Abstract
The RNA-dependent RNA polymerase 3Dpol is required for the elongation of positive- and negative-stranded picornavirus RNA. During the course of investigating the effect of the transgenic expression of viral genes on the host immune response, we evaluated the viral load present in the host after infection. To our surprise, we found that 3D transgenic expression in genetically susceptible FVB mice led to substantially lower viral loads after infection with Theiler's murine encephalomyelitis virus (TMEV). As a result, spinal cord damage caused by chronic viral infection in the central nervous system was reduced in FVB mice that expressed 3D. This led to the preservation of large-diameter axons and motor function in these mice. The 3D transgene also lowered early viral loads when expressed in FVB-Db mice resistant to persistent TMEV infection. The protective effect of 3D transgenic expression was not altered in FVB-Rag−/−.3D mice that are deficient in T and B cells, thus ruling out a mechanism by which the overexpression of 3D enhanced the adaptive immune clearance of the virus. Understanding how endogenously overexpressed 3D polymerase inhibits viral replication may lead to new strategies for targeting therapies to all picornaviruses.
Picornavirus infection is a major contributor to worldwide disease. Diseases such as poliomyelitis and hand-foot-and-mouth disease can be fatal. Other picornaviruses, such as rhinovirus, are partly responsible for upper respiratory tract infections. There are no drugs to treat picornavirus illness. However, some therapies show promise in vitro and in animal models. Winthrop compounds, which bind to hydrophobic sites on the surface of the virion that are important in viral attachment to the host and uncoating, decreased the number of upper respiratory symptoms following challenge with coxsackie virus 21 (45). A similar pocket binding drug, pleconaril (VP63843), showed 95% inhibition against 215 non-polio enteroviruses (39). A phase II trial of this drug against enteroviral meningitis decreased disease duration compared to the placebo (1). However, these drugs have side effects and were less effective in larger studies. Another limitation is that mutant viruses arose that sterically inhibited binding by substituting a bulky amino acid in the binding pocket (21). The administration of small interfering RNA (siRNA) has shown some promise in controlling picornavirus infections; however, the development of a delivery system is a major hurdle (8-10, 44).
The picornavirus Theiler's murine encephalomyelitis virus (TMEV) is a member of the Cardiovirus genus. TMEV is divided into two groups based on disease in mice after intracerebral injection (12, 15). The highly virulent GDVII subgroup causes fatal encephalitis, and the lowly virulent Theiler's original (TO) subgroup, which includes BeAn and DA strains, causes a persistent infection in the white matter of the central nervous system (CNS), leading to chronic inflammation and demyelination in genetically susceptible mice. Chronic inflammation and demyelination leads to secondary axonal dysfunction and paralysis. Therefore, the BeAn and DA strains of TMEV infection in mice are used as animal models of demyelinating diseases such as multiple sclerosis (4, 11, 13).
Inbred strains of mice differ in their susceptibility to TMEV (14, 18). Resistance to persistent infection depends on the haplotype of the major histocompatibility complex (H-2). Mice of the H-2b,d,k haplotype are resistant to persistent infection, whereas mice of the H-2f,p,q,r,s,v haplotype are susceptible to persistent infection (32). Resistance to persistent infection has been further defined to the D locus of H-2 (33, 35). The mice used in this paper all are on an FVB/NJ background. Due to the prominent pronuclei in their fertilized eggs and large litter size, FVB/NJ mice commonly are used for transgenic injection (43). These mice are of the H-2q haplotype and are susceptible to persistent TMEV infection. Persistent infection leads to chronic spinal cord inflammation and demyelination in all inbred mice that are genetically susceptible to TMEV. FVB-Db mice contain the H-2Db transgene, which confers resistance to persistent TMEV infection (2). FVB mice and FVB-Db mice have similar early acute disease in the brain at 7 days postinfection (dpi); however, unlike FVB mice, FVB-Db mice control the virus in the brain and spinal cord by day 45 and do not develop demyelination.
Picornaviruses perform multiple tasks inside host cells for successful viral replication, with very few gene products being responsible for these tasks. The single-stranded RNA picornavirus genome has, on average, 7,500 nucleotides and produces a single polyprotein that is cleaved by its own virus-encoded proteases. One of these proteins, the RNA-dependent RNA-polymerase 3Dpol, is required for the elongation of positive- and negative-stranded viral RNA. 3Dpol oligomerizes, which favors elongation and binding to RNA (16). 3Dpol forms a membranous replication complex with VPg and precursor proteins 3AB and 3CD to initiate VPg uridylylation, which serves as a primer for positive- and negative-strand RNA replication by 3Dpol (25, 40, 42). The stimulatory effect of 3AB on RNA replication by 3Dpol is inhibited by increasing concentrations of 3D (26, 31).
During the course of investigating the effect of the transgenic expression of viral genes on the host immune response to TMEV, we evaluated the viral load present in the host after infection. Mice expressing the 3D transgene were used as control mice, since previous studies have shown that the 3D protein was ignored by T and B cells in the immune system (3, 22, 29). To our surprise, we found that the 3D transgenic mice substantially reduced TMEV in vivo. Therefore, we set out to study the effect of 3D overexpression in a transgenic mouse model of TMEV infection.
MATERIALS AND METHODS
Mice.
FVB/NJ (001800) and B6.129S7-Rag1tm1Mom/J (002216) mice were obtained from Jackson Laboratory (Bar Harbor, ME). B6.129S7-Rag1tm1Mom/J mice were backcrossed to FVB/NJ mice for nine generations to make Rag1−/− mice on the FVB/NJ background. FVB-Db mice were provided by Larry Pease (19). A total of 646 mice were used for this study. All animal protocols followed National Institutes of Health guidelines and were approved by the Mayo Foundation Institutional Animal Care and Use Committee.
Cell lines and primary cells.
SQSV and L2 cells were grown in Dulbecco's minimum essential medium (DMEM) containing l-glutamine, glucose, sodium pyruvate, 10% heat-inactivated fetal bovine serum, and penicillin-streptomycin.
Generation of 3D transgenic mice.
Transgenic 3D mice were made as described previously (29). Transgenic mice were generated by cloning nucleotides 6586 to 7968, which code for the entire 3D polymerase in the DA strain of TMEV (GenBank accession no. M20301), into the eukaryotic expression vector pUB6, which contains an upstream human ubiquitin c promoter (Invitrogen, Carlsbad, CA). 3D TMEV DNA was amplified by PCR from pDAFL3 and directionally cloned into pUB6 using a BamHI site on the 5′ end of the cloned fragment and an EcoRV site on the 3′ end. The construct was cloned while maintaining the His Tag included in the vector, thereby allowing the identification of 3D by this marker. This vector then was cut with BglII and PvuII to yield a fragment of 2,935 bp. The resulting fragment encoded all 462 amino acids of the viral RNA polymerase, which then was gel purified and sequenced before injection into embryos (see Fig. S1 in the supplemental material). The sequence revealed a base pair substitution at nucleotide 7620. This substitution did not change the codon, leaving the amino acid sequence identical to that of the sequenced viral 3D polymerase. The gel-purified DNA was injected into FVB embryos for implantation into pseudopregnant females. All embryo injections and implantations were done at the Mayo Clinic Transgenic Core Facility under the direction of Chella David. Tail samples from the offspring were used to obtain genomic DNA for the determination of transgene integration. DNA samples were screened using primers for the TMEV 3D gene as well as the ubiquitin c promoter region. Thirty-nine potential founder mice containing 3Dpol were screened for transgene integration, six mice were positive, and two were chosen for breeding based on the highest copy of transgene integration. These mice were used to establish two lines. Line 1 bred well and constitutes the majority of the data in this study. Line 2 did not breed well and thus could not be maintained; however, we obtained pathological data from this line at the 45-day time point. Line 2 showed a greater reduction of virus-induced demyelination by the insertion of the 3D transgene than line 1 (see Fig. 3C). The less-effective line 1, which was used for these experiments, nevertheless profoundly affected TMEV-induced infection and demyelination. All mice used in the experiments were screened by PCR for the presence of the 3D transgene prior to their use in subsequent assays. FVB-3D transgenic mice then were crossed to FVB-Db transgenic mice to obtain FVB-Db.3D mice or FVB-Rag−/− mice to obtain FVB-Rag−/−.3D mice. All of these transgenic lines behaved similarly to the wild-type controls prior to infection with TMEV.
