Abstract
Retroelements are important evolutionary forces but can be deleterious if left uncontrolled. Members of the human APOBEC3 family of cytidine deaminases can inhibit a wide range of endogenous, as well as exogenous, retroelements. These enzymes are structurally organized in one or two domains comprising a zinc-coordinating motif. APOBEC3G contains two such domains, only the C terminal of which is endowed with editing activity, while its N-terminal counterpart binds RNA, promotes homo-oligomerization, and is necessary for packaging into human immunodeficiency virus type 1 (HIV-1) virions. Here, we performed a large-scale mutagenesis-based analysis of the APOBEC3G N terminus, testing mutants for (i) inhibition of vif-defective HIV-1 infection and Alu retrotransposition, (ii) RNA binding, and (iii) oligomerization. Furthermore, in the absence of structural information on this domain, we used homology modeling to examine the positions of functionally important residues and of residues found to be under positive selection by phylogenetic analyses of primate APOBEC3G genes. Our results reveal the importance of a predicted RNA binding dimerization interface both for packaging into HIV-1 virions and inhibition of both HIV-1 infection and Alu transposition. We further found that the HIV-1-blocking activity of APOBEC3G N-terminal mutants defective for packaging can be almost entirely rescued if their virion incorporation is forced by fusion with Vpr, indicating that the corresponding region of APOBEC3G plays little role in other aspects of its action against this pathogen. Interestingly, residues forming the APOBEC3G dimer interface are highly conserved, contrasting with the rapid evolution of two neighboring surface-exposed amino acid patches, one targeted by the Vif protein of primate lentiviruses and the other of yet-undefined function.
Retroelements have invaded the genomes of all organisms, from bacteria to primates, in amounts that often greatly exceed the sum of host protein-coding sequences. As such, they are essential evolutionary forces, but they must be tightly regulated to avoid deleterious consequences that would result from their uncontrolled spreading. The seven members of the human APOBEC3 family of cytidine deaminases partake in this process as innate immunity effectors that inhibit a large panel of retroelements, from endogenous retrotransposons to exogenous retroviruses (reviewed in references 14 and 22).
APOBEC3 proteins are characterized by the presence of one or two domains comprising the canonical zinc-coordinating motif (HXEX24-30PCX2-4C), essential for catalytic activity (33). APOBEC3G (A3G) contains two such domains, only the C terminal of which is endowed with editing potential (26, 43), whereas its N-terminal counterpart is enzymatically inactive but has RNA binding properties (32, 43). A3G can block the propagation of Vif-defective (Δvif) human immunodeficiency virus type 1 (HIV-1) (52) but is inactivated by the wild-type virus through Vif-induced proteasomal degradation (18, 40, 53, 55, 65). In the absence of Vif, the A3G antiviral effect requires both zinc-coordinating motifs, the N terminal for recruitment into particles and the C terminal for lethal editing of the viral DNA. A3G interacts via its N terminus with the nucleocapsid (NC) moiety of Gag, possibly through an RNA intermediate, and is thereby recruited at sites of viral assembly at the plasma membrane (4, 7, 56). Once the virus is in target cells, A3G interferes with reverse transcription at several steps (24, 25, 41), among which is the C-to-U editing of the nascent minus-strand viral DNA that critically depends on a functional C-terminal zinc-coordinating motif (27, 38, 66). It is noteworthy that the editing activity of A3G is not essential for its blockade of at least two other retroelements, the hepatitis B virus (58) and the Alu endogenous retrotransposon (16, 29).
In cells, A3G partakes in high-molecular-mass ribonucleoproteic complexes together with close to 100 other proteins (16, 21, 36). A3G binds single-stranded polynucleotides (32, 43) and has been found to associate with 7SL, Alu, and hYs RNAs (16, 62). A3G can also form oligomers (7, 10, 15, 33, 43), and the small angle of the X-ray scattering profile of these complexes is compatible with the volume of a homodimer (63). Mutating the N-terminal zinc-binding motif of A3G abrogates its RNA binding and self-association potential (43).
The recent structure determination of the A3G C-terminal domain has shed light on determinants dictating single-stranded DNA binding and editing (11, 28, 49). In contrast, the lack of structural data on its N-terminal counterpart, as well as the absence of enzymatic activity of this domain, have precluded reaching a similar understanding of the specific roles played by the zinc-binding motif and other features of this region of A3G. As a step toward investigating these issues, we conducted a large-scale mutagenesis/functional study of the A3G N-terminal module, mapping determinants crucial for RNA binding; oligomerization; and activity against an exogenous retrovirus, Δvif HIV-1, and an endogenous retroelement, Alu. We then integrated these data in a structural model. Our results reveal that residues important for the restriction of both retroelements form a surface predicted to mediate contacts between two A3G monomers and to form a groove that can potentially accommodate single-stranded polynucleotides. Furthermore, the results of phylogenetic analyses indicate that this region is highly conserved, in contrast with two adjacent patches of amino acids subjected to positive selection, one corresponding to the region targeted by Vif, the lentiviral antagonist of A3G, and the other of yet-unknown function.
MATERIALS AND METHODS
Constructs.
HIV-1 viruses encoding all but the Vif protein were produced from the pR9Δvif plasmid (60). The plasmids required for the LINE-1-driven Alu retrotransposition assay were a kind gift from J. V. Moran (pALU_pA+_neoTET and pJM101_L1.RP_NoNeo) (19, 29). pCMV plasmids coding for APOBEC proteins fused to a C-terminal hemagglutinin (HA) tag are described in reference 34. Point mutations were introduced using Quikchange site-directed mutagenesis (Stratagene). Each mutant was verified by performing a sequencing reaction. The C-terminal segment 198 to 384 of A3G was amplified from pCMV-A3G-HA by PCR using primers A3G198-384_F, 5′ GCGTATCGATAAGCTTGGCCAAGGATGGATCCACCCACATTCAC 3′, and 009, 5′ GCACTGGAGTGGCAACTTCC 3′, and introduced back into pCMV-A3G-HA with HindIII and XbaI to replace the wild-type protein. The plasmid coding for C-terminally myc-tagged A3G under the cytomegalovirus promoter was obtained by D. Kabat through the AIDS Research and Reference Reagent Program (pCDNA3.1.A3G-myc-His). The Vpr construct with residues 1 to 14 deleted (Vpr14-88) was cloned from the wild-type HIV-1 plasmid pR9 (60) by PCR using primers Vpr_BF, 5′ CCGGGGGGAAGCTTGGCCACCATGCCATACAATGAATGGACA 3′, and Vpr_ER-pcs, 5′ CCGGGGGGAAGCTTTTCCACCTACGATCGGATAATTCTGGCTTACTCTCCTCTGTCGAGTAAC 3′, and a proofreading DNA polymerase (PfuTurbo, Stratagene). The Vpr_ER-pcs primer contains the HIV protease-dependent cleavage site SQNYPIV. The PCR fragment obtained was next introduced into the HindIII site of pCMV-A3G-HA to obtain plasmid pCMV-Vpr14-88-A3G-HA. The plasmid coding for Vif C133S is described in reference 34 (pRRLsin.cPPT.PGK.GFP-2A-Vif-myc).
