Abstract
The human cytomegalovirus (HCMV) open reading frame UL48 encodes a 253-kDa tegument protein that is closely associated with the capsid and was recently shown to have ubiquitin-specific protease activity (J. Wang, A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson, J. Virol. 80:6003-6012, 2006). Here, we examined the cleavage specificity of this deubiquitinase (DUB) and replication characteristics of an active-site mutant virus. The purified catalytic domain of the UL48 DUB (1 to 359 amino acids), corresponding to the herpes simplex virus UL36USP DUB (L. M. Kattenhorn, G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh, Mol. Cell 19:547-557, 2005), efficiently released ubiquitin but not ubiquitin-like modifications from a hemagglutinin peptide substrate. Mutating the active-site residues Cys24 or His162 (C24S and H162A, respectively) abolished this activity. The HCMV UL48 and HSV UL36USP DUBs cleaved both Lys48- and Lys63-linked ubiquitin dimers and oligomers, showing more activity toward Lys63 linkages. The DUB activity of the full-length UL48 protein immunoprecipitated from virus-infected cells also showed a better cleavage of Lys63-linked ubiquitinated substrates. An HCMV (Towne) mutant virus in which the UL48 DUB activity was destroyed [UL48(C24S)] produced 10-fold less progeny virus and reduced amounts of viral proteins compared to wild-type virus at a low multiplicity of infection. The mutant virus also produced perceptibly less overall deubiquitination than the wild-type virus. Our findings demonstrate that the HCMV UL48 DUB contains both a ubiquitin-specific carboxy-terminal hydrolase activity and an isopeptidase activity that favors ubiquitin Lys63 linkages and that these activities can influence virus replication in cultured cells.
Human cytomegalovirus (HCMV) is a member of the betaherpesvirus subfamily. In healthy individuals, HCMV infection is asymptomatic and causes latent or persistent infections. However, primary infections of newborns and reactivation from latent infection in immunocompromised and immunosuppressed individuals cause severe disease (26). The virion of HCMV consists of an icosahedral capsid containing a 235-kb linear genome embedded within a tegument layer of proteins that interface it with an enclosing envelope (14, 26). Although some of the tegument proteins are dissociated from the capsid as it passes through the plasma membrane (32), most are delivered to the cell, and some have early functions that help the virus establish infection, including regulating viral gene expression and modifying host cell antiviral responses (2, 6, 13, 19, 24, 37, 43). Tegument proteins are also required during late stages of replication, including capsid egress from the nucleus (27) and envelopment (5, 8, 33-35). Recently, a herpesvirus group-common tegument protein, the largest protein encoded by each of the herpesvirus genomes, was discovered to contain a cysteine protease activity within the first 500 amino acids of its amino end. It is a ubiquitin (Ub)-specific protease (USP) that removes Ub from protein substrates (20), but its function during herpesvirus replication is presently unknown.
The attachment of Ub monomers and polymers to the target has important roles in regulating cellular processes including protein degradation, protein and vesicle trafficking, cell cycle control, signal transduction, gene transcription, and receptor endocytosis (7). Ubiquitination occurs via an enzymatic cascade, which includes a single ATP-dependent Ub-activating enzyme (E1), dozens of Ub-conjugating proteins (E2), and hundreds of Ub ligases (E3s) (16). Ub is synthesized as a precursor whose carboxy end must be removed by specific Ub carboxy-terminal hydrolases (UCHs) before it can be transferred to a target substrate. This posttranslational modification can be reversed by USPs, which catalyze the hydrolysis of the isopeptide bond linking the C-terminal Gly-Gly residues of Ub to a Lys residue in the substrate. Thus, deubiquitinating proteases (DUBs) can have either UCH or USP activity or both. Most of the known DUBs, including the herpesvirus enzyme, are cysteine proteases characterized by a Cys-His-Asp catalytic triad (1, 15, 22, 36, 41).
The herpesvirus DUB was discovered as a ∼40-kDa fragment of the 336-kDa VP1/2 tegument protein of herpes simplex virus type 1 (HSV-1) (20). It is encoded by the UL36 open reading frame of HSV-1 and was designated UL36USP. It bears no homology to known DUBs but is conserved in the UL36 equivalents of other herpesviruses (20). Studies with recombinant UL36USP and the corresponding domains from murine cytomegalovirus (MCMV) and Epstein-Barr virus verified their DUB activities and established them as the prototype of a new family of viral DUBs (21, 30, 31). In HCMV, UL48 is the homolog of HSV-1 UL36 and encodes a 253-kDa high-molecular-mass tegument protein. Although this protein is essential for replication, as demonstrated by the lethal effect of deleting the gene (8, 9, 12, 44), the DUB catalytic activity per se is not absolutely required. This was established by showing that mutants of HCMV encoding pUL48 with no DUB activity (i.e., active Cys24 and His162 replaced in point mutants [C24I and H162A, respectively]) are still replication competent (39) and has been generalized to other herpesviruses (3, 4, 18, 23). It has also been determined that the full-length pUL48 present in virions is catalytically active and able to cleave polyubiquitin (poly-Ub) chains linked through their Lys48 residues (39), but more information about the substrate specificity of this enzyme will be helpful if not necessary to understand its biological role during replication.
In this study, we have used the cloned catalytic domain of HCMV pUL48 and full-length pUL48 immunoprecipitated from virus-infected cells to investigate the substrate specificity of this enzyme in vitro. We have also constructed an HCMV (strain Towne) mutant virus with no pUL48 DUB activity (i.e., Cys24 nucleophile changed to Ser24), and compared it with the wild-type virus to evaluate the impact of this viral DUB on the progression of infection, viral protein synthesis, and patterns of ubiquitinylation in virus-infected cells.
MATERIALS AND METHODS
Cell culture and virus infectivity assays.
Primary human foreskin fibroblast (HF) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a 5% CO2 humidified incubator at 37°C. For infection experiments, HF cells were infected with virus at the specified multiplicities of infection (MOIs). At the times indicated, the growth medium was collected, combined with lysates prepared from the cell layer by freezing and thawing three times, clarified by centrifugation, and stored at −70°C until assayed for infectivity.
Virus infectivity assays were performed as described previously (17). In brief, the diluted samples were used to inoculate monolayers of HF cells in a 24-well plate. After adsorption, fresh warm culture medium was added. At 24 h postinfection, the growth medium was aspirated, and the cells were fixed with cold methanol for 10 min. The cell layer was then washed three times with phosphate-buffered saline (PBS) and incubated with anti-IE1 rabbit polyclonal antibody diluted in PBS at 37°C for 1 h, followed by incubation with phosphatase-conjugated anti-rabbit immunoglobulin G in PBS at 37°C for 1 h. Finally, the cell layer was rinsed with PBS and treated with detection solutions at room temperature for 1 h according to the manufacturer's directions. The number of cells staining positive for HCMV IE1 was counted in at least three fields per well under a light microscope.