FIG. 3.
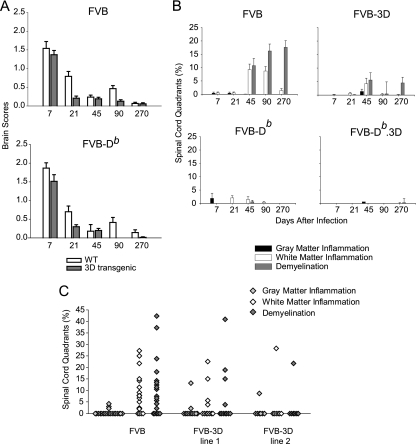
Pathology of the brain and spinal cord in 3D transgenic mice. (A) Time course of brain scores in mice after infection with TMEV. Seven areas of the brain (cerebellum, brain stem, cortex, hippocampus, striatum, corpus callosum, and meninges) were graded based on a four-point scale as described in Materials and Methods. Scores from all areas were averaged and are plotted as shown. Bars represent the means ± SEM of each group. WT, wild type. (B) Percentages of spinal cord quadrants containing gray matter inflammation, white matter inflammation, or demyelination. Data are expressed as percentages of quadrants with the pathological abnormality as a function of all spinal cord quadrants. Note that the 45-day time point contains combined data from lines 1 and 2 of FVB-3D transgenic mice. (More than 50 mice were studied in all groups for brain and spinal cord. See the supplemental material for actual numbers.) (C) Data from transgenic line 1 and line 2 at the 45-day time point shown in panel B are shown separately and compared to results for the nontransgenic FVB mice. Each point represents one mouse.
Virus and infection.
The Daniels strain of TMEV was used for all experiments (24). Mice were injected intracranially with 2 × 105 PFU of TMEV in a volume of 10 μl.
RNA isolation.
Total RNA was extracted from brain and spinal cord as described previously (38). Briefly, the tissues were frozen and stored in liquid nitrogen. Tissue samples were homogenized in TRIzol (Invitrogen, Carlsbad, CA), and total RNA was isolated according to the manufacturer's recommendations. Total RNA concentrations were determined by a spectrophotometer and then equilibrated to 50 ng/μl and stored at −80°C.
Reverse transcription-PCR (RT-PCR) for analysis of viral RNA transcripts.
A 238-bp region of the DA strain of TMEV that codes for the VP2 capsid protein was used to quantify viral transcripts in the brain and spinal cord of infected mice using primers VP2-F and VP2-R. The housekeeping gene GAPDH was quantified to control for differences in total mRNA amount and integrity using primers GAPDH-F and GAPDH-R. Primer pair sequences for VP2 and GAPDH are listed in Table S1 in the supplemental material.
Total RNA was reverse transcribed and amplified in one step using a LightCycler instrument (Roche, Indianapolis, IN). The reactions were done in 20-μl capillaries containing 10 μl of 2× Quantifast SYBR green master mix (Qiagen, Valencia, CA), 0.2 μM of each forward and reverse primer, 20 U of Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA), 0.2 μg of total RNA, and molecular-grade water. Reaction conditions for VP2 and GAPDH were the following: reverse transcription at 55°C for 10 min, followed by denaturation at 95°C for 2 min and then 40 cycles of amplification, which included melting at 95°C for 10 s, annealing/extension at 60°C for 30 s, and data acquisition at 82°C for 6 s. The reaction conditions for melting curve analysis were 95°C for 10 s, 60°C for 5 s, and ramping up to 95°C at 0.1°C/s with continuous data acquisition.
Standards were generated for VP2 by cloning the amplicon of the gene into pcDNA3.1 (Invitrogen, Carlsbad, CA), and 10-fold serial dilutions of a known quantity of the plasmid were run alongside unknown samples. PCR product curves were linear across serial 10-fold dilutions, and the melting curve analysis indicated the synthesis of a single homogenous product at the expected melting temperature. Data were expressed as the total amount of VP2 transcript in the brain or spinal cord calculated as follows: [(copy number obtained from standard curve) × (total amount of RNA from tissue)]/amount of RNA used in reaction. To set the baseline sensitivity of the assay to 1, the total copy number obtained from the nonspecific amplification of an uninfected mouse brain or spinal cord then was divided by total viral transcripts in infected mice.
Viral plaque assay.
SQSV cells were infected with TMEV at a multiplicity of infection of 1 in a 12-well plate containing 1 × 105 cells per well. Cells were washed in Dulbecco's phosphate-buffered saline (PBS) (Mediatech) and harvested after 8, 16, and 24 h by scraping the cells off in 1 ml DMEM (Invitrogen, Carlsbad, CA). The cells went through three freeze-thaw cycles, were vortexed, and were centrifuged for 1 min at 16,000 × g. Tenfold serial dilutions of the supernatant were made in DMEM containing 2% fetal bovine serum (FBS). A volume of 250 μl of each dilution was plated in triplicate on L2 cells, which were plated at a density of 1 × 105/well in 12-well plates the day before. The cells were incubated at 37°C for 1 h before the addition of 1 ml of DMEM containing 2% FBS and 0.5% low-melting-point agarose. After 72 h at 37°C, the cells were fixed and stained with cresyl violet, and plaques were counted. Replicates from the same experiment were used to quantify viral RNA transcripts by quantitative real-time RT-PCR to obtain a correlation between the two assays.
Immune staining for virus antigen in the CNS.
Immunocytochemistry was performed on paraffin-embedded sections as previously described (27). Slides were deparaffinized in xylene and rehydrated through an ethanol series (absolute, 95, 70, and 50%). Virus antigen staining used polyclonal antiserum to TMEV (36), which reacts strongly with the capsid proteins of TMEV. Following incubation with a secondary biotinylated antibody (Vector Laboratories, Burlingame, CA), immunoreactivity was detected using the avidin-biotin immunoperoxidase technique (Vector Laboratories). The reaction mixture was developed using Hanker-Yates reagent with hydrogen peroxide as the substrate (Polysciences, Warrington, PA). Slides were lightly counterstained with Gill III hematoxylin. We examined the distribution of virus antigen at 0 and 7 days in the brain and at 0, 45, and 270 days in the spinal cord. Each area of the brain was scored for the number of antigen-positive cells per ×40 high-power field in areas of maximal pathology. Five mice were used for each strain except for FVB-Db, for which four mice were used. The uninfected control groups used two mice per strain. For the spinal cord, we scored every spinal quadrant for the presence or absence of virus antigen-positive cells in either the gray or white matter in every animal. We examined, on average, eight spinal cord sections with representative samples from the cervical, thoracic, and lumbar areas. Five randomly selected mice per strain and experimental group were used, and two mice were used for the uninfected control group. The data were expressed as the percentages of spinal cord quadrants showing virus antigen-positive cells in the white matter of the spinal cord.
Spinal cord morphometry.
Following perfusion with Trump's fixative (phosphate-buffered 4% formaldehyde with 1% glutaraldehyde, pH 7.4), spinal cords were cut into 1-mm coronal blocks. Every third block was osmicated and embedded in glycol methacrylate. Two-micrometer sections were prepared and stained with a modified erichrome-cresyl violet stain (30). Morphological analysis was performed on 12 to 15 sections per mouse as previously described (34). Each quadrant from every coronal section from each mouse was graded for the presence or absence of gray matter disease, meningeal inflammation, and demyelination. The score was expressed as the percentage of spinal cord quadrants examined with the pathological abnormality. A maximum score of 100 indicated a particular pathological abnormality in every quadrant of all spinal cord sections of a given mouse. All grading was performed on coded sections without knowledge of the experimental group. Additional spinal cord blocks were embedded in paraffin for immunocytochemistry. Spinal cords at 270 dpi were embedded in araldite plastic and stained with para-phenylene diamine (PPD) to detect myelinated and demyelinated axons.
