Abstract
Flavivirus assembles into an inert particle that requires proteolytic activation by furin to enable transmission to other hosts. We previously showed that immature virus undergoes a conformational change at low pH that renders it accessible to furin (I. M. Yu, W. Zhang, H. A. Holdaway, L. Li, V. A. Kostyuchenko, P. R. Chipman, R. J. Kuhn, M. G. Rossmann, and J. Chen, Science 319:1834-1837, 2008). Here we show, using cryoelectron microscopy, that the structure of immature dengue virus at pH 6.0 is essentially the same before and after the cleavage of prM. The structure shows that after cleavage, the proteolytic product pr remains associated with the virion at acidic pH, and that furin cleavage by itself does not induce any major conformational changes. We also show by liposome cofloatation experiments that pr retention prevents membrane insertion, suggesting that pr is present on the virion in the trans-Golgi network to protect the progeny virus from fusion within the host cell.
Maturation, by which a noninfectious immature virus particle is converted to an infectious virion, is an essential step in the replication cycle of many viruses. It often involves the proteolytic processing of a precursor protein coupled with the conformational transformation of the virion. The assembly pathway for flaviviruses is well established, and the structures of the entire virion as well as individual glycoproteins representing different stages of the life cycle have been determined. Thus, flaviviruses are an excellent system for investigating the dynamic aspects of maturation in enveloped viruses.
Flavivirus maturation requires furin (21), a cellular protease located primarily in the trans-Golgi network (TGN) (16). Immature particles, containing heterodimers of the precursor membrane protein (prM) and the envelope protein (E), bud into the endoplasmic reticulum (ER) and then are transported through the cellular secretory pathway to the extracellular environment (12, 23). The cleavage of prM by furin generates the membrane-anchored protein M and the soluble product pr. In the mature, infectious virion, the pr peptide is absent, and the virus undergoes membrane fusion in the endosome at low pH. In contrast, immature particles produced from furin-deficient LoVo cells, or cells grown in the presence of protease inhibitors or acidotropic reagents to prevent furin cleavage, contain prM and are significantly less infectious (21).
Crystal structures of the E protein (9, 14, 18, 19, 29) show that each polypeptide chain contains three domains: the structurally central amino-terminal domain (DI), the dimerization domain (DII) containing the fusion loop, and the carboxy-terminal immunoglobulin-like domain (DIII). In the presence of lipids, low pH induces rearrangements of the E proteins, resulting in the formation of homotrimers with the fusion loops and the C-terminal membrane anchors located at the same end (4, 15). The conformational change of the E proteins presumably facilitates the merging of the viral membrane with the endosomal membrane, thereby mediating the entry of virus through membrane fusion. The structures of the entire virion in the immature (26, 27) and the mature (10, 17, 25) forms have been studied by cryoelectron microscopy (cryoEM). The mature virion, approximately 500 Å in diameter, has a smooth surface on which 90 E dimers form a closely packed protein shell with a herringbone pattern (10). The immature particles (26, 27), with an external diameter of approximately 600 Å, contain 60 prominent spikes, each consisting of a trimer of prM-E heterodimers. The difference in the E protein organization is striking, indicating that the maturation process requires major positional exchanges of the E proteins (including their trans-membrane components). This raises the question of whether the structure of the immature form is physiologically relevant.