Cell culture and transfection.
HEK293T and CD4+, HIV-1 long terminal repeat-LacZ-containing, HeLa-derived P4.2 cells (9) were regularly passaged in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin), while HeLa HA cells were passaged in minimum essential medium supplemented with nonessential amino acids (Gibco) in addition to 10% fetal calf serum, antibiotics, and 2 mM glutamine. HEK293T cells were transfected by using Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. HeLa HA cells were transfected with Fugene 6 using a previously described protocol (29).
Single-round HIV-1 infectivity assay.
Viruses were produced in duplicates by transfection of HEK293T cells seeded in six-well plates, using the pR9Δvif plasmid, in the presence or of or without APOBEC proteins. To determine the activity levels of the mutants independently of their expression levels (see Fig. 2C), we produced Δvif HIV-1 viruses in the presence of wild-type A3G at three or four different doses and compared the restriction activity levels of the mutants to that of the corresponding dose of the wild type. At 12 h posttransfection, the medium was changed, and it was collected 24 h later. Cell-free virions were prepared by filtration through a 0.45-μM-pore-size nitrocellulose membrane (Spin-X; Corning) and concentrated in a microcentrifuge for 1.5 h at 13,000 rpm at 4°C. The pellet was resuspended in 20 μl of phosphate-buffered saline, and the amount of physical particles was monitored by reverse transcriptase activity assay (1). The infectious titer was determined by duplicate infections on the P4.2 indicator cell line (39). The specific titer was calculated by dividing the infectious titer by the amount of physical particles as determined by reverse transcriptase activity. The expression level of APOBEC proteins was evaluated by Western blot analysis of 2 μg of cell extract collected at the time of virion harvesting. To evaluate the amount of APOBEC protein inside viral particles, an equivalent of 1.5 × 106 cpm of virions was run on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. APOBEC proteins were detected with a mouse HA-specific, peroxidase-conjugated 3F10 monoclonal antibody (Roche). The viral capsid was detected by using mouse p24-specific antibody (B. Chesebro and H. Chen through the AIDS Research and Reference Reagent Program).
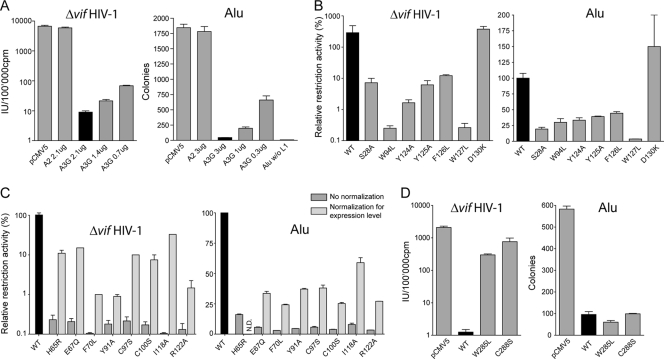
FIG. 2.
Functional analysis of wild-type (black bars) and mutant A3Gs against Δvif HIV-1 and Alu through single-round assays. (A) Dose-dependent inhibition of Δvif HIV-1 and Alu replication, with A2 serving as negative control, as described in Materials and Methods. Transfection with the Alu reporter in the absence of a LINE-1 helper plasmid (Alu w/o L1; w/o, without) was performed to control the LINE-1 dependency of neomycin resistance. (B to D) Relative restriction activities of stably expressed N-terminal domain mutants (B) and of N-terminal domain mutants with low steady-state levels before (dark gray) or after (light gray) normalization for expression levels (C), all expressed as a percentage of the wild-type activity. (D) Impact of C-terminal domain mutants on Δvif HIV-1 and Alu replication. Means and standard errors are representative of the results for independent duplicates from a single experiment (A and D) or from one to three different experiments (B and C). pCMV5, empty vector; WT, wild type; IU, infectious units; cpm, counts per minute; N.D., not done.
Alu retrotransposition assay.
The LINE-1-driven Alu retrotransposition assay was performed as previously described (29). Briefly, 3 × 105 to 5 × 105 HeLa HA cells were seeded into T75 flasks and transfected 15 h later with pALU_pA+_neoTET, pJM101_L1.RP_NoNeo, and pCMV5 or an APOBEC-encoding plasmid. Three days posttransfection, the cells were selected with G418 at 2 mg/ml for 2 days and then at 0.5 mg/ml for 4 to 5 more days. Colonies were then fixed with ethanol and stained with crystal violet and counted, and the counts averaged from duplicates. The expression levels of APOBEC proteins were evaluated by Western blot analysis of lysate from cells transfected in parallel and collected at the time of selection. Evaluation of the expression-independent activity of the mutants was performed by transfecting wild-type A3G at three different doses and comparing the activity levels of the mutants to that of the corresponding dose of the wild type.
Immunoprecipitation.
Transfected HEK293T cells were lysed in radioimmunoprecipitation assay buffer supplemented with antiprotease cocktail set I (Calbiochem) and 100 U/ml of RNase inhibitor (RNAseIN; Promega). After a short spin to remove cell debris, a 50- to 100-μg amount of cell lysate was adjusted to 500 μl with lysis buffer. Ten microliters was retrieved and loaded on a SDS-PAGE gel, and 20 μl was used for reverse transcription-PCR (RT-PCR). Forty microliters of anti-HA affinity matrix (Roche) was added to the rest of the mix, which was swirled for 2 to 3 h at 4°C and then extensively washed using radioimmunoprecipitation assay buffer supplemented with NaCl at up to 500 mM. After the last wash, half of the beads were incubated in β-mercaptoethanol and SDS-containing buffer for 3 min at 95°C before being run on a SDS-PAGE gel. RNA was extracted from the other half and analyzed by RT-PCR. For myc-tagged A3G, immunoprecipitations were performed similarly, using anti-c-Myc affinity matrix (Santa Cruz), and the protein was detected by Western blotting with a mouse anti-c-Myc antibody (Roche).