Brain pathology.
Brain pathology was assessed at 7, 21, 45, 90, and 270 dpi using our previously described technique (28). Following perfusion with Trump's fixative, we made two coronal cuts in the intact brain at the time of removal from the skull (one section through the optic chiasm and a second section through the infundibulum). As a guide, we used the Atlas of the Mouse Brain and Spinal Cord corresponding to sections 220 and 350, page 6 (41). This resulted in three blocks that then were embedded in paraffin. This allowed for the systematic analysis of the pathology of the cortex, corpus callosum, hippocampus, brain stem, striatum, and cerebellum. Resulting slides then were stained with hematoxylin and eosin. Pathological scores were assigned without knowledge of experimental group to the following areas of the brain: cortex, corpus callosum, hippocampus, brainstem, striatum, and cerebellum. Each area of the brain was graded on a four-point scale as follows: 0, no pathology; 1, no tissue destruction and only minimal inflammation; 2, early tissue destruction (loss of architecture) and moderate inflammation; 3, definite tissue destruction (demyelination, parenchymal damage, cell death, neurophagia, and neuronal vacuolation); and 4, necrosis (complete loss of all tissue elements with associated cellular debris). Meningeal inflammation was assessed and graded as follows: 0, no inflammation; 1, one cell layer of inflammation; 2, two cell layers of inflammation; 3, three cell layers of inflammation; 4, four or more cell layers of inflammation. The area with maximal tissue damage was used for the assessment of each brain region.
Assessment of functional disease using an accelerated rotarod assay.
The Rotamex rotarod (Columbus Instruments, Columbus, OH) measures balance, coordination, and motor control and was used to assess neurologic function. The rotarod consists of a suspended rod powered by a variable-speed motor capable of running at a fixed speed or accelerating at a constant rate. Mice were trained and tested according to the protocol established previously (17). Prior to injection with TMEV, each mouse received 3 days of training using a constant speed protocol. This consisted of three 3-min trials during 3 days (12 rpm on day 1, 13 rpm on day 2, and 14 rpm on day 3). Mice then were tested using an accelerated assay (initial speed of 10 rpm, accelerating at 10 rpm until the mouse fell off). The maximum speed was 70 rpm at 6 min. Rotarod performance was measured at 21, 45, 90, and 180 dpi. All subsequent trials consisted of 1 day of accelerated speed testing (three trials consisting of an initial speed of 10 rpm and accelerating at 10 rpm until the mouse fell off). The speed (in revolutions per minute) at the time of fall was recorded for each mouse, and data were expressed as the percent preservation of the PBS-infected FVB control group function. Ratios to calculate the percent preservation at 0, 21, 45, 90, and 180 dpi were calculated by dividing the trial means of each mouse's performance by the average trial means of the PBS-infected FVB control group for each time point after infection.
Axonal counts.
Animals were sacrificed for the analysis of spinal cord pathology and axonal counts. Mice were anesthetized with an overdose of sodium pentobarbital, and tissues were fixed by the intracardiac perfusion of Trump's fixative. Spinal cords were removed and cut into 1-mm blocks, and every third block was postfixed and stained with osmium tetroxide and embedded in araldite plastic (Polysciences, Warrington, PA). A mid-thoracic section was selected for axonal count analysis, since this would be the most representative of both ascending and descending tracks from the brainstem neurons. One-micron-thick cross-sections were cut from this block and stained with 4% PPD to visualize the myelin sheaths. An Olympus Provis AX70 microscope fitted with a DP70 digital camera and a 60× oil objective captured six sample images from the mid-thoracic spinal cord section according to the sampling scheme shown in Fig. 7A. We sampled approximately 400,000 μm2 of white matter from each animal. Images were captured from regions of normal-appearing white matter containing a relative absence of demyelination. The calculation of absolute axon numbers was performed as previously reported (7).
FIG. 7.
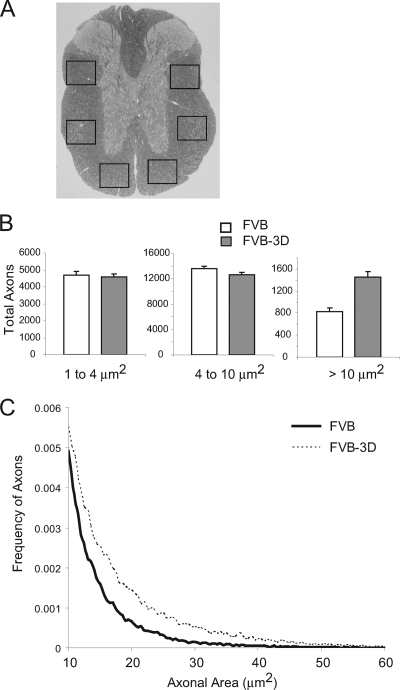
Axon counts at 270 dpi in FVB (n = 29) and FVB-3D mice (n = 24). (A) Cross-section of the spinal cord. Boxes show the six areas where axons were counted. (B) Data show the total number of axons per mouse (means ± SEM) as a function of the size of axon diameter (1 to 4 μm, small; 4 to 10 μm, medium; >10 μm, large). (C) Histogram of axons greater than 10 μm in size. The lines represent the relative frequency of large-diameter axons using 0.5-μm2 intervals as bins. Note the tail of large-diameter axons in infected FVB-3D mice but not in infected FVB mice.
Irradiation of mice.
Mice were exposed to 600 rads the day prior to infection with TMEV.
Statistics.
Data were analyzed using either the Student's t test for equal variance and normally distributed data or the Mann-Whitney rank sum test for data that were not. For comparisons of more than one group, analysis of variance was used for equal variance and normally distributed data, in which the Holmes-Sidak or Tukey test was used for pair-wise multiple comparisons, and Kruskal-Wallis analysis of variance on ranks for data that were not equally variant or normally distributed, in which Dunn's test was used for pair-wise multiple comparisons. The log-rank test was used for the analysis of Kaplan-Meier survival curves. The level for significance was set at P < 0.05 for all tests.
RESULTS
Characterization of 3D transgenic FVB mice.
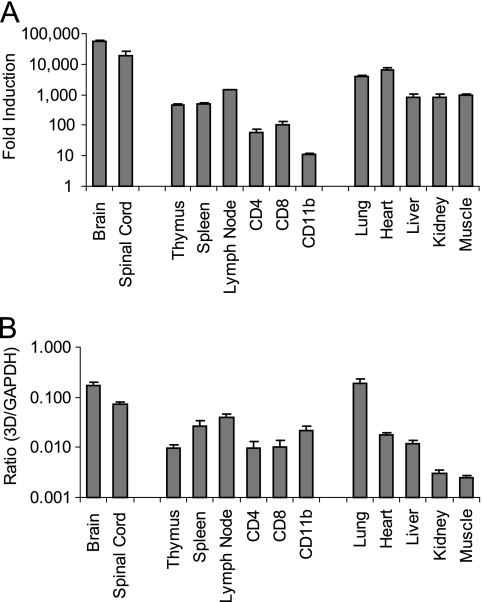
Our 3D transgenic mice express the entire TMEV 3D polymerase under the control of a ubiquitin promoter. Previously, we quantified 3D transgenic mRNA transcripts in both lines of mice in the brain, and similar levels of transcript were found (29). To further asses the expression levels in different tissues and cells, we quantified the transgenic 3D transcripts in line 1 by real-time quantitative RT-PCR (Fig. 1; also see Table S2 in the supplemental material). Total RNA was isolated from the brain, spinal cord, thymus, spleen, lymph nodes, kidney, lungs, heart, and muscle. Transcripts of the transgene were detected in all tissues. The highest numbers of 3D transcripts were seen in the CNS, which showed a 5.5 × 104-fold increase above the background level in the brain and a 1.9 × 104-fold increase in the spinal cord. This is equivalent to 16.7% in the brain and 7.3% in the spinal cord of the high expression levels seen with GAPDH mRNA in each tissue. We also isolated RNA from CD4, CD8, and CD11b cells sorted by fluorescence-activated cell sorting from the spleen of mice, which showed small numbers of transcripts compared to those of other organs and tissues.