Previously, we showed that at low pH, the immature virus undergoes a major conformational change in which the E proteins are rearranged into a configuration similar to that of the mature virus (24). The prM protein in the low-pH form of the immature virus, unlike the neutral-pH form, is accessible to furin cleavage (11, 24). Given that furin is abundant in the TGN, it is likely that the cleavage of the immature virus takes place in the TGN. However, it is not clear how virions are stabilized in the TGN after furin cleavage, where the pH value is similar to that of the endosome. It seems possible that pr remains associated with the virion in the TGN to prevent fusion, which is consistent with the observation that pr peptides comigrate with the virion in sucrose gradient sedimentation at pH 5.5 (24). However, in that experiment, approximately 30% of pr was found in fractions distinct from the virus (24). To further investigate the retention of pr at low pH, we report here the structure of furin-cleaved immature particles at pH 6.0 as determined by cryoEM. We show that at acidic pH the cleaved immature virus particles have essentially the same structure as that of the uncleaved immature particles, demonstrating that pr peptides are associated with virions at low pH and that furin cleavage by itself does not induce any major conformational changes. We further show that the presence of pr prevents the virus from interacting with liposomes at low pH, suggesting that pr retention inhibits membrane fusion in the TGN.
MATERIALS AND METHODS
Purification and in vitro furin cleavage of immature dengue particles.
Immature dengue particles were generated and purified as previously described (8, 24). C6/36 mosquito cells were infected with dengue virus 2 strain New Guinea C at a multiplicity of infection of 0.1 for 2 h at 30°C. At 22 h postinfection, the medium was replaced with fresh minimal essential medium containing 2% fetal bovine serum (FBS) and 20 mM NH4Cl. Virus particles were harvested at 48 to 60 h postinfection and precipitated using 8% polyethylene glycol 8000. Particles were pelleted through a 20% sucrose cushion and further purified in a 10 to 30% potassium-tartrate gradient centrifugation. Purified virus sample was concentrated in NTE buffer (12 mM Tris at pH 8, 120 mM NaCl, and 1 mM EDTA) using an Amicon Ultra-4 100-kDa MWCO concentrator (Millipore). The concentration of the virus was estimated by comparing the intensity of the band corresponding to the E protein to those of known concentrations of bovine serum albumin (BSA) on Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
In vitro furin cleavage was carried out as described previously (24). Briefly, purified immature dengue particles in NTE buffer at approximately 1 mg/ml E protein were mixed with 50 mM morpholinepropanesulfonic acid (MES) at a 1:1 (vol/vol) ratio to adjust the pH to 6.0. Furin (New England Biolabs Inc.) was added at a ratio of 0.2 U per μg of E protein, and the mixture was incubated at 30°C for 16 h in the presence of 3 mM CaCl2. For back neutralization, the sample was mixed with buffer containing 100 mM Tris at pH 8.0 and 120 mM NaCl at a 1:1 (vol/vol) ratio.
Infectivity studies using an immunofluorescence assay.
Purified immature viruses were incubated at pH 6.0 with or without furin at 30°C for 16 h and then back neutralized. The samples were used to infect confluent baby hamster kidney (BHK) cells. At 36 h postinfection, cells were fixed with methanol and incubated with the monoclonal anti-E antibody followed by fluorescein isothiocyanate-conjugated secondary antibody. Fluorescent images were taken using an inverted epifluorescent microscope equipped with a digital charge-coupled-device camera (Nikon).
CryoEM image processing and three-dimensional reconstruction.
The furin-cleaved immature dengue particles were flash frozen on holey carbon grids in liquid ethane. Charge-coupled-device images were recorded with a CM200 FEG transmission electron microscope (Philips) using a nominal magnification of 50,000 and an electron dose of approximately 25 e− per Å2. A total of 1,612 particles were boxed and normalized using the program ROBEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt) from 45 images of 1.8- to 3.8-μm defocus levels. Phases, but not amplitudes, were corrected by an appropriate calculation of the contrast transfer function. The low-pH structure of uncleaved immature dengue virus (24) was used as a starting model for image reconstruction. The origins and orientation parameters (x, y, θ, φ, and w) were determined using the program PFT (2), and electron density maps were calculated using the program EM3DR (3, 7). Due to the heterogeneity of the sample, only 635 boxed particles were used for the reconstruction to achieve a resolution of 22 Å using a Fourier shell correlation cutoff of 0.5.
To calculate the difference map (see Fig. 2E), the electron density maps of the immature and mature virions were calculated to 22 Å using the same pixel size. The difference map was generated in ROBEM after scaling and normalization.
