Abstract
Peach latent mosaic viroid (PLMVd) is a small, single-stranded, circular RNA pathogen that infects Prunus persica trees. As with all other known viroids, the PLMVd genome does not encode any proteins. Consequently, it must interact with host cellular factors in order to ensure its life cycle. With the objective of identifying cellular proteins that interact with PLMVd, Northwestern hybridizations were performed using partially purified peach leaf extracts. Mass spectrometric analysis of the detected RNA-protein complexes led to the identification of six putative RNA-binding proteins. One of these was found to be elongation factor 1-alpha (eEF1A), and because of its known involvement in the replication and translation of various RNA viruses, further characterizations were performed. Initially, the existence of this interaction received support from an experiment that immunoprecipitated the eEF1A from a crude extract of infected peach leaves, coupled with reverse transcription-PCR detection of the PLMVd. Subsequently, eEF1A interaction with PLMVd strands of both polarities was confirmed in vitro by electrophoresis mobility shift assays, fluorescence spectroscopy, and the prediction of an altered PLMVd RNase mapping profile in the presence of the protein. The potential contribution of eEF1A to the molecular biology of PLMVd, including for viroid replication, is discussed.
Viroids are small (250 to 400 nucleotides [nt]), single-stranded, circular pathogenic RNAs that infect higher plants, frequently causing significant losses in agriculture. In infected cells, viroids replicate in a DNA-independent manner via a rolling circle mechanism that follows either a symmetric or an asymmetric mode. In the symmetric mode, the infecting circular monomer, which is assigned a plus [(+)] polarity by convention, is replicated into linear concatemeric minus [(−)] strands which are then spliced and ligated, yielding minus-strand circular monomers. By using the latter RNA molecules as templates, the same three steps are then repeated in order to produce the progeny. In contrast, in the asymmetric mode, the linear concatemeric (−) strands serve directly as the template for the synthesis of the linear concatemeric (+) strands.
There are 34 different viroid species known to belong to either the Pospiviroidae or Avsunviroidae family. Members of the Pospiviroidae family share a conserved central region, have a nuclear localization, and replicate via an asymmetric rolling circle mechanism. Conversely, the members of the Avsunviroidae are localized in the chloroplasts and replicate through a symmetric rolling circle mechanism involving hammerhead self-cleavage activities (14).
Peach latent mosaic viroid (PLMVd) is the causative agent of an economically important disease that leads to the death of infected Prunus persica trees, often in epidemic proportions (13). Peach latent mosaic is distributed worldwide and is transmitted by contaminated land tools, grafting, aphids (Myzus persicae), and even pollen, as recently reported (1, 22). The disease generally remains latent (i.e., asymptomatic) in trees for 5 to 7 years before symptoms begin to appear. These symptoms include bud necrosis, delayed shoot development, and branches becoming necrotic and dying off. Fruit symptoms consist of discolored areas on the skin. PLMVd also infects apricot, pear, nectarine, plum, citrus, almond, and cherry trees. Since PLMVd possesses a hammerhead structure and replicates in chloroplasts, it belongs to the Avsunviroidae family. This infectious RNA is 335 to 351 nt in size, depending on the sequence variant, and folds into a branched secondary structure (7, 22).
Considering that the viroid genome does not encode any protein, these infectious agents need to interact with cellular factors in order to ensure their life cycle. For example, to be infectious, viroids have to pass through many critical steps. First, they must be imported into the subcellular organelle that contains the RNA polymerase that supports their replication. Second, they must then be exported out of this replication site and transported both from cell to cell and within the vascular system. Finally, they need to penetrate other parts of the host plant in order to ensure the propagation of the infection. Consequently, it is hypothesized that various host proteins could be implicated not only in these different steps but also in other central mechanisms, such as the viroid's replication and its associated pathogenic activities (14).
Some proteins have been reported to bind viroids both in vitro and in vivo. For example, it was shown that RNA polymerase II can replicate viroids from the Pospiviroidae family in vitro (17), an observation that is supported by the demonstration that α-amanitin inhibits the synthesis of potato spindle tuber viroid (PSTVd) in infected cells (32). Similarly, the hop stunt viroid was shown to bind the cucumber phloem lectin protein PP2 of phloem exudates (18). This interaction has been proposed to play an important role in hop stunt viroid transport within the vascular system of the plant. Lastly, a screening assay performed with the goal of identifying RNA-binding proteins from a cDNA expression library revealed an interaction between PSTVd and the host tomato protein Virp1 (19, 27, 29), an interaction that recently was demonstrated to be important in the infectivity of this viroid (25). All of these studies were performed with viroids belonging to the Pospiviroidae family; to date, there is only one report in the literature of a protein interacting in vivo with a species belonging to the Avsunviroidae family. Specifically, the PARBP33 protein was shown to interact with the avocado sunblotch viroid, enhancing its self-cleavage activity in experiments conducted in vitro (10).
As yet, no protein has been reported to interact with PLMVd. Self-cleavage assays have shown that neither the activity of the PLMVd-derived hammerhead motifs nor that of the full-length viroid was modulated by the presence of PARBP33 under the conditions tested, as was observed for avocado sunblotch viroid (A. Dubé and J. P. Perreault, unpublished data). With the objective of identifying any protein that interacts with PLMVd, Northwestern hybridization was performed with partially purified peach leaf protein extracts. Mass spectrometric analysis of the resulting RNA-protein complexes led to the identification of six putative RNA-binding proteins, one of which was elongation factor 1-alpha (eEF1A). The interaction of this protein was confirmed in vitro, and further characterization of the PLMVd-eEFIA complex was undertaken.
MATERIALS AND METHODS
Protein fractionation.
All of the purification steps were performed at 4°C. Initially, 10 g of uninfected peach leaves (cv. Siberian C) were cut into small pieces, ground into a fine powder under liquid nitrogen, and then homogenized in 50 ml of PA buffer [50 mM Tris-HCl (pH 7.8), 1 mM EDTA, 50 mM (NH4)2SO4, and 10% glycerol] containing 1% polyvinylpyrrolidone, 1 mM benzamidine-phenylmethylsulfonyl fluoride, 1× CLAP (i.e., a protease inhibition cocktail; a 1,000× solution is composed of 10 μg/ml each of chymostatin, leupeptin, antipain, and pepstatin), and 1 mM dithiothreitol (DTT). Cell debris were removed by centrifugation at 10,000 × g at 4°C for 15 min. The supernatants were cleared using syringes containing cheese cloth, and the flowthrough was then filtered through Stericups (Millipore). The resulting extracts were purified using 5-ml HiTrap heparin HP columns (GE Healthcare) and an AKTA fast-performance liquid chromatography (FPLC) system (GE Healthcare).
All solutions used with the FPLC were passed through 0.22-μm filters prior to use. The column was washed with PB buffer [50 mM Tris-HCl (pH 7.8), 1 mM EDTA, 50 mM (NH4)2SO4, 10% glycerol, and 2.5% Triton X-100 (vol/vol)], and the extracts were then loaded on the column. The column was eluted with PB buffer containing 150, 400, 600, and finally 1,500 mM (NH4)2SO4. Fractions (2 ml) were collected, and those containing proteins were dialyzed against 4 liters of Tris-HCl (pH 7.8), 1 mM EDTA, 1 mM DTT, 40 mM NaCl, and 10% glycerol overnight at 4°C. The resulting protein-containing fractions were analyzed by electrophoresis through 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels (29:1 ratio of acrylamide to bisacrylamide) and then visualized by Coomassie blue staining. The protein concentrations were determined using the Bradford assay.
Synthesis of PLMVd transcripts and RNA probes.
The synthesis and purification of both (+) and (−) PLMVd transcripts were performed as described previously (7). Briefly, in vitro transcription reactions were performed using digested recombinant plasmid pPD1, which contains two tandemly repeated PLMVd sequences cloned into the PstI restriction site of pBluescript II KS, as the template. In this construct, the insert is flanked by T3 and T7 promoters for the production of (+) and (−) polarity transcripts, respectively. The transcription reactions were performed for 3 h at 37°C in a final volume of 100 μl containing 80 mM HEPES-KOH (pH 7.5), 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, a 5 mM concentration of each deoxynucleoside triphosphate, 0.004 U/μl pyrophosphatase (Roche Diagnostics), 37 U RNA Guard (Amersham Biosciences), and 10 μg of purified T7 or T3 RNA polymerase. For random internal labeling, 30 μCi of [α-32P]UTP (3,000 Ci/mmol; New England Nuclear) was added to the transcription reaction mixtures. During transcription, RNA molecules of both polarities self-cleaved, yielding 338-nt linear monomeric species.