FIG. 1.
Levels of 3D mRNA transcripts in 3D transgenic mice. (A) Viral transcripts were quantified by real-time RT-PCR by using SYBR green. Primers specific for the 3D transgene were used. Cycle threshold values in nontransgenic FVB mice were used as a reference point for nonspecific amplification. Data are expressed as the mean induction (n-fold) ± SEM (n = 3) from the nonspecific amplification of nontransgenic FVB for each tissue. (B) Ratio of 3D increase (n-fold) to GAPDH increase (n-fold). GAPDH transcripts were quantified by real-time RT-PCR. GAPDH increase (n-fold) in each mouse was relative to the value for the same reference point as that used to quantify 3D transcripts. Data are expressed as mean ratios ± SEM (n = 3).
3D transgenic expression reduces viral load in FVB and FVB-Db mice.
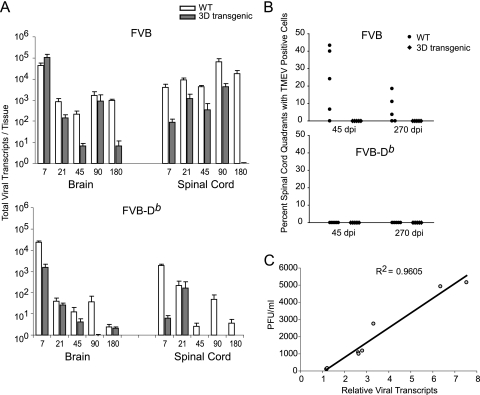
We infected 3D transgenic FVB mice (FVB-3D), which express 3D under the control of a human ubiquitin promoter, and wild-type FVB mice with the DA strain of TMEV. After infection, total RNA was extracted from the brain and spinal cord to quantitate viral transcripts by real-time quantitative PCR expressed as total viral RNA transcripts per tissue (Fig. 2A; also see Table S3 in supplemental material). In the brain, both FVB and FVB-3D mice contained about 4.5 logs of viral transcripts by 7 dpi. Although viral transcripts in FVB and FVB-3D mice decreased over the next 173 days, the drop in transcripts was significantly faster in FVB-3D mice. By 180 dpi, FVB mice had 2.5 logs more viral transcripts than did FVB-3D mice (P = 0.016).
FIG. 2.
Levels of TMEV in the brain and spinal cord of 3D transgenic mice. (A) Viral transcripts were quantified by real-time RT-PCR by using SYBR green. Primers specific for VP2 were used. Viral transcripts calculated from nonspecific amplification in uninfected FVB mice were divided by the viral transcripts of infected mice to set the baseline sensitivity of the assay to 1. White bars show values of infected nontransgenic mice, and dark bars show values of 3D transgenic mice. Data are shown as means ± SEM of five mice per group. WT, wild type. (B) Percentages of spinal cord quadrants containing TMEV-positive cells in the white matter are shown after 45 and 270 dpi by the same immunoperoxidase staining technique as that used for the brain. Each point represents one mouse. (C) Correlation of PFU from plaque assay and relative viral transcripts by real-time RT-PCR after the infection of SQSV cells with TMEV for 8, 16, and 24 h. The same VP2-specific primers were used as those for panel A. Each point represents data obtained from separate replicate samples for both assays.
Following TMEV infection in the brain of mice, the virus spreads to the spinal cord. The amount of virus that persists in the spinal cord after infection depends on both the virus's intrinsic ability to replicate and the ability of the immune system to clear virus. We studied the effect of 3D on TMEV in the spinal cord. Unlike the brain at day 7, the levels of viral transcripts in the spinal cord of FVB-3D mice already were significantly lower than those in FVB mice (4,100 ± 1,500 in FVB and 90 ± 37 in FVB-3D; P = 0.008). While FVB mice continued to maintain more than 4,100 viral transcripts through 180 dpi, FVB-3D mice never had more than 4,200 viral transcripts during any of the time points tested. FVB-3D mice continued to have significantly less transcript than FVB mice at 21 (P = 0.008), 45 (P = 0.003), and 180 dpi (P = 0.016). At 90 days, viral transcripts in both FVB and FVB-3D mice rose to the highest levels in the spinal cord of all time points tested; however, mean transcripts in FVB-3D mice still were 1.32 logs lower than mean transcripts in FVB mice, although this was not statistically significant. At 180 dpi, FVB mice contained 18,000 viral transcripts, while FVB-3D transcripts dropped from 4,200 at 90 dpi to undetectable amounts at 180 dpi.
While analyzing the rise and fall throughout the persistent phase of infection, we found that FVB mice had a significant rise in viral transcripts in both the brain (P = 0.016) and spinal cord (P = 0.008) from 45 to 90 days, but the rise in 3D transgenic mice was minimal and not statistically significant during the same period of infection. Furthermore, 3D transgenic mice had a statistically significant drop from 21 to 45 days in the brain (P = 0.008) and from 90 to 180 days in the spinal cord (P = 0.016), both of which were not significant in nontransgenic FVB mice during the same period of infection.
Since the H-2 haplotype is known to independently alter virus persistence in inbred mice, we asked whether the 3D transgene effect also was seen in a haplotype that normally confers resistance to persistent viral infection. To study this, we crossed FVB-3D mice with FVB mice that contain the H-2Db transgene to obtain FVB-Db.3D mice. Unlike what we observed with the 3D transgene in susceptible FVB mice at 7 dpi, we did see significantly lower levels of viral transcripts in resistant FVB-Db mice containing the 3D transgene (P < 0.001). The levels of viral transcripts in the brain of both FVB-Db and FVB-Db.3D mice continued to decrease equally with time after 21 dpi, and as expected, there were no significant differences between the groups in the remaining time points. In the spinal cord, the FVB-Db.3D mice had very few viral transcripts (6.2 ± 2.0) by 7 dpi compared to the FVB-Db mice (1,900 ± 300) (P = 0.008). At 45, 90, and 180 dpi, we did not detect any viral transcripts above the background level in the FVB-Db.3D mice, while the FVB-Db mice still contained a few viral transcripts; this finding was significantly different from that for FVB-Db.3D mice (P = 0.008 at 45 days, P = 0.016 at 90 days, and P = 0.016 at 180 days).
To evaluate whether our method of quantifying VP2 viral transcripts correlates with the presence of active and cytopathic virus, we infected a cell line derived from mouse fibroblasts (SQSV) with TMEV. Virus was harvested at 8, 16, and 24 h after infection to quantitate cytopathic virus by plaque assay and VP2 transcripts by real-time quantitative RT-PCR (Fig. 2C). We found a direct correlation between the two assays, with a Pearson's correlation coefficient of 0.96 (P < 0.001).
To further assess the viral load in the brain and spinal cord, we checked for the presence of viral protein by immunohistochemistry in mice using an anti-TMEV rabbit antibody at 7 dpi in the brain and 45 and 270 dpi in the spinal cord. In the brain, most of the virus was detected in the hippocampus. There were no significant differences between FVB versus FVB-3D and FVB-Db versus FVB-Db.3D mice in all areas of the brain studied (see Table S4 in supplemental material). In the spinal cord, there was a statistical difference (P = 0.032) in the percentage of spinal cord quadrants containing virus, with the FVB-3D mice being negative for viral staining in all mice tested (n = 5) and FVB mice having 22.8% ± 8.66% (n = 5) virus-positive quadrants (Fig. 2B; also see Table S5 in supplemental material). At 270 days, virus still was detected in three of five FVB mice, whereas all five FVB-3D mice tested were negative. Viral staining was negative in spinal cord quadrants of all FVB-Db and FVB-Db.3D mice studied (n = 5 for both FVB-Db and FVB-Db.3D). Taken together, these data indicate that the expression of the 3D transgene affects virus propagation in FVB and FVB-Db mice.