FIG. 2.
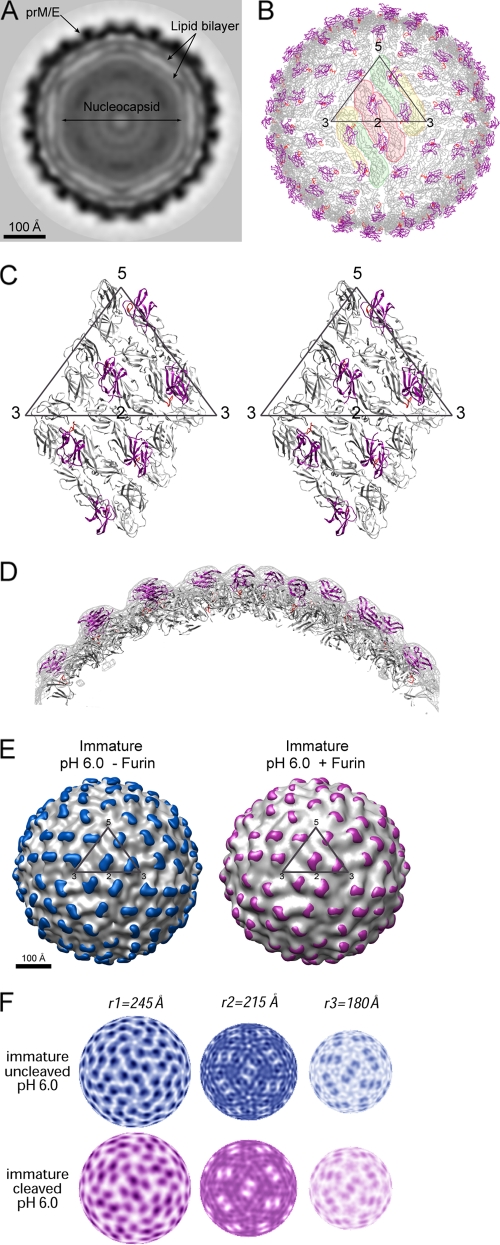
Structure of furin-cleaved immature virus at pH 6.0. (A) A central cross-section viewed along an icosahedral threefold axis. The darkness of the shading is proportional to the magnitude of the cryoEM electron density. Viral components are labeled. (B) Pseudoatomic structure of the virion upon fitting the crystal structures of prM-E into the cryoEM map. The E and pr proteins are shown in gray and magenta, respectively. The fusion loop is colored in red. An icosahedral asymmetry unit is outlined in a triangle. The E dimer on the icosahedral twofold axis is shaded pink, whereas the two monomers of the general-position dimer are shaded yellow and green. (C) Stereo diagram showing the interactions of pr (magenta) with E (gray). (D) A cross-section through the cryoEM density showing the overall quality of the fitting. The EM density is shown in gray, and protein molecules are colored as described for panel C. (E) Structure comparison of the immature virus with and without furin cleavage. The difference density calculated by subtracting the density of the mature virion is shaded in color (blue, before furin cleavage; magenta, after furin cleavage). An icosahedral asymmetric unit is outlined in a black triangle. (F) Comparison of the radial density sections of the immature particles with and without furin treatment. Maps at radii corresponding to the outer shell (r1), inner shell (r2), and membrane region (r3), respectively, are shown.
Fitting of the prM-E structure into the cryoEM density map of the furin-cleaved immature virus at low pH.
A model containing a dimer of prM-E was extracted from the pseudoatomic structure of the low-pH, uncleaved immature virus (24) and used to fit into the 22-Å reconstruction map of furin-cleaved particles using EMfit (20). First, the molecular twofold axis of [prM-E]2 was superimposed onto one of the icosahedral twofold axes of the virion. The search model then was rotated and translated along the icosahedral axis to obtain the best fit. A map in which the densities within 6 Å of each Cα atom of the fitted dimer were set to zero was used for fitting [prM-E]2 in the general position of the icosahedral asymmetric unit. The pseudoatomic structure of the entire virion then was generated by applying icosahedral symmetry (Table 1; also see Fig. 2).
