in the liver, the effect of these proteins on HBV RNA and DNA synthesis was investigated in the human Huh7 hepatoma cell line.
Transfection of the HBV DNA (4.1-kbp) construct into Huh7 cells supports HBV transcription and replication (Fig. 1A and C, lane 1). Expression of increasing levels of PGC1α activates, whereas that of SHP inhibits, 3.5-kb HBV RNA synthesis and viral replication in a dose-dependent manner (Fig. 1). The effects of PGC1α and SHP on HBV DNA and RNA synthesis were quantitatively similar, indicating that the effects of PGC1α and SHP on transcription were reflected in the levels of observed viral replication (Fig. 1). Interestingly, increasing the levels of PGC1α enhanced viral biosynthesis at all levels of SHP expression in a similar manner (Fig. 1D). However, SHP inhibition was relatively ineffective at the lower levels examined and was readily apparent only at the highest level of SHP expression (Fig. 1D), suggesting that viral replication in Huh7 cells is relatively insensitive to SHP-mediated inhibition. The observation that PGC1α and SHP modulate HBV biosynthesis in Huh7 cells is consistent with the suggestion that nuclear receptors control viral transcription in these hepatoma cells.
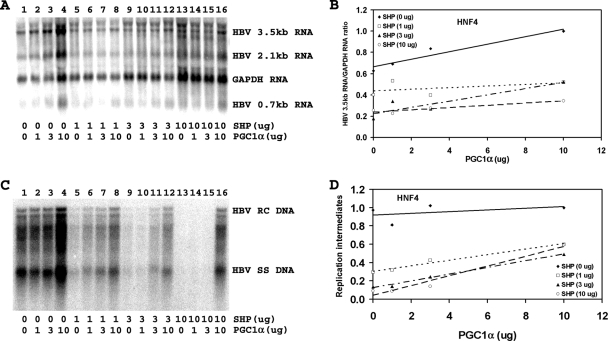
PGC1α and SHP modulate HNF4α-dependent HBV biosynthesis in human embryonic kidney 293T cells.
In an attempt to investigate the relative importance of various nuclear receptors in determining the level of HBV biosynthesis in liver cells, the effect of PGC1α and SHP on HBV transcription and replication was examined in nonhepatoma cells where viral synthesis was governed by a single nuclear receptor. Transfection of the HBV DNA (4.1-kbp) construct with the HNF4α expression vector into 293T cells supports HBV transcription and replication (Fig. 2A and C, lane 1). Expression of increasing levels of PGC1α activates, whereas that of SHP inhibits, both 3.5-kb HBV RNA synthesis and viral replication in a dose-dependent manner (Fig. 2). The effects of PGC1α and SHP on HNF4α-mediated HBV DNA and RNA synthesis in 293T cells were quantitatively similar, indicating that the effects of PGC1α and SHP on transcription were reflected in the levels of observed viral replication (Fig. 2). Increasing the levels of PGC1α modestly enhanced viral biosynthesis at all levels of SHP expression, although this effect appeared greatest at the highest level of SHP expression (Fig. 2D). HNF4α-dependent viral biosynthesis was highly sensitive to SHP inhibition, especially at the lowest levels of PGC1α expression (Fig. 2D). These observations demonstrate that the effects of PGC1α and SHP on HBV biosynthesis in Huh7 cells (Fig. 1) are quantitatively different from those observed in 293T cells expressing HNF4α (Fig. 2). This finding suggests that HNF4α may not have a significant role in governing HBV biosynthesis in Huh7 cells.
FIG. 2.
Effect of PGC1α and SHP expression on HBV biosynthesis in the human embryonic kidney cell line 293T expressing HNF4α. Cells were transfected with the HBV DNA (4.1-kbp) construct plus the HNF4α expression vector (lane 1) or the HBV DNA (4.1-kbp) construct plus the HNF4α, PGC1α, and SHP expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the 3.5-kb HBV RNA results from three independent experiments. Trend lines were calculated using linear regression analysis. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediate results from three independent experiments. Trend lines were calculated using linear regression analysis.