3D transgene reduces brain and spinal cord pathology in FVB mice.
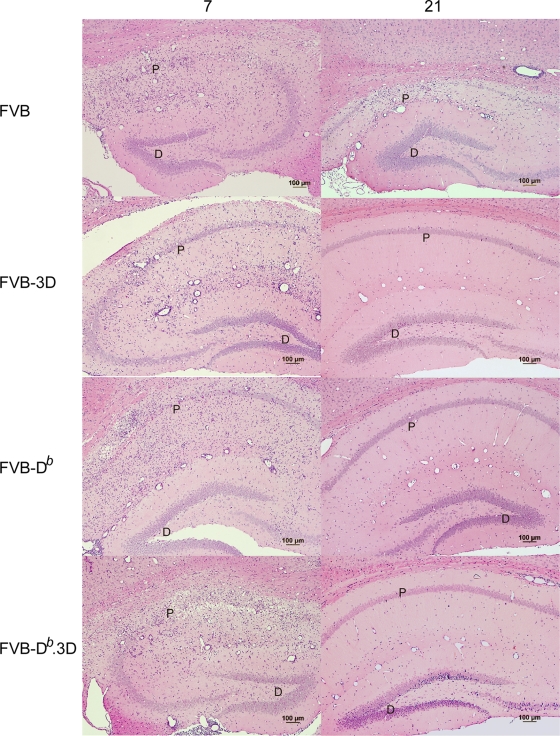
Because the 3D transgene affects the propagation of virus in both FVB and FVB-Db mice, we wanted to determine whether the transgenic expression of 3D also resulted in less brain and spinal cord pathology. We graded six different areas of the brain using a four-point scale based on the amount of inflammation and tissue destruction in each area (see Materials and Methods). For analysis, we combined the brain pathology scores from each area of the brain and graphed the results as the means ± standard errors of the means (SEM) for each experimental mouse group. At 7 dpi, there was no significant difference in the brain pathology scores between FVB and FVB-3D mice (Fig. 3A; also see Table S6 in the supplemental material). This indicated that no intrinsic problem existed with the ability of the virus to induce disease in FVB versus FVB-3D mice. However, there was a significant reduction (P = 0.02) in the amount of brain pathology by 21 dpi in the FVB-3D transgenic mice compared to that of FVB mice. There was the destruction of the pyramidal layer in the hippocampus at days 7 and 21 in FVB, but the restoration of this layer by day 21 in FVB-3D mice was much like that seen in resistant FVB-Db mice (Fig. 4). By 45 dpi, brain pathology had subsided in both FVB and FVB-3D mice, and there were no statistical differences between the groups at 45, 90, and 270 dpi. We also studied the effect of the 3D transgene on brain pathology scores in FVB-Db mice. FVB-Db.3D mice had modestly lower brain pathology scores than FVB-Db mice at 7 and 21 days, even though they were not statistically different. Brain pathology had subsided in both groups by 45 days, and there were no differences in brain pathology scores at 45, 90, and 270 dpi.
FIG. 4.
Hippocampal injury in brain 7 and 21 dpi. Hematoxylin and eosin staining in the hippocampus of mice show the destruction of the pyramidal cell layer in FVB, FVB-3D, FVB-Db, and FVB-Db.3D mice at day 7. By 21 days, damage in the pyramidal layer was recovered in FVB-3D mice similarly to the degree of recovery seen in FVB-Db and FVB-Db.3D mice (P, pyramidal cell layer; D, dentate gyrus). Note the absence of pathology in the dentate gyrus of the hippocampus.
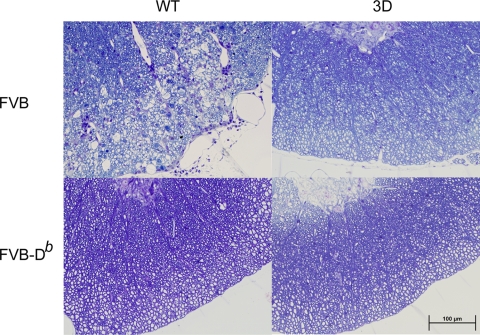
Since Theiler's virus causes a chronic demyelinating disease of the spinal cord in FVB mice, we expected to see white matter inflammation and demyelination by 45 days in FVB mice and none or very minimal inflammation in FVB-Db mice, which are resistant to persistent infection in the spinal cord (Fig. 3B; also see Table S7 in the supplemental material). Both FVB and FVB-3D mice had very little white matter inflammation at 7 and 21 days in the spinal cord, and they had no demyelination. By 45 days, FVB-3D had significantly lower levels of white matter inflammation (P = 0.015) and demyelination (P = 0.035) than FVB mice, with FVB-3D mice containing only 4.20% ± 2.04% of spinal quadrants with white matter inflammation and 5.58% ± 2.67% with demyelination, whereas the FVB mice had white matter inflammation in 9.27% ± 2.06% and demyelination in 10.74% ± 2.69% of spinal cord quadrants. When data from both lines were analyzed separately at this time point, we found that line 2 had significantly less white matter inflammation (P = 0.038) and demyelination (P = 0.038) than nontransgenic FVB mice, while line 1 did not show a statistical significance (Fig. 3C; also see Tables S7 and S8 in the supplemental material). FVB mice continued to demyelinate after 45 days, while demyelination had not increased from the previous time point in FVB-3D mice. At 90 days, FVB-3D had significantly less inflammation (P < 0.001) and demyelination (P = 0.015) than FVB mice, which had white matter inflammation in 8.72% ± 1.68% and demyelination in 16.23% ± 2.62% of spinal cord quadrants, while FVB-3D mice had only 0.29% ± 0.29% with white matter inflammation and 0.44% ± 0.44% with demyelination. Slides of spinal cord sections show the typical inflammation and demyelination seen in FVB mice at 90 days, in contrast to the absence of pathological abnormalities in FVB-3D, FVB-Db, and FVB-Db.3D mice (Fig. 5). By 270 days, no significant differences were seen in quadrants containing white matter inflammation between the groups, as white matter inflammation subsided in FVB mice. However, there was a statistical difference in the percent of spinal cord quadrants containing demyelinated axons (P < 0.001), as the extent of demyelination in FVB mice increased to 17.6% ± 2.50% and FVB-3D mice maintained the low levels of demyelination seen at 45 dpi. As expected, due to the clearance of the virus in resistant mice, we detected very little or no white matter inflammation and demyelination in either FVB-Db or FVB-Db.3D mice at 7, 21, 45, 90, and 270 dpi.
FIG. 5.
Spinal cord sections of mice at 90 dpi. Plastic-embedded sections were stained with a modified erichrome-cresyl violet stain. Lateral columns of cord show inflammation and demyelination in FVB mice but not in FVB-3D, FVB-Db, or FVB-Db.3D mice. All pictures were taken in a similar region of the spinal cord with a ×40 objective. WT, wild type.
3D transgene protects FVB mice from functional deficits.
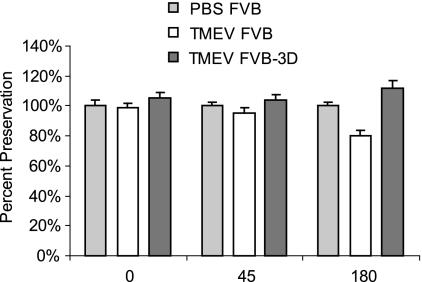
To test whether the 3D transgene protected mice from functional deficits, we infected FVB (n = 10) and FVB-3D (n = 25) mice and assayed them by rotarod at 0, 45, and 180 dpi (Fig. 6). Infected mice were compared to age-matched PBS-infected FVB mice (n = 10). There were no statistical differences between TMEV- and PBS-infected FVB mice until 180 dpi, at which point the TMEV-infected FVB mice had significantly decreased function (80% ± 3%; P = 0.002), whereas the TMEV-infected FVB-3D mice showed a preservation of function (112% ± 5%; not significant) relative to that of PBS-infected control mice (100% ± 3%).