The transcription reactions were stopped by the addition of RNase-free RQ1 DNase (Promega) and then incubated for 30 min at 37°C. One volume of stop buffer (0.03% [wt/vol] each of bromophenol blue and xylene cyanol, 10 mM EDTA [pH 7.5], and 97.5% [vol/vol] deionized formamide) was then added, and the resulting mixtures were denatured for 2 min at 65°C prior to being fractionated by denaturing (8 M urea) 5% polyacrylamide gel electrophoresis (PAGE; 19:1 ratio of acrylamide to bisacrylamide) using 45 mM Tris-borate (pH 7.5) and 1 mM EDTA as the buffer (1× TBE buffer). Nonradioactive transcripts were detected by UV shadowing, while radioactive ones were detected by autoradiography. The bands corresponding to the 338-nt full-length fragments of both polarities were excised, and the RNAs were eluted overnight in 500 mM NH4OAc, 1 mM EDTA, and 0.1% SDS, ethanol precipitated, purified on Sephadex G-50 spin columns (Amersham), and lyophilized. After the samples were dissolved in ultrapure water, the RNA concentrations were determined either by absorbance spectrophotometry at 260 nm or by Cerenkov counting, and the samples were then stored dry at −20°C. Both the (+) and (−) strand-specific riboprobes used for the Northwestern hybridizations were synthesized and purified in an analogous manner, except that a StripEZ transcription kit (Ambion) was used and the transcription reactions were performed in the presence of 50 μCi of [α-32P]UTP (3,000 Ci/mmol; New England Nuclear). The same procedure was used for the synthesis of both the tRNAAla and the CU-based polymer, except that the DNA templates were prepared by PCR filling reactions using Pwo DNA polymerase (Roche Diagnostic), an oligonucleotide that included the T7 RNA promoter complementary sequence followed by either the tRNA sequence (5′-TGGAGATGCAGGGAATCGAACCCTGTGCCTCTCGCATGCAAAGCGAGCGCTCTACCATATGAGCTACATCCCCTATAGTGAGTCGTATTA-3′) or the CU sequence [5′-(AG)27CCCTATAGTGAGTCGTATTA-3′], and an oligonucleotide corresponding to the T7 RNA promoter sequence. For hY3-delta ribozyme, the following two oligonucleotides were used in the filling reaction mix: delta-forward (5′-TAATACGACTCACTATAGGGCTGGTCCGAGTGCAGTGGTGTTTACAACTAATTGATCACAACCCAGTTACAGATTGGGCCCACGGAATCGGTAGGCTTCGGCCTACGGGTAG-3′) and delta-reverse (5′-AAAGGCTAGTCAAGTGAAGCAGTGGGAGTGGAGAAGGAACAAAGAGGGCCCACGTGGAGGAGCCGCCAGCAGCTACCCGTAGGCCGAAGCCT-3′).
Northwestern hybridizations.
The SDS-PAGE gels used for the analysis of the protein fractions were always set up with duplicate samples being loaded on the gel: one-half of the gel was used for Coomassie blue staining, and the other half was used for Northwestern hybridization. After transfer of the proteins from the nonstained half of the gel onto nitrocellulose membranes, the membranes were washed three times at room temperature in 25 ml of Northwestern buffer (10 mM Tris-HCl [pH 7.4], 50 mM NaCl, 1 mM EDTA, 400 μg/ml Ficoll 400, 400 μg/ml polyvinylpyrrolidone, 400 μg/ml bovine serum albumin [BSA], and fresh 1 mM DTT) for 20 min at room temperature. Then, after a preincubation of 1 h in 25 ml of Northwestern buffer which served as a renaturation step, yeast tRNA (20 μg/ml) and 5,000 cpm of either (+) or (−) PLMVd radioactive RNA probe were added to the buffer solution, and the hybridization was performed at room temperature for 1 h. The membranes were washed three times in Northwestern buffer for 10 min and then were revealed by autoradiography. The bands containing the proteins interacting with PLMVd were cut out of the stained half of the SDS-PAGE gel and were sequenced by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (WEMB Biochem). The resulting peptide sequences were compared with all known plant sequences found in the NCBI database.
Immunoprecipitation and reverse transcription-PCR (RT-PCR) detection of the eEF1A-PLMVd complex.
Protein extraction from healthy and PLMVd-infected peach leaves was performed as described earlier. These protein extracts (1 ml) were mixed with 10 μl of maize eEF1A antiserum (kindly provided by Brenda Hunter, University of Arizona) and incubated for 1 h at 4°C under agitation. Twenty microliters of resuspended protein A/G Plus-agarose (Santa Cruz Biotechnology) was then added, and the mixtures were incubated for another 1 h at 4°C. The beads were precipitated by centrifugation at 1,000 × g for 5 min, and the supernatants were discarded by aspiration. The pellets were washed four times with 1 ml of phosphate-buffered saline (PBS). After the last wash, the pellets were resuspended in 100 μl PBS, and then Trizol extractions (Invitrogen) were performed in order to isolate eEF1A-binding RNAs.
The presence of PLMVd bound to the immunoprecipitated eEF1A was detected by RT-PCR. Briefly, RT reactions were carried out using SuperScript III (Invitrogen) as recommended by the manufacturer. The antisense primer PLM250-275 (5′-ATCACTTCTGGAGGGGACCGGGTTTG-3′) was used for the detection of the (+) polarity strands. After the reactions, 10 μg of RNase A was added and the mixtures were incubated for 10 min at 37°C. Five microliters of the RT reaction mixture was used for subsequent PCR amplification using purified Pfu DNA polymerase as described below. For the PCRs, the antisense primer PLM250-275 and the sense primer PLM63-86 (5′-TAATGACCTCTCAGCCCCTCCACC-3′) were used. Forty cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C were performed, and PCR products were detected by 1% agarose gel electrophoresis.
Cloning of eEF1Α from Prunus persica.
Total peach RNA samples were extracted from 200 mg of frozen (−80°C) healthy peach leaves (cv. Siberian C), using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA quality was verified by electrophoresis on 1% agarose gels, and quantities were determined by spectrophotometry at 260 nm. The resulting samples were used for RT-PCR amplifications. RT reactions were performed using SuperScript II enzyme (Invitrogen) for 2 h in the presence of an oligo(dT) antisense primer {Smart-3 [5′-CATGGAATTCGGATCC(T)30VN-3′, where V indicates A, C, or G and N indicates A, C, G, or T]}. According to the eEF1Α gene sequences from Arabidopsis thaliana (GenBank accession no. AAK82537, AAL31193, CAA34455, and AAL86336), Oryza sativa (GenBank accession no. O64937), and Solanum lycopersicum (GenBank accession no. CAA37212), a degenerate sense primer (1-EF1alpha [5′-ATGGGTAANGAGAAGWNNCACATNANCATTGTGG-3′, where W indicates A or T and N indicates A, C, G, or T]) was designed for Prunus persica eEF1Α cDNA amplification. PCR amplifications of the cDNAs were performed in a final volume of 50 μl containing 2 μM of each DNA primer, 1 mM MgSO4, a 0.2 mM concentration of each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.8), 25 mM KCl, 5 mM (NH4)2SO4, and 1 μg of purified Pfu polymerase. The mixture was subjected to 30 amplification cycles (1 min at 94°C, 1 min at 54°C, and 1 min 30 s at 72°C), followed by a final extension of 5 min at 72°C, using an Eppendorf Mastercycler gradient thermal cycler (Eppendorf).
In order to obtain the 5′ portion of the mRNA, a 5′ rapid amplification of cDNA ends (5′-RACE) strategy was performed using a RACE kit (Invitrogen). The antisense oligonucleotides used were EF1a-RACE2 (5′-CATCTGCTTGACACCAAGAGTAAACGCAAGC-3′) and EF1a-RACE3 (5′-GCATGCTCACGGGTCTGACCATCCTTGGAAATACCAGC-3′), which were used in a strategy of nested PCR amplification. The eEF1A sequence obtained was then used to design oligonucleotides (sense EF1a-NdeI [5′-CATATGGGCAAAGAAAAGTTTCACATC-3′] and antisense EF1a-XhoI [5′-CTCGAGCTTCTTCTTTGCTGCAGCCTTGG-3′]) specific for the production of a full-length cDNA that was cloned into the pET21b vector (Novagen).