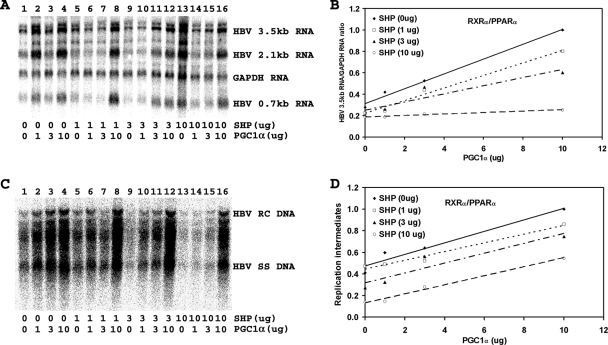
PGC1α and SHP modulate RXRα/PPARα-dependent HBV biosynthesis in human embryonic kidney 293T cells.
Transfection of the HBV DNA (4.1-kbp) construct with the RXRα and PPARα expression vectors into 293T cells supports HBV transcription and replication (Fig. 3A and C, lane 1). Expression of increasing levels of PGC1α activates, whereas that of SHP inhibits, 3.5-kb HBV RNA synthesis and viral replication in a dose-dependent manner (Fig. 3). Increasing the levels of PGC1α enhanced viral biosynthesis at all levels of SHP expression in a similar manner (Fig. 3D). However, SHP inhibition was relatively ineffective at the lower levels examined and was readily apparent only at the highest level of SHP expression (Fig. 3D), suggesting that RXRα/PPARα-mediated viral replication in 293T cells is relatively insensitive to SHP-mediated inhibition. These observations indicate that the effects of PGC1α and SHP on HBV biosynthesis in Huh7 cells (Fig. 1) are quantitatively more similar to those seen in 293T cells expressing RXRα/PPARα than to those seen in 293T cells expressing HNF4α (Fig. 2 and 3). These findings suggest that RXRα/PPARα may have a more important role than HNF4α in governing HBV biosynthesis in Huh7 cells.
FIG. 3.
Effect of PGC1α and SHP expression on HBV biosynthesis in the human embryonic kidney cell line 293T expressing RXRα/PPARα. Cells were transfected with the HBV DNA (4.1-kbp) construct plus the RXRα and PPARα expression vectors (lane 1) or the HBV DNA (4.1-kbp) construct plus the RXRα, PPARα, PGC1α, and SHP expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the 3.5-kb HBV RNA results from three independent experiments. Trend lines were calculated using linear regression analysis. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediate results from three independent experiments. Trend lines were calculated using linear regression analysis. All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear receptors RXRα and PPARα.
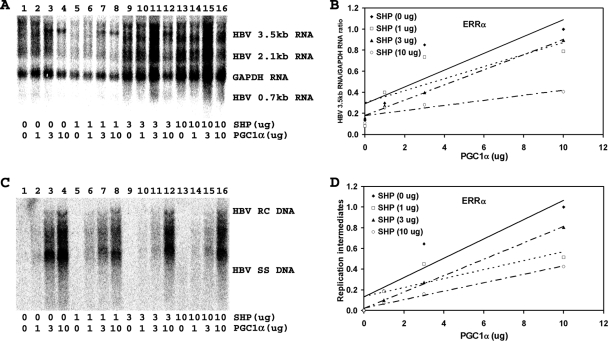
PGC1α and SHP modulate ERR-dependent HBV biosynthesis in human embryonic kidney 293T cells.
There are three isoforms of ERR, with ERRα being the most highly expressed one in the liver (3). The ERRβ and ERRγ isoforms are expressed at very low levels in the liver, suggesting that these isoforms probably do not contribute to liver-specific gene expression to a major extent (3). Transfection of the HBV DNA (4.1-kbp) construct with the ERR expression vectors into 293T cells supports limited HBV transcription and replication (Fig. 4 to 6, compare panel A, lane 1, and panel C, lane 1). Expression of increasing levels of PGC1α activates, whereas that of SHP inhibits, 3.5-kb HBV RNA synthesis and viral replication in a dose-dependent manner (Fig. 4 to 6). The effects of PGC1α and SHP on HBV DNA and RNA synthesis were quantitatively similar, indicating that the effects of PGC1α and SHP on transcription were reflected in the levels of observed viral replication (Fig. 4 to 6). Increasing the levels of PGC1α enhanced viral biosynthesis at all levels of SHP expression (Fig. 4D, 5D, and 6D). Additionally, ERR-dependent viral biosynthesis was sensitive to SHP inhibition, with ERRβ displaying the greatest sensitivity to SHP expression (compare Fig. 4D, 5D, and 6D). Most notably, the effects of PGC1α and SHP on HBV biosynthesis in 293T cells expressing ERRα and ERRγ (Fig. 4 and 6) were quantitatively very similar to those observed in Huh7 cells (Fig. 1). These observations suggest that ERRα or ERRγ may be the major transcription factor contributing to HBV biosynthesis in Huh7 cells.