FIG. 6.
Rotarod assay of TMEV-infected mice. Mice were placed on an accelerating rotarod, and the speed at which the mice fell was recorded. Data shown are the percentages at which the mice fell at each time compared the that of age-matched PBS-infected FVB controls (n = 10 for FVB, 25 for FVB-3D, and 10 for PBS controls).
3D transgene protects FVB mice from dropout of large-diameter axons.
We have reported that the loss of large-diameter axons (greater than 10 μm2) correlates with functional deficits measured by rotarod assay (7). Since function is preserved in FVB mice expressing the 3D transgene, we compared the number of axons in FVB and FVB-3D mice 270 days after infection. Axons were counted from six different regions in the spinal cord for each animal (Fig. 7A). As expected, the FVB-3D mice had maintained significantly more large-diameter axons than FVB mice (P < 0.001) (Fig. 7B). In FVB-3D mice, 1,449 ± 101 (n = 24) axons were greater than 10 μm2, whereas FVB mice contained only 830 ± 63 (n = 29) axons greater than 10 μm2. Figure 7C shows the reduced frequency of large-diameter axons in FVB mice compared to that of FVB-3D as distributed in 0.5-μm2 increments. There was no significant difference found in the number of small axons 1 to 4 μm2 in size and medium axons between 4 to 10 μm2 in infected FVB and FVB-3D mice at 270 dpi.
The effect of viral propagation in 3D transgenic mice is not the consequence of enhanced adaptive immunity to the virus.
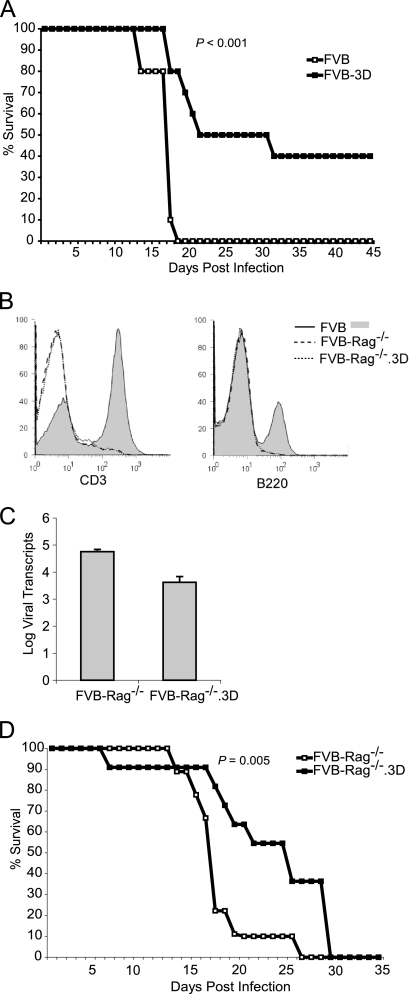
Previously, we have shown that resistant mice that express the immunodominant viral transgene VP2 under the same ubiquitin promoter as that of our 3D transgenic mice become susceptible to persistent infection (29). Because the VP2 transgene was ubiquitously expressed in many tissues, including the thymus, VP2 peptide-specific T cells were thought to have been deleted by negative selection in the thymus and/or activation-induced cell death in the periphery. This demonstrated that T-cell responses can be altered by transgenic expression and can affect viral clearance and CNS pathology. However, unlike VP2, 3D is not thought to contain any immunodominant epitopes. We then hypothesized that the expression of the 3D transgene, containing epitopes with weaker affinities for T-cell receptors (TCR), have an opposite effect on the adaptive immune system from that of stronger immunodominant epitopes, like those in VP2. Because positive selection in the thymus requires moderate, not strong or weak, affinity interactions with TCR, 3D expression could provide moderate stimulation to T cells that otherwise would die due to the lack of stimulation in wild-type mice. These 3D-selected T cells could have the potential to interact with immunodominant VP2 peptides. The change in the T-cell repertoire then would lead to a stronger anti-TMEV immune response, thus clearing the virus. We therefore set out to determine if the mechanism for the reduction of virus in the 3D transgenic mice was due to clearance by the adaptive immune response. To study this, we immunocompromised FVB and FVB-3D mice by irradiating them the day before injection with TMEV virus (Fig. 8A). All of the wild-type mice died (n = 10), whereas 8 of 10 FVB-3D mice survived by day 19, and 4 of 10 FVB-3D mice survived past 45 days. Kaplan-Meier survival curves of FVB and FVB-3D mice showed a significant difference by a log-rank test (P < 0.001). The irradiated FVB mice had an average survival time of 17.3 ± 0.6 days, whereas the irradiated FVB-3D mice survived significantly longer, 31.1 ± 4.1 days. Because immunocompromising the mice by irradiation may not completely eliminate the adaptive immune system or may have an effect on other cell types, we crossed the FVB-3D transgenic mice with FVB mice containing the Rag-1 knockout. Rag-1 knockout causes B- and T-cell development arrest at an early age, resulting in a mouse that is deficient in T and B cells (20) (Fig. 8B). This experiment definitively addresses whether the mechanism by which 3D affects viral infection is via the adaptive immune response. At day 14 after infection, FVB-Rag−/−.3D mice contained 3.6 ± 0.21 log viral transcripts, whereas wild-type FVB-Rag−/− mice had 4.8 ± 0.08 log viral transcripts in the spinal cord, which was statistically different (P = 0.004) (Fig. 8C). FVB.Rag−/−.3D mice (n = 11) survived significantly longer than FVB-Rag−/− mice (n = 9) after infection with TMEV (P = 0.005 by log-rank test) (Fig. 8D). By day 15, FVB-Rag−/− mice were moribund from viral encephalitis, with a median survival time of 18 days, whereas the FVB-Rag−/−.3D mice survived for a median of 26 days.
FIG. 8.
Survival of infected immunodeficient FVB mice. (A) Survival curve of FVB and FVB-3D mice irradiated (600 rads) 1 day before TMEV infection (n = 10 for each group). (B) Flow cytometry analysis of splenocytes from FVB, FVB-Rag−/−, and FVB-Rag−/−.3D mice using anti-CD3 and anti-B220 antibodies. Note the absence of CD3+ T cells and B220+ B cells in the FVB-Rag−/− and FVB-Rag−/−.3D mice but not in FVB mice. (C) Viral transcripts in the spinal cord of FVB-Rag−/− and FVB-Rag−/−.3D mice at 14 dpi. (D) Survival curve of FVB-Rag−/− (n = 9) and FVB-Rag−/−.3D mice (n = 11) after infection with TMEV.
DISCUSSION
We found that endogenously expressed 3Dpol slows viral propagation in mice that are susceptible to persistent TMEV infection. This effect of the 3D transgene in these mice leads to lower pathology scores in the brain and spinal cord, resulting in the preservation of axons and functional abilities in FVB mice after TMEV infection.