TABLE 1.
Fitting of [prM-E]2 into the cryoEM map of furin-cleaved prM particle at low pH using the EMfit program
| Mola | sumfb (%) | −denc (%) | Clashf (%) | centxd (Å) | centyd (Å) | centzd (Å) | θ1e | q2e | q3e |
|---|---|---|---|---|---|---|---|---|---|
| 2f | 75.4 | 0.0 | 0.0 | 0.0 | 0.0 | 217.8 | 2.5 | 0.0 | 0.0 |
| Quasi-2f | 73.2 | 0.6 | 0.0 | 40.1 | −28.5 | 207.7 | 231.2 | 21.2 | 131.2 |
The [prM-E]2 dimer of uncleaved prM particles (24) was used to fit into the twofold (2f) and quasi-two-fold (Quasi-2f) locations of furin-cleaved low-pH prM particles. Mol, molecule.
Average density of Cα atoms expressed as a percentage of the highest density in the map.
Percentage of atoms in negative density.
The refined coordinates of the center of the mass of [prM-E]2 in the map.
Τhe refined Eulerian angles (in degrees) required to rotate the atomic coordinates of [prM-E]2 from a standard orientation into position.
Clash is the percentage of atoms in one molecule that are closer than 6 Å to another symmetry-related molecule when Cα atoms are used for fitting.
Liposome cofloatation.
Liposomes were prepared by mixing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (Avanti Polar Lipids), and 1-cholesterol (Sigma) at a molar ratio of 1:1:1. The lipids were dried under vacuum and suspended in either the NTE buffer or low-pH buffer (50 mM MES at pH 5.5, 120 mM NaCl). The mixture was frozen and thawed five times, followed by extrusion through two 100-nm-pore-size polycarbonate membranes with a syringe-type mini-extruder (Avanti Polar Lipids) 21 times.
Virus particles were mixed with liposomes at the corresponding pH values at an approximately 1:25 molar ratio. After a 30-min incubation at 25°C, the mixture was acidified with 250 mM MES at pH 5.5, 120 mM NaCl and incubated for 10 min at 37°C. The sample was adjusted to a final volume of 2 ml of 20% (wt/wt) sucrose in 50 mM MES at pH 5.5 and 120 mM NaCl and was layered onto a 1-ml 60% (wt/wt) sucrose cushion. The gradients were completed with a layer of 0.8 ml of 15% (wt/wt) sucrose and a layer of 0.2 ml buffer without sucrose. Centrifugation was carried out for 2 h at 38,000 rpm at 4°C in a Beckman SW60Ti rotor, and fractions of 400 μl were collected. The amount of viral RNA in each fraction was determined by quantitative reverse transcription-PCR (qRT-PCR).
Viral RNA purification and qRT-PCR.
Viral RNA was purified from 140 μl of each fraction using a QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer's recommendations and was eluted in 60 μl of nuclease-free water. The qRT-PCR was performed using the SuperScript III Platinum SYBR green One-Step qRT-PCR kit (Invitrogen) with primers 5′-CCAATGCGTTCAATCGGCT-3′ and 5′-CCAATGCGTTCAATCGGCT-3′, which bind to nucleotides 7475 to 7495 and 7612 to 7630 of the dengue genome, respectively. The amplification reactions were carried out in a 25-μl reaction mixture containing a 1-μl aliquot of purified viral RNA from each fraction. All reactions were performed in triplicate. Cycling conditions were 20 min at 50°C and 5 min at 95°C, followed by 40 cycles of 5 s at 95°C and 1 min at 60°C. Purified viral RNA from untreated virus in serial dilution was used to generate a standard curve for relative quantification.