RT-PCR analysis.
RNA was extracted using an RNeasy mini kit (Qiagen). After DNase treatment (Sigma), RNA was reverse transcribed using random hexamers and Superscript III reverse transcriptase as recommended by the manufacturer (Invitrogen). For each sample, a control reaction lacking the enzyme was included to check for contaminating DNA. We amplified the resulting cDNAs by PCR using GoTaq green master mix (Promega) and specific primers for 7SL (7SL_BF2, 5′ TCCGCACTAAGTTCGGCATC 3′, and 7SL_ER2, 5′ AGTGGCTATTCACAGGCGCG 3′), hY1 (hY1_F2, 5′ CTGGTCCGAAGGTAGTGAG 3′, and hY1_R2, 5′ CTAGTCAAGTGCAGTAGTG 3′), hY3 (hY3_F2, 5′ GCTGGTCCGAGTGCAGTGGTGTTTAC 3′, and hY3_R2, 5′ AGGCTAGTCAAGTGAAGCAGTG 3′), and Alu (AluWBF, 5′ TCACGCCTGTAATCCCAGCA 3′, and AluR2, 5′ GATCTCGGCTCACTGCAAG 3′). The linearity of the PCR was verified by parallel amplification of serially diluted human genomic DNA.
Structural model.
The N-terminal domain of the ABC3G_HUMAN sequence (residues 12 to 194; UniProtKB [59] code Q9HC16) was defined as the target sequence, while the recently resolved human A3G C-terminal domain (28) (PDB [3] ID 3E1U) served as a template. The two A3G domains show a good sequence identity (36%). Sequence alignment was done with the align2d function of the MODELLER program (50), as well as with the CLUSTALW (13) and T-COFFEE (44) algorithms, and subsequently modified based on structural alignments with CHIMERA (47).
Homology models were built with the MODELLER program (50); 1,000 models were calculated and scored with the MODELLER objective function and the ANOLEA energy (42) using the default 5-residue-window averaging. ANOLEA estimates the folding free energy of each amino acid in a three-dimensional structure. The model with a MODELLER objective function of less than 1,100 and the best ANOLEA energy for the nonloop regions was chosen for further refinement. Residues 140 to 146 were treated with the MODELLER loop refinement routine because of their poor alignment with the template structure. Again, 1,000 models were calculated, and the best conformation according to its ANOLEA score was used in the refined A3G monomer model. A zinc ion was added to the pseudoactive site of the model before it was energy minimized with the CHARMM program (6) and the CHARMM22 all-atom force field (37). The minimization consisted of 100 steps of steepest descent. A harmonic restraint of 5 kcal/mol/Å2 toward their initial position was imposed on all heavy atoms except for the zinc site residues His65, Glu67, Cys97, and Cys100, which were restrained to display the same distances to the zinc ion as in the X-ray structure. Two copies of the final N-terminal model were structurally aligned to chains A and C of the APOBEC2 (A2) tetramer (48) (PDB ID 2NYT) in order to create a putative A3G head-to-head dimer structure. The side-chain conformations of residues 27 and 124 to 127 at the dimerization interface were reassigned using SCWRL3 (8) and minimized again with CHARMM in order to remove clashes. The FoldX program (23) was used to perform an in silico alanine scan by mutating each residue to an alanine and estimating the change of folding energy. Molecular visualization and structural alignments were done using CHIMERA (47).
RESULTS
Common determinants on the A3G N-terminal domain are required for the inhibition of Δvif HIV-1 and Alu.
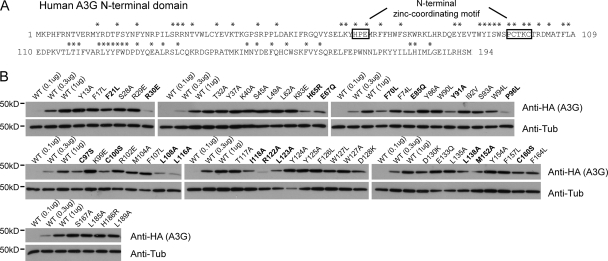
To initiate a structure-function study of the human A3G N-terminal domain, we first individually mutated 55 residues in this region (Fig. 1A) (67), concentrating on positions that are highly conserved among primate orthologues or subjected to positive selection during primate evolution (46, 51) and opting for conservative amino acid substitutions in most cases (5). Thirty-six of the resulting mutants exhibited wild-type steady-state levels of expression, while 19 appeared less stable (Fig. 1B). We then tested these derivatives for their efficiency at blocking Δvif HIV-1 infection and Alu retrotransposition in single-round assays (Fig. 2). Although the magnitudes of the blockades obtained with wild-type A3G differed between the two systems, in both cases it was dose dependent, with the inactive A2 serving each time as a negative control (Fig. 2A). Remarkably, all mutants exhibited parallel phenotypes for both classes of retroelements: of the well-expressed mutants, six (S28A, W94L, Y124A, Y125A, F126L, and W127L) exhibited marked decreases in Δvif HIV-1 and Alu restriction, whereas one, the D130K mutant, had instead a reproducible increase in inhibitory potency (Fig. 2B). The W127A mutant, slightly more stable than the W127L mutant described previously (31), was also defective (not illustrated). In contrast, mutations at several positions subjected to positive selection during primate evolution (Tyr13, Arg29, Glu85, Lys99, Arg102, and Asp128) did not affect activity against Δvif HIV-1 or Alu (data not shown). The activity levels of the 19 less stable mutants were normalized for the expression levels. Eleven of them (F21L, R30E, E85Q, P96L, L108A, L116A, L123A, L138A, M152A, C160S, and F164L) then appeared as active as the wild type against both Δvif HIV-1 and Alu (not illustrated), while eight, including derivatives carrying changes in zinc-coordinating residues (H65R, E67Q, C97S, and C100S), were defective (Fig. 2C). This result highlights the importance of an intact N-terminal zinc-coordinating motif for inhibiting both retroelements, contradicting a previous report that this motif is dispensable for Alu restriction (16). We also tested two C-terminal domain mutants, one at a position homologous to the N-terminal Trp94 (W285L) and the other with a change at a zinc-binding position (C288S). These two residues have been shown to contribute to the catalytic activity of recombinant C-terminal domain A3G (11, 12). Both mutants were defective for Δvif HIV-1 restriction but fully competent for Alu inhibition (Fig. 2D), consistent with the previously noted importance of editing for blocking the former but not the latter (16, 29, 38).