Expression and purification of eEF1Α.
The eEF1Α (C-terminal His6-tag fusion) insert was expressed in Escherichia coli BL21(DE3). Cells were grown in 1 liter of LB medium containing 100 μg/ml ampicillin at 37°C to an optical density at 600 nm of 0.5. The cultures were chilled on ice for 30 min, and expression was induced by adding ethanol and IPTG (isopropyl-β-d-thiogalactopyranoside) to final concentrations of 2% and 0.4 mM, respectively, and then incubating cultures at 18°C for 20 h. The cells were harvested by centrifugation at 5,000 × g for 10 min, and the resulting cell pellets were frozen at −80°C for 10 min and then placed on ice for 10 min. The last two steps were repeated three times, and the bacteria then were resuspended in 50 ml buffer A (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 10% sucrose). After the addition of lysozyme (50 μg/ml) and Triton X-100 (0.1%) to the bacteria, the cells were lysed by sonication. The resulting lysates were clarified by centrifugation for 45 min at 10,000 × g at 4°C.
The supernatants were then passed over a 5-ml Ni-nitrilotriacetic acid affinity column (Qiagen), and the column was washed with buffer A containing 0.1% Triton X-100 and 5 mM imidazole. The elutions were performed with buffer B (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 10% glycerol) containing increasing concentrations of imidazole (50, 100, 200, 500, and 1,000 mM). Protein-containing fractions were pooled and dialyzed against 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 2 mM DTT, and 10% glycerol overnight at 4°C. The resulting protein samples were loaded on a 1-ml heparin-Sepharose 6 fast-flow column (Amersham Biosciences), and the column was washed with buffer HS (50 mM Tris-HCl [pH 7.8] and 10% glycerol) containing 50 mM (NH4)2SO4. The column was then eluted with HS buffer containing 150, 400, 600, or 1,500 mM (NH4)2SO4. The resulting fractions were dialyzed as described previously, and the resulting proteins were concentrated using Amicon Ultra-4 5K filters (Millipore). Protein concentrations were determined by Bradford assay, and the purity was evaluated by 10% SDS-PAGE followed by Coomassie blue staining.
In order to confirm the presence of eEF1A, Western blots were performed. Briefly, protein aliquots were fractionated in 10% SDS-PAGE gels and then transferred to nitrocellulose membranes. The membranes were blocked with 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) containing 5% dried milk overnight at 4°C. The blocked membranes were incubated with the primary antibody, mouse Tetra His (Qiagen), diluted 1:1,500 in 4 ml of 1× PBS containing 1% dried milk, for 2 h at 25°C. The membranes were then washed three times in 1× PBS before adding a rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (Promega), diluted 1/10,000 in 1× PBS containing 1% dried milk, and were incubated for 1 h at 25°C. Finally, the proteins were detected using an ECL detection kit according to the manufacturer's protocol (Perkin Elmer).
EMSA.
The binding of the eEF1Α protein from Prunus persica to the PLMVd RNAs of both polarities was characterized by electrophoretic mobility shift assays (EMSAs). Initially, the labeled PLMVd RNAs were heated at 65 to 70°C and then rapidly put on ice in order to ensure proper folding. This condition gives a secondary structure identical to the one obtain previously by our laboratory (7). PLMVd RNA (5 nM; 500 cpm) and yeast tRNA (0.1 μg; 50 nM) were incubated together in binding buffer (10 mM Tris-HCl [pH 7.5], 5 mM magnesium acetate, 100 mM NH4Cl, and 0.5 mM DTT) for 5 min at 4°C. Increasing amounts of either eEF1A protein (100 to 1,000 nM) or control protein (2,000 nM of BSA) were then added, and the mixtures were incubated for 15 min at 4°C. After the incubation, 5 μl of loading buffer (1× TBE, 50% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol) was added prior to electrophoresis through 5% native polyacrylamide gels (29:1 acrylamide/bisacrylamide ratio) in 1× TBE buffer overnight at 4°C. The gels were exposed to phosphor screens and were then revealed using a Storm scanner (Molecular Dynamics). Analyses were performed using both Image Quant and Prism software.
GDP binding assays.
To make radioactive GDP, 150 μCi of [α-32P]GTP (3,000 Ci/mmol; New England Nuclear) and 10 μM of nonradiolabeled GTP were added to purified Saccharomyces cerevisiae RNA triphosphatase (Cet1), which possesses an ATPase activity, in a buffer containing 10 mM Tris-HCl (pH 7.5) and 10 mM NaCl. After an incubation of 30 min at 30°C, the proteins were removed by phenol-chloroform extraction. For the binding assays, 5 μl (∼500,000 cpm) of the radiolabeled GDP and various concentrations (10 μM to 1 mM) of nonradioactive GDP were added to 30 μl of purified eEF1A protein (75 nM) in EMSA buffer (see above). Also, 50 μg (70 nM) of BSA was added to favor the precipitation in 20% trichloroacetic acid. The volume was completed to 100 μl, and the sample was incubated on ice for 30 min. The proteins then were precipitated by centrifugation at 1,300 rpm for 15 min. Supernatants were discarded, and the pellets were washed with 100 μl of 20% trichloroacetic acid and then spun for 2 min. Finally, the supernatants were removed and the radioactivity was determined by a scintillation counter.
Fluorescence spectroscopy.
Real-time RNA binding experiments were performed by fluorescence spectroscopy, using an F-2500 fluorescence spectrophotometer (Hitachi). RNA solutions (25-μl samples of 10 μM solutions) were injected at predetermined times into 80 μl of 0.8 μM eEF1A protein solution in binding buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM NaCl, and 5 mM MgCl2. The samples were excited at 290 nm, and the fluorescence was monitored as a function of time at 329 nm. The slope of each spectrum was calculated in order to determine the relative association rate (μM−1 s−1).
5′- and 3′-end labeling.
Purified 338-nt PLMVd RNA, opened at position 1 for strands of both (+) and (−) polarity, was labeled according to previously reported procedures (7). The RNA contained two point mutations [U9→A and A330→U, in the (+) polarity strand] that prevent hammerhead self-cleavage from taking place and result in the synthesis of full-length PLMVd RNA strands. Transcripts (25 pmol) were dephosphorylated in a final volume of 10 μl, using 10 U of Antarctic phosphatase according to the manufacturer's recommended protocol (New England Biolabs). The reactions were terminated by heating for 8 min at 65°C. Subsequently, dephosphorylated RNAs (5 pmol) were 5′-end labeled in the presence of 3.2 pmol [γ-32P]ATP (6,000 Ci/mmol; New England Nuclear) and 3 U of T4 polynucleotide kinase according to the manufacturer's recommended protocol (USB). The reactions were performed at 37°C for 60 min. For 3′-end labeling, purified 338-nt PLMVd transcripts (20 pmol) were incubated with 10% dimethyl sulfoxide, 40 μCi of [α-32P]CMP, 3′-monophosphate (3,000 mCi/mmol; New England Nuclear), and 40 U of purified T4 RNA ligase in a final volume of 10 μl containing 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 1 mM ATP, and 10 mM DTT. After 60 min of incubation at 37°C, the reactions were terminated by the addition of 1 volume formamide dye buffer, and the mixtures were fractionated through denaturing 5% PAGE gels. After autoradiography, the bands containing the appropriate 5′- and 3′-end-labeled RNAs were excised and the RNA recovered as described above.
Enzymatic probing of PLMVd strands.
For enzymatic probing, trace amounts of snap-cooled 5′- or 3′-end-labeled 338-nt PLMVd (<1 nM) were dissolved in 3 μl of water, and yeast tRNA was added to a final concentration of 0.1 μg/μl. The resulting RNA mixtures were added to buffer containing 10 mM Tris-HCl (pH 7.5), 0.5 mM DTT, and 100 mM NH4Cl in either the presence or absence of 0.5 μM eEF1A. Uniquely in the case of the reaction performed in the presence of RNase V1, the buffer also contained 5 mM MgCl2. The mixtures were incubated for 1 min at 25°C in the presence of either 0.2 U of RNase T1 (Roche), 0.1 U of RNase PhyM (IRL), 1 pg/μl RNase A (USB), or 0.045 U of RNase V1 (Pierce Molecular Biology) and were then quenched by the addition of 40 μl of 40 mM EDTA.