Expression plasmids.
Plasmids encoding the glutathione S-transferase (GST) fusion proteins GST-Ub-hemagglutinin (HA), GST-SUMO-1-HA, GST-ISG15-HA, or GST-Ufm1-HA were provided by Chin Ha Chung (Seoul National University, Seoul, Republic of Korea). A mammalian expression plasmid encoding HA-Ub was provided by Gary S. Hayward (Johns Hopkins University School of Medicine, Baltimore, MD). A plasmid encoding residues 1 to 359 of HCMV UL48, fused to an N-terminal His6 purification tag (UL48 DUB), was prepared in pDEST17 using Gateway technology (Invitrogen). Plasmids expressing the C24S (TGC to TCC) or H162A (CAC to GCC) mutant version of UL48 DUB [UL48 DUB(C24S) and UL48 DUB(H162A), respectively] were made by using the Stratagene QuikChange site-directed mutagenesis protocol. A plasmid expressing the GST-UL48 DUB(C24S) fusion protein was constructed in a pGEX derivative by using Gateway technology. A DNA fragment encoding the first 533 amino acids of HSV UL36 was amplified by PCR from HSV-1 genomic DNA isolated from virus-infected cells and cloned by using the pENTR vector (Invitrogen). An expression plasmid encoding His6-UL36 DUB was generated on plasmid pDEST17 by using Gateway technology. Mammalian expression plasmids expressing Myc6-tagged UL48 (wild type and C23S mutant) were produced with plasmid pCS3-MT (17) by using Gateway technology.
UCH or Ub-like protein (Ubl) carboxy-terminal hydrolase assays.
Recombinant GST-Ub-HA, GST-SUMO-1-HA, GST-ISG15-HA, and GST-Ufm1-HA fusion proteins were expressed in Escherichia coli cells and partially purified by using glutathione-Sepharose 4B beads (Peptron) according to the manufacturer's instructions. Wild-type and mutant UL48 DUB proteins were expressed in E. coli cells and partially purified by immobilized metal affinity chromatography (see below). A total of 3 μg of the purified GST-Ub-HA, GST-SUMO-1-HA, GST-ISG15-HA, or GST-Ufm1-HA protein was mixed with 100 or 200 ng of wild-type or mutant UL48 DUB. PBS and 2 mM dithiothreitol were added to reach a final volume of 40 μl. The reaction mixtures were incubated at 37°C for 24 h. After the termination of the reaction using protein sample buffer, the reaction products were separated by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE). Proteins were then visualized by Coomassie blue staining.
Protein purification by immobilized metal affinity chromatography.
E. coli BL21 cells containing the expression plasmids were grown at 37°C in 200 ml LB containing appropriate antibiotics to an optical density at 600 nm of ∼0.5, induced for protein expression at 30°C with isopropyl-β-d-thiogalactopyranoside (0.4 mM; Sigma), and harvested 4 h later. The cell pellets were suspended in 10 ml lysis buffer (20 mM Tris-HCl [pH 8.0], 5 mM imidazole, 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) and sonicated for 1 min by using a microtip probe (1 s on and 3 s off). The suspension was centrifuged at 14,000 rpm for 30 min at 4°C, and the supernatant was collected. Recombinant proteins were partially purified over Ni-nitrilotriacetic acid resin (Invitrogen) according to the manufacturer's procedure. The resin-bound protein was washed with 60 mM imidazole, and the bound proteins were then eluted with a 2-ml step gradient of imidazole (100 to 500 mM).
In vitro cleavage assays of di-Ub and poly-Ub.
K48- and K63-linked diubiquitin (di-Ub) or poly-Ub proteins used in this study were purchased from Boston Biochem (catalog no. UC-200 for K48-di-Ub, UC-300 for K63-di-Ub, UC220 for K48-poly-Ub, and UC320 for K63-poly-Ub) or Biomol International (catalog no. UW8860 for K48-poly-Ub and UW9570 for K63-poly-Ub). For di-Ub cleavage assays, the reactions were conducted by using a 20-μl volume containing 1 μg of di-Ub and 20 ng of wild-type or C24S mutant HCMV UL48 DUB in PBS with 1 mM dithiothreitol. For poly-Ub cleavage assays, 1 or 2 μg of poly-Ub was incubated with 20 to 30 ng of HCMV UL48 DUB or HSV UL36 DUB or 250 ng of His-OTUB1 (catalog no. E-522; Boston Biochem). The reaction mixtures were incubated at 37°C. After the termination of the reaction by using SDS sample buffer at different time points, the reaction products were separated by SDS-15% PAGE (Gradi-Gel II; Elpis Biotech, Republic of Korea) and stained with Coomassie blue or detected by immunoblotting.
In vitro GST pull-down assays.
GST and GST fusion proteins were generated in E. coli cells. The standard procedure for the GST pull-down assays used was described previously (29).
T-BAC mutagenesis.
An HCMV (strain Towne) bacterial artificial chromosome (BAC) clone (T-BAC) (25) was used as the template for mutagenesis. In this BAC clone, a 9-kb portion of the genome (US1 to US12) is replaced by a fragment containing both the F plasmid sequence and a green fluorescent protein (GFP) expression cassette (25). The 2.8-kb BamHI-SphI restriction fragment containing the UL48 C24S mutation was subcloned into transfer vector pGS284, a derivative of pCV442 (ampicillin resistant and sucrose sensitive) (25). The mutations were transferred into the T-BAC UL48 sequence by homologous recombination (17). In brief, the mutant UL48 sequences were transferred into the T-BAC by cross-streaking E. coli cells (S17-pir; strain GS111) transformed with the transfer vector onto RecA+ E. coli cells (DH10B; strain GS243) hosting wild-type HCMV T-BAC (chloramphenicol resistant). The resulting exoconjugates were selected from intersection regions by growing cells on LB plates containing ampicillin plus chloramphenicol. Several exoconjugate colonies from each dish were incubated in LB broth containing chloramphenicol for 6 h at 37°C. Cultures were serially diluted and spread onto pairs of LB plates, one containing chloramphenicol and one containing chloramphenicol plus 5% sucrose and no NaCl. T-BAC DNAs in colonies isolated from sucrose plates, which subsequently grew on plates containing chloramphenicol but not on plates containing ampicillin, were examined by restriction enzyme digestion and DNA sequencing to validate the gross integrity of their genome and confirm the intended UL48 sequence.