FIG. 4.
Effect of PGC1α and SHP expression on HBV biosynthesis in the human embryonic kidney cell line 293T expressing ERRα. Cells were transfected with the HBV DNA (4.1-kbp) construct plus the ERRα expression vector (lane 1) or the HBV DNA (4.1-kbp) construct plus the ERRα, PGC1α, and SHP expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the 3.5-kb HBV RNA results from three independent experiments. Trend lines were calculated using linear regression analysis. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediate results from three independent experiments. Trend lines were calculated using linear regression analysis.
FIG. 5.
Effect of PGC1α and SHP expression on HBV biosynthesis in the human embryonic kidney cell line 293T expressing ERRβ. Cells were transfected with the HBV DNA (4.1-kbp) construct plus the ERRβ expression vector (lane 1) or the HBV DNA (4.1-kbp) construct plus the ERRβ, PGC1α, and SHP expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the 3.5-kb HBV RNA results from three independent experiments. Trend lines were calculated using linear regression analysis. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediate results from three independent experiments. Trend lines were calculated using linear regression analysis.
Relative importance of individual nuclear receptors to HBV biosynthesis in the human hepatoma Huh7 cells.
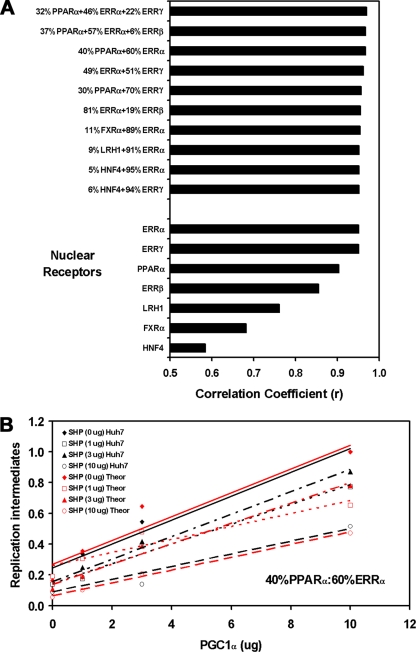
Qualitative comparison of the effects of PGC1α and SHP on HBV biosynthesis in both the human hepatoma Huh7 cells and the human embryonic kidney 293T cells in the presence of nuclear receptors indicated that ERRα or ERRγ might be controlling the majority of viral RNA and DNA synthesis in the Huh7 cells (Fig. 1 to 6). However, in an attempt to more precisely define the relative contributions of the various nuclear receptors to viral biosynthesis in Huh7 cells, the effects of PGC1α and SHP on viral replication in Huh7 cells were compared quantitatively to their effects on viral biosynthesis in 293T cells expressing individual nuclear receptors (Fig. 7A). Direct comparison of the individual replication profiles (Fig. 1 to 6) (27a) indicated that the effects of PGC1α and SHP on viral replication in Huh7 cells were most similar to those observed in 293T cells expressing ERRα (Correlation coefficient r = 0.95) and could be ordered as follows: ERRα (r = 0.95), ERRγ (r = 0.95), RXRα/PPARα (r = 0.90), ERRβ (r = 0.85), LRH1 (r = 0.76), RXRα/FXRα (r = 0.68), and HNF4α (r = 0.58). As it is likely that more than one nuclear receptor is contributing to viral biosynthesis in Huh7 cells, optimal theoretical combinations of transcription factors were identified, and the similarity of their replication profiles was compared with the effects of PGC1α and SHP on viral replication in Huh7 cells (Fig. 7). Six pair-wise combinations displayed correlation coefficients, ranging from r values of 0.95 to 0.97. In all of these combinations, ERRα plus ERRγ contributed a minimum of 81% of the total activity when PPARα was not considered. Depending on the combination of factors, PPARα could contribute up to 40% of the total activity (Fig. 7B). The additional nuclear receptors HNF4α, RXRα/FXRα, LRH1, and ERRβ appear to contribute to viral replication to a very limited extent only, based on this analysis. This conclusion is not altered when combinations of three or more factors are evaluated (Fig. 7A). Therefore, it appears that HBV transcription and replication in Huh7 cells is governed primarily by ERRα plus ERRγ, with a potentially significant contribution also coming from PPARα. The other nuclear receptors examined do not appear to contribute greatly to viral biosynthesis in Huh7 cells (Fig. 7).