In order to prepare a guanosine ladder, the RNA was also incubated in the presence of RNase T1 at 65°C for 10 min. For alkaline hydrolysis, the PLMVd RNA (<1 nM) was dissolved in a mixture of 9 μl of water and 1 μl of 2 N NaOH and then incubated at room temperature for 1 min. In that case, the reactions were quenched by the addition of 10 μl of 1 M Tris-HCl (pH 7.5). In all cases, the resulting mixtures were purified by phenol-chloroform extraction and ethanol precipitation in the presence of glycogen and were then dissolved in loading buffer (10 μl of 50% formamide and 10 mM EDTA solution), fractionated in denaturing 8% PAGE gels, and visualized by exposure of the gels to phosphorimaging screens.
Nucleotide sequence accession number.
The complete eEF1A sequence from Prunus persica was submitted to the NCBI database (GenBank accession no. FJ267653).
RESULTS
Isolation of PLMVd-binding proteins.
In order to isolate host proteins that have the ability to bind PLMVd transcripts, we decided to use total peach leaf extracts, primarily because this approach permits the identification of proteins important in all steps of the viroid life cycle. An alternative would have been the use of a chloroplast preparation, as this is the organelle in which PLMVd is proposed to replicate; however, this would restrict the number of putative proteins that might be identified. Protein extracts were therefore prepared from freshly harvested healthy peach leaves. The resulting extracts were purified by FPLC using a heparin-Sepharose column in order to enrich the extract in nucleic acid binding proteins. The column was successively eluted with 150, 400, 600, and 1,500 mM ammonium sulfate solutions. Most proteins eluted at the 150 and 400 mM ammonium sulfate concentrations. The most abundant protein in the total extract possessed an electrophoretic mobility equivalent to that of a 50-kDa protein, a molecular mass that corresponds to a chloroplast-encoded large subunit of rubisco, a protein that does not bind to the column (Fig. 1A).
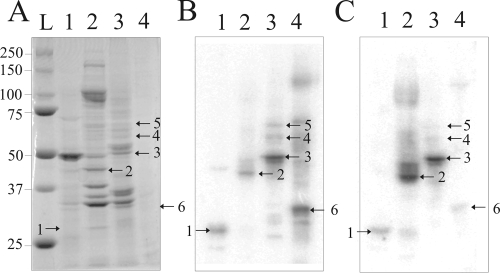
FIG. 1.
Detection of Prunus persica proteins interacting with PLMVd by Northwestern blot analysis. (A) Coomassie blue staining of the 10% PAGE gel. (B and C) Proteins were transferred onto nitrocellulose membranes, which were then probed using the (−) (B) and (+) (C) PLMVd strands. The blots show total protein extract from peach tree leaves (lanes 1) and purified protein fractions obtained after fractionation on a heparin-Sepharose column and subsequent elution with 150 mM (lanes 2), 400 mM (lanes 3), and 600 mM (lanes 4) ammonium sulfate. In panel A, lane L is a molecular weight marker. The numbering of the bands corresponds to that used in Table 1.
In order to identify proteins with the potential to bind PLMVd RNA species, Northwestern hybridizations were performed. Briefly, protein fractions were separated by PAGE and then transferred onto nitrocellulose membranes for further hybridization, using radiolabeled unit-length PLMVd transcripts of either (+) or (−) polarity. Typical autoradiograms using the probe of each polarity are illustrated in Fig. 1B and C. Significantly, irrespective of the polarity of the probe used, virtually identical autoradiograms were obtained, albeit with different band intensities. This might be interpreted as an indication that PLMVd strands of both polarities are folded into similar global secondary structures, thereby resulting in equivalent interactions with the host proteins. In the total extract, a single protein with a migration pattern corresponding to that of an ∼30-kDa protein was consistently detected (Fig. 1B and C, band 1). The same band was detected in the 150 mM ammonium sulfate column fraction, although in a reduced amount (the most abundant band detected corresponded to an ∼45-kDa protein and was numbered band 2). These two proteins were the only ones consistently detected in the 150 mM ammonium sulfate fraction. In the 400 mM ammonium sulfate fraction, three proteins, ranging from 50 to 70 kDa, were consistently detected (bands 3, 4, and 5, with the most abundant being protein 3). Finally, a single band corresponding to an ∼35-kDa protein was detected in the 600 mM ammonium sulfate fraction (band 6).
Northwestern hybridizations were also performed using other probes, corresponding to either the PSTVd RNA species, as a model viroid of the Pospiviroidae, or a model hepatitis delta virus ribozyme, as an unrelated but highly structured RNA species. In the latter case, no RNA-protein complex was detected using a peach leaf protein extract. Conversely, the PSTVd RNA probe led to the detection of two bands in the peach leaf extract, specifically bands 2 and 3, suggesting that the corresponding proteins have the ability to bind both viroid species. Finally, Northwestern hybridizations using the PLMVd probe of (+) polarity and protein extracts isolated from both tomato and tobacco plants were also performed (data not shown). In both cases, bands 2 and 3 were observed, but never band 1, 4, 5, or 6.
Identification of peach proteins with potential to bind PLMVd.
The six bands on the Coomassie blue-stained gels located at the positions corresponding to the six proteins identified in the PLMVd Northwestern blots were excised and used for identification of the derived peptides by LC-MS/MS analysis. Because the genome of Prunus persica has been sequenced only partially, the NCBI database containing all known plant sequences was used for the identification of the various proteins. Up to eight different peptides homologous to a unique plant protein were retrieved for each protein band (Table 1). The percent coverage between a peptide and a given protein species ranged from 1.6% to 15%.
TABLE 1.
LC-MS/MS results
| Band no. | FPLC fraction | Molecular mass (kDa) | Protein name | No. of peptides | % Coveragea |
|---|---|---|---|---|---|
| 1 | Total extract | 30 | β-1,3-Glucanase | 1 | 3 |
| 2 | 150 mM HS buffer | 45 | Aminomethyltransferase | 4 | 11.5 |
| 3 | 400 mM HS buffer | 50 | eEF1A | 6 | 11.8 |
| 4 | 400 mM HS buffer | 60 | Putative chaperone | 8 | 15 |
| 5 | 400 mM HS buffer | 67 | Dynamin | 1 | 1.6 |
| 6 | 600 mM HS buffer | 35 | L5 ribosomal protein | 2 | 7.6 |
Represents the percentage of total amino acids sequenced for each protein compared to its total length.
Band 1, which was detected only in the total extract, led to the detection of only one peptide, representing 3% coverage, of β-1,3-glucanase, a hydrolytic 30-kDa (as determined by the autoradiogram) enzyme responsible for β-1,3-glucan degradation (35). The tomato homologue of this enzyme has been shown to be overexpressed upon citrus exocortis viroid infection, and in another study, the tobacco homologue was shown to be able to increase the susceptibility to a viral pathogen (3, 15). Confidence in the accurate identification of the protein responsible for band 2 was significantly higher, as four peptides, covering 11.5% of the enzyme aminomethyltransferase (Table 1), were detected. This 45-kDa protein is a component of the glycine cleavage system which oxidatively decarboxylates glycine (36). The detection of this protein might be considered suspicious, as it has no known RNA binding domain; however, the γ-aminobutyrate-aminotransferase from tomato was reported to nonspecifically bind PSTVd (28). Six peptides with sequences homologous to that of eEF1A, together covering 11.8% of the open reading frame and encoding a protein of 50 kDa, were detected for the protein corresponding to band 3. The primary function of eEF1A is as a GTP-dependent carrier of aminoacylated tRNAs to the A site of the ribosome during translation. The eEF1A protein is also known to bind a variety of different nucleic acids, including short double-stranded RNA molecules (34). The protein corresponding to band 4 led to the identification of eight sequenced peptides homologous to a putative chaperone (15% coverage). That corresponding to band 5 led to the identification of only one peptide, corresponding to 1.6% of a dynamin-like GTP-binding protein. This low homology could be explained by the fact that this protein family is not highly conserved among different species. The dynamin family cluster includes proteins implicated in membrane division, vesicle formation, cytokinesis, and endocytosis (9). Finally, the protein corresponding to band 6 provided two peptides, corresponding to 7.6% coverage, of the L5 ribosomal protein, which is a component of the large subunit of the ribosome known to bind 5S RNA (31).
Immunoprecipitation of eEF1A-PLMVd complex.
