Abstract
The human genome contains more than half a million human endogenous retrovirus (HERV) long terminal repeats (LTRs) that can be regarded as mobile regulatory modules. Many of these HERV LTRs have been recruited during evolution as transcriptional control elements for cellular gene expression. We have cloned LTR sequences from two HERV families, HERV-H and HERV-L, differing widely in their activity and tissue specificity into a murine leukemia virus (MLV)-based promoter conversion vector (ProCon). Various human cell lines were infected with the HERV-MLV hybrid vectors, and cell type-specific expression of the reporter gene was compared with the promoter specificity of the corresponding HERV LTRs in transient-transfection assays. Transcription start site analysis of HERV-MLV hybrid vectors revealed preferential use of the HERV promoter initiation site. Our data show that HERV LTRs function in the context of retroviral vectors in certain cell types and have the potential to be useful as cell type-specific promoters in vector construction.
About 8 to 9% of the human genome consists of human endogenous retroviruses (HERVs) and long terminal repeat (LTR) retroelements (22, 24). These sequences are thought to be relicts of germ line infections that became genetically fixed during primate evolution (for a review, see references 13, 26, and 44). Since then, they have amplified and spread throughout the primate genome by reinfection and/or retrotransposition.
In contrast to HERV protein coding sequences, which accumulated numerous inactivating mutations or deletions, HERV LTRs have preserved their promoter activity and still contain active regulatory elements, such as enhancer sequences, transcription factor binding sites, or polyadenylation signals. We have analyzed more than 100 arbitrarily isolated HERV LTR sequences, including 5′, 3′, and solitary LTRs, in a transient-transfection assay and found that about one-third of these LTRs are still active and may drive gene expression (2, 36; S. Weinhardt et al., unpublished data). Thus, HERVs and other LTR retrotransposons represent mobile regulatory modules that may contribute to the transcriptional regulation of cellular genes (5, 18, 28, 33, 47). There are a number of bona fide examples for the recruitment of HERV LTRs as transcriptional control elements for cellular genes (for a review, see references 16, 23, and 28), among them LTRs belonging to the multicopy families HERV-H (21, 41) and HERV-L (8, 10). In many cases, LTRs are used as alternative promoters/enhancers that confer differential tissue specificities to genes, thus increasing their transcriptional potential. One of the best-studied examples for HERV LTR-mediated tissue-specific regulation is the insertion of a HERV-E element upstream of an ancestral amylase gene that acts as a parotid gland-specific enhancer (35, 46). Interestingly, human genes initiated within HERV LTRs appear to have greater tissue specificity than genes lacking HERV promoters (5). Currently, about 5.8% of human genes are thought to be controlled by HERV promoters (5, 33).
In general, HERV LTRs appear to be active in a tissue-specific manner. Using a retrovirus-specific microarray, we have established a comprehensive HERV expression profile of 19 different human tissues (12, 38, 39). Some HERVs are ubiquitously expressed, whereas others are highly specific and transcriptionally active only in a few tissues. In addition, we and others have shown that isolated HERV LTRs maintain their promoter specificity in transient-transfection assays, suggesting that cell type specificity is mediated by the presence of transcription factor binding sites within the LTR and the availability of corresponding transcription factors in the cell and does not depend on additional cellular sequences located upstream or downstream of the LTR. For example, cloned HERV-H LTRs show a similar promoter activity in various human cell lines in transient-transfection assays as suggested by the endogenous transcription patterns of HERV-H proviruses in human tissues and cell lines (11, 14, 36, 38). To further test this assumption and to investigate the effect of reintegration on the cell type specificity of HERV promoters, we cloned three LTR sequences from two different HERV families into a modified Moloney murine leukemia virus (MLV)-based retroviral vector (pLXSNEGFP), which contains the enhanced green fluorescent protein (EGFP) gene under the transcriptional control of the retroviral LTR and the neomycin resistance gene under the control of the simian virus 40 promoter (19). pLXSNEGFP belongs to the family of ProCon vectors that allow cloning of promoter sequences by replacing the U3 region of the MLV 3′LTR (Fig. 1A). After reverse transcription, the promoter sequences are duplicated and transferred to the 5′ LTR, thus driving the transcription of the transgene in the infected cells (31, 34).
FIG. 1.