Pulsed-field gel electrophoresis.
The recombinant T-BAC DNAs were digested with restriction endonucleases and subjected to electrophoresis in a 1% agarose (Bio-Rad) gel containing 0.5× TBE (1× TBE is 89 mM Tris-HCl [pH 7.4], 89 mM boric acid, and 25 mM EDTA [pH 8.0]) by using a Chef-DRII apparatus (Bio-Rad) for 5 h at 6 V/cm, with switching times ramped at 0.1 s. The DNA fragments were stained with ethidium bromide (0.5 μg/ml) in 0.5× TBE buffer.
Electroporation and production of virus stocks.
Electroporation was used to introduce the T-BAC DNAs into HF cells. Each reaction mixture contained 5 × 106 HF cells suspended in 250 μl of medium containing 10% serum, 2 μg of the appropriate T-BAC DNA, 1 μg of plasmid pCMV71 encoding pp71 to enhance activating the major immediate-early promoter, and 1 μg of plasmid pEGFP-C1 to monitor electroporation efficiency. Electroporations were done in 0.4-cm cuvettes at 250 V and 960 μF, and the cells were transferred onto T-25 plates. When the surviving cells reached confluence, they were transferred at a dilution of 1:2 into new flasks. Cultures were grown at 37°C, and the spread of GFP fluorescence was monitored.
Antibodies.
Rabbit anti-peptide antibodies for UL48 were raised against the synthetic peptide N′-SSSPKKTPEKRRKDLSGS-C′ (pUL48 residues 278 to 295). Anti-HA rat monoclonal antibody (MAb) 3F10 conjugated with peroxidase was purchased from Roche. Mouse MAb 6E1 for the major immediate-early IE1 (72-kDa) protein and rabbit polyclonal antibody raised against purified IE1, which detects both IE1 and IE2 (86 kDa), were previously described (17). Mouse MAbs against p52 (UL44) and pp28 (UL99) were purchased from Advanced Biotechnologies, Inc. Mouse MAbs against Ub (P4D1) and β-actin were purchased from Santa Cruz and Sigma, respectively.
Immunoprecipitation of the UL48 proteins from virus-infected cells.
Subconfluent HF cells in a 150-mm dish were mock infected or infected with the recombinant viruses at an MOI of 3. At 80 h after incubation, cells were harvested and sonicated in 1 ml coimmunoprecipitation assay buffer (50 mM Tris-Cl [pH 7.4], 50 mM NaF, 5 mM sodium phosphate, and 0.1% Triton X-100 containing protease inhibitors [Sigma]) by using a microtip probe (Vibra cell; Sonics and Materials, Inc.) for 10 s (pulse on for 1 s and pulse off for 3 s). The clarified cell lysates were incubated with anti-UL48 antibodies for 16 h at 4°C. Sixty microliters of a 50% slurry of protein A- and G-Sepharose (Amersham) was then added. After incubation for 2 h at 4°C, the beads were pelleted, washed seven times with coimmunoprecipitation assay buffer, and used as the source of infected-cell pUL48 in Ub cleavage assays.
Immunoblot analysis.
For the detection of viral antigens, the virus-infected cells were harvested in PBS containing 5 mM N-ethylmaleimide, denatured in protein sample buffer, boiled for 5 min, and fractionated via SDS-8% PAGE. For the Ub cleavage assays, the reaction products were separated via SDS-gradient PAGE (Gradi-Gel II). Standard detection procedures for the enhanced chemiluminescence system (Roche) and X-ray film were followed.
RESULTS
Recombinant HCMV UL48 DUB has UCH activity.
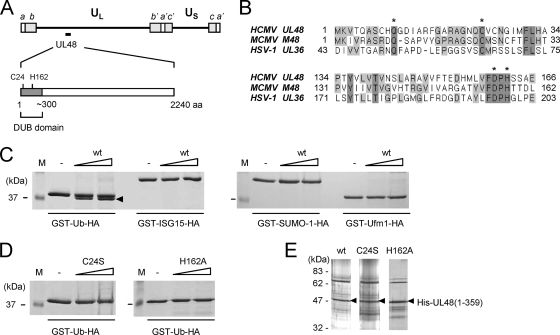
Based on the sequence homology among eight different human herpesviruses, four amino acids (Gln52, Cys65, Asp197, and His199) that may directly contribute to enzymatic proteolysis were identified in the N-terminal DUB domain of HSV-1 US36 (20). The corresponding residues of HCMV UL48 and MCMV M48 also contained within the highly conserved ∼300-amino-acid N-terminal sequences are aligned (Fig. 1A and B).
FIG. 1.
UCH activity of HCMV UL48 DUB. (A) The HCMV genome consisting of unique long (UL) and unique short (US) sequences bound by terminal repeats (a/a′, b/b′, and c/c′) and the amino-terminal region (∼300 amino acids) of UL48 exhibiting DUB activity are illustrated. The Cys and His catalytic residues (see below) are indicated. (B) Amino acid sequences containing the predicted catalytic residues (Gln, Cys, Asp, and His) are aligned for the DUB domains of HCMV UL48, MCMV M48, and HSV-1 UL36 (20). Amino acid numbers within each sequence are indicated, conserved amino acids are shaded, and the predicted catalytic residues are indicated by asterisks. (C) In vitro assays for the C-terminal hydrolase activity of UL48 DUB toward Ub or three Ubl-HA fusions. Three micrograms of the purified GST-Ub-HA or GST-Ubl-HA fusion protein was incubated at 37°C for 24 h with 100 or 200 ng of bacterially expressed wild-type (wt) UL48 DUB or with no DUB added. The reaction products were separated by SDS-12% PAGE and stained with Coomassie blue. The arrowhead indicates GST-Ub. Lane “M” contains molecular mass marker proteins, and the 37-kDa marker is shown. (D) GST-Ub-HA was assayed as described above (C) but using UL48 DUB(C24S) or UL48 DUB(H162A). (E) Samples equivalent to the 100-ng amount of each UL48 DUB used in the assays shown in C and D were subjected to SDS-PAGE followed by silver staining.