CryoEM map accession numbers.
The cryoEM map of furin-cleaved immature dengue particles at low pH has been deposited with the European Bioinformatics Institute (accession number 5117). The coordinates for the fitted prM/E molecules into the cryoEM map have been deposited under the accession number 3IYA.
RESULTS
In vitro maturation of dengue virus.
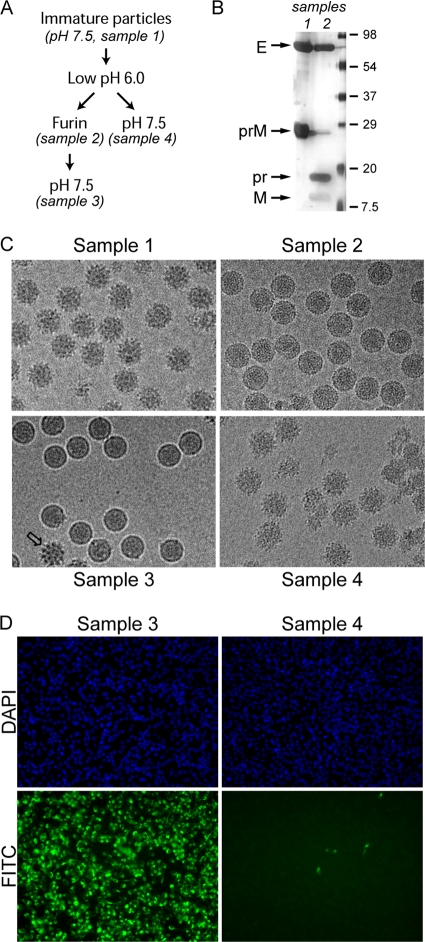
To characterize the conformational changes associated with maturation, prM-containing particles obtained by adding the acidotropic reagent NH4Cl to the growth media (sample 1) (Fig. 1A) were subjected to in vitro furin cleavage at mildly acidic pH (sample 2), followed by neutralization (sample 3). Immature particles subjected to low-pH incubation and neutralization in the absence of furin were included as controls (sample 4). As determined by SDS-PAGE, approximately 95% of prM was cleaved by furin at pH 6.0 (Fig. 1B), generating two proteolytic products, pr (91 residues) and M (75 residues). Consistently with previous data (26, 27), cryoEM micrographs showed that at neutral pH, prM-containing particles have a spiky surface, with each protrusion corresponding to a trimer of prM-E heterodimers (Fig. 1C, sample 1). Upon furin cleavage at pH 6.0, the particles were converted to a round shape with a smooth surface (Fig. 1C, sample 2). At pH 7, these particles (Fig. 1C, sample 3) became slightly smaller, with a shape and size similar to those of mature virions harvested from infected cells (13). In contrast, particles with intact prM reverted to the spiky form after back neutralization (Fig. 1C, sample 4). A few spiky particles were present in sample 3 (Fig. 1C), presumably due to incomplete prM cleavage. To test the infectivity of the in vitro-matured particles, confluent BHK cells were infected with viruses, and the resultant E protein expression was analyzed by immunofluorescence (Fig. 1D). Whereas E proteins are readily detectable in cells infected with furin-cleaved sample 3, particles that underwent the same pH treatment without furin cleavage were noninfectious (sample 4). These data show that the maturation of prM-containing particles can be induced in vitro. The shape and infectivity of the in vitro-matured samples are indistinguishable from those of mature virus particles harvested from infected cells (23).
FIG. 1.
Maturation of dengue immature particles by in vitro furin cleavage. (A) Flow chart of pH and furin treatments of prM-containing particles. (B) Samples 1 and 2 were analyzed by SDS-PAGE silver staining to examine the extent of furin cleavage. The capsid protein was not visible by silver stain, likely due to the absence of cysteine residues (6). (C) EM of the virus particles subjected to pH changes and furin treatment as outlined for panel A. (D) The infectivity of low-pH-treated particles with and without furin cleavage was analyzed by an immunofluorescence assay. Virus-infected BHK cells were probed with a monoclonal anti-E antibody and detected using a fluorescein isothiocyanate (FITC)-conjugated secondary antibody. Nuclei of all cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). The cells were visualized with an epifluorescent microscope. The samples were infected with the same amount of virus particles.