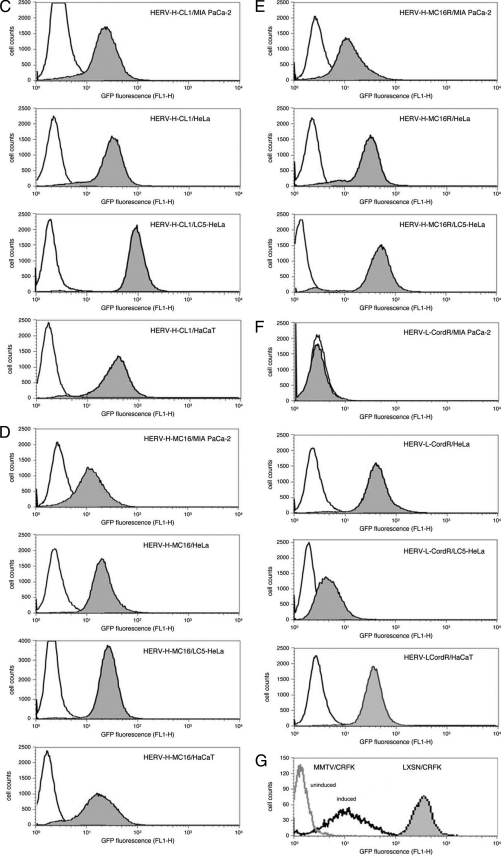
Transcriptional activity of HERV promoters in different cell lines. (A) HERV-MLV hybrid vector constructs. HERV U3 or U3-R sequences were inserted into the 3′ LTR of the MLV-based promoter conversion vector pLXSNEGFP to replace the MLV U3 region. HERV-H U3 regions were isolated by PCR from plasmid pBL-HERV-H-CL1 (36) using primers PHfor (5′-CCGACACCGCGGTGTCAGGCCTCTGAGCCCAA-3′) and PHCL1rev (5′-CGCGCCACGCGTGAAGGGAGATAGGGGTGGGGC-3′), and from plasmid pBL-HERV-H-MC16 (36) using primers PHfor and PHMC16rev (5′-CGCGCCACGCGTGAAGGGAGATGGGGTGGGAC-3′). The HERV-H-MC16 U3-R region was amplified from plasmid pBL-HERV-H-MC16 using primers PHfor and PHMC16Rrev (5′-CGCGCCACGCGTGTGAGCAACATGGCTGTTTATT-3′). The HERV-L U3-R region (6) was amplified from plasmid pBL-HERV-L using primers PLfor (5′-CCGGCCCGCGGTGATGATTAATACCAAGT-3′) and PLRrev (5′-CCGGCCACGCGTGATATATATTGGGGTTTAT-3′). All primers carry the MluI or SacII restriction site (shown in boldface type) as an extension, and the for and rev subscripts stand for forward and reverse, respectively. PCR products were digested with SacII and MluI and ligated into the vector pLXSNEGFP. The vector was transfected into the packaging cell line PA317, and the resultant particles were used to infect different cell lines. After reverse transcription and integration in the genome of the target cells, the EGFP gene is under the control of the HERV promoter. SV40, simian virus 40. (B) Relative luciferase activity in cell lines transiently transfected with pBL constructs containing the U3-R region of HERV-H-CL1, HERV-H-MC16, and HERV-L-Cord. All human cell lines used for transfection were either purchased from the American Type Culture Collection (ATCC) or authenticated by the German Collection of Microorganisms and Cell Cultures (DSMZ). The luciferase activity of the promoterless reporter plasmid was assigned a value of 1.0. Results were standardized with the cotransfected plasmid pRL-TK containing the thymidine kinase promoter upstream of the Renilla luciferase gene. The data are the means plus standard deviations (error bars) of at least three independent experiments. (C to G) Promoter activity of HERV-MLV hybrid vectors. MIA PaCa-2, HeLa, LC5-HeLa, and HaCaT cells were infected with LXSN-HERV-H-CL1 (C), LXSN-HERV-H-MC16 (D), LXSN-HERV-H-MC16R (E), and LXSN-HERV-L-CordR (F) and cloned by selection in medium containing G418. Expression of EGFP was monitored by FACS with a FACSCalibur flow cytometer (Becton-Dickinson) and analyzed with FlowJo FACS analysis software (Treestar) measuring the fluorescence of at least 50,000 cells (gray area). Uninfected cells were used as a negative control (white area). (G) For positive controls, CRFK cells were infected with the original MLV-based vector LXSNEGFP (gray area) and with LXSN-MMTV, a derivative containing the glucocorticoid-inducible, heterologous MMTV promoter. LXSN-MMTV-infected cells in the presence of dexamethasone (induced) and absence of dexamethasone (uninduced).
For HERV LTRs, we selected members of the class I family HERV-H, which are transcribed in many different tissues (11, 14, 36, 38, 49), and members of the class III family HERV-L, which are expressed only in skin, thyroid gland, and reproductive organs (38). Both HERV families represent a huge reservoir of regulatory sequences for gene expression. The HERV-H family, which is distantly related to gammaretroviruses including MLV, comprises more than 1,000 proviral copies per haploid human genome and a similar number of solitary LTRs (17, 26). The foamy virus-related HERV-L family (3, 6) consists of about 200 full-length elements and 6,000 solitary LTRs (26). To identify LTR sequences as appropriate promoters, the U3-R region of a number of HERV-H and HERV-L LTRs was cloned into the firefly luciferase expressing vector pBL and tested in various human cell lines using the Dual-Luciferase reporter assay (Promega) as described previously (36).