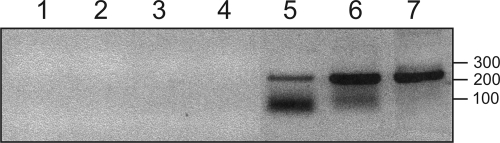
We decided to further characterize the interaction between eEF1A and PLMVd because this protein possesses an RNA binding motif and because a relatively good peptide coverage of the protein was obtained (>10%). With the goal to provide additional physical support for the eEF1A-PLMVd complex in vivo, protein extracts were prepared from PLMVd-infected peach leaves and then the eEF1A was immunoprecipitated using a maize polyclonal antiserum. The latter was shown to be efficient for peach eEF1A protein detection by Western blotting, as well as for immunoprecipitation of the peach eEF1A protein alone (data not shown). The RNA within the pellets was retrieved by Trizol extraction, and the PLMVd RNA strands of (+) polarity were detected after RT-PCR amplification. No PLMVd was detected in the extract of the healthy leaves (Fig. 2, lane 3). Similarly, no PCR product was observed from the extract of the infected leaves if the eEF1A antiserum was omitted during the immunoprecipitation step (Fig. 2, lane 4). Conversely, two bands of DNA were detected when the eEF1A antiserum was incubated in the presence of the extract from the infected leaves (Fig. 2, lane 5). The upper band had the same electrophoretic position as the PCR products from RT-PCR amplifications using either an RNA extract from infected leaves or PLMVd strands prepared by runoff transcription (Fig. 2, lanes 6 and 7). Subsequently, a Southern blot hybridization was performed to confirm the PLMVd identity of this PCR product (data not shown). In the later experiment, no hybridization of the smaller band could be detected, indicating that this was a nonspecific PCR product, which might have occurred since the amount of immunoprecipitated PLMVd was very small. These results confirm that at least a portion of PLMVd retrieved within the infected leaves was bound to the immunoprecipitated eEF1A. More importantly, this experiment added physical support for the presence of the eEF1A-PLMVd complex in vivo.
FIG. 2.
Detection of PLMVd immunoprecipitated with eEF1A. eEF1A proteins were immunoprecipitated from healthy and infected peach leaf extracts, RNAs were extracted, and then PLMVd was amplified by RT-PCR and analyzed by 1% agarose electrophoresis. Lane 1, control RT-PCR amplification performed without nucleic acid (only water); lane 2, RT-PCR amplification performed using the healthy leaves, but without immunoprecipitation; lane 4, RT-PCR amplification performed after immunoprecipitation in the absence of eEF1A antiserum, using PLMVd-infected leaves; lanes 3 and 5, RT-PCR amplifications performed after immunoprecipitation in the presence of eEF1A antiserum, using healthy and PLMVd-infected leaves, respectively; lanes 6 and 7, RT-PCR amplifications performed using RNA extract from PLMVd-infected leaves and PLMVd synthesized by runoff transcription, respectively. Adjacent to the gel is a DNA molecular size marker, in bp.
Cloning, expression, and purification of eEF1Α.
In order to permit further characterization, the peach eEF1A protein was initially cloned. The eEF1A cDNA from Prunus persica was amplified by RT-PCR (see Materials and Methods). The resulting eEF1A Prunus persica sequence (NCBI accession no. FJ267653) corresponds to an mRNA of 1,771 nt that includes 5′- and 3′-untranslated regions of 88 and 339 nt, respectively. The open reading frame region is 1,344 nt long and encodes a 447-amino-acid protein that is 96.8% homologous to the Arabidopsis thaliana gene product, 97.5% homologous to the Oryza sativa gene product, and 95.7% homologous to that from Zea mays (Fig. 3). The differences lie along the length of the sequence and are not clustered in any one region. Moreover, there are two fewer amino acids at the carboxy-terminal end of the coding sequence of the Prunus persica eEF1A protein than at that of the protein from Arabidopsis.
FIG. 3.
Alignment of eEF1A amino acid sequences from Prunus persica (GenBank accession no. FJ267653), Arabidopsis thaliana (accession no. CAA34455), Zea mays (accession no. AAF42978), and Oryza sativa (accession no. BAA23657). The amino acid sequence of Prunus persica eEF1A was determined using Expasy Translated Tool, while the sequence alignments were performed using AliBee multiple-alignment software.
The Prunus persica eEF1A gene was subcloned into an expression vector in order to permit the subsequent production of a His6-tagged recombinant protein in E. coli. After expression, this protein was purified using a nickel column followed by a second, heparin-containing column (Fig. 4A). The presence of the His-tagged eEF1A protein was confirmed by Western blot hybridization using a His tag antibody (Fig. 4B). The final protein's purity was estimated to be near 96%, and circular dichroism analysis showed that the protein was folded (data not shown). Finally, the structural integrity of the recombinant eEF1A protein was verified by determining its capacity to bind radioactive GDP. Since the elongation factor possesses a GTP binding site that involves amino acids from several distinct regions of the protein (16), this is an accurate means of verifying that it is correctly folded. The experiment was performed using a constant concentration of eEF1A and increasingly concentrated mixtures of radioactive [α-32P]GDP and nonradioactive GDP and resulted in a binding curve that was fitted by a single-binding-site equation with a dissociation constant (Kd) of 25 μM (data not shown). This result indicates that a large proportion, if not all, of the enzyme adopts a tertiary structure that has the ability to bind GDP.
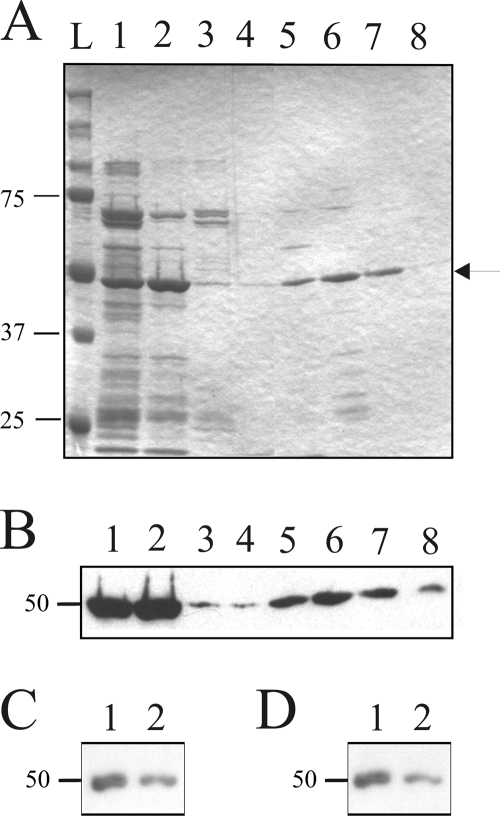
FIG. 4.
Expression and characterization of recombinant Prunus persica eEF1A. (A) Coomassie blue staining of an SDS-PAGE gel containing His6-tagged eEF1A purification fractions. Lanes 1 and 2, eluates from the Ni-nitrilotriacetic acid column, using 200 mM and 500 mM imidazole, respectively; lanes 3 and 4, flowthrough and wash fractions of the proteins from the heparin column purifications of the samples in lanes 1 and 2, respectively; lanes 5 to 8, fractions collected from the elutions performed in the presence of 150 mM, 400 mM, 600 mM, and 1,500 mM ammonium sulfate, respectively. The lane labeled L is a molecular size marker. The arrow indicates the position of recombinant eEF1A. (B) Western blot hybridization performed using an anti-His6 antibody on an SDS-PAGE gel similar to that presented in panel A. (C and D) Autoradiograms of Northwestern hybridizations performed using the fractions corresponding to the elutions performed in the presence of 400 mM and 600 mM ammonium sulfate (lanes 1 and 2, respectively), using the (−) and (+) PLMVd strands as probes, respectively.
eEF1Α interacts with PLMVd.
In order to confirm the proposed interaction between Prunus persica eEF1A and PLMVd, Northwestern hybridizations were performed using the purified recombinant protein and PLMVd RNA strands of either (+) or (−) polarity as probes. A unique, sharp band corresponding to a 50-kDa protein, the expected size of eEF1A, was obtained with the probes of both polarities (Fig. 4C and D).