FIG. 7.
Theoretical evaluation of the nuclear receptor combinations governing HBV biosynthesis in Huh7 cells. (A) Correlation coefficients were determined for optimal combinations of nuclear receptors based on their effects on viral replication in 293T cells compared with that in Huh7 cells in the presence of the different levels of the PGC1α and SHP coregulators. The combinations of nuclear receptors are reported in a descending order, with respect to their correlation coefficient values. Data used to determine the correlation coefficient values for RXRα/FXRα and LRH1 are included in the companion study (27a). 32% RXRα/PPARα plus 46% ERRα plus 22% ERRγ, r = 0.969; 37% RXRα/PPARα plus 57% ERRα plus 6% ERRβ, r = 0.968; 40% RXRα/PPARα plus 60% ERRα, r = 0.967; 49% ERRα plus 51% ERRγ, r = 0.962; 30% RXRα/PPARα plus 70% ERRγ, r = 0.957; 81% ERRα plus 19% ERRβ, r = 0.955; 11% RXRα/FXRα plus 89% ERRα, r = 0.954; 9% LRH1 plus 91% ERRα, r = 0.952; 5% HNF4α plus 95% ERRα, r = 0.952; 6% HNF4α plus 94% ERRγ, r = 0.952; 100% ERRα, r = 0.951; 100% ERRγ, r = 0.950; 100% RXRα/PPARα, r = 0.903; 100% ERRβ, r = 0.855; 100% LRH1, r = 0.761; 100% RXRα/FXRα, r = 0.682; 100% HNF4α, r = 0.585. (B) Example of the theoretical optimal best fit of the levels of viral replication obtained with RXRα/PPARα plus ERRα in 293T compared with that in Huh7 cells in the presence of the different levels of the PGC1α and SHP coregulators. Trend lines were calculated using linear regression analysis.
DISCUSSION
HBV transcription of the 3.5-kb pregenomic RNA is regulated by several nuclear receptors (Fig. 1 to 6) (27a, 40). As this transcript is reverse transcribed by the viral polymerase, regulation of transcription from the nucleocapsid promoter is an important determinant of HBV biosynthesis (40, 42). In addition, nuclear receptor activity is tightly regulated in the liver, as this is an important tissue with respect to whole-body energy and metabolic homeostasis (1, 23). Therefore, it appears that HBV biosynthesis is intimately linked to the gene regulation programs within the hepatocyte that adjust in response to changing physiological conditions and environmental demands (20, 37). The activities of nuclear receptors are modulated by a variety of coregulators in the liver (1, 23). However, PGC1α is particularly important for maintaining glucose homeostasis by modulating HNF4α-mediated regulation of liver gluconeogenesis (34). Similarly, SHP has a role in reducing the flux of cholesterol to bile acids within the liver. When bile acid levels within the hepatocytes increase, they activate FXR, which in turn increases transcription of the SHP gene (11, 24). In this manner, toxic levels of bile acids are prevented from accumulating within the liver. SHP negatively regulates a variety of nuclear receptors, restricting the level of expression of responsive genes (1).
Based on these findings, two basic issues relating to the role of nuclear receptors in controlling HBV biosynthesis are apparent. First, the relative importance of the various nuclear receptors regulating HBV biosynthesis under specific physiological conditions needs to be established. Second, the importance of coregulators in modulating nuclear receptor-mediated HBV biosynthesis has to be determined if the effects of altered liver metabolism on viral transcription and replication are to be understood. In an attempt to address these issues, the effects of PGC1α and SHP on viral biosynthesis were initially examined in the human hepatoma Huh7 cell line. A distinct pattern of activation of HBV biosynthesis by PGC1α was observed in the presence of different amounts of SHP (Fig. 1). Although increasing amounts of PGC1α displayed a relatively consistent positive effect on viral biosynthesis, relatively high levels of SHP were required to mediate any major effect on HBV replication (Fig. 1C and D). Most importantly, PGC1α and SHP had a similar effect on viral biosynthesis in the human embryonic 293T cell line in the presence of ERRα and ERRγ (Fig. 4C and D, 6C and D, and 7A). In contrast, none of the other nuclear receptors displayed a pattern of activation by PGC1α and inhibition by SHP that was similar to their effect in Huh7 cells (Fig. 2, 3, 5, and 7). Indeed, each nuclear receptor displayed a unique pattern of responsiveness to PGC1α-mediated activation and SHP-mediated inhibition (Fig. 2 to 6). Together, these observations suggest that viral biosynthesis in Huh7 cells is primarily governed by ERRα plus ERRγ or combinations of transcription factors with similar responsiveness to PGC1α and SHP (Fig. 4, 6, and 7). In addition, it suggests that this approach may be used to examine the roles of nuclear receptors in controlling HBV biosynthesis in additional hepatoma cell lines and possibly in vivo in hepatocytes under various physiological conditions.