HERV-H-MC16 and HERV-H-CL1 proved to be the most active, representing type I and type Ia subgroups of HERV-H LTRs, respectively. The HERV-H-CL1 LTR was found to be active in all cell lines tested so far, with the highest transcriptional activity in LC5-HeLa cells and astrocytes (U373) and lower activities in pancreatic cells (MIA PaCa-2), epidermal keratinocytes (HaCaT), and breast cancer cells (MCF7) as shown in Fig. 1B and described previously (36). The HERV-H-MC16 LTR displays almost consistent transcription levels in MIA PaCa-2, HeLa, LC5-HeLa, HaCaT, and U373 cells. In addition, we selected the 5′ LTR of the HERV-L provirus identified by Cordonnier et al. (6) because of its high specificity for keratinocytes (Fig. 1B) (S. Weinhardt, unpublished data). To further examine the tissue specificity of the selected LTRs, transgenic mice were established carrying the HERV-H-CL1 and the HERV-L-Cord U3-R region with the luciferase or EGFP gene as a reporter gene. Preliminary data suggest that the HERV-H-CL1 LTR initiates transcripts only in testes but not in other organs, whereas the HERV-L LTR is not active in any murine tissue (37; Weinhardt, unpublished), confirming previous data indicating that the activity of various HERV-H LTRs is restricted in murine cell lines compared to human cells (11). Thus, the promoter activity of these HERV LTRs appears to be primarily limited to human cells.
For construction of HERV-MLV hybrid vectors, the HERV LTR sequences were amplified by PCR and inserted into the 3′ U3-deleted MLV vector (Fig. 1A). Plasmids pLXSN-HERV-H-CL1 and pLXSN-HERV-H-MC16 contain the 327-bp U3 region of HERV-H-CL1 and the 255-bp U3 region of HERV-H-MC16, respectively. To compare the infectivity and expression efficiency of constructs with and without a HERV R region, we also inserted a 323-bp fragment comprising the U3 and R regions of HERV-H-MC16 (pLXSN-HERV-H-MC16R). A 397-bp fragment containing the complete U3-R region of HERV-L was used for construction of pLXSN-HERV-L-CordR, since the exact border between U3 and R has not yet been determined.
The HERV-MLV hybrid constructs and the original MLV-derived pLXSNEGFP vector, as well as pLXSN-MMTV containing the inducible mouse mammary tumor virus (MMTV) promoter instead of the MLV U3 region in the 3′ LTR (34), were transfected into the amphotropic murine packaging cell line PA317. Virus titers were determined by infection of the feline kidney epithelial cell line CRFK. Human cell lines previously used for analysis of HERV promoter activity in transient luciferase assays were infected with supernatants obtained from vector-producing PA317 cells as described previously (30). Infected cells were cloned by neomycin selection. To verify the promoter conversion, DNA was analyzed by PCR using primers specific for the HERV U3, MLV U3 and MLV R regions in combination with a primer complementary to a sequence in the EGFP gene. Sequence analysis of the amplification products revealed that the HERV promoter was present in the correct configuration within the 5′ LTR of the vector provirus in all cell lines investigated (data not shown).
Fluorescence-activated cell sorting (FACS) analysis was performed to determine the promoter activity of the HERV-MLV constructs and the proportion of cells expressing EGFP (Fig. 1C to F). For comparability, only cell clones harboring a single integrated provirus were used. As expected from the results of transient-transfection assays, HERV-H promoters were active in all cell lines investigated so far but showed slightly differential EGFP expression depending on the cell type. In contrast, the HERV-L promoter displayed a high degree of cell type specificity and was active only in HaCaT and HeLa cells. The original MLV-based vector LXSNEGFP showed nearly the same activity in all cell lines (shown in Fig. 1G for CRFK cells), which was about threefold higher than the highest activity found for a HERV-MLV hybrid vector, i.e., LXSN-HERV-H-CL1 in LC5-HeLa cells (Fig. 1C). A MMTV promoter containing hybrid vector derived from LXSNEGFP (Fig. 1G) was not active in CRFK cells but could be stimulated by the glucocorticoid dexamethasone (20, 34), giving EGFP expression levels comparable to those of LXSN-HERV-H-MC16 and LXSN-HERV-H-MC16R in MIA PaCa-2 cells (Fig. 1D and E). The activity of the HERV promoters was not influenced by dexamethasone (data not shown). In summary, these data suggest a slightly reduced promoter activity of heterologous retroviral promoters compared to the primary MLV U3 region irrespective whether they are of murine or human origin.