One major observation we noticed in FVB-3D mice was lower levels of viral transcripts in the cord at all time points after infection. Past studies using FVB mice expressing TMEV transgenes have led to the tolerance of T cells to dominant viral peptides resulting in increased viral transcripts (29). We concluded that there were two possibilities for our findings: either 3D directly interfered with viral replication, or 3D affected the adaptive immune system to clear the virus more efficiently. Interestingly, transgenic 3D still lowered the levels of viral transcripts in FVB-Db mice, which are resistant to persistent TMEV infection. We suspected that if the 3D transgene worked by changing the T-cell repertoire capable of recognizing more dominant viral peptide(s) or altering the viral peptides being presented, thus allowing the better clearance of the virus in susceptible mice, there would not be lower levels of viral transcripts in mice expressing the 3D transgene on a resistant background. Despite this, there was still a significant decrease of viral transcripts in FVB-Db mice containing the 3D transgene in the spinal cord, arguing that the effect of 3D on TMEV was not caused by a change in the adaptive immune response. Also, the fact that the viral load already was lower at the acute phase of infection in FVB-Db mice, before time for the development of protective T- and B-cell responses in the CNS, suggested that 3D did not affect the adaptive immune response to clear the virus. To completely rule this out as a mechanism, we did studies of mice lacking an immunocompetent adaptive immune system. Whole-body gamma irradiation depletes peripheral lymphocytes in mice. Following irradiation, infected FVB-3D mice lived longer than the infected wild-type FVB mice. The limitations of whole-body irradiations are the incomplete inactivation of all peripheral lymphocytes, effects on other cell types, and the fact that lymphocytes get replenished 4 weeks after irradiation (37). To overcome these limitations, we crossed FVB-3D mice with FVB-Rag−/− mice. Infected FVB-Rag−/− mice expressing the 3D transgene lived significantly longer than the infected control FVB-Rag−/− mice. Furthermore, FVB-Rag−/−.3D mice had significantly fewer TMEV RNA transcripts in the spinal cord than FVB-Rag−/− mice 14 days after infection. The immunosuppression of these mice did not allow us to study the effects of the adaptive immune response during the persistence phase of the infection. However, the difference we observed during the acute phase of infection in the absence of any adaptive immunity strongly suggests that the effect of 3D transgenic expression on TMEV propagation is caused by a mechanism(s) other than increased antiviral immunity due to the alteration of T- and/or B-cell repertoires.
An important aspect of this research was to exclude the possibility that the random insertion of the 3D transgene interfered with TMEV replication due to the disruption of an endogenous gene at or around the insertion site instead of the action of 3D itself. Fortunately, we had two lines available for study that yielded a pathological analysis of spinal cord sections 45 days after TMEV infection. Both lines expressing 3D resulted in decreased virus-induced demyelination following TMEV infection. Furthermore, the line that we were unable to continue, line 2, had an even greater reduction in virus-induced pathology than that of line 1. Line 1, which resulted in the majority of the data in the paper, showed the clear inhibition of TMEV infection by the transgenic expression of 3D. Because the FVB-3D lines behaved similarly to decrease spinal cord inflammation and demyelination, we are confident the result was not an effect of fortuitous insertion that decreased viral replication.
Despite ruling out a role for the adaptive immune response to explain the phenotype of FVB-3D mice, the mechanism of how transgenic 3D inhibits viral replication in vivo is not known. We have considered several hypotheses. (i) Endogenously expressed 3D may have interfered with virus-expressed proteins. There is evidence that 3D oligomerizes with itself and other picornavirus proteins to efficiently transcribe the RNA genome. Transgenic 3D may lose its ability to interact with a small subset of proteins or RNA itself and therefore have a dominant-negative effect on viral 3D polymerase. (ii) Even if 3D has not lost any function, the change in the relative stoichiometry of 3D molecules to viral precursor molecules like 3AB and 3CD may inhibit the processing of these proteins and slow RNA replication in the cytoplasm. (iii) The presence of endogenous 3D polymerase residing in the cytoplasm of the host as the virus enters may allow a head start of the host response to double-stranded RNA by type I interferons. Type I interferons are released by many cells infected with a virus that uses double-stranded RNA to replicate, thus they are considered early response cytokines of the innate immune system. The antiviral action of type I interferons is both autocrine and paracrine, inhibiting viral replication within the cell and cells in the surrounding environment. Past studies have shown the expression of type I interferons after TMEV infection and their critical role in controlling infection (5, 6, 23). Because Rag−/− mice have relatively normal type I interferon responses, the experiments crossing Rag−/− mice to 3D transgenic mice do not rule out this mechanism of action. (iv) Transgenic 3D may have indirectly affected virus replication by up- or downregulating other genes in the host cells. These genes may initiate a cascade of events to inhibit virus replication or enhance innate immune clearance. It is a paradox that the primary function of the 3D polymerase, when expressed and processed normally in the picornavirus genome, is to replicate positive- and negative-stranded RNA; however, when expressed transgenically in mice, 3D inhibits replication. An important question is whether the effect of the transgenic expression of 3D influences viral infection with other picornaviruses. Preliminary data using encephalomyocarditis virus infection shows a dramatic reduction in viral transcripts in infected mice expressing 3D compared to that of infected nontransgenic mice (see Fig. S2 in supplemental material). These results need to be repeated with other picornaviruses and nonpicornaviruses to determine the applicability of the findings. Understanding how endogenously expressed 3D polymerase inhibits viral replication may lead to the identification of new small molecules for potential therapies that can be used for all picornaviruses.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (NS RO1 32129 and NS RO1 24180), the National Multiple Sclerosis Society (R63172 and CA 1011A8), and the Multiple Sclerosis Research Foundation of Canada (CMS-05). We also acknowledge support from the Applebaum Foundation.
Footnotes
Published ahead of print on 16 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abzug, M. J., G. Cloud, J. Bradley, P. J. Sanchez, J. Romero, D. Powell, M. Lepow, C. Mani, E. V. Capparelli, S. Blount, F. Lakeman, R. J. Whitley, and D. W. Kimberlin. 2003. Double blind placebo-controlled trial of pleconaril in infants with enterovirus meningitis. Pediatr. Infect. Dis. J. 22:335-341. [DOI] [PubMed] [Google Scholar]
- 2.Azoulay, A., M. Brahic, and J. F. Bureau. 1994. FVB mice transgenic for the H-2Db gene become resistant to persistent infection by Theiler's virus. J. Virol. 68:4049-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron, K., X. Zhang, B. Seal, M. Rodriguez, and M. K. Njenga. 2001. Antigens to viral capsid and non-capsid proteins are present in brain tissues and antibodies in sera of Theiler's virus-infected mice. J. Virol. Methods 91:11-19. [DOI] [PubMed] [Google Scholar]
- 4.Dal Canto, M. C., and H. L. Lipton. 1977. Multiple sclerosis. Animal model: Theiler's virus infection in mice. Am. J. Pathol. 88:497-500. [PMC free article] [PubMed] [Google Scholar]
- 5.Delhaye, S., S. Paul, G. Blakqori, M. Minet, F. Weber, P. Staeheli, and T. Michiels. 2006. Neurons produce type I interferon during viral encephalitis. Proc. Natl. Acad. Sci. USA 103:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiette, L., C. Aubert, U. Muller, S. Huang, M. Aguet, M. Brahic, and J. F. Bureau. 1995. Theiler's virus infection of 129Sv mice that lack the interferon alpha/beta or interferon gamma receptors. J. Exp. Med. 181:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe, C. L., J. D. Adelson, and M. Rodriguez. 2007. Absence of perforin expression confers axonal protection despite demyelination. Neurobiol. Dis. 25:354-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun, E. J., Y. R. Nam, J. Ahn, H. Tchah, C. H. Joo, Y. Jee, Y. K. Kim, and H. Lee. 2008. Antiviral potency of a siRNA targeting a conserved region of coxsackievirus A24. Biochem. Biophys. Res. Commun. 376:389-394. [DOI] [PubMed] [Google Scholar]