The suggestion that HBV biosynthesis in Huh7 cells might be primarily governed by ERRα or ERRγ is somewhat unexpected. Of the ERR isoforms, only ERRα is abundant in the liver (3). Therefore, it appears likely that only the ERRα isoform might have a significant role in HBV biosynthesis in vivo during natural infection. However, ERRα is expressed at relatively high levels in a broad spectrum of tissues (3), whereas HBV transcription is highly restricted in vivo (13). This suggests that ERRα alone cannot explain the transcriptional control of tissue specificity, restricting HBV biosynthesis. The dependence of ERRα on additional liver-enriched nuclear receptors for activity at the nucleocapsid promoter might explain the possible role of ERRα in the regulation of HBV transcription in vivo. In this context, it is noteworthy that RXRα/PPARα is a liver-enriched nuclear receptor that may also contribute significantly to the level of viral transcription and replication in Huh7 cells (Fig. 7) (3). In nonhepatoma cells, it has been shown that RXRα/PPARα can support HBV biosynthesis (Fig. 3) (40), and PPARα agonists have also been shown to activate viral replication in vivo (32). However, the absence of PPARα in vivo does not affect the level of viral biosynthesis under normal physiological conditions (32). In vivo, it is the neonatal loss of HNF4α which results in the complete loss of viral biosynthesis (21). In this case, it may be the loss of the direct effect of HNF4α on 3.5-kb pregenomic HBV RNA synthesis that accounts for the absence of viral replication. Alternatively, it is possible that the loss of expression of additional nuclear receptors, which are developmentally dependent on HNF4α expression, prohibits HBV biosynthesis (18). Regardless of the explanation, it is apparent that ERRα or RXRα/PPARα alone, or ERRα in combination with RXRα/PPARα, does not control viral biosynthesis in the liver, and therefore, it appears that the factors governing HBV transcription in Huh7 cells might be significantly different from those performing the same role in vivo.
To analyze further the nuclear receptors that might control viral biosynthesis in Huh7 cells, the possibility that multiple nuclear receptors might be governing HBV transcription and replication in Huh7 cells was considered (Fig. 7). No theoretical combination of nuclear receptors, including significant activity derived from HNF4α, RXRα/FXRα, or LRH1, was identified that displayed a pattern of activation by PGC1α and inhibition by SHP that corresponded to that observed in Huh7 cells. This suggests that these nuclear receptors contribute little to HBV biosynthesis in Huh7 cells.
Utilizing the effect of PGC1α and SHP on nuclear receptor-mediated viral biosynthesis, ERRα and ERRγ, and possibly RXRα/PPARα, have been identified as potentially the most important nuclear receptors for HBV biosynthesis in Huh7 cells. To characterize further the nuclear receptors governing viral transcription and replication, it will be necessary to reduce or eliminate individual nuclear receptors from specific cell lines, possibly using small interfering RNAs, with a view to establishing further their contribution to HBV biosynthesis. Although this approach may be successful, it is possible that elimination of one nuclear receptor may lead to viral biosynthesis being governed by one or more of the remaining nuclear receptors capable of supporting HBV transcription. This might be addressed by determining the effect of PGC1α and SHP on viral biosynthesis in these modified Huh7 cells. However, it is unclear that the nuclear receptors observed to control HBV biosynthesis in Huh7 cells are particularly relevant in vivo (3, 12). More importantly, the characterization of the effect of coregulators on viral biosynthesis in hepatoma cells and nonhepatoma cells expressing individual nuclear receptors should permit the roles of glucagon and insulin signaling through PGC1α plus bile acid and proinflammatory signaling through SHP to be defined in a comprehensive manner. Understanding the relative importance of the various nuclear receptors and their coregulators in vivo is likely to permit the identification of suitable targets for additional HBV antiviral therapies.