Interestingly, the activities of LXSN-HERV-H-CL1 and LXSN-HERV-H-MC16 nearly exactly reflect the expression activity and cell specificity patterns of the corresponding pBL clones containing HERV-H LTR in transient luciferase assays (Fig. 1B). No essential differences between HERV-H-MLV hybrid vectors containing or lacking the HERV R region were found. Only in HeLa-LC5 cells, a slightly higher activity (about twofold) of LXSN-HERV-H-MC16R compared to LXSN-HERV-H-MC16 was observed (Fig. 1E). An enhancing effect of the R region on HERV promoters has been described previously for some class II HERVs, HERV-K(HML-4) and HERV-K(HML-2), and may be due to additional transcription factor binding sites within the R region or posttranscriptional events, such as stabilizing effects on the mRNA (2). Furthermore, the use of an additional transcription initiation site within the HERV R region or the differences in spacing between promoter and transcription start site generated by a second R region may play a role.
As expected, LXSN-HERV-LCordR containing the HERV-L promoter shows similar EGFP expression levels but a higher cell type specificity than those of the HERV-H-MLV hybrid vectors. LXSN-HERV-L is inactive in the pancreas cell line MIA PaCa-2 but highly active in HeLa and HaCaT cells. Compared with pBL plasmids containing the luciferase gene under the control of HERV-L LTR sequences, LXSN-HERV-LCordR displays the same cell type specificity (Fig. 1B and F). Remarkably, little or no activity was observed for LXSN-HERV-LCordR- and HERV-L LTR-containing pBL plasmids in the HeLa subclone LC5-HeLa, although both were highly active in the original HeLa cells. LC5-HeLa cells represent a subclone from L132 cells (29), a cell line that was originally thought to be derived from embryonic lung tissue but was subsequently found to have been established via HeLa cell contamination (CCL-5 cells [American Type Culture Collection]). Although expressing keratin, LC5-HeLa cells have lost some features typical for HeLa cells and show instead fibroblast-like characteristics (29). The endogenous HERV transcription profile of LC5-HeLa cells as established by microarray analysis does not show HERV-L expression, which is typical for the cervix (38) as well as HeLa cells (data not shown). Accordingly, transfection with three different HERV-L LTRs cloned in the luciferase-containing pBL vector did not result in expression of the reporter gene in LC5-HeLa cells, but HERV-L LTR promoter activity could be induced by treatment with phorbol ester (O. Diem, unpublished data). Thus, slight alterations of a cell type, possibly caused by selection of a certain karyotype and/or long-term cultivation, may lead to differential HERV activities in those cells.
Taken together, these data suggest that cell type specificity of HERV promoters may be conferred on retroviral vectors. Interestingly, our results are also in good agreement with previous findings obtained with a retrovirus-specific microarray used to investigate the endogenous expression patterns and tissue specificities of different HERVs (38). The data confirm the high specificity of HERV-L elements, the expression of which is restricted essentially to skin, thyroid gland, and tissues involved in reproduction, such as uterus, cervix, placenta, and testes, in contrast to the ubiquitous expression of HERV-H elements (11, 14, 36, 38). Therefore, we conclude that reintegration does not alter cell type specificity in principle, even though expression levels should also be influenced by the genomic context.
To investigate whether adaptive evolution of the HERV LTR sequences may have occurred during infection and selection of infected cell lines, the sequences of five LXSN-HERV-H-CL1-infected cell clones, five LXSN-HERV-H-MC16-infected cell clones, three LXSN-HERV-H-MC16R-infected cell clones, and five LXSN-HERV-L-CordR-infected cell clones were compared with the original HERV LTR sequences of the vectors. No sequence variations could be detected, suggesting that mutations must be a rare event at least in nonreplicative vectors.
In our experiments, infected cell clones have been selected by neomycin resistance. Therefore, only integrations within active chromosomal regions have been investigated, so the HERV promoter, like the internal simian virus 40 promoter driving neomycin resistance gene expression, is unlikely to be silenced by methylation or inactivated chromatin. Our data should therefore particularly reflect the availability of transcription factors and their interaction with the HERV promoter in a given cell type. The longer U3 regions of HERVs compared to MLV, especially of HERV-L, may contain more potential binding sites for transcription modulating factors that confine activity in certain cells and thus may increase cell type specificity. The interaction of HERV-H and HERV-L LTRs with cellular factors was investigated previously, and some potential transcription factor binding sites were identified (1, 7, 9, 10, 32, 42). Binding sites for the transcription factor Sp1 have been detected in HERV-H and HERV-L LTRs but appear to act in a different context. HERV-H type Ia LTRs (represented by HERV-H-CL1) contain three Sp1 binding sites that probably act synergistically (32, 42). In several active HERV-L LTRs (including HERV-L-Cord), at least one Sp1 binding site was identified within the U3 region, although at different locations in LTRs with diverse cell type specificities (9, 10; Weinhardt, unpublished). Sp1 is ubiquitously expressed in many different cells, and Sp1 binding sites are commonly present in LTRs. However, several alternatively spliced transcripts encoding different isoforms of Sp1 are known, which are associated with different cell types or stages (45). The differential tissue specificities of HERV-L and HERV-H LTRs may be due to binding of different Sp1 variants or additional, as-yet-unknown, interacting cellular factors, e.g., cell type-dependent repressors.