FIG. 1.
Large-scale mutagenesis of A3G N-terminal domain residues and effects on steady-state levels of the proteins. (A) Amino acid sequence of human A3G positions 1 to 194, with mutated residues analyzed for restriction activity marked with an asterisk. (B) Steady-state levels of wild-type (WT) A3G in HEK293T cells transiently transfected with threefold dilutions of the wild type and the highest dose (1 μg) of mutant plasmid DNA, analyzed by Western blotting using HA-specific antibodies. Mutants with steady-state levels lower by threefold or more than that of the wild type are highlighted in bold. anti-Tub, antitubulin.
Functionally defective N-terminal A3G mutants are also impaired for RNA binding and oligomerization.
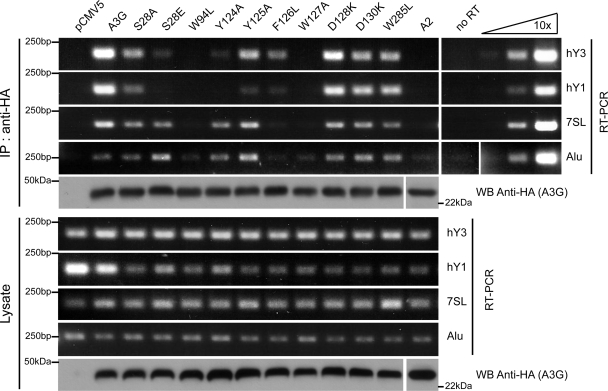
Interaction with RNA seems to govern two properties of A3G, namely, binding to the HIV-1 NC, which leads to recruitment into nascent particles, a prerequisite for viral inactivation, and oligomerization (4, 15, 56). Furthermore, an intact N-terminal zinc-coordinating motif is required for the self-association of A3G, as well as RNA binding (43). Therefore, we probed the RNA binding ability of A3G N-terminal mutants by RT-PCR analysis of immunoprecipitates recovered from transiently transfected HEK293T cells (Fig. 3). Contrary to the results for A2, wild-type A3G was found to associate with hY1, hY3, 7SL, and Alu RNAs, as previously described (16, 62). In contrast, functionally defective N-terminal mutants had, to various degrees, a decreased affinity for one or more of these RNAs. Although the background level was relatively high for Alu (Fig. 3, lane marked “no RT”), interaction with this RNA was reduced to negligible levels for the two most inactive mutants, the W94L and W127A mutants. The D130K and W285L mutants did not show a significant change of affinity for any RNA tested.
FIG. 3.
Small cellular RNA binding abilities of A3G N-terminal mutants. Extracts of HEK293T cells transiently transfected with plasmids expressing HA-tagged forms of A2 and wild-type and mutant A3G were either first immunoprecipitated (IP) with HA-specific antibodies (IP: anti-HA) or directly analyzed (Lysate) by Western blotting (for HA) or semiquantitative RT-PCR with primers specific for indicated small RNAs. Right panel shows results for 10-fold dilutions of human genomic DNA. “No RT” indicates control sample run in the absence of reverse transcriptase enzyme. WB, Western blotting.
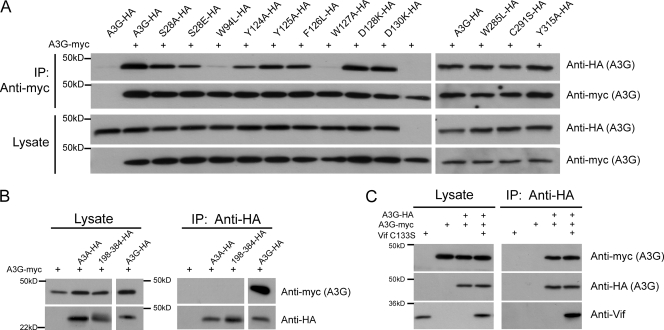
To evaluate the oligomerization potential of the A3G mutants, we expressed them as HA-tagged derivatives together with myc-tagged wild-type A3G and performed HA- and myc-specific Western blot analyses of antimyc-mediated immunoprecipitations (Fig. 4). While wild-type HA-tagged A3G efficiently coimmunoprecipitated with its myc-tagged counterparts, N-terminal mutants were impaired for oligomerization to degrees largely corresponding to their RNA binding and antiviral activity defects (Fig. 4A, left). In contrast, three mutations in the C-terminal domain of A3G (W285L, C288S, and Y315A), in spite of being at positions homologous to those in three N-terminal oligomerization-defective mutants (W94L, C97S, and Y124A) (45), had no impact on A3G self-association (Fig. 4A, right). Neither APOBEC3A (A3A) nor a version of A3G with an N-terminal deletion (A3G198-384) (12) could bind wild-type A3G (Fig. 4B). Interestingly, most of the A3G oligomerization determinants identified in this work have aromatic groups (Trp94, Tyr124, Tyr125, Phe126, and Trp127), most of which do not have counterparts in A3A and in the C-terminal domain of A3G. Unfortunately, A3G derivatives lacking the C-terminal domain were unstable and could not be tested for RNA binding or self-association.
FIG. 4.
A3G homodimerization is affected by N-terminal domain mutations but not by the presence of Vif C133S. (A) Indicated HA-tagged wild-type A3G and mutants were transiently coexpressed in HEK293T cells along with myc-tagged wild-type A3G (+). Cell lysate (Lysate) and immunoprecipitated material (IP: antimyc) were loaded onto SDS-PAGE gels, followed by Western blot analysis using HA- and myc-specific antibodies. A3G-HA alone served as control for nonspecific binding (first lane). (B) HA-tagged A3A, A3G, and A3G198-384 were cotransfected with A3G-myc (+) in HEK293T cells, and lysates were either directly loaded onto SDS-PAGE gels (Lysate) or used for immunoprecipitation using anti-HA-coated beads (IP: anti-HA). A3G-myc alone served as control for nonspecific binding. (C) Cells transiently expressing wild-type myc- and HA-tagged A3G in the presence or absence of Vif C133S were lysed and used for immunoprecipitation with anti-HA-coated beads. Vif C133S and A3G-myc alone served as controls for nonspecific binding. +, present.