Structure of the furin-cleaved immature particle at mildly acidic pH.
The structure of furin-treated dengue immature particles at low pH was determined to 22-Å resolution by cryoEM and image reconstruction (Fig. 2). An equatorial cross-section of the reconstruction shows a multilayer structure comprised of the nucleocapsid, the lipid bilayer, and the glycoproteins prM and E (Fig. 2A). The density in the nucleocapsid region appears to be amorphous, which is consistent with earlier studies indicating that the core lacks a defined structure (10, 26, 28). The lipid bilayer is situated between radii of 165 and 205 Å, as previously observed in dengue virus reconstructions (25, 26). Fitting the prM-E crystal structure (11) into the electron density outside the membrane shows that the E proteins are arranged in the same herringbone pattern as that of a mature virion (Fig. 2B). Each E dimer is associated with two pr peptides, which are located at equivalent positions covering the fusion loops along the E dimer interface (Fig. 2B and C). The electron density outside the membrane shows excellent agreement with the structure of prM-E heterodimer (Fig. 2D).
Within the limits of current resolution, the structure of the cleaved particles at low pH essentially is identical to that of the uncleaved form at the same pH (24) (Fig. 2E). Electron density maps of the two structures were similar (Fig. 2F). For example, at the radial section of 215 Å, both structures have patterns consistent with the herringbone arrangement of the E proteins in the mature virus. At a radius corresponding to the membrane region (180 Å; designated r3), both maps show three groups of densities with elongated shapes centered at the icosahedral twofold axis, which correspond to the transmembrane helices of the M and E proteins (25). Furthermore, the difference map between the uncleaved and cleaved forms contained no significant densities compared to the height of densities in the original maps, demonstrating that there is no evident conformational change upon furin cleavage. It therefore also is apparent that the occupancy of the pr peptides in the cleaved form must approach 100%.
Retention of pr on the virus at low pH prevents membrane insertion.
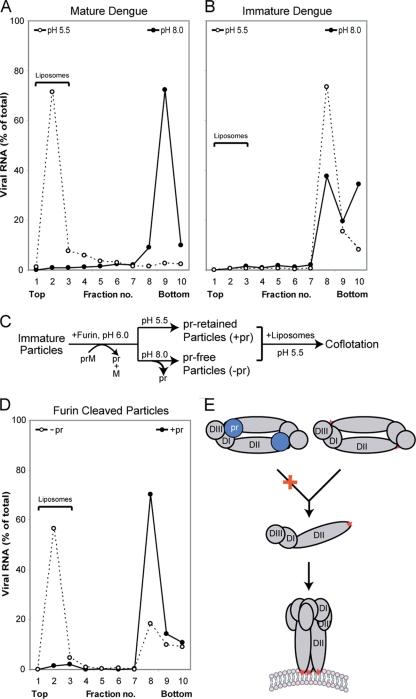
Liposome cofloatation experiments were performed to test if the presence of pr on the viral particle prevents interactions of the virus with the membrane at low pH, a step preceding fusion. As expected, mature dengue virus cofloated with liposomes at acidic pH but not at neutral pH (Fig. 3A). Immature virus containing intact prM did not interact with lipids at either pH (Fig. 3B). To test the function of pr in preventing lipids interactions, immature particles treated with furin at pH 6.0 were either kept at low pH or neutralized to pH 8.0 to release pr (Fig. 3C) (24). The particles then were mixed with liposomes at pH 5.5, followed by sucrose density gradients. Similarly to mature virus, neutralized, pr-free particles cofloated with liposome at pH 5.5 (Fig. 3D). In contrast, particles kept at low pH did not interact with liposomes and remained in the bottom fractions of the sucrose gradients, demonstrating that the retention of pr at the virion surface at acidic pH prevents membrane insertion. The membrane fusion of flaviviruses involves the dissociation of the E dimers and the subsequent formation of homotrimers (1, 15, 22). As the pr peptide makes contact with both E monomers within a dimer (Fig. 2C), it is likely that pr stabilizes the E dimer and thereby inhibits subsequent membrane insertion (Fig. 3E).