To determine transcription initiation sites of HERV-H- and HERV-L-MLV hybrid vectors (Fig. 1A), rapid amplification of 5′ cDNA ends (5′RACE) of vector constructs integrated in HeLa, LC5-HeLa, or HaCaT cells was performed as described previously (27). Nested PCR was carried out with forward primers specific for the 5′RACE adapter. Both reverse primers were located within the EGFP gene to avoid amplification of endogenous HERV-H or HERV-L elements. Several independent clones derived from each PCR product were sequenced.
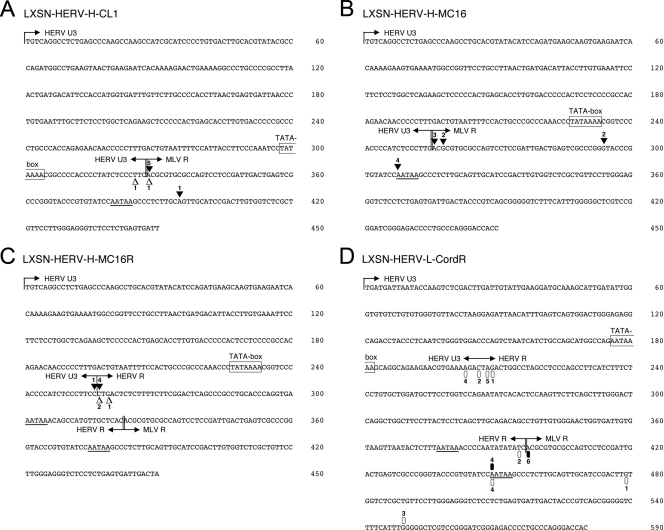
Generally, the transcription start of HERV-H promoter-containing vectors was found to be more precisely defined than that of LXSN-HERV-LCordR. In six of eight cases, LXSN-HERV-H-CL1 used exactly the predicted initiation site at the transition between the HERV-H U3 region and the MLV R region in infected HeLa and LC5-HeLa cells (Fig. 2A).
FIG. 2.
Mapping of transcription start sites in different HERV-MLV hybrid LTRs by 5′RACE. Total RNA was extracted from HeLa and LC5-HeLa cells infected with LXSN-HERV-H-CL1, LXSN-HERV-H-MC16, or LXSN-HERV-H-MC16R, as well as two different HaCaT cell clones infected with LXSN-HERV-L-CordR, all showing high EGFP expression by FACS analysis. Nested PCR was carried out using the FirstChoice RLM-RACE kit (Ambion) with forward primers specific for the 5′RACE adapter and reverse primers specific for the reporter gene EGFP, GFPrev-outer (5′-TAAGTTGCTGGCCAGCTTACCTC-3′) and 3′GFPrev-inner (5′-TTACCTCCCGGTGGTGGGTCG-3′). Several independent clones derived from each PCR product were sequenced. Symbols represent 5′ ends of individual RACE clones obtained from different cell lines, and the numbers indicate that the clones were found multiple times. Potential boundaries between U3 and R regions are marked by vertical lines and arrows. Boundaries between HERV and MLV sequences are marked by vertical double lines and by arrows. The TATA sequences are boxed. The polyadenylation signals are underlined. (A) Transcription initiation sites of LXSN-HERV-H-CL1. White triangles represent the results of 5′RACE in infected HeLa cells, and black triangles represent RACE clones obtained from infected LC5-HeLa cells. (B) Transcription initiation sites of LXSN-HERV-H-MC16 in infected LC5-HeLa cells. (C) Transcription initiation sites of LXSN-HERV-H-MC16R containing in addition the HERV R region. White triangles represent the results of 5′RACE in infected HeLa cells, and black triangles represent RACE clones obtained from infected LC5-HeLa cells. (D) Transcription initiation sites of LXSN-HERV-L-CordR. Black and white ovals represent the results of 5′RACE in two different HaCaT cell clones.
The transcription initiation sites of LXSN-HERV-H-MC16 in LC5-HeLa cells are mainly located in two regions (Fig. 2B), one around the beginning of the MLV R region and a second exactly at the first A of the MLV polyadenylation signal. The use of the latter as an additional transcription start site was also observed in LXSN-HERV-L-CordR-infected cells (Fig. 2D). Further potential transcription start sites within the MLV R region of LXSN-HERV-H-CL1 and LXSN-HERV-H-MC16 did not appear several times in independent experiments and might therefore be due to failure during the RACE procedure.