- 9.Kim, S. M., K. N. Lee, J. Y. Park, Y. J. Ko, Y. S. Joo, H. S. Kim, and J. H. Park. 2008. Therapeutic application of RNA interference against foot-and-mouth disease virus in vitro and in vivo. Antivir. Res. 80:178-184. [DOI] [PubMed] [Google Scholar]
- 10.Kim, Y. J., J. Ahn, S. Y. Jeung, D. S. Kim, H. N. Na, Y. J. Cho, S. H. Yun, Y. Jee, E. S. Jeon, H. Lee, and J. H. Nam. 2008. Recombinant lentivirus-delivered short hairpin RNAs targeted to conserved coxsackievirus sequences protect against viral myocarditis and improve survival rate in an animal model. Virus Genes 36:141-146. [DOI] [PubMed] [Google Scholar]
- 11.Lehrich, J. R., B. G. Arnason, and F. H. Hochberg. 1976. Demyelinative myelopathy in mice induced by the DA virus. J. Neurol. Sci. 29:149-160. [DOI] [PubMed] [Google Scholar]
- 12.Lipton, H. L. 1980. Persistent Theiler's murine encephalomyelitis virus infection in mice depends on plaque size. J. Gen. Virol. 46:169-177. [DOI] [PubMed] [Google Scholar]
- 13.Lipton, H. L. 1975. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 11:1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipton, H. L., and M. C. Dal Canto. 1979. Susceptibility of inbred mice to chronic central nervous system infection by Theiler's murine encephalomyelitis virus. Infect. Immun. 26:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorch, Y., A. Friedmann, H. L. Lipton, and M. Kotler. 1981. Theiler's murine encephalomyelitis virus group includes two distinct genetic subgroups that differ pathologically and biologically. J. Virol. 40:560-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 17.McGavern, D. B., L. Zoecklein, K. M. Drescher, and M. Rodriguez. 1999. Quantitative assessment of neurologic deficits in a chronic progressive murine model of CNS demyelination. Exp. Neurol. 158:171-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melvold, R. W., D. M. Jokinen, R. L. Knobler, and H. L. Lipton. 1987. Variations in genetic control of susceptibility to Theiler's murine encephalomyelitis virus (TMEV)-induced demyelinating disease. I. Differences between susceptible SJL/J and resistant BALB/c strains map near the T cell beta-chain constant gene on chromosome 6. J. Immunol. 138:1429-1433. [PubMed] [Google Scholar]
- 19.Mendez-Fernandez, Y. V., A. J. Johnson, M. Rodriguez, and L. R. Pease. 2003. Clearance of Theiler's virus infection depends on the ability to generate a CD8+ T cell response against a single immunodominant viral peptide. Eur. J. Immunol. 33:2501-2510. [DOI] [PubMed] [Google Scholar]
- 20.Mombaerts, P., J. Iacomini, R. S. Johnson, K. Herrup, S. Tonegawa, and V. E. Papaioannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869-877. [DOI] [PubMed] [Google Scholar]
- 21.Nikolova, I., and A. S. Galabov. 2003. Development of resistance to disoxaril in coxsackie B1 virus-infected newborn mice. Antivir. Res. 60:35-40. [DOI] [PubMed] [Google Scholar]
- 22.Njenga, M. K., C. Marques, and M. Rodriguez. 2004. The role of cellular immune response in Theiler's virus-induced central nervous system demyelination. J. Neuroimmunol. 147:73-77. [DOI] [PubMed] [Google Scholar]
- 23.Njenga, M. K., L. R. Pease, P. Wettstein, T. Mak, and M. Rodriguez. 1997. Interferon alpha/beta mediates early virus-induced expression of H-2D and H-2K in the central nervous system. Lab. Investig. 77:71-84. [PubMed] [Google Scholar]
- 24.Ohara, Y., S. Stein, J. L. Fu, L. Stillman, L. Klaman, and R. P. Roos. 1988. Molecular cloning and sequence determination of DA strain of Theiler's murine encephalomyelitis viruses. Virology 164:245-255. [DOI] [PubMed] [Google Scholar]
- 25.Pathak, H. B., S. K. Ghosh, A. W. Roberts, S. D. Sharma, J. D. Yoder, J. J. Arnold, D. W. Gohara, D. J. Barton, A. V. Paul, and C. E. Cameron. 2002. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J. Biol. Chem. 277:31551-31562. [DOI] [PubMed] [Google Scholar]
- 26.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 27.Pavelko, K. D., K. M. Drescher, D. B. McGavern, C. S. David, and M. Rodriguez. 2000. HLA-DQ polymorphism influences progression of demyelination and neurologic deficits in a viral model of multiple sclerosis. Mol. Cell Neurosci. 15:495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavelko, K. D., C. L. Howe, K. M. Drescher, J. D. Gamez, A. J. Johnson, T. Wei, R. M. Ransohoff, and M. Rodriguez. 2003. Interleukin-6 protects anterior horn neurons from lethal virus-induced injury. J. Neurosci. 23:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavelko, K. D., L. R. Pease, C. S. David, and M. Rodriguez. 2007. Genetic deletion of a single immunodominant T-cell response confers susceptibility to virus-induced demyelination. Brain Pathol. 17:184-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce, M., and M. Rodriguez. 1989. Erichrome stain for myelin on osmicated tissue embedded in glycol methacrylate plastic. J. Histotechnol. 12:35-36. [Google Scholar]
- 31.Richards, O. C., and E. Ehrenfeld. 1998. Effects of poliovirus 3AB protein on 3D polymerase-catalyzed reaction. J. Biol. Chem. 273:12832-12840. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, M., and C. S. David. 1985. Demyelination induced by Theiler's virus: influence of the H-2 haplotype. J. Immunol. 135:2145-2148. [PubMed] [Google Scholar]
- 33.Rodriguez, M., and C. S. David. 1995. H-2 Dd transgene suppresses Theiler's virus-induced demyelination in susceptible strains of mice. J. Neurovirol. 1:111-117. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, M., A. J. Dunkel, R. L. Thiemann, J. Leibowitz, M. Zijlstra, and R. Jaenisch. 1993. Abrogation of resistance to Theiler's virus-induced demyelination in H-2b mice deficient in beta 2-microglobulin. J. Immunol. 151:266-276. [PubMed] [Google Scholar]
- 35.Rodriguez, M., J. Leibowitz, and C. S. David. 1986. Susceptibility to Theiler's virus-induced demyelination. Mapping of the gene within the H-2D region. J. Exp. Med. 163:620-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez, M., J. L. Leibowitz, and P. W. Lampert. 1983. Persistent infection of oligodendrocytes in Theiler's virus-induced encephalomyelitis. Ann. Neurol. 13:426-433. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez, M., A. K. Patick, and L. R. Pease. 1990. Abrogation of resistance to Theiler's virus-induced demyelination in C57BL mice by total body irradiation. J. Neuroimmunol. 26:189-199. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez, M., K. Pavelko, and R. L. Coffman. 1995. Gamma interferon is critical for resistance to Theiler's virus-induced demyelination. J. Virol. 69:7286-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotbart, H. A., and A. D. Webster. 2001. Treatment of potentially life-threatening enterovirus infections with pleconaril. Clin. Infect. Dis. 32:228-235. [DOI] [PubMed] [Google Scholar]
- 40.Shen, M., Z. J. Reitman, Y. Zhao, I. Moustafa, Q. Wang, J. J. Arnold, H. B. Pathak, and C. E. Cameron. 2008. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex. J. Biol. Chem. 283:875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidman, R. L., J. B. Angevine, and E. T. Pierce. 1971. Atlas of the mouse brain and spinal cord. Harvard University Press, Cambridge, MA.
- 42.Strauss, D. M., and D. S. Wuttke. 2007. Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase. J. Virol. 81:6369-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taketo, M., A. C. Schroeder, L. E. Mobraaten, K. B. Gunning, G. Hanten, R. R. Fox, T. H. Roderick, C. L. Stewart, F. Lilly, C. T. Hansen, et al. 1991. FVB/N: an inbred mouse strain preferable for transgenic analyses. Proc. Natl. Acad. Sci. USA 88:2065-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, E. L., T. M. Tan, V. Tak Kwong Chow, and C. L. Poh. 2007. Inhibition of enterovirus 71 in virus-infected mice by RNA interference. Mol. Ther. 15:1931-1938. [DOI] [PubMed] [Google Scholar]
- 45.Woods, M. G., G. D. Diana, M. C. Rogge, M. J. Otto, F. J. Dutko, and M. A. McKinlay. 1989. In vitro and in vivo activities of WIN 54954, a new broad-spectrum antipicornavirus drug. Antimicrob. Agents Chemother. 33:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.