To investigate whether the DUB domain of UL48 contains UCH activity, we cloned a sequence encoding the 359 N-terminal amino acids of UL48 into an expression vector that added a His6 tag to its N-terminal end (UL48 DUB). We also generated point mutants of this construct in which the active-site Cys24 was replaced with Ser (C24S) or the active-site His162 was replaced with Ala (H162A). UL48 DUB and mutants were expressed in E. coli cells and partially purified over Ni-nitrilotriacetic acid resin. We also expressed and affinity purified carboxy-HA-tagged Ub, ISG15, SUMO-1, and Ufm1 as fusion proteins having GST at their amino ends (i.e., GST-Ub-HA, GST-ISG15-HA, GST-SUMO-1-HA, and GST-Ufm1-HA). When bacterially expressed UL48 DUB was incubated with the GST fusion proteins, it cleaved GST-Ub-HA to produce GST-Ub (Fig. 1C) but did not cleave GST-ISG15-HA, GST-SUMO-1-HA, or GST-Ufm1-HA (Fig. 1C). When C24S and H162A mutant UL48 DUBs were tested, no activity was detected, consistent with the critical catalytic role of these residues (20, 39) (Fig. 1D). Protein staining showed that the assay mixtures contained comparable amounts of wild-type and mutant UL48 DUBs (Fig. 1E). These results demonstrate that UL48 DUB has Ub-specific UCH activity.
UL48 DUB has higher USP activity toward K63-linked Ub than K48-linked Ub.
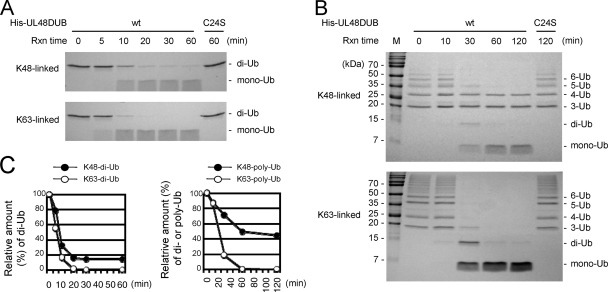
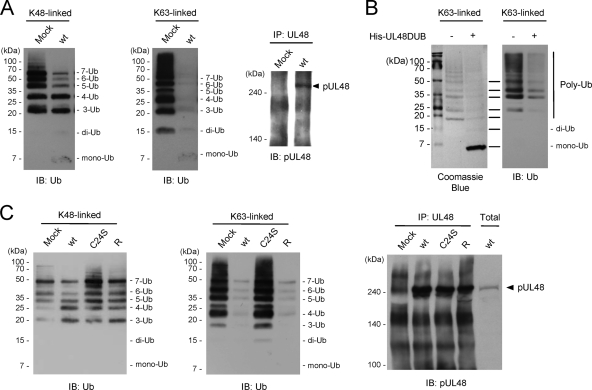
We next investigated whether UL48 DUB shows linkage selectivity in its USP activity and first tested Ub dimer substrates (di-Ub). Wild-type or C24S mutant UL48 DUBs were incubated with K48- or K63-linked di-Ub in a time course reaction, and the products were detected by SDS-PAGE and protein staining. The results showed that wild-type UL48 DUB cleaved both K48- and K63-linked di-Ub into monomers, but the C24S mutant showed no activity during the 1-h incubation period (Fig. 2A). Notably, UL48 DUB cleaved K63-linked di-Ub about twofold faster than it cleaved K48-linked di-Ub (Fig. 2C, left).
FIG. 2.
UL48 DUB cleaves both K48- and K63-linked Ub. (A) K48- or K63-linked di-Ub (Boston Biochem) was incubated with wild-type (wt) (first six lanes) or C24S mutant UL48 DUB at 37°C for the indicated times. Reactions were stopped by adding protein sample buffer to the mixture, and mixtures were subjected to SDS-15% PAGE, followed by Coomassie blue protein staining. Rxn time, reaction time. (B) K48- or K63-linked poly-Ub (Boston Biochem) was incubated with wild-type or C24S mutant UL48 DUB, and the products were analyzed as described above (A) for the times indicated. A Gradi-Gel II gel was used for SDS-PAGE, and staining was performed with Coomassie blue. The positions of mono-Ub to hexaubiquitin are on the right, and molecular mass markers are indicated on the left. (C) Data in A and B were quantified. Shown here are graphs of the calculated values for the di-Ub (left) and poly-Ub (right) remaining.
We then tested the linkage specificity of UL48 DUB USP activity toward poly-Ub substrates by using K48-linked or K63-linked poly-Ub (Boston Biochem) (chains of three to seven Ubs). The results showed that UL48 DUB also cleaved both K48- and K63-linked poly-Ub (Fig. 2B). Consistent with the results obtained using di-Ub substrates, it cleaved K63-linked poly-Ub more efficiently than K48-linked poly-Ub. In the experiment shown, about 80% of K63-linked poly-Ub was converted to monomeric Ub during 30 min of incubation, whereas only 30% of K48-linked poly-Ub was cleaved during the same incubation time. Additionally, although most K63-linked poly-Ub was converted to monomeric Ub within 60 min, nearly 40% of K48-linked poly-Ub remained uncleaved after 120 min of incubation (Fig. 2C, right).
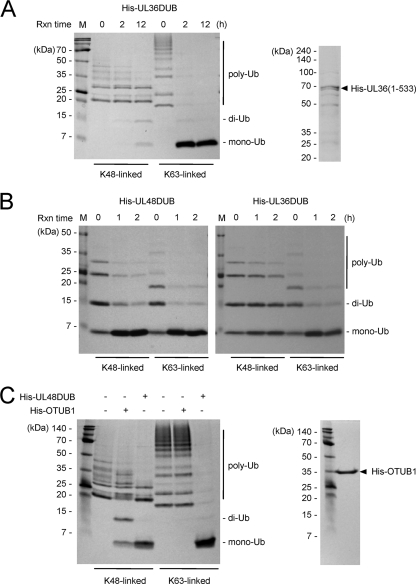
We also tested the linkage selectivity of the corresponding HSV-1 UL36 DUB by using the same poly-Ubs. Like UL48 DUB, UL36DUB cleaved both substrates and showed higher USP activity for K63-linked poly-Ub than for K48-linked poly-Ub (Fig. 3A). Similar results were obtained when the experiment was repeated using poly-Ub substrates from a different source (i.e., Biomol International). UL48 DUB and UL36 DUB cleaved both K48- and K63-linked poly-Ub, and both DUBs showed higher USP activity for K63 linkages than for K48 linkages (Fig. 3B). The deubiquitinating enzyme otubain 1 (OTUB1) was recently shown to preferentially cleave K48-linked poly-Ub over K63-linked poly-Ub (10). In our control experiments, OTUB1 (purchased from Boston Biochem) cleaved K48-linked poly-Ub but not K63-linked poly-Ub under the same conditions where UL48 DUB efficiently cleaved both K48-linked and K63-linked poly-Ub chains (Fig. 3C). Overall, our results demonstrate that UL48 DUB cleaves both K48- and K63-linked di- or poly-Ub in vitro, but it has higher USP activity for K63 linkages than for K48 linkages.