Acknowledgments
We are grateful to Anastasia Kralli (The Scripps Research Institute, La Jolla, CA) for the plasmids pSG5-mERRα, pcDNA3-2xFLAG-mERRβ, pSV5-mERRγ, and pcDNA3-HA-hPGC1α, Eric F. Johnson (The Scripps Research Institute, La Jolla, CA) for the plasmids pCMVHNF4 and pCMVPPARα-G, Ronald M. Evans (Salk Institute, La Jolla, CA) for the plasmid pRS-hRXRα, and David Mangelsdorf (Southwestern Medical Center, Dallas, TX) for the plasmid pCMX-SHP. We thank Harel Dahari for his assistance with the statistical analysis.
This work was supported by Public Health Service grant AI30070 from the National Institutes of Health.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Bavner, A., S. Sanyal, J. A. Gustafsson, and E. Treuter. 2005. Transcriptional corepression by SHP: molecular mechanisms and physiological consequences. Trends Endocrinol. Metab. 16:478-488. [DOI] [PubMed] [Google Scholar]
- 2.Bonnelye, E., J. M. Vanacker, T. Dittmar, A. Begue, X. Desbiens, D. T. Denhardt, J. E. Aubin, V. Laudet, and B. Fournier. 1997. The ERR-1 orphan receptor is a transcriptional activator expressed during bone development. Mol. Endocrinol. 11:905-916. [DOI] [PubMed] [Google Scholar]
- 3.Bookout, A. L., Y. Jeong, M. Downes, R. M. Evans, and D. J. Mangelsdorf. 2005. Tissue-specific expression patterns of nuclear receptors. NURSA www.nursa.org/10.1621/datasets.02001.
- 4.Chang, C., K. Jeng, C. Hu, S. J. Lo, T. Su, L.-P. Ting, C.-K. Chou, S. Han, E. Pfaff, J. Salfeld, and H. Schaller. 1987. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 6:675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D., G. Lepar, and B. Kemper. 1994. A transcriptional regulatory element common to a large family of hepatic cytochrome P450 genes is a functional binding site of the orphan receptor HNF-4. J. Biol. Chem. 269:5420-5427. [PubMed] [Google Scholar]
- 6.Choi, Y. H., M. J. Park, K. W. Kim, H. C. Lee, Y. H. Choi, and J. Cheong. 2004. The orphan nuclear receptor SHP is involved in monocytic differentiation, and its expression is increased by c-Jun. J. Leukoc. Biol. 76:1082-1088. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Dubois, M. F., C. Pourcel, S. Rousset, C. Chany, and P. Tiollais. 1980. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc. Natl. Acad. Sci. USA 77:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forman, B. M., J. Chen, and R. M. Evans. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. USA 94:4312-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glebe, D., and S. Urban. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 13:22-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 12.Guidotti, L. G., C. M. Eggers, A. K. Raney, S. Y. Chi, J. M. Peters, F. J. Gonzalez, and A. McLachlan. 1999. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J. Virol. 73:10377-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, S., R. T. Stravitz, P. Dent., and P. B. Hylemon. 2001. Down-regulation of cholesterol 7α-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J. Biol. Chem. 276:15816-15822. [DOI] [PubMed] [Google Scholar]
- 15.Hu, X. L., H. S. Margolis, R. H. Purcell, J. Ebert, and B. H. Robertson. 2000. Identification of hepatitis B virus indigenous to chimpanzees. Proc. Natl. Acad. Sci. USA 97:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida, H., K. Ueda, K. Ohkawa, Y. Kanazawa, A. Hosui, F. Nakanishi, E. Mita, A. Kasahara, Y. Sasaki, M. Hori, and N. Hayashi. 2000. Identification of multiple transcription factors, HLF, FTF, and E4BP4, controlling hepatitis B virus enhancer II. J. Virol. 74:1241-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knutti, D., A. Kaul, and A. Kralli. 2000. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell. Biol. 20:2411-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyrmizi, I., P. Hatzis, N. Katrakili, F. Tronche, F. J. Gonzalez, and I. Talianidis. 2006. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 20:2293-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford, R. E., D. Chavez, K. M. Brasky, R. B. Burns III, and R. Rico-Hesse. 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc. Natl. Acad. Sci. USA 95:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., C. E. Oropeza, K. H. Kaestner, and A. McLachlan. 2009. Limited effects of fasting on hepatitis B virus (HBV) biosynthesis in HBV transgenic mice. J. Virol. 83:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, L., C. E. Oropeza, Jr., B. Sainz, S. L. Uprichard, F. J. Gonzalez, and A. McLachlan. 2009. Developmental regulation of hepatitis B virus biosynthesis by hepatocyte nuclear factor 4α. PLoS ONE 4:e5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., Y. H. Xie, Y. Y. Kong, X. Wu, L. Zhu, and Y. Wang. 1998. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which finds and activates enhancer II of hepatitis B virus. J. Biol. Chem. 273:29022-29031. [DOI] [PubMed] [Google Scholar]