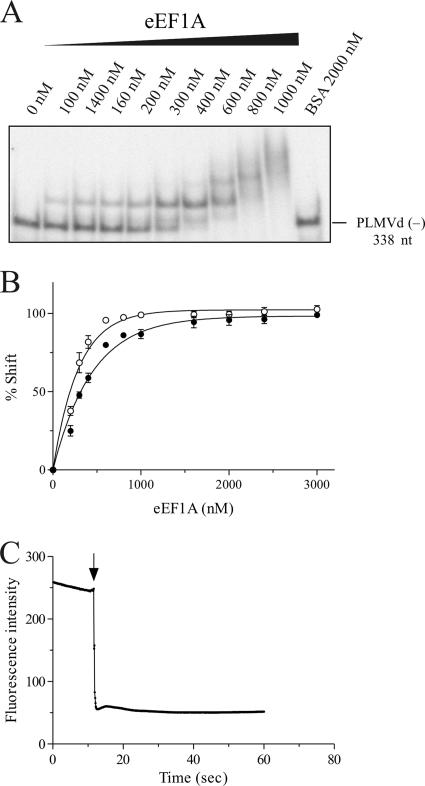
Physical evidence for the interaction between eEF1A and PLMVd was obtained by EMSA. These experiments were always performed in the presence of an excess of yeast tRNA in order to eliminate any nonspecific interactions. Radiolabeled PLMVd transcripts were incubated in the presence of increasing concentrations of recombinant elongation factor and were then fractionated in native PAGE gels. PLMVd (−) strands were shifted by the presence of eEF1A (Fig. 5A). Similar results were obtained for the PLMVd strand of (+) polarity (data not shown). In the presence of large amounts of eEF1A, more than one RNA-protein complex was detected. It is most likely that under these conditions, either the elongation factor binds to several sites on PLMVd or eEF1A forms a dimer. The fraction of shifted PLMVd RNA was determined and is illustrated as a function of the concentration of eEF1A in Fig. 5B. eEF1A had an apparent Kd of 386 nM ± 49 nM for the PLMVd (+) polarity strand and of 210 nM ± 31 nM for the (−) polarity strand (Table 2). This is a stronger level of binding than that observed for either tRNAAla or an RNA species composed of 27 consecutive CU dinucleotide repeats, used as controls (Table 2). These two controls were performed in order to compare the binding of the PLMVd to the eEF1A recombinant protein with the binding of the elongation factor with a structured (tRNAAla) and a nonstructured (CU-based polymer) RNA species. The binding of PLMVd to eEF1A was virtually identical when the experiment was repeated in the presence of different concentrations of GTP (data not shown). According to this experiment, the binding of eEF1A is not GTP dependent. However, this does not completely exclude the possibility that eEF1A was purified with bound GTP and that only the eEF1A-GTP complexes bound PLMVd.
FIG. 5.
EMSAs and fluorescence analysis of PLMVd-eEF1A interaction. (A) Autoradiogram of native polyacrylamide gel obtained using the PLMVd strand of (−) polarity and increasing concentrations of eEF1A. (B) Graphic representation of the percentage of shifted complexes as a function of the eEF1A concentration. Closed circles indicate the PLMVd strand of (+) polarity, while open circles indicate the PLMVd strand of (−) polarity. (C) Fluorescence of eEF1A as a function of time. The arrow indicates the point of injection of the PLMVd (−) RNA. The calculated slope corresponds to the ka of protein binding to the RNA.
TABLE 2.
Dissociation constants (Kd) and association rates (ka) for the interaction of eEF1A with various RNAsb
| RNA species | EMSA Kd (nM) | Fluorescence spectroscopy ka (μM−1 s−1) |
|---|---|---|
| (−) PLMVd | 210 ± 31 | 420 ± 29 |
| (+) PLMVd | 386 ± 49 | 342 ± 6 |
| tRNAAla | >900 | 250 ± 30 |
| CU27 | NDa | 178 ± 19 |
ND, not determined.
Data are means ± standard deviations.
In order to provide further physical support for the EMSA data, the binding experiments were followed by fluorescence spectroscopy. The eEF1A protein includes three tryptophan residues that fluoresce following excitation at 290 nm. If the binding of the RNA species to eEF1A modifies the fluorescence signal, then it is possible to estimate its association rate. A typical example is illustrated in Fig. 5C for the binding of PLMVd strands of (−) polarity. In these experiments, the binding of the PLMVd strands of both polarities to eEF1A resulted in higher association constants (ka) than were observed for both the tRNAAla and the CU-based polymer (Table 2). Thus, this may contribute to explaining why PLMVd would be bound preferentially by eEF1A compared to other RNA species.
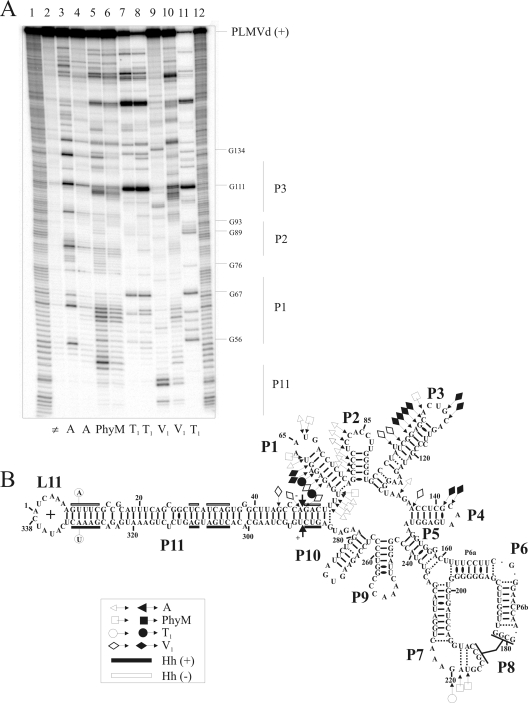
Finally, additional support for the eEF1A-PLMVd interaction was provided by enzymatic mapping of the RNA component. PLMVd strands were prepared using a mutated version of the hammerhead sequence that prevents self-cleavage from occurring, without affecting its secondary structure, thereby resulting in the synthesis of full-length transcripts (338 nt) starting from position 1. The 5′ or 3′ end, located in the left-hand terminal loop (loop L11) (Fig. 6), was 32P labeled and then probed in either the presence or absence of an excess of eEF1A. Yeast tRNA was added to the trace amounts of PLMVd as a competitor in order to avoid any nonspecific binding. Three ribonucleases (RNases that preferentially hydrolyze the 3′-phosphodiester bond of specific ribonucleotides located in single-stranded regions; the three used for this study were RNase A, which cleaves after both single-stranded C and U; PhyM, which cleaves after single-stranded A and U; and T1, which cleaves after single-stranded G) and RNase V1, which cleaves the phosphodiester bonds of nucleotides located in double-stranded regions irrespective of their identity, were used. A typical example of an autoradiogram of a sequencing gel probing 5′-end-labeled PLMVd (+) transcripts is illustrated in Fig. 6A. Several regions showing different banding patterns, when examined in either the absence or presence of eEF1A, are observable. For example, both RNase A and RNase PhyM readily hydrolyzed the U53, U54, U56, U63, and U66 residues of the P1 stem-loop region of (+) polarity in the absence of eEF1A, but only did so at a reduced level in its presence (Fig. 6A, compare lanes 3 and 4 with lanes 5 and 6, respectively). With RNase T1, the neighboring G58 and G60 residues were hydrolyzed to a greater extent in the presence of eEF1A than in its absence (Fig. 6A, compare lanes 7 and 8). Finally, the banding pattern obtained with RNase V1 in the right-hand P11 stem (positions A48 and G49) was less cleaved in the presence of eEF1A, suggesting that this domain became single stranded in most of the PLMVd molecules (lanes 9 and 10).
FIG. 6.
Enzymatic probing of PLMVd with ribonucleases in either the presence or the absence of eEF1A. (A) Typical autoradiogram of an 8% PAGE gel used for analysis of probing performed with a 5′-end-labeled PLMVd strand of (+) polarity. Lanes 1 and 12, identical ladders obtained by alkaline hydrolysis of PLMVd strands; lane 2, nonhydrolyzed RNA sample. The remaining lanes contained RNA samples that were hydrolyzed by RNase A (lanes 3 and 4), RNase PhyM (lanes 5 and 6), RNase T1 (lanes 7 and 8), and RNase V1 (lanes 9 and 10) in either the absence (lanes 3, 5, 7, and 9) or the presence (lanes 4, 6, 8, and 10) of eEF1A. Lane 11 contains PLMVd strands hydrolyzed by RNase T1 at 65°C in order to provide a guanosine ladder. (B) Compilation of mapping differences resulting from the addition of eEF1A on the model secondary structure of the PLMVd (+) strand. Opened and closed symbols indicate a decrease and increase in the susceptibility to a given RNase, respectively. The legend in the figure shows the symbols used to indicate cleavage by the various RNases (▵, RNase A; □, RNase PhyM; ○, RNase T1; and ⋄, RNase V1). The two mutated nucleotides preventing self-cleavage are circled on the left-hand extremity of the P11 stem. The black and white boxes indicate the conserved nucleotides for both (+) and (−) hammerhead sequences, respectively.