FIG. 3.
Association of pr inhibits membrane insertion. Virus particles were mixed with liposomes at pH 5.5 and analyzed by sedimentation in sucrose gradients. Fractions from the gradients were analyzed by qRT-PCR to determine the amount of viral RNA. Liposomes float to the top of the gradient, whereas virus particles sediment to the bottom. (A) Mature dengue virus harvested from infected cells. (B) Immature dengue particles. (C) Flow chart of in vitro furin treatment for samples used in panel D. (D) Furin-treated particles with (−pr) and without (+pr) neutralization. (E) Conformational changes of the glycoproteins necessary to promote membrane fusion. E proteins are shown in gray, and the three domains are labeled (DI, DII, and DIII). The fusion loops are indicated by a red star. The presence of pr (blue) likely prevents E dimer dissociation at low pH.
DISCUSSION
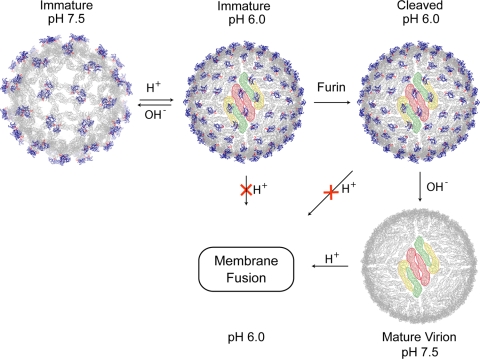
Flaviviruses enter cells by endocytosis, in which the low pH induces membrane fusion. On the other hand, the progeny viruses must be stable at the low-pH environment of the secretory pathway to prevent premature fusion. Previously, Heinz and colleagues noted this apparently paradoxical role of acidic pH in flavivirus activation and suggested that after furin has cleaved prM, pr continues to protect virions in the TGN (21). Here, we have provided direct structural and functional evidence that pr is retained at low pH to prevent membrane fusion. The structures of dengue virus representing four different states in the maturation pathway (Fig. 4) directly support the previously proposed mechanism (11, 24) of how flaviviruses are processed and stabilized. Inside an infected cell, immature virus particles bud into the ER as spiky virions. The acidic environment of the TGN induces a major conformational change, resulting in the exposure of the furin cleavage site. The prM protein is cleaved by furin in the TGN, and the proteolytic product pr remains associated to prevent fusion until the progeny virion is released to the extracellular milieu.
FIG. 4.
Conformational metamorphosis of dengue viruses. Structures of the virion at different stages of the life cycle are shown. The E and pr proteins are shown in gray and blue, respectively, and the fusion loops are in red. An E protein raft, consisting of three parallel dimmers, is shaded in color.
Previously, it had been assumed that proteolytic cleavage induces the conformational changes of the virus, which transforms an inert virion into an infectious one. However, studies of flaviviruses (21, 24, and this work) show that no major conformational changes occur in the virions upon furin cleavage. It is possible that cleavage causes the local refolding of the M protein, which unfortunately cannot be observed at the current resolution of the cryoEM reconstructions. Changes in pH were shown to have a greater effect on the virus structure than that of the proteolytic cleavage (Fig. 4). Particles containing intact prM undergo a pH-dependent, reversible transformation that involves large-scale motions of the E and prM proteins. Once prM is cleaved, there is no further rearrangement of E proteins when the pH is adjusted back to neutral. The presence of pr peptides at low pH with or without the covalent linkage stabilizes the metastable E-E dimer, thereby preventing membrane fusion. It seems that the cleavage of prM by furin merely breaks the linkage between the pr peptide and the M protein and enables the release of pr at neutral pH. The resulting mature virus undergoes pH-induced membrane fusion in the next infection cycle.