FIG. 3.
Comparison of HCMV UL48 DUB and HSV-1 UL36 DUB USP activities. (A) K48- or K63-linked poly-Ub (Boston Biochem) was incubated with UL36 DUB for the indicated times and analyzed as described in the legend of Fig. 2B. The rightmost single lane shows a sample of the UL36 DUB [His-UL36(1-533)] preparation separated by SDS-PAGE and stained with Coomassie blue. Rxn time, reaction time. (B) K48- or K63-linked poly-Ub (Biomol International) was incubated withUL48 DUB and UL36 DUB for the indicated times, and the reaction products were analyzed as described above (A). (C) K48- or K63-linked poly-Ub (Boston Biochem) was incubated with OTUB1 or UL48 DUB for 12 h and analyzed as described above (A). A sample of the His-OTUB1 used, following SDS-PAGE and Coomassie blue staining, is shown to the right. Molecular mass markers, positions of Ub substrates and products, and times of incubation are indicated and abbreviated as described in the legends in preceding figures.
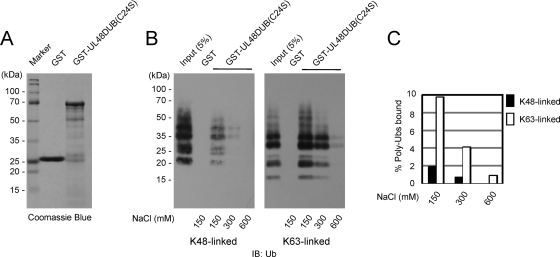
We further investigated whether UL48 DUB has different binding affinities for K48- and K63-linked Ub polymers. In in vitro GST pull-down assays using purified GST-UL48 DUB(C24S), the UL48 DUB domain bound directly to both K48-linked and K63-linked poly-Ub chains but showed about a fivefold-higher binding affinity for K63-linked poly-Ub chains than K48-linked poly-Ub (Fig. 4).
FIG. 4.
Comparison of UL48 DUB binding affinities for K48- and K63-linked poly-Ub. (A) Samples of the GST and GST-UL48 DUB(C24S) proteins purified from E. coli cells were subjected to SDS-PAGE followed by Coomassie blue staining. (B) GST pull-down assay. The GST and GST-UL48 DUB(C24S) proteins immobilized on glutathione-Sepharose beads were incubated with K48-linked or K63-linked poly-Ub (Boston Biochem) under different salt conditions as indicated. The poly-Ub chains bound were fractionated by SDS-PAGE and detected by immunoblotting (IB) with anti-Ub antibody. Five percent of the input poly-Ub chains are also shown (left lane in each panel). (C) The amounts of poly-Ub bound under different salt conditions were quantified. Shown here are graphs of the calculated values for K48-linked (black bars) and K63-linked (white bars) Ub polymers bound.
Generation of mutant virus expressing pUL48(C24S) and its revertant.
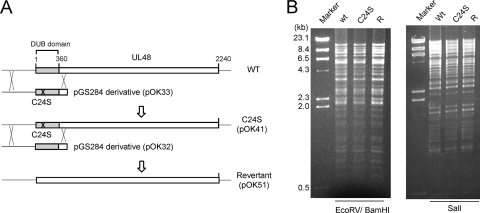
In order to test the DUB activity of the full-length UL48 protein (pUL48), we recovered it from virus-infected cells. Three viruses were prepared from a Towne HCMV bacmid (T-BAC), as described in Materials and Methods: wild-type virus encoding normal pUL48, a mutant virus encoding pUL48 with the C24S change [pUL48(C24S)], and a revertant virus made by changing the UL48(C24S) mutation back to the wild type.
The C24S mutation was introduced into the UL48 gene of the T-BAC clone by homologous recombination (Fig. 5A and see Materials and Methods) and confirmed by sequencing a PCR-amplified fragment of the bacmid DNA that includes the site. A revertant of the UL48(C24S) T-BAC clone was made by homologous recombination between it and a wild-type UL48 DNA fragment cloned into pGS284 (Fig. 5A). DNA sequencing of an appropriate PCR-amplified fragment verified the change of mutant Ser24 back to wild-type Cys24. No gross deletions or rearrangements in the mutant or the revertant genomes were detected by comparing their endonuclease restriction fragment patterns with those of the wild-type T-BAC following pulsed-field gel electrophoresis and staining with ethidium bromide (Fig. 5B).
FIG. 5.
Preparation of UL48(C24S) mutant T-BAC and its revertant. (A) Scheme for making the C24S mutant and its revertant T-BAC clones. Plasmids containing either a 2.8-kb DNA fragment of wild-type (WT) UL48 (pOK32) or the same fragment mutated to encode the C24S mutation (pOK33) were used for homologous recombination with the parent T-BAC clone as diagrammed. These resulted in the mutant (pOK41) and revertant (pOK51) T-BAC clones. (B) Restriction endonuclease DNA fragment patterns obtained following EcoRV/BamHI or SalI digestion of wild-type (wt), C24S mutant, and revertant T-BAC DNAs. Shown here are the fragments separated by pulsed-field gel electrophoresis and stained with ethidium bromide. The sizes of molecular mass marker fragments of λ-HindIII are shown in the left lane.
Virus production was initially noted by the spread of GFP fluorescence (i.e., the ectopic GFP gene in T-BAC) and viral cytopathic effects, which began after about 10 days and included most cells within 3 or 4 weeks. The spread of virus was comparable for the wild-type, mutant, and revertant (data not shown), consistent with previous findings that the DUB activity of HCMV pUL48 and its homologs is not essential for the transfected viral genome to replicate and produce infectious virus in permissive cells (3, 4, 18, 23).
Analysis of the USP activity of the UL48 protein from virus-infected cells.
We next examined the USP activity of pUL48 recovered from virus-infected cells. pUL48 was immunoprecipitated with anti-UL48 antibodies from cells infected with the wild-type virus and incubated with K48-linked or K63-linked poly-Ubs. The resulting mixtures were analyzed by SDS-PAGE and immunoblotting, which showed that immunoprecipitated pUL48 cleaved both substrates. In agreement with the results obtained using bacterially expressed UL48 DUB, pUL48 from infected cells cleaved K63-linked poly-Ub more efficiently than K48-linked poly-Ub (Fig. 6A). The unexpected faint bands of monoubiquitin (mono-Ub) and di-Ub bands in the K63 immunoblot are due to the weak reactivity of these anti-Ub MAbs with mono-Ub (42), as evidenced by the relative intensities of the same reaction products stained with Coomassie blue or detected by immunoblotting (Fig. 6B, right).