Several experiments performed with either the 5′- or 3′-end-labeled PLMVd transcripts and various migration times were required in order to obtain a complete compilation of the differences in RNase mapping caused by eEF1A binding. Importantly, the data obtained in the absence of eEF1A were in good agreement with the previously reported extensive mapping of the PLMVd (+) polarity strand that was used to determine its secondary structure (7). The binding of eEF1A resulted in several mapping differences that can be classified into two types. First, there were nucleotides located in single-stranded regions that became partially resistant to RNase hydrolysis, suggesting that either they were protected by the elongation factor within the RNA-protein complex or they were involved in a modified structure that resulted from the binding of eEF1A (e.g., along the P1 stem). Second, there were nucleotides located in double-stranded regions that were hydrolyzed solely in the presence of eEF1A (except for the case of RNase V1), an observation which can only result from a modification of the secondary structure, specifically going from being double to single stranded (e.g., P11 stem). These two types of modifications are not necessarily independent. For example, it is possible that the binding of eEF1A occurred with the P1 stem region and resulted in the unfolding of the P11 stem while, concurrently, the P3 stem became more susceptible to RNase V1 hydrolysis. In the case of the (−) strand, all of the observed RNase probing modifications occurred in the region of the junction between P10 and P11, that is, adjacent to the self-cleavage site. Since the secondary structure of that strand has not yet been reported, interpretation is more ambiguous. Importantly for the probing of the (+) strand, the observed differences are concentrated in two distinct regions (P11-P1-P2-P3 and P7-P8), in agreement with the possibility that more than one eEF1A protein can bind to a molecule of PLMVd, as suggested by the EMSA experiments.
DISCUSSION
Today, there is no known approach that would identify all proteins interacting with an RNA species, such as a viroid, that requires many different host partners in order to ensure its complete life cycle. The combination of partial FPLC purification of peach extracts, Northwestern hybridization, and mass spectrometry described here identified six putative interacting proteins in a single experiment (i.e., a single set of conditions), whereas all other approaches previously reported (e.g., RNA ligand screening and in vivo cross-linking) yielded only one (10, 29). However, there are at least two obvious limits to this strategy. The first is the fact that the use of a Northwestern blot implies performing denaturing PAGE. Some proteins may remain in inactive conformations and therefore cannot be identified by this technique. The second limit is associated with the observation that neither a plastid polymerase nor a component of the RNA silencing machinery was isolated in our experiments. However, one possible explanation for that observation is that the strength of the interaction between the viroid and the protein was too weak to resist all of the washing steps. Moreover, some proteins may interact with the viroid only when complexed with another protein(s), or only in the presence of required cofactors, species which were not preserved during the preparation and purification procedure. In other words, there are several factors that can influence the recruitment of a given protein by the viroid and its subsequent identification by this method.
Since the localization of some proteins, e.g., the putative chaperone, remains unknown, we cannot conclude that no chloroplast proteins were identified. Moreover, even if the majority of proteins are retrieved from the cytoplasm, we cannot argue that these interactions are not specific since there is a cytoplasm phase in the life cycle of PLMVd. The observation that all six of the proteins identified here interact with PLMVd strands of both polarities was not surprising, since it is believed that both strands fold into nearly identical structures and share many features (6).
Based on the facts that the LC-MS/MS experiment identified peptides covering over 10% of a putative candidate and that this candidate possessed an RNA binding motif, it was considered that this protein, the elongation factor eEF1A, was a suitable candidate protein for further significant characterization. Moreover, eEF1A is a very abundant protein, representing 1 to 5% of the total soluble protein in the cell (5). In fact, it is so abundant that it is not unreasonable to suggest that the detection of the PLMVd-eEF1A complex might simply be the result of a nonspecific interaction. It is clear that physical support for the existence of this interaction provided by the further characterization reported here increases the likelihood that the five other proteins proposed to interact with PLMVd all represent valid, specific interactions between the protein in question and PLMVd. The eEF1A cDNA from Prunus persica was cloned, and its sequence did not show any significant differences compared to eEF1A proteins from other plants. The recombinant eEF1A protein was expressed in E. coli and then highly purified. Several analyses of the purified recombinant protein suggest that it folds into a proper tertiary structure and specifically binds to PLMVd strands of both polarities in vitro. Experiments using immunoprecipitation of the eEF1A-PLMVd complex from crude extracts of peach leaves by use of eEF1A antibodies, coupled with RT-PCR detection of PLMVd, led to detection of small amounts of PLMVd in the precipitated fraction. However, there are several limitations to this approach, including the fact that the elongation factor is very abundant and that only a small fraction of it was probably interacting with the viroid, thereby suggesting that this part of the data must be considered with caution. Moreover, several factors, including the quality of the leaves, the time of harvest, and the amount of PLMVd (i.e., level of replication), to name only these examples, influenced the reproducibility of this experiment (i.e., whether PLMVd was detected after the immunoprecipitation of eEF1A protein). Alternatively, we tried introducing an aptamer at various sites within the PLMVd species in order to retrieve the eEF1A-PLMVd complex. However, such chimerical molecules could not be detected even 1 year after the inoculation, suggesting that they did not replicate in peach leaves (A. Dubé, O. Parisi, H. Jijakli, and J. P. Perreault, unpublished data). Importantly, the detection of PLMVd with the immunoprecipitation of eEF1A supports the idea that the complex is formed under cellular conditions and can be considered the first step in the direction of characterizing this complex in vivo.
Clearly, the next interesting issue raised by these results, which warrants study, is determining the biological consequences associated with the formation of the PLMVd-eEF1A complex. Although this is not the goal of the current study, it is tempting to speculate about this issue. It is unlikely that the formation of this complex will sufficiently reduce the amount of eEF1A so that it will interfere with its normal cellular function (e.g., reducing translation), due to its abundance. The potential binding site located on the PLMVd strand of (+) polarity, as determined by nuclease probing, might be useful in the development of a hypothesis to explain eEF1A's role in the viroid's life cycle. First, these data suggest that eEF1A binds the P1 stem and that this binding is associated with the opening of the right-hand, double-stranded region of the P11 stem. This region includes both the hammerhead cleavage and self-ligation sites [i.e., positions C289 and C49 for the (+) and (−) strands, respectively (8, 23)] (Fig. 5B), as well as the host polymerase initiation site [positions A50/C51 and U286 for the (+) and (−) strands, respectively (12, 30)]. The modification observed in the nuclease mapping experiments in this region may correlate with that observed for the PLMVd (−) strand, although it is more limited in the latter case.
Several experiments were performed to investigate the potential implication of eEF1A in either the self-cleavage or self-ligation reaction (F. Bolduc and J. P. Perreault, unpublished data); however, no significant variation of either the self-cleavage or self-ligation level was detected, regardless of the conditions tested. Alternatively, it might be possible that the binding of eEF1A to, or near, the P1 stem favors the initiation of replication. eEF1A has been shown to be involved in the replication of several RNA viruses either by interacting directly with the RNA species (e.g., it interacts with the 3′ ends of turnip yellow mosaic virus [24], brome mosaic virus [2], and West Nile virus [11]) or by interacting directly with the viral RNA-dependent RNA polymerase (e.g., as observed for poliovirus [21] and tobacco mosaic virus [TMV] [37]). However, since the chloroplastic polymerase responsible for PLMVd replication remains to be identified and there is no model enzymatic assay yet available, such involvement cannot be verified at present. Second, the interaction in the L7 loop (Fig. 5B), which is located in close proximity to the P8 pseudoknot, is in agreement with the previous demonstration that an elongation factor binds to such a motif within TMV (38). Since the contribution of eEF1A to various cellular mechanisms, including translation, cytoskeleton formation, and the protein exportation mechanism, to name only a few examples, has been described (20, 26), the formation of the PLMVd-eEF1A complex might also have an impact on several cellular mechanisms (e.g., RNA transport and RNA stability). Interestingly, the facts that PLMVd interacts with the homologous elongation factors from tobacco and tomato plants and that PSTVd, the prototype member of the Pospiviroidae family, can interact with peach eEF1A indicate that this interaction does not represent a PLMVd host specificity. It has already been proposed that many RNA viruses, including hepatitis delta virus, require interaction with the host's eEF1A protein during their life cycles (33, 37, 38). It is likely that viroids obey this rule.