- 23.Lin, J., C. Handschin, and B. M. Spiegelman. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1:361-370. [DOI] [PubMed] [Google Scholar]
- 24.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 25.Mangelsdorf, D. J., E. S. Ong, J. A. Dyck, and R. M. Evans. 1990. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature 345:224-229. [DOI] [PubMed] [Google Scholar]
- 26.McLachlan, A., D. R. Milich, A. K. Raney, M. G. Riggs, J. L. Hughes, J. Sorge, and F. V. Chisari. 1987. Expression of hepatitis B virus surface and core antigens: influences of pre-S and precore sequences. J. Virol. 61:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muerhoff, A. S., K. J. Griffin, and E. F. Johnson. 1992. The peroxisome proliferator-activated receptor mediates the induction of CYP4A6, a cytochrome P450 fatty acid omega-hydroxylase, by clofibric acid. J. Biol. Chem. 267:19051-19053. [PubMed] [Google Scholar]
- 27a.Ondracek, C. R., V. C. Reese, C. N. Rushing, C. E. Oropeza, and A. McLachlan. 2009. Distinct regulation of hepatitis B virus biosynthesis by peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner in human hepatoma cell lines. J. Virol. 83:12545-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oropeza, C. E., L. Li, and A. McLachlan. 2008. Differential inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by small heterodimer partner. J. Virol. 82:3814-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks, D. J., S. G. Blanchard, R. K. Bledsoe, G. Chandra, T. G. Consler, S. A. Kliewer, J. B. Stimmel, T. M. Willson, A. M. Zavacki, D. D. Moore, and J. Lehmann. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365-1368. [DOI] [PubMed] [Google Scholar]
- 30.Ramiere, C., C. Scholtes, O. Diaz, V. Icard, L. Perrin-Cocon, M. A. Trabaud, V. Lotteau, and P. Andre. 2008. Transactivation of the hepatitis B virus core promoter by the nuclear receptor FXRα. J. Virol. 82:10832-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raney, A. K., J. L. Johnson, C. N. A. Palmer, and A. McLachlan. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71:1058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raney, A. K., E. F. Kline, H. Tang, and A. McLachlan. 2001. Transcription and replication of a natural hepatitis B virus nucleocapsid promoter variant is regulated in vivo by peroxisome proliferators. Virology 289:239-251. [DOI] [PubMed] [Google Scholar]
- 33.Raney, A. K., and A. McLachlan. 1991. The biology of hepatitis B virus, p. 1-37. In A. McLachlan (ed.), Molecular biology of the hepatitis B virus. CRC Press, Boca Raton, FL.
- 34.Rhee, J., Y. Inoue, J. C. Yoon, P. Puigserver, M. L. Fan, F. J. Gonzalez, and B. M. Spiegelman. 2003. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. USA 100:4012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shlomai, A., N. Paran, and Y. Shaul. 2006. PGC-1α controls hepatitis B virus through nutritional signals. Proc. Natl. Acad. Sci. USA 103:16003-16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Summers, J., P. M. Smith, M. Huang, and M. Yu. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sureau, C., J.-L. Romet-Lemonne, J. I. Mullins, and M. Essex. 1986. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 47:37-47. [DOI] [PubMed] [Google Scholar]
- 40.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren, K. S., J. L. Heeney, R. A. Swan, Heriyanto, and E. J. Verschoor. 1999. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus). J. Virol. 73:7860-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Will, H., W. Reiser, T. Weimer, E. Pfaff, M. Buscher, R. Sprengle, R. Cattaneo, and H. Schaller. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon, J. C., P. Puigserver, G. X. Chen, J. Donovan, Z. D. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, C. B. Newgard, and B. M. Spiegelman. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131-138. [DOI] [PubMed] [Google Scholar]