Individual mutations of two aspartates to lysine at positions 128 and 130 of A3G impair recognition by HIV-1 Vif (31, 39). Owing to the close proximity of these residues to the patch at residues 124 to 127 that is involved in oligomerization, we investigated whether Vif could prevent A3G complex formation. For this, we performed the myc-/HA-tagged A3G coimmunoprecipitations in the presence or absence of HIV-1 Vif C133S, which binds A3G but fails to trigger its proteasomal degradation (35). As reported previously (35), Vif C133S did not affect the antiviral potency of A3G (data not shown). In line with this result, it did not interfere with A3G oligomerization, in spite of associating with the cytidine deaminase (Fig. 4C). Of note, the mutants D128K and D130K that do not bind Vif had full oligomerization and RNA binding abilities (Fig. 3 and 4A), and the oligomerization-defective A3G W94L and W127A mutants were efficiently degraded by wild-type Vif (data not shown) (31). Altogether, these data indicate that distinct A3G domains dictate oligomerization and recognition by HIV-1 Vif.
Functional rescue of inactive A3G N-terminal domain mutants by forced packaging into HIV-1 virions.
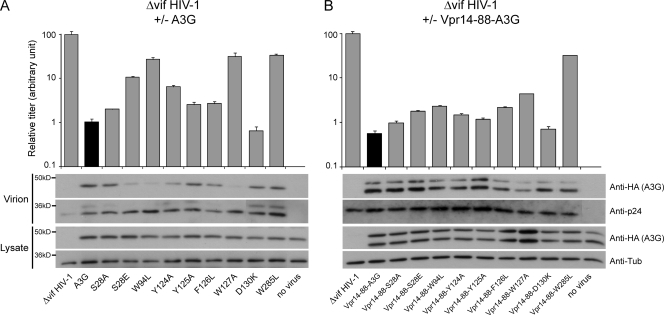
The incorporation of A3G into assembling HIV-1 particles is essential for its antiviral activity against this pathogen (43). As a remarkable correlate, the functional defects of well-expressed A3G N-terminal domain mutants with impaired RNA binding and antiviral activities were inversely proportional to their packaging efficiency (Fig. 5A), in agreement with the RNA-dependent association of A3G to the HIV-1 NC (4, 56). This is notably exemplified by the S28A and S28E mutants, whose impact on Δvif HIV-1 infectivity closely paralleled their packaging efficiencies. In contrast, the C-terminal W285L mutant was efficiently encapsidated in spite of its restriction defect. To further probe the contribution of the packaging defect of A3G N-terminal mutants to their functional impairment, we forced their encapsidation by fusion with a fragment of the HIV-1 Vpr protein that was sufficient for recruitment into viral particles but devoid of cytotoxicity (amino acids 14 to 88) (2). A peptide linker that is sensitive to the viral protease was placed between the two parts of the resulting fusion proteins, allowing the release of the A3G moiety following virion maturation to avoid subsequent influence of Vpr on the A3G activity. Vpr-mediated particle incorporation significantly rescued the antiviral phenotype of the packaging-defective A3G mutants (Fig. 5B). Although the activities of the least-defective mutants were not fully restored, the antiviral effects of the S28E, W94L, and W127A mutants increased approximately 10 times in this context. This indicates that N-terminal residues that are essential for A3G packaging do not subsequently play a major role in HIV-1 inhibition. In contrast, the W285L catalytic mutant remained inactive even when fused to Vpr.
FIG. 5.
Encapsidation and antiviral activities of packaging-defective A3G N-terminal domain mutants are rescued by fusion with Vpr14-88. Top, relative infectivity levels of Δvif HIV-1 exposed or not to wild-type or mutant A3G. Infectivity of viruses produced in the absence of A3G was given the arbitrary value of 100. Values represent means and standard errors of the results from independent duplicates. Bottom, Western blot analyses of cellular and viral extracts using HA-, tubulin- (anti-Tub), or p24-specific antibodies. (A) Results with native A3G. (B) Results with Vpr14-88-A3G fusion proteins.
Modeling a head-to-head dimer of A3G.
A clear interpretation of our functional and biochemical data is complicated by the absence of any structural information on the N-terminal domain of A3G, due at least in part to the insolubility of the full-length protein at high concentration (11). It would be particularly interesting to determine whether determinants of restriction are part of a single domain or are dispersed in different areas. Taking advantage of the recently published crystal structure of the A3G C-terminal domain (residues 197 to 384) (28) and of the good sequence identity between the two domains (36%), we modeled the structure of the N-terminal domain (residues 13 to 194) using the MODELLER program (50). As the sequence identity between these structures is significantly higher than that with A2 (27%), the model presented herein may be considered more reliable than earlier A3G models based solely on A2 (30, 67).