FIG. 6.
Infected-cell UL48 protein cleaves poly-Ub. (A) K48- or K63-linked poly-Ub (1 μg; Boston Biochem) was incubated at 37°C for 2 h with pUL48 prepared from HCMV-infected cells by immunoprecipitation (IP) with anti-UL48 antibodies (“wt”) (see Materials and Methods). A sample from noninfected cells was similarly prepared (mock). After centrifugation, the supernatants of the reaction mixtures were subjected to SDS-PAGE (Gradi-Gel II), followed by immunoblotting (IB) with anti-Ub antibody (left two panels). The pellets were resuspended in protein sample buffer and subjected to SDS-PAGE and immunoblotting with anti-UL48 antibody (right two lanes). (B) K63-linked poly-Ub (Boston Biochem) was incubated without (−) or with (+) wild-type UL48 DUB for 2 h as described in the legend of Fig. 2B. The reaction products were separated by SDS-PAGE (Gradi-Gel II) and either stained with Coomassie blue (left) or detected by immunoblotting with anti-Ub antibody (right). The positions of mono-Ub, di-Ub, and poly-Ub are indicated on the right, and sizes of molecular mass markers are indicated on the left. (C) K48- or K63-linked poly-Ub was incubated with wild-type, C24S mutant, or revertant (R) pUL48 prepared from the virus-infected cells by immunoprecipitation or with similarly prepared material from noninfected cells (mock), and the samples were analyzed as described above (A) (left two panels). The relative amounts of immunoprecipitated pUL48 were determined as described above (A). pUL48 in wild-type virus-infected cells, collected and directly solubilized in protein sample buffer 72 h after infection at an MOI of 3 (“Total,” rightmost lane), served as a size marker for immunoprecipitated pUL48.
We similarly compared the activities of wild-type, C24S mutant, and revertant pUL48 from virus-infected cells. The results showed no activity for immunoprecipitated C24S mutant pUL48, but pUL48 from both wild-type and revertant virus-infected cells was active and more so for K63-linked poly-Ub than K48-linked poly-Ub (Fig. 6C, left and center). The amounts of immunoprecipitated wild-type, mutant, and revertant pUL48 proteins were comparable, and all comigrated with pUL48 in infected cells (Fig. 6C, right). Overall, these results confirm that pUL48 expressed in virus-infected cells contains USP activity that is eliminated by mutating the catalytic Cys24 and show that this activity cleaves K63-linked poly-Ub better than K48-linked poly-Ub.
The DUB activity of UL48 contributes to efficient viral growth in cultured cells at low MOIs.
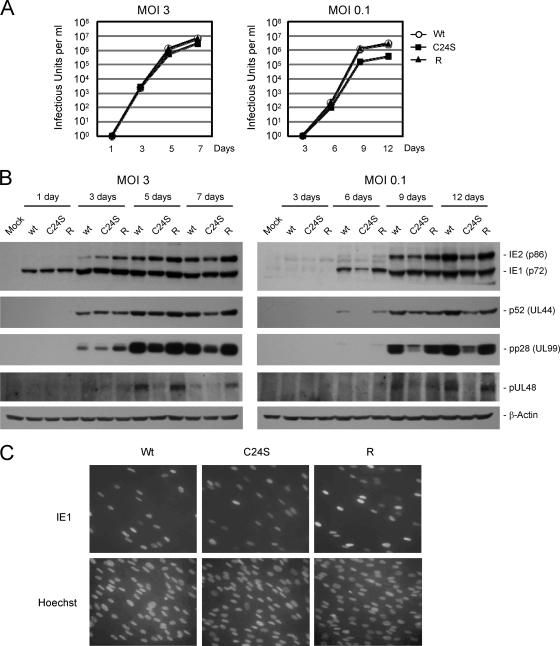
To compare the relative infectivities of the wild-type, C24S mutant, and revertant viruses, two survey experiments were performed. First, HF cells were infected with virus at an MOI of 3 or 0.1. The total amounts of virus produced (intracellular and extracellular combined) were collected at various time points after infection, and titration assays were conducted as described in Materials and Methods. At an MOI of 3, the growth kinetics of the UL48(C24S) mutant virus were similar to those of wild-type and revertant viruses (Fig. 7A, left). However, at an MOI of 0.1, mutant virus infection produced about 10-fold-fewer progeny virions than did wild-type and revertant viruses (Fig. 7A, right). The results of immunoblotting assays showed that the slightly reduced growth of the mutant virus at a low MOI correlated with the low-level accumulation of viral immediate-early (IE1-p72 and IE2-p86), delayed-early (p52 [UL44]), and late (pp28 [UL99 and pUL48]) proteins in infected cells (Fig. 7B). The use of comparable amounts of input viruses was confirmed by indirect immunofluorescence assay (Fig. 7C).
FIG. 7.
Analysis of the growth curves of wild-type, C24S mutant, and revertant viruses and accumulation of viral proteins in infected cells. (A) HF cells in 12-well plates were infected with wild-type (Wt), UL48(C24S) mutant, or revertant (R) virus at an MOI of 3 or 0.1. The time course results shown represent the total amounts of virus produced at the indicated sampling times. The results indicated are the averages of data from two independent assays. (B) HF cells in 60-mm dishes were infected with recombinant viruses at an MOI of 3 or 0.1. Total-cell extracts were prepared at the indicated time points and were subjected to SDS-8% PAGE, followed by immunoblotting for IE1, IE2, p52 (UL44), pp28 (UL99), and pUL48 (left, MOI of 3; right, MOI of 0.1). (C) HF cells were infected with virus at an MOI of 1. At 8 h after infection, cells were fixed in methanol, and an immunofluorescence assay was performed with anti-IE1 MAb 6E1. DNA was stained with Hoechst dye.