To investigate the influence of an additional HERV R region upstream of the MLV R region on the selection of transcription initiation sites, we performed 5′RACE of LXSN-HERV-H-MC16R and LXSN-HERV-LCordR containing two R regions derived from HERV and MLV sequences. All transcription start sites of LXSN-HERV-H-MC16R in HeLa and LC5-HeLa cells were found to cluster around the boundary between the HERV U3 and HERV R regions (Fig. 2C). The transcription initiation sites of the MLV R region were not used by LXSN-HERV-H-MC16R.
For LXSN-HERV-LCordR, two different clones of infected HaCaT cells were analyzed. In contrast to HERV-H-MLV hybrid vectors, LXSN-HERV-LCordR appears to possess multiple transcription initiation sites in a region spanning about 300 bp of HERV and MLV sequences (Fig. 2D). The transition between the HERV-L U3 and R regions in HERV-L elements has not yet been defined. In one of the two analyzed clones of infected HaCaT cells, the majority of transcription start sites of LXSN-HERV-LCordR cluster within 7 bp of the HERV-L sequence, suggesting that this might be the natural U3/R boundary of HERV-L elements. In both clones, the transcription start site located at the border between the HERV-L R region and the MLV R region is also used, as well as the minor initiation site starting with the first A of the polyadenylation signal. As sequence variations between the LTRs could be excluded, the differential use of transcription start sites in different clones of the same cell line infected with the same vector suggests a possible influence of vector integration sites on initiation of transcription. Notably, the promoter activity of the HERV-L LTR was significantly lower in cells in which only the MLV start site was used than in the cells preferentially initiating at the HERV start site (data not shown).
In contrast to exogenous retroviruses, which mostly have a strong TATA box and a single major transcription start site, multiple initiation sites have been found in several endogenous retroviruses and retroviral elements (15, 27, 43). Multiple transcription initiation sites are thought to be characteristic for promoters with a weak TATA box but several Sp1 binding sites. The majority of human genes possess highly variable transcription start sites, reflecting the dynamic nature of transcription (48). This led to the assumption that endogenous retroviruses residing in the genome gradually approximate cellular genes and assume more flexibility in transcriptional control than their exogenous counterparts (27). Thus, HERV LTRs probably resemble cellular transcription units more than they resemble promoters of exogenous retroviruses.
Cell type-specific promoters and enhancers are a prerequisite for the construction of targeted retroviral expression vectors and their controlled use in gene therapy. HERV LTRs represent a huge reservoir of regulatory sequences in the human genome that are easy to isolate and characterize. They have a number of features that make them advantageous for the construction of therapeutic vectors. They have adapted themselves to their hosts over millions of years, and thus, pathogenic sequences have largely been eliminated during evolution. Recombination of HERV elements with HERV-derived vectors will not create completely new types of retroviruses, as would be the case with vectors based on animal retroviruses or human exogenous retroviruses, such as lentiviruses. On the other hand, homologous recombination of HERV sequences with HERV-based vectors might be utilized for targeted gene transfer. In contrast to cellular promoters, which often depend on additional signal structures located at some distance upstream or downstream, the regulatory elements of retroviruses are concentrated in a small and clearly defined region to maintain transcriptional independence regardless of the integration site in the host genome. Furthermore, many HERV LTRs are characterized by multiple Sp1 binding sites that may protect against inactivation by de novo methylation (4, 40).
In our study, all of the HERV-H hybrid vectors are replication defective. Replication-competent retroviral vectors, however, could be used for example for selection of cell type-specific and efficient HERV promoters, when the original MLV promoter is replaced by a mixture of arbitrarily amplified HERV LTR sequences. The HERV promoter sequence, which replicates most efficiently in a certain cell type, could then be isolated after several replication cycles. Recently, synthetic promoters have been inserted in place of the MLV promoter in replication-competent retroviral vectors (25, 30), and such viruses have been shown to be replication competent for a number of replication cycles before losing the heterologous promoter. HERV LTRs may prove to be even more stable in the context of replicating vectors, since they are of retroviral origin and yet can still restrict expression in a cell type-specific manner. Considering that a multitude of HERV LTRs have already been recruited during evolution as control elements for gene expression, HERV LTRs should be a valuable source of new cell type-specific regulatory sequences and represent promising candidates for the construction of retroviral vectors for use in human gene therapy.