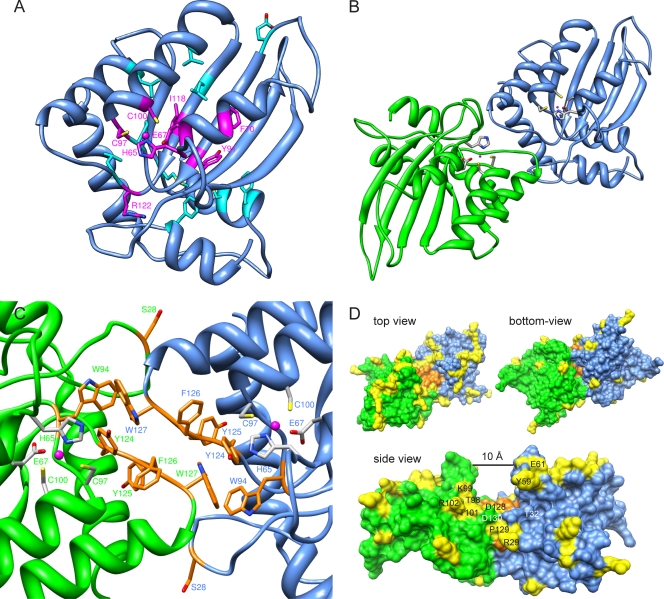
Our model is shown in detail in Fig. 6. It has a good ANOLEA score profile (data not shown), with a pattern comparable to that of the X-ray structure of the C-terminal domain, indicating good reliability of the predicted structure. Like the C-terminal domain, the N-terminal region of A3G forms a central anti-parallel β-sheet surrounded by α-helices. The positions of the loops delimited by residues 22 to 34 and 56 to 64 differ significantly and the regions containing residues 123 to 134 and 140 to 146 differ to a lesser extent from models based on A2. In our model, the N-terminal residues found to affect the expression of A3G upon mutation (Fig. 1B) are either linked to the zinc ion (His65, Glu67, Cys97, and Cys100) or have an important structural function (Fig. 6A), as suggested by an in silico alanine scan of the model (see Table S1 in the supplemental material). Out of those mutants, the ones that are defective regardless of their expression level (Fig. 2C) are modified at positions involved in zinc binding (His65, Glu67, Cys97, and Cys100) or seem to play a prominent role in stabilizing the structure of the protein (Phe70, Tyr91, and Ile118) (Fig. 6A; also see Table S1 in the supplemental material). Although Arg122 was not predicted to influence the overall structure of A3G, its high degree of conservation among cytidine deaminases, including A2, correlates with its important functional role. Since zinc often stabilizes protein domains (57), the disruption of this motif in the A3G N terminus might alter function indirectly, by altering local conformation. For A2, a single-domain cytidine deaminase, two monomers assemble in a long anti-parallel β-sheet with one zinc-coordinating domain at each extremity (48). Two of those dimers interact to form a linear tetramer. Since most of the residues important for A3G self-association resided in regions homologous to the ones mediating A2 tetramer formation and since various experiments have suggested that A3G could form, among different heterogeneous complexes, a homodimer (7, 10, 15, 33, 43, 63), we built a structural model of an A3G head-to-head dimer by structural alignment of two A3G N-terminal domains to two A2 monomers (Fig. 6B). In this model, the residues found to be important for A3G functionality (Ser28, Trp94, Tyr124, Tyr125, Phe126, and Trp127) partake in stabilizing the dimer, notably, by forming numerous π-π interactions between the five aromatic residues and their cognate copies from the other monomer (Fig. 6C). Trp127 makes extensive contacts with Trp94 and Tyr124 of the other monomer. In agreement, the W94L and W127A mutants were the two mutants with the weakest binding affinity for wild-type A3G (Fig. 5A). Although they do not make direct contacts with Ser28 on the other monomer, Asp128 and -130 are in close proximity to it, an observation that parallels the observation of a reduced capacity of S28E, in comparison to that of S28A, to oligomerize with wild-type A3G (Fig. 5A). Two additional residues participating in intersubunit contacts on A2 (48) are present on a helix corresponding to the most C-terminal helix in our model. Nine mutations in this helix (F157L, N176D, K180A, Y181A, I183A, L184A, I187A, E191Q, and I192A) did not affect A3G self-association in coimmunoprecipitation experiments (data not shown). Although this helix is close to the putative dimer interface, it displays less-extensive interactions with the other monomer than the loop containing residues 124 to 127 or the corresponding helix in the A2 tetramer, in agreement with the results of our mutational analysis.
FIG. 6.
Homology dimer model of A3G. (A) A3G N-terminal domain model based on homology with the crystal structure of the A3G C terminus. Residues affecting protein steady-state level when mutated are indicated as follows. Magenta, mutants are functionally defective even after normalization for expression levels (Fig. 2C); light blue, mutants are as active as the wild type after normalization for expression levels (Phe21, Glu85, Pro96, Leu108, Leu116, Leu123, Leu138, Met152, Cys160, and Phe164). (B and C) Nitrogen, oxygen, sulfur, and zinc atoms are depicted in blue, red, yellow, and magenta, respectively. (B) Top view of the dimer model, showing two N-terminal domains of A3G (one green and the other blue) in a head-to-head complex, as modeled from the A2 tetramer structure. Zinc-binding residues (His65, Glu67, Cys97, and Cys100) are shown in gray. (C) The predicted dimer interface in detail, with residues important for A3G oligomerization and Δvif/Alu restriction shown in orange. Intersubunit contacts of the aromatic side chains allow the formation of energetically favorable π-π interactions. (D) Space-filling visualization of A3G dimer. As in panel C, the surface of the residues important for oligomerization is depicted in orange. Yellow surfaces correspond to the residues found to be under positive selection in primate A3Gs. The side view evidences the 10-Å groove formed by the two monomers at the dimer interface, as well as the residues under positive selection that are close to the Vif-binding site comprising residues 128 to 130. Thr32 and Asp130 (labeled in white) are two conserved residues that both influence recognition by HIV-1 Vif.
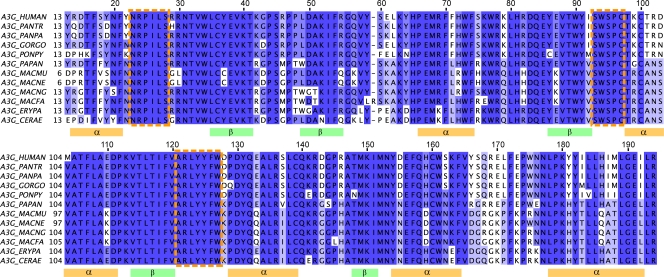
The dimer interface forms a groove with a width of approximately 10 Å (Fig. 6D, side view), compatible with the diameter of single-stranded but not double-stranded polynucleotidic chains, in agreement with in vitro data (64). The aromatic residues Trp94 and Tyr124 to Trp127 located at the bottom could interact with the nucleotide bases. The corners of the groove are delimited by two negatively charged residues, Asp128 and Asp130, which do not have extensive interactions with the other monomer, in agreement with our coimmunoprecipitation data (Fig. 5A). Primate A3Gs have an unusually high rate of residues under positive selection, characteristic of rapid evolution (46, 51). Mapping these residues onto the model reveals that the large majority occur on one side of the dimeric protein, leaving the other side with only a few such residues (Fig. 6D, top view and bottom view, respectively). At the dimer interface, a patch of amino acids subjected to positive selection is centered on the Vif-binding site, previously demonstrated to comprise residues 128 to 130 (Fig. 6D, side view) (31, 67). Alignment of Old World monkey and hominid A3Gs shows that conserved residues overlap with secondary structure elements, further supporting the validity of our model (Fig. 7). In the loop domains, the three largest clusters of conserved residues encompass one or more of the six amino acids identified as important for restriction (Ser28, Trp94, Tyr124, Tyr125, Phe126, and Trp127).