The effect of furin cleavage on flavivirus structure observed in this study is reminiscent of studies of influenza hemagglutinin (HA). In influenza virus, the proteolytic cleavage of the envelope glycoprotein precursor HA0 generates a metastable conformation (HA) that mediates membrane fusion. The structures of HA0 and HA essentially are superimposable, except for 19 residues located at the cleavage site (5). Upon cleavage, the N terminus of HA2 (the fusion peptide) inserts into a pocket, burying ionizable residues in the cavity. Therefore, for both dengue and influenza viruses, corresponding to class II and class I fusion mechanisms, respectively, proteolytic cleavage does not induce the large-scale refolding of the fusion proteins. However, cleavage is essential to prime the proteins for membrane fusion by breaking a covalent peptide bond either in the fusion protein itself (HA) or in an accompanying protein (prM), making it possible for the fusion protein to refold into the fusogenic conformation in a subsequent step.
Acknowledgments
We are grateful to Wei Zhang and Sheemei Lok for helpful discussions.
The work was supported by an NIH grant (AI076331 to M.G.R.). J.C. is an HHMI investigator.
I.Y. and J.C. designed the research; I.Y., H.A.H., and P.R.C. performed the research; R.J.K. and M.G.R. contributed reagents and tools; and J.C., I.Y., M.G.R., and R.J.K. wrote the paper.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 6.Chuba, P. J., and S. Palchaudhuri. 1986. Requirement for cysteine in the color silver staining of proteins in polyacrylamide gels. Anal. Biochem. 156:136-139. [DOI] [PubMed] [Google Scholar]
- 7.Fuller, S. D., S. J. Butcher, R. H. Cheng, and T. S. Baker. 1996. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol. 116:48-55. [DOI] [PubMed] [Google Scholar]
- 8.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 9.Kanai, R., K. Kar, K. Anthony, L. H. Gould, M. Ledizet, E. Fikrig, W. A. Marasco, R. A. Koski, and Y. Modis. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 80:11000-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, L., S. M. Lok, I. M. Yu, Y. Zhang, R. J. Kuhn, J. Chen, and M. G. Rossmann. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830-1834. [DOI] [PubMed] [Google Scholar]
- 12.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th edn. ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 13.Lok, S. M., V. Kostyuchenko, G. E. Nybakken, H. A. Holdaway, A. J. Battisti, S. Sukupolvi-Petty, D. Sedlak, D. H. Fremont, P. R. Chipman, J. T. Roehrig, M. S. Diamond, R. J. Kuhn, and M. G. Rossmann. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312-317. [DOI] [PubMed] [Google Scholar]
- 14.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 16.Molloy, S. S., L. Thomas, J. K. VanSlyke, P. E. Stenberg, and G. Thomas. 1994. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 13:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 18.Nybakken, G. E., C. A. Nelson, B. R. Chen, M. S. Diamond, and D. H. Fremont. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80:11467-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136:190-200. [DOI] [PubMed] [Google Scholar]
- 21.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70:8142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wengler, G. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, I. M., W. Zhang, H. A. Holdaway, L. Li, V. A. Kostyuchenko, P. R. Chipman, R. J. Kuhn, M. G. Rossmann, and J. Chen. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834-1837. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, Y., B. Kaufmann, P. R. Chipman, R. J. Kuhn, and M. G. Rossmann. 2007. Structure of immature West Nile Virus. J. Virol. 81:6141-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Y., V. A. Kostyuchenko, and M. G. Rossmann. 2007. Structural analysis of viral nucleocapsids by subtraction of partial projections. J. Struct. Biol. 157:356-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]