Acknowledgments
This work was supported in part by a grant from the Bayerische Forschungsstiftung, Forschungsverbund FORGEN 2.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Anderssen, S., E. Sjøttem, G. Svineng, and T. Johansen. 1997. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology 234:14-30. [DOI] [PubMed] [Google Scholar]
- 2.Baust, C., W. Seifarth, U. Schön, R. Hehlmann, and C. Leib-Mösch. 2001. Functional activity of HERV-K-T47D-related long terminal repeats. Virology 283:262-272. [DOI] [PubMed] [Google Scholar]
- 3.Bénit, L., J. B. Lallemand, J. F. Casella, H. Philippe, and T. Heidmann. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 73:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boumber, Y. A., Y. Kondo, X. Chen, L. Shen, Y. Guo, C. Tellez, M. R. Estecio, S. Ahmed, and J. P. Issa. 2008. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 4:e1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conley, A. B., J. Piriyapongsa, and I. K. Jordan. 2008. Retroviral promoters in the human genome. Bioinformatics 24:1563-1567. [DOI] [PubMed] [Google Scholar]
- 6.Cordonnier, A., J. F. Casella, and T. Heidmann. 1995. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J. Virol. 69:5890-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Parseval, N., H. Alkabbani, and T. Heidmann. 1999. The long terminal repeats of the HERV-H human endogenous retrovirus contain binding sites for transcriptional regulation by the Myb protein. J. Gen. Virol. 80:841-845. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, C. A., P. Medstrand, and D. L. Mager. 2003. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. USA 100:12841-12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, C. A., M. T. Romanish, L. E. Gutierrez, L. N. van de Lagemaat, and D. L. Mager. 2006. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 366:335-342. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, C. A., L. N. van de Lagemaat, G. J. Baillie, and D. L. Mager. 2005. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene 364:2-12. [DOI] [PubMed] [Google Scholar]
- 11.Feuchter, A., and D. Mager. 1990. Functional heterogeneity of a large family of human LTR-like promoters and enhancers. Nucleic Acids Res. 18:1261-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank, O., M. Giehl, C. Zheng, R. Hehlmann, C. Leib-Mösch, and W. Seifarth. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 79:10890-10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gifford, R., and M. Tristem. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291-315. [DOI] [PubMed] [Google Scholar]
- 14.Goodchild, N. L., D. A. Wilkinson, and D. L. Mager. 1993. Recent evolutionary expansion of a subfamily of RTVL-H human endogenous retrovirus-like elements. Virology 196:778-788. [DOI] [PubMed] [Google Scholar]
- 15.Günzburg, W. H., F. Heinemann, S. Wintersperger, T. Miethke, H. Wagner, V. Erfle, and B. Salmons. 1993. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature 364:154-158. [DOI] [PubMed] [Google Scholar]
- 16.Jern, P., and J. M. Coffin. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709-732. [DOI] [PubMed] [Google Scholar]
- 17.Jern, P., G. O. Sperber, and J. Blomberg. 2004. Definition and variation of human endogenous retrovirus H. Virology 327:93-110. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, I. K., I. B. Rogozin, G. V. Glazko, and E. V. Koonin. 2003. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19:68-72. [DOI] [PubMed] [Google Scholar]
- 19.Klein, D., S. Indraccolo, K. von Rombs, A. Amadori, B. Salmons, and W. H. Gunzburg. 1997. Rapid identification of viable retrovirus-transduced cells using the green fluorescent protein as a marker. Gene Ther. 4:1256-1260. [DOI] [PubMed] [Google Scholar]
- 20.Klein, R., B. Ruttkowski, S. Schwab, T. Peterbauer, B. Salmons, W. H. Gunzburg, and C. Hohenadl. 2008. Mouse mammary tumor virus promoter-containing retroviral promoter conversion vectors for gene-directed enzyme prodrug therapy are functional in vitro and in vivo. J. Biomed. Biotechnol. 2008:683505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalski, P. E., J. D. Freeman, and D. L. Mager. 1999. Intergenic splicing between a HERV-H endogenous retrovirus and two adjacent human genes. Genomics 57:371-379. [DOI] [PubMed] [Google Scholar]
- 22.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 23.Leib-Mösch, C., W. Seifarth, and U. Schön. 2005. Influence of human endogenous retroviruses on cellular gene expression, p. 123-143. In E. D. Sverdlov (ed.), Retroviruses and primate evolution. Landes Bioscience, Georgetown, TX.
- 24.Li, W. H., Z. Gu, H. Wang, and A. Nekrutenko. 2001. Evolutionary analyses of the human genome. Nature 409:847-849. [DOI] [PubMed] [Google Scholar]
- 25.Logg, C. R., A. Logg, R. J. Matusik, B. H. Bochner, and N. Kasahara. 2002. Tissue-specific transcriptional targeting of a replication-competent retroviral vector. J. Virol. 76:12783-12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mager, D. L., and P. Medstrand. 2003. Retroviral repeat sequences, p. 57-63. In D. Cooper (ed.), Nature encyclopedia of the human genome. Nature Publishing Group, London, United Kingdom.