FIG. 7.
Alignment of hominid and Old World monkey A3Gs. The alignment shows high conservation in predicted secondary structures deduced from the model (orange and green rectangles), as well as in loops containing the residues involved in restriction activity (orange-dashed boxes), as seen by the results in Fig. 2B. Alignment was performed using MUSCLE (20). Dark blue, >80% homology; blue, >60% homology; light blue, >40% homology; white, <40% homology; PANTR, Pan troglodytes; PANPA, Pan paniscus; GORGO, Gorilla gorilla; PONPY, Pongo pygmaeus; PAPAN, Papio anubis; MACMU, Macaca mulatta; MACNE, Macaca nemestrina; MACNG, Macaca nigra; MACFA, Macaca fascicularis; ERYPA, Erythrocebus patas; CERAE, Cercopithecus aethiops.
DISCUSSION
In the present work, we combined functional analyses and structural modeling to delineate a structural region of the A3G N terminus that is involved in mediating oligomerization and RNA binding and is essential for A3G packaging into HIV-1 virions, inhibition of Δvif HIV-1 infectivity, and blockade of Alu retrotransposition. This structural region notably encompasses a series of aromatic residues, Trp94, Tyr124, Tyr125, Phe126, and Trp127, that are predicted to mediate intersubunit contacts through a series of π-π interactions, as is often found at protein-protein interfaces (17). The dimer interface is further characterized by the formation of a putative groove that has the width and electrostatic potential to bind single-stranded but not double-stranded polynucleotides. Although our data do not formally demonstrate the presence of a dimer rather than a higher-order structure, recent evidence indicates that the major form of oligomer detected in experimental settings comparable to ours is a dimer (30). In that report, Huthoff et al. similarly proposed an A3G head-to-head dimer model, mainly based on analysis of mutants with the two N-terminal mutations Y124A and W127A and homology with the structure of A2. The two models show striking similarities, notably revealing the importance of residues 124 to 127 in establishing intermonomeric contacts. However, interesting differences emerge between the two models, probably stemming from the use here of the now-available structure of the more closely related A3G C terminus as a template, while that of the more distant A2 was utilized by Huthoff et al. For instance, their study did not predict the putative RNA-accommodating groove created at the dimer interface, due to a different positioning of the loop containing residues 22 to 34 which buried the zinc-coordinating motif and placed Ser28 far from the other monomer. Also, a more twisted dimer positioned Asp128 at the dimerization interface, resulting in contacts with Arg122 of the other monomer, a possibility difficult to reconcile with the absence of both any oligomerization phenotype of the D128K mutant and the accessibility of this residue to Vif (Fig. 4A). Finally, Huthoff et al. found that positively charged amino acids located at the edge of the dimer interface (Arg24, Arg30, and Arg136) might engage in stabilizing interactions with RNA. However, Arg24 (54) and Arg30 (Fig. 1B; also see Table S1 in the supplemental material) were both found to contribute to protein stability, suggesting that these residues influence RNA binding only indirectly. In addition, we observed that charge-inverting mutations at four other positions on the same interface (Arg29, Lys63, Lys99, and Arg102) did not reduce A3G restriction activity or RNA binding (not illustrated).
Our functional analyses confirm that the C-terminal catalytic site of A3G promotes Δvif HIV-1 but not Alu inhibition. In contrast, they indicate that A3G requires an intact dimerization domain to act against both classes of retroelements. In the case of HIV-1, however, the implicated residues seem necessary mainly for gaining access to budding viral particles. Indeed, the defective phenotype of all derivatives impaired for packaging due to mutations at these positions could be rescued by forcing their incorporation into virions by fusion with Vpr (Fig. 5B). Thus, the N-terminal module of A3G does not appear to play a major role in other aspects of the antiviral effect of the cytidine deaminase on HIV-1. In contrast, in the absence of a clearer understanding of how A3G blocks Alu retrotransposition, the role of the N-terminal domain in this process can only remain speculative. Some degree of correlation between Alu RNA binding ability and the Alu inhibition potential of the mutants studied here (Fig. 3) suggests a model whereby A3G blocks retrotransposition of this element by sequestering its RNA away from the LINE trans-acting proteins. However, this simple model is not supported by data obtained with A3A, since in the experimental system described here, this cytidine deaminase exhibited far greater levels of anti-Alu activity than A3G but no obvious affinity for its RNA, at least based on the results of RT-PCR analysis of A3A-specific immunoprecipitates (not illustrated).
The potential RNA-accommodating groove formed by the dimer interface (Fig. 6D) is consistent with the RNase sensitivity of A3G oligomers (45). Furthermore, the fact that mutations abrogating dimerization and perturbing association with small cellular RNAs also prevented packaging into HIV-1 virions indicates that these three events are linked. The RNase sensitivity of the A3G-HIV-1 NC interaction was previously interpreted as indicating that an RNA intermediate is involved in linking the two partners (4, 56). Alternatively, it could be that NC does not recognize A3G as a monomer but as a dimer, the stabilization of which is RNA dependent. However, the apparent promiscuity of A3G packaging, which can be recruited into HIV-1 virions via either the cognate retroviral NC or its severe acute respiratory syndrome coronavirus homologue (61), makes a protein-protein-only interaction less likely, unless it is primarily structural motifs rather than specific amino acid sequences that are at play.
Several studies have pointed to the role of Asp128 and Asp130 in binding Vif (31, 39). A recent report indicated that Thr32 might be involved as well (54). On the A3G monomer, Asp128/Asp130 and Thr32 are located on opposite sides of the dimerization-mediating aromatic stretch. However, in our dimer model, Thr32 is situated close to Asp128 of the adjacent monomer (Fig. 6D, side view). Together with the results of coimmunoprecipitation studies indicating that Vif does not prevent A3G oligomerization (Fig. 4C), this strongly suggests that Vif can recognize a cytidine deaminase dimer. From a therapeutic perspective, since Vif binding and A3G function involve distinct regions, possible side effects of a Vif-targeting drug on A3G activity may be limited.