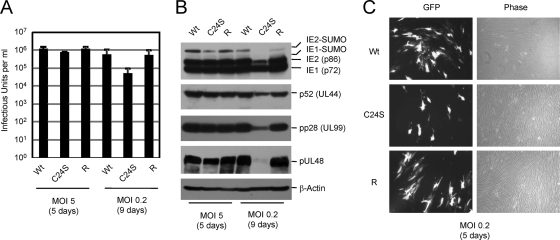
In the second experiment, separately produced virus stocks were used, the MOIs were increased to 5 and 0.2, and virus production in the culture medium was measured only on days 5 (MOI of 5) and 9 (MOI of 2). Results were similar to those for the first experiment. The amount of infectious virus released into the culture medium for mutant virus infection was comparable to those for the wild-type and revertant viruses at a high MOI, whereas it was lower by 10-fold than those in wild-type and revertant virus infections at a low MOI (Fig. 8A). Consistently, a similar reduction in the level of mutant virus production at a low MOI correlated with a reduced amount of viral proteins (Fig. 8B). Furthermore, we found that when cells were infected at a low MOI, the spread of GFP signals in the mutant virus infection was slower than those for wild-type and revertant virus infections (Fig. 8C). Overall, the analysis of the UL48(C24S) mutant virus suggests that although UL48 DUB activity is not essential for viral growth, it may contribute to viral growth in cultured cells at low MOIs.
FIG. 8.
Virus production by the UL48(C24S) mutant and its revertant. (A and B) HF cells were infected with wild-type (Wt), UL48(C24S) mutant, or revertant (R) virus at an MOI of 5 or 0.2 as described in the legend of Fig. 7. Samples in culture medium were taken on days 5 (MOI of 5) and 9 (MOI of 0.2), diluted, and assayed for virus titer as described in the legend of Fig. 7A. Total-cell extracts were prepared and analyzed by immunoblotting as described in the legend of Fig. 7B. (C) The GFP images (left) and corresponding phase-contrast images (right) of cells were taken on day 5 after infection with viruses at an MOI of 0.2.
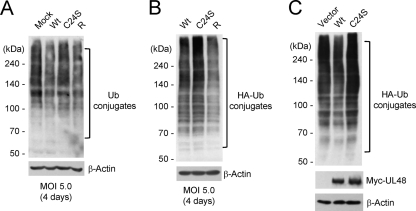
Effects of wild-type and UL48(C24S) mutant virus infections on the level of cellular Ub conjugates.
We examined whether the level of cellular Ub conjugates is changed by virus infection or differently affected by the wild-type and mutant viruses. The results showed that compared to mock-infected cells, the level of endogenous Ub conjugates appeared to be slightly reduced in cells infected with wild-type and revertant viruses but not in cells infected with the mutant virus (Fig. 9A). Consistently, when cells were first transfected with HA-Ub and then infected with virus, the level of HA-Ub conjugates in mutant virus-infected cells was higher than those in wild-type and revertant virus-infected cells (Fig. 9B). Furthermore, similar results were obtained when cells were cotransfected with HA-Ub and UL48 (wild-type and mutant) plasmids (Fig. 9C), suggesting that the small difference in the levels of cellular Ub conjugates observed between wild-type and mutant viruses indeed reflects the difference between wild-type and mutant UL48 DUB activities. Collectively, these results suggest that UL48 DUB activity may affect the level of Ub conjugates in virus-infected cells.
FIG. 9.
Comparison of the levels of Ub conjugates in cells infected with wild-type, C24S mutant, and revertant viruses. (A) HF cells were mock infected or infected with wild-type (Wt), C24S mutant, and revertant (R) virus at an MOI of 5. At 4 days after infection, cells were washed with PBS containing 5 mM N-ethylmaleimide. Total-cell extracts were prepared by boiling the samples in protein sample buffer and were subjected to SDS-6% PAGE followed by immunoblotting with anti-Ub antibody. (B) HF cells were first transfected with plasmids expressing the HA-Ub fusion protein for 24 h. The cells were then divided into three parts, infected with the indicated viruses, and analyzed as described above (A), with anti-HA antibody. (C) 293T cells were cotransfected with plasmid expressing HA-Ub and Myc6-UL48 (wild type or C24S mutant) for 48 h. Total-cell extracts were prepared, and immunoblotting was performed as described above (A) with anti-HA and anti-UL48 antibodies.
DISCUSSION
We demonstrate that UL48 DUB contains UCH activity that is specific for Ub but not for Ubl proteins such as ISG15, SUMO-1, and Ufm1. Since UL48 DUB also contained USP activity (39; this study), UL48 DUB falls into a DUB family that has both USP and UCH activities. We also show that UL48 cleaves both K48- and K63-linked di- and poly-Ubs but has higher Ub-depolymerizing activity for K63-linked polymers than for K48-linked polymers. This linkage-selective activity of UL48 was evident when an assay was conducted with the immunoprecipitated UL48 protein prepared from HCMV-infected cells. Recently, M48, an MCMV equivalent of UL48, was shown to cleave both K48- and K63-linked di-Ub, but it cleaved K48-linked di-Ub five times faster than it cleaved K63-linked di-Ub (31). Whether this difference implicates distinct functions between UL48 and M48 is not clear.
In this study, like UL48 DUB, HSV-1 UL36 DUB also cleaved both K48- and K63-linked Ub polymers and showed higher cleavage activity for K63-linked polymers than for K48-linked polymers. This result is in contrast to a previously reported observation that UL36 DUB cleaved only K48-linked polymers of His-tagged Ub but not K63-linked polymers (20), possibly due to the different poly-Ub substrates used. Importantly, we confirmed the K63-favoring linkage-selective activity of UL48 DUB and UL36 DUB using Ub polymers without a tag and different Ub polymer sources. Overall, our data suggest that the DUBs of herpesviruses appear to be conserved to react to both K48- and K63-linked Ubs. However, it should be noted that the in vivo Ub linkage specificity of these proteins may be influenced by their interactions with target or other partner proteins in infected cells.
About 100 cellular DUBs are encoded by the human genome, and they have been classified into five families based on their DUB domain structures: USPs, UCHs, ovarian tumor-related proteases, Machado-Joseph disease protein domain proteases, and JAB1/PAB1/MPN domain-containing metalloenzymes (28). However, the function of most of these families is unknown, and information about their cleavage specificity is limited. Although Ub precursor processing is thought to be performed by many DUBs (1), whether they generally cleave both K48 and K63 linkages or target specific linkages is largely unknown (1, 22, 36). In our control reactions, otubain 1 cleaved only K48 linkages, as expected, and cellular isopeptidase T (USP5), like UL48 DUB and the ovarian tumor-related protease domain-containing protease A20 (11, 40), cleaved both K48-linked and K63-linked poly-Ubs (data not shown). It is expected that specific DUBs cleave specific linkages more efficiently than others. Given the functional difference between K48-linked and K63-linked Ub polymers, the UL48 DUB activity is likely to have multiple impacts on cellular Ub biology.