eEF1A binding to viral structures has been shown to be GTP dependent. For example, GTP has been shown previously to be essential for the binding of the 3′-terminal tRNA-like structure of TMV (38). However, to our knowledge, the GTP-dependent behavior of eEF1A binding to RNA structures other than tRNA-like structures has not yet been reported. For example, the GTP dependence of the binding of the pseudoknot structure of West Nile virus was not verified (4). In the case of eEF1A binding to PLMVd, we performed an experiment that aimed to address this question, and it seems not to be the case. However, we have to keep in mind that it is not excluded that a fraction of the purified eEF1A may have retained GTP essential for PLMVd interaction. Future work should be directed toward elucidating the biological consequences of PLMVd-eEF1A complex formation. It is of primary importance to confirm the formation of the protein-PLMVd complex for each putatively identified protein and then to study the biological implications of those interactions. For example, the identified putative chaperone, which had the best coverage score (15%), has no known specific activity. Consequently, it could be interesting to further investigate this protein. It is important to understand that the life cycle of PLMVd remains largely elusive, and therefore there is no solid base from which to reject a potential interacting protein yet. Northwestern blotting is a risky but interesting strategy for the identification of proteins that interact with a viroid, and further characterization of these interactions should contribute to revealing the significance of each viroid RNA-host protein complex.
Acknowledgments
We acknowledge Catherine Grenier for technical assistance during the initial stage of this project, François Bolduc for technical assistance, and Dan Thompson for kindly providing all of the peach leaves.
This work was supported by a grant from the Natural Sciences and Engineering Research Council (NSERC, Canada; grant 155219-07) to J.-P.P. The RNA group is supported by grants from the Université de Sherbrooke and the Canadian Institutes of Health Research (CIHR; grant PRG-80169). A.D. was the recipient of a predoctoral fellowship from NSERC. J.-P.P. holds the Canada Research Chair in Genomics and Catalytic RNA. M.B. and J.-P.P. are members of the Centre de Recherche Clinique Étienne-Lebel.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Barba, M., E. Ragozzino, and F. Faggioli. 2007. Pollen transmission of peach latent mosaic viroid. J. Plant Pathol. 89:287-289. [Google Scholar]
- 2.Bastin, M., and T. C. Hall. 1976. Interaction of elongation factor 1 with aminoacylated brome mosaic virus and tRNAs. J. Virol. 20:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beffa, R. S., R. M. Hofer, M. Thomas, and F. Meins, Jr. 1996. Decreased susceptibility to viral disease of [beta]-1,3-glucanase-deficient plants generated by antisense transformation. Plant Cell 8:1001-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning, K. S., J. Humphreys, W. Hobbs, G. B. Smith, and J. M. Ravel. 1990. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem. 265:17967-17973. [PubMed] [Google Scholar]
- 6.Bussiere, F., J. Lehoux, D. A. Thompson, L. J. Skrzeczkowski, and J. Perreault. 1999. Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J. Virol. 73:6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bussiere, F., J. Ouellet, F. Cote, D. Levesque, and J. P. Perreault. 2000. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cote, F., and J. P. Perreault. 1997. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol. 273:533-543. [DOI] [PubMed] [Google Scholar]
- 9.Danino, D., and J. E. Hinshaw. 2001. Dynamin family of mechanoenzymes. Curr. Opin. Cell Biol. 13:454-460. [DOI] [PubMed] [Google Scholar]
- 10.Daros, J. A., and R. Flores. 2002. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, W. G., J. L. Blackwell, P. Y. Shi, and M. A. Brinton. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81:10172-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado, S., A. E. Martinez de Alba, C. Hernandez, and R. Flores. 2005. A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 79:12934-12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desvignes, J. 1986. Peach latent mosaic and its relation to peach mosaic and peach yellow mosaic virus diseases. Acta Hortic. 193:51-57. [Google Scholar]
- 14.Ding, B., and A. Itaya. 2007. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant-Microbe Interact. 20:7-20. [DOI] [PubMed] [Google Scholar]
- 15.Domingo, C., V. Conejero, and P. Vera. 1994. Genes encoding acidic and basic class III beta-1,3-glucanases are expressed in tomato plants upon viroid infection. Plant Mol. Biol. 24:725-732. [DOI] [PubMed] [Google Scholar]
- 16.Dreher, T. W., O. C. Uhlenbeck, and K. S. Browning. 1999. Quantitative assessment of EF-1alpha.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 274:666-672. [DOI] [PubMed] [Google Scholar]
- 17.Fels, A., K. Hu, and D. Riesner. 2001. Transcription of potato spindle tuber viroid by RNA polymerase II starts predominantly at two specific sites. Nucleic Acids Res. 29:4589-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez, G., and V. Pallas. 2004. A long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with Hop stunt viroid RNA. J. Virol. 78:10104-10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gozmanova, M., M. A. Denti, I. N. Minkov, M. Tsagris, and M. Tabler. 2003. Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 31:5534-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross, S. R., and T. G. Kinzy. 2005. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12:772-778. [DOI] [PubMed] [Google Scholar]
- 21.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 22.Hassen, I. F., S. Massart, S. Roussel, J. Kummert, H. Fakhfakh, M. Marrakchi, and M. H. Jijakli. 2006. Genomic structure of new Tunisian peach latent mosaic viroid variants. Commun. Agric. Appl. Biol. Sci. 71:1257-1265. [PubMed] [Google Scholar]
- 23.Hernandez, C., and R. Flores. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 89:3711-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi, R. L., J. M. Ravel, and A. L. Haenni. 1986. Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. EMBO J. 5:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalantidis, K., M. A. Denti, S. Tzortzakaki, E. Marinou, M. Tabler, and M. Tsagris. 2007. Virp1 is a host protein with a major role in potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 81:12872-12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khacho, M., K. Mekhail, K. Pilon-Larose, A. Pause, J. Cote, and S. Lee. 2008. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell 19:5296-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniataki, E., A. E. Martinez de Alba, R. Sagesser, M. Tabler, and M. Tsagris. 2003. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9:346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez de Alba, A. E. 2000. Isolation and characterisation of viroid-binding proteins. Universidad del Pais Vasco, Bilbao, Spain.
- 29.Martinez de Alba, A. E., R. Sagesser, M. Tabler, and M. Tsagris. 2003. A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 77:9685-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motard, J., F. Bolduc, D. Thompson, and J. P. Perreault. 2008. The peach latent mosaic viroid replication initiation site is located at a universal position that appears to be defined by a conserved sequence. Virology 373:362-375. [DOI] [PubMed] [Google Scholar]
- 31.Perederina, A., N. Nevskaya, O. Nikonov, A. Nikulin, P. Dumas, M. Yao, I. Tanaka, M. Garber, G. Gongadze, and S. Nikonov. 2002. Detailed analysis of RNA-protein interactions within the bacterial ribosomal protein L5/5S rRNA complex. RNA 8:1548-1557. [PMC free article] [PubMed] [Google Scholar]
- 32.Rackwitz, H. R., W. Rohde, and H. L. Sanger. 1981. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature 291:297-301. [DOI] [PubMed] [Google Scholar]
- 33.Sikora, D., V. S. Greco-Stewart, P. Miron, and M. Pelchat. 2009. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology 390:71-78. [DOI] [PubMed] [Google Scholar]
- 34.Slobin, L. I. 1983. Binding of eucaryotic elongation factor Tu to nucleic acids. J. Biol. Chem. 258:4895-4900. [PubMed] [Google Scholar]
- 35.Thimmapuram, J., T. S. Ko, and S. S. Korban. 2001. Characterization and expression of beta-1,3-glucanase genes in peach. Mol. Genet. Genomics 265:469-479. [DOI] [PubMed] [Google Scholar]
- 36.Walker, J. L., and D. J. Oliver. 1986. Glycine decarboxylase multienzyme complex. Purification and partial characterization from pea leaf mitochondria. J. Biol. Chem. 261:2214-2221. [PubMed] [Google Scholar]
- 37.Yamaji, Y., T. Kobayashi, K. Hamada, K. Sakurai, A. Yoshii, M. Suzuki, S. Namba, and T. Hibi. 2006. In vivo interaction between tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347:100-108. [DOI] [PubMed] [Google Scholar]
- 38.Zeenko, V. V., L. A. Ryabova, A. S. Spirin, H. M. Rothnie, D. Hess, K. S. Browning, and T. Hohn. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76:5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]