Considering that A3G dimerization and RNA binding are important for inhibiting retroelements as distant as HIV-1 and Alu, it is not surprising that the residues involved in this process are highly conserved (Fig. 7). Notably, the subjection of residues located at the corner of the dimer interface to positive selection (Fig. 6D, side view) (46, 51) is consistent with their recognition by the A3G antagonist Vif (31, 67). In contrast, there is no obvious explanation for the finding that another group of residues lined up on one side of the protein (Fig. 6D, top view versus bottom view) similarly underwent rapid evolution. While it is tempting to postulate that these positions might be important for target recognition, the normal levels of activity of the corresponding mutants against both HIV-1 and Alu suggest that these elements at least were not responsible for the selective pressure exerted at these positions, leaving the nature of the underlying evolutionary forces undefined.
Supplementary Material
Acknowledgments
This work was supported by grants from the Strauss Foundation and the Infectigen Association to D.T. and the Swiss National Science Foundation to D.T. and O.M.
Special thanks to Nadine Zangger for sequence alignments and Sujana Nylakonda and Nicola MacKinnon for technical assistance. The following reagents were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID: pCDNA3.1.A3G-myc-His from David Kabat and mouse p24-specific antibody from Bruce Chesebro and Hardy Chen. For computational resources, we thank the Vital-IT team at the Swiss Institute of Bioinformatics. The modeling part of this work was performed within the Protein Modeling Facility (PMF) of the University of Lausanne. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Footnotes
Published ahead of print on 23 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ao, Z., Z. Yu, L. Wang, Y. Zheng, and X. Yao. 2008. Vpr14-88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS ONE 3:e1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, H., K. Henrick, H. Nakamura, and J. L. Markley. 2007. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35:D301-D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., and B. R. Cullen. 2008. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA 14:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordo, D., and P. Argos. 1991. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 217:721-729. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. Charmm: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187-217. [Google Scholar]
- 7.Burnett, A., and P. Spearman. 2007. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 81:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canutescu, A. A., A. A. Shelenkov, and R. L. Dunbrack, Jr. 2003. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12:2001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 10.Chelico, L., E. J. Sacho, D. A. Erie, and M. F. Goodman. 2008. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. J. Biol. Chem. 283:13780-13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, K. M., E. Harjes, P. J. Gross, A. Fahmy, Y. Lu, K. Shindo, R. S. Harris, and H. Matsuo. 2008. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452:116-119. [DOI] [PubMed] [Google Scholar]
- 12.Chen, K. M., N. Martemyanova, Y. Lu, K. Shindo, H. Matsuo, and R. S. Harris. 2007. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 581:4761-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317-353. [DOI] [PubMed] [Google Scholar]
- 15.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 16.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho, K. I., D. Kim, and D. Lee. 2009. A feature-based approach to modeling protein-protein interaction hot spots. Nucleic Acids Res. 37:2672-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 19.Dewannieux, M., C. Esnault, and T. Heidmann. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41-48. [DOI] [PubMed] [Google Scholar]
- 20.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goila-Gaur, R., and K. Strebel. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerois, R., J. E. Nielsen, and L. Serrano. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320:369-387. [DOI] [PubMed] [Google Scholar]
- 24.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, F., S. Cen, M. Niu, Y. Yang, R. J. Gorelick, and L. Kleiman. 2007. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 81:11322-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hache, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920-10924. [DOI] [PubMed] [Google Scholar]
- 27.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 28.Holden, L. G., C. Prochnow, Y. P. Chang, R. Bransteitter, L. Chelico, U. Sen, R. C. Stevens, M. F. Goodman, and X. S. Chen. 2008. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456:121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulme, A. E., H. P. Bogerd, B. R. Cullen, and J. V. Moran. 2007. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huthoff, H., F. Autore, S. Gallois-Montbrun, F. Fraternali, and M. H. Malim. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huthoff, H., and M. H. Malim. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwatani, Y., H. Takeuchi, K. Strebel, and J. G. Levin. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 80:5992-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 34.Jost, S., P. Turelli, B. Mangeat, U. Protzer, and D. Trono. 2007. Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J. Virol. 81:10588-10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi, M., A. Takaori-Kondo, Y. Miyauchi, K. Iwai, and T. Uchiyama. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 280:18573-18578. [DOI] [PubMed] [Google Scholar]
- 36.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105-29119. [DOI] [PubMed] [Google Scholar]
- 37.MacKerell, A. D., D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102:3586-3616. [DOI] [PubMed] [Google Scholar]
- 38.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 39.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 40.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 41.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melo, F., and E. Feytmans. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141-1152. [DOI] [PubMed] [Google Scholar]
- 43.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 44.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 45.Opi, S., H. Takeuchi, S. Kao, M. A. Khan, E. Miyagi, R. Goila-Gaur, Y. Iwatani, J. G. Levin, and K. Strebel. 2006. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 80:4673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortiz, M., G. Bleiber, R. Martinez, H. Kaessmann, and A. Telenti. 2006. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology 3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 48.Prochnow, C., R. Bransteitter, M. G. Klein, M. F. Goodman, and X. S. Chen. 2007. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445:447-451. [DOI] [PubMed] [Google Scholar]
- 49.Rausch, J. W., L. Chelico, M. F. Goodman, and S. F. Le Grice. 2009. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J. Biol. Chem. 284:7047-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 51.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 53.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 54.Shirakawa, K., A. Takaori-Kondo, M. Yokoyama, T. Izumi, M. Matsui, K. Io, T. Sato, H. Sato, and T. Uchiyama. 2008. Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat. Struct. Mol. Biol. 15:1184-1191. [DOI] [PubMed] [Google Scholar]
- 55.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 56.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 57.Takahasi, K., M. Yoneyama, T. Nishihori, R. Hirai, H. Kumeta, R. Narita, M. Gale, Jr., F. Inagaki, and T. Fujita. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428-440. [DOI] [PubMed] [Google Scholar]
- 58.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 59.Uniprot Consortium. 2009. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 37:D169-D174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, S. M., and C. T. Wang. 2009. APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein. Virology 388:112-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, T., C. Tian, W. Zhang, K. Luo, P. T. Sarkis, L. Yu, B. Liu, Y. Yu, and X. F. Yu. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 81:13112-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wedekind, J. E., R. Gillilan, A. Janda, J. Krucinska, J. D. Salter, R. P. Bennett, J. Raina, and H. C. Smith. 2006. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J. Biol. Chem. 281:38122-38126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 65.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, K. L., B. Mangeat, M. Ortiz, V. Zoete, D. Trono, A. Telenti, and O. Michielin. 2007. Model structure of human APOBEC3G. PLoS ONE 2:e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.