Consistent with the results using bacterially expressed UL48 DUB, pUL48 expressed in HCMV-infected cells also cleaved K63-linked poly-Ub with higher kinetics than K48-linked poly-Ub. It was previously reported that detergent-disrupted extracellular virus particles recovered from HCMV-infected cells cleaved K48-linked poly-Ub but not K63-linked poly-Ub (39). These two studies employed different methods to prepare pUL48 and therefore might have evaluated its DUB activity with different sensitivities. This difference may also indicate that the UL48 DUB activity for K63 linkage is masked in virions.
Results of our genetic analysis with C24S mutant virus are consistent with data from other studies showing that the DUB activities of UL48 (39) and its homologs (3, 4, 18, 23) are not essential for viral growth in permissive cultured cells. We found that UL48(C24S) mutant virus is also viable in other semipermissive cell types such as U373 cells and U2-OS cells (data not shown). However, growth curve analysis demonstrates that the mutant virus is detectably impaired for growth at low MOIs. This correlated with a reduced accumulation of viral immediate-early, early, and late proteins and a slow spread of the GFP signals in cells infected with mutant virus, compared to the wild type, at low MOIs. We also observed that the mutant virus produces smaller plaques than the wild-type virus at low MOIs and that UL48 did not alter the steady-state level of IE2 or UL44 in cotransfected cells (data not shown). Therefore, the growth defect of the mutant virus at low MOIs may be attributed to the slow spread of the virus to neighboring cells, consistent with the slower progression of cytopathic effects and the reduced level of production of extracellular virus particles observed for the UL48(C24I) and UL48(H162A) mutant viruses (39).
The possibility that the replication of the UL48(C24S) mutant virus may be enabled by complementation from the viral genome or by a cellular activity is unresolved. If such complementation results from a compensating change or function within the viral genome, it is external to the UL48 gene, since the C24S mutant and wild-type UL48 differ only by the intended point mutations. Alternatively, mutant viral DUB activity may be partially compensated for by a cellular activity. If so, it is plausible that this factor is not as abundant in highly differentiated cells as it is in less-differentiated cell types (e.g., cultured HFs). Such a difference could reconcile the modest replication defect of the UL48(C24S) mutant virus in HF cells with the relatively stronger defects observed in animals infected with counterpart mutants of Marek's disease virus (18) and pseudorabies virus (4).
The function of herpesviral DUBs remains undetermined. They may increase the stability of target proteins during the progression of infection by inhibiting K48-linked polyubiquitination, which leads to the proteasomal degradation of target proteins. However, the fact that UL48 DUB efficiently reacts to K63-linked Ub chains also suggests that UL48 may modulate cellular signaling mediated by K63-linked Ub chains. In this regard, it is notable that the DUB activity reacting to K63-linked Ub chains plays important roles in the regulation of both innate and adaptive immune responses (38). It will also be intriguing to test whether Ub binding activity rather than DUB activity affects viral growth in culture. Identifying cellular (or viral) substrates for UL48 DUB activity or its cellular binding partners and evaluating their interactions will be necessary to understand the enigmatic role of the DUB domain of UL48 in viral pathogenesis.
Acknowledgments
We thank Chin Ha Chung (Seoul National University) for GST-Ub/Ubl-HA constructs.
This work was supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MEST) (Ubiquitome Research Program 20090065395) to J.-H.A. and by USPHS grant AI082246 to W.G.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Amerik, A. Y., and M. Hochstrasser. 2004. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695:189-207. [DOI] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottcher, S., H. Granzow, C. Maresch, B. Mohl, B. G. Klupp, and T. C. Mettenleiter. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottcher, S., C. Maresch, H. Granzow, B. G. Klupp, J. P. Teifke, and T. C. Mettenleiter. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt, W. J., and S. Boppana. 2004. Human cytomegalovirus virion proteins. Hum. Immunol. 65:395-402. [DOI] [PubMed] [Google Scholar]
- 6.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciechanover, A. 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6:79-87. [DOI] [PubMed] [Google Scholar]
- 8.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edelmann, M. J., A. Iphofer, M. Akutsu, M. Altun, K. di Gleria, H. B. Kramer, E. Fiebiger, S. Dhe-Paganon, and B. M. Kessler. 2009. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418:379-390. [DOI] [PubMed] [Google Scholar]
- 11.Evans, P. C., H. Ovaa, M. Hamon, P. J. Kilshaw, S. Hamm, S. Bauer, H. L. Ploegh, and T. S. Smith. 2004. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 378:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, W. 2008. Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 325:187-204. [DOI] [PubMed] [Google Scholar]
- 15.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Huh, Y. H., Y. E. Kim, E. T. Kim, J. J. Park, M. J. Song, H. Zhu, G. S. Hayward, and J.-H. Ahn. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 82:10444-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 104:20025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalejta, R. F., and T. Shenk. 2003. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J. Virol. 77:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 21.Kattenhorn, L. M., R. Mills, M. Wagner, A. Lomsadze, V. Makeev, M. Borodovsky, H. L. Ploegh, and B. M. Kessler. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187-11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J. H., K. C. Park, S. S. Chung, O. Bang, and C. H. Chung. 2003. Deubiquitinating enzymes as cellular regulators. J. Biochem. (Tokyo) 134:9-18. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 28.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 29.Park, J. J., Y. E. Kim, H. T. Pham, E. T. Kim, Y. H. Chung, and J. H. Ahn. 2007. Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J. Gen. Virol. 88:3214-3223. [DOI] [PubMed] [Google Scholar]
- 30.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmolke, S., P. Drescher, G. Jahn, and B. Plachter. 1995. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J. Virol. 69:1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo, J. Y., and W. J. Britt. 2007. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol. 81:6536-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo, J. Y., and W. J. Britt. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soboleva, T. A., and R. T. Baker. 2004. Deubiquitinating enzymes: their functions and substrate specificity. Curr. Protein Pept. Sci. 5:191-200. [DOI] [PubMed] [Google Scholar]
- 37.Stamminger, T., M. Gstaiger, K. Weinzierl, K. Lorz, M. Winkler, and W. Schaffner. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J. Virol. 76:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, S. C. 2008. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8:501-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694-699. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson, K. D., and M. Hochstrasser. 1998. The ubiquitinating enzymes. Plenum Press, New York, NY.
- 42.Winborn, B. J., S. M. Travis, S. V. Todi, K. M. Scaglione, P. Xu, A. J. Williams, R. E. Cohen, J. Peng, and H. L. Paulson. 2008. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283:26436-26443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]