- 27.Maksakova, I. A., and D. L. Mager. 2005. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J. Virol. 79:13865-13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medstrand, P., L. N. van de Lagemaat, C. A. Dunn, J. R. Landry, D. Svenback, and D. L. Mager. 2005. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet. Genome Res. 110:342-352. [DOI] [PubMed] [Google Scholar]
- 29.Mellert, W., A. Kleinschmidt, J. Schmidt, H. Festl, S. Emler, W. K. Roth, and V. Erfle. 1990. Infection of human fibroblasts and osteoblast-like cells with HIV-1. AIDS 4:527-535. [DOI] [PubMed] [Google Scholar]
- 30.Metzl, C., D. Mischek, B. Salmons, W. H. Gunzburg, M. Renner, and D. Portsmouth. 2006. Tissue- and tumor-specific targeting of murine leukemia virus-based replication-competent retroviral vectors. J. Virol. 80:7070-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mrochen, S., D. Klein, S. Nikol, J. R. Smith, B. Salmons, and W. H. Gunzburg. 1997. Inducible expression of p21WAF-1/CIP-1/SDI-1 from a promoter conversion retroviral vector. J. Mol. Med. 75:820-828. [DOI] [PubMed] [Google Scholar]
- 32.Nelson, D. T., N. L. Goodchild, and D. L. Mager. 1996. Gain of Sp1 sites and loss of repressor sequences associated with a young, transcriptionally active subset of HERV-H endogenous long terminal repeats. Virology 220:213-218. [DOI] [PubMed] [Google Scholar]
- 33.Piriyapongsa, J., N. Polavarapu, M. Borodovsky, and J. McDonald. 2007. Exonization of the LTR transposable elements in human genome. BMC Genomics 8:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saller, R. M., F. Öztürk, B. Salmons, and W. H. Günzburg. 1998. Construction and characterization of a hybrid mouse mammary tumor virus/murine leukemia virus-based retroviral vector. J. Virol. 72:1699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuelson, L. C., K. Wiebauer, C. M. Snow, and M. H. Meisler. 1990. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol. Cell. Biol. 10:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schön, U., W. Seifarth, C. Baust, C. Hohenadl, V. Erfle, and C. Leib-Mösch. 2001. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology 279:280-291. [DOI] [PubMed] [Google Scholar]
- 37.Schönfeld, R. M. 2003. Generation of transgenic mice to evaluate promoter activity and specificity of two human endogenous retrovirus long terminal repeats. Ph.D. dissertation. Ludwig-Maximilians-University, Munich, Germany.
- 38.Seifarth, W., O. Frank, U. Zeilfelder, B. Spiess, A. D. Greenwood, R. Hehlmann, and C. Leib-Mösch. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seifarth, W., B. Spiess, U. Zeilfelder, C. Speth, R. Hehlmann, and C. Leib-Mosch. 2003. Assessment of retroviral activity using a universal retrovirus chip. J. Virol. Methods 112:79-91. [DOI] [PubMed] [Google Scholar]
- 40.Šenigl, F., J. Plachý, and J. Hejnar. 2008. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J. Virol. 82:7818-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sin, H. S., J. W. Huh, D. S. Kim, D. W. Kang, D. S. Min, T. H. Kim, H. S. Ha, H. H. Kim, S. Y. Lee, and H. S. Kim. 2006. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch. Virol. 151:1985-1994. [DOI] [PubMed] [Google Scholar]
- 42.Sjøttem, E., S. Anderssen, and T. Johansen. 1996. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3′ to the TATA box. J. Virol. 70:188-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strazzullo, M., B. Majello, L. Lania, and G. La Mantia. 1994. Mutational analysis of the human endogenous ERV9 proviruses promoter. Virology 200:686-695. [DOI] [PubMed] [Google Scholar]
- 44.Sverdlov, E. E. 2005. Retroviruses and primate genome evolution. Landes Bioscience, Georgetown, TX.
- 45.Thomas, K., J. Wu, D. Y. Sung, W. Thompson, M. Powell, J. McCarrey, R. Gibbs, and W. Walker. 2007. SP1 transcription factors in male germ cell development and differentiation. Mol. Cell. Endocrinol. 270:1-7. [DOI] [PubMed] [Google Scholar]
- 46.Ting, C. N., M. P. Rosenberg, C. M. Snow, L. C. Samuelson, and M. H. Meisler. 1992. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 6:1457-1465. [DOI] [PubMed] [Google Scholar]
- 47.van de Lagemaat, L. N., J. R. Landry, D. L. Mager, and P. Medstrand. 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 19:530-536. [DOI] [PubMed] [Google Scholar]
- 48.Wakaguri, H., R. Yamashita, Y. Suzuki, S. Sugano, and K. Nakai. 2008. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 36:D97-D101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson, D. A., J. D. Freeman, N. L. Goodchild, C. A. Kelleher, and D. L. Mager. 1990. Autonomous expression of RTVL-H endogenous retroviruslike elements in human cells. J. Virol. 64:2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]