Abstract
B lymphocytes converted into lymphoblastoid cell lines (LCLs) by an Epstein-Barr virus that expresses a conditional EBNA3C require complementation with EBNA3C for growth under nonpermissive conditions. Complementation with relatively large EBNA3C deletion mutants identified amino acids (aa) 1 to 506 (which includes the RBP-Jκ/CSL [RBP-Jκ] binding domain) and 733 to 909 to be essential for LCL growth, aa 728 to 732 and 910 to 992 to be important for full wild-type (wt) growth, and only aa 507 to 727 to be unimportant (S. Maruo, Y. Wu, T. Ito, T. Kanda, E. D. Kieff, and K. Takada, Proc. Natl. Acad. Sci. USA 106:4419-4424, 2009). When mutants with smaller deletions were used, only aa 51 to 400 and 851 to 900 were essential for LCL growth; aa 447 to 544, 701 to 750, 801 to 850, and 901 to 992 were important for full wt growth; and aa 4 to 50, 401 to 450, 550 to 707, and 751 to 800 were unimportant. These data reduce the EBNA3C essential residues from 68% to 40% of the open reading frame. Point mutations confirmed RBP-Jκ binding to be essential for wt growth and indicated that SUMO and CtBP binding interactions were important only for full wt growth. EBNA3C aa 51 to 150, 249 to 311, and 851 to 900 were necessary for maintaining LCL growth, but not RBP-Jκ interaction, and likely mediate interactions with other key cell proteins. Moreover, all mutants null for LCL growth had fewer S+G2/M-phase cells at 14 days, consistent with EBNA3C interaction with RBP-Jκ as well as aa 51 to 150, 249 to 311, and 851 to 900 being required to suppress p16INK4A (S. Maruo, Y. Wu, S. Ishikawa, T. Kanda, D. Iwakiri, and K. Takada, Proc. Natl. Acad. Sci. USA 103:19500-19505, 2006). We have confirmed that EBNA3C upregulates TCL1 and discovered that EBNA3C upregulates TCL1 through RBP-Jκ, indicating a central role for EBNA3C interaction with RBP-Jκ in mediating cell gene transcription.
Epstein-Barr virus (EBV) is etiologically associated with Burkitt's lymphoma, Hodgkin's lymphoma, other B- and T-cell lymphomas, nasopharyngeal carcinoma, some gastric carcinomas, and B-lymphocyte proliferative disease in immunodeficient patients (for a review, see reference 53). Latent EBV infection induces primary B lymphocytes to continuously proliferate as lymphoblastoid cell lines (LCLs) (21, 50). In proliferating lymphocytes, EBV expresses nuclear proteins EBNA-1, -2, -3A, -3B, -3C, and -LP; integral membrane proteins LMP-1, -2A, and -2B; small nonpolyadenylated RNAs EBER1 and EBER2; and BamA rightward transcripts, which encode polyadenylated RNAs and microRNAs. EBNA-1, -2, -3A, -3C, and -LP and LMP1 are essential for peripheral B-lymphocyte conversion to LCLs, whereas EBNA3B is dispensable and LMP2A is critical for conversion of less mature B lymphocytes to LCLs (for a review, see reference 28).
The experiments described here were undertaken to identify EBNA3C residues necessary for maintaining LCL growth so as to better understand the proteins through which EBNA3C enables LCL growth. Substantial evidence indicates that EBNA3C regulation of transcription through the cell transcription factor RBP-Jκ/CSL (RBP-Jκ) is essential for maintaining LCL growth (42, 43). EBNA3A, EBNA3B, and EBNA3C have a domain near their N termini through which they associate with large fractions of the cellular RBP-Jκ (25, 41, 51, 55, 62). Since EBNA2 activates promoters through RBP-Jκ (17, 20), EBNA3A, EBNA3B, and EBNA3C likely limit EBNA2 access to RBP-Jκ and differentially modulate EBNA2 effects (14, 51, 54, 60, 62). Indeed, EBNA3A and EBNA3C deletion or point mutants that are deficient in association with RBP-Jκ or in inhibition of EBNA2 activation of the EBV Cp promoter are unable to maintain LCL growth, whereas EBNA3B is unimportant for LCL growth (40-43). EBNA3A, EBNA3B, and EBNA3C also have domains that can activate and domains that can repress transcription when positioned near a promoter (4, 8, 13, 39, 51). Importantly, EBNA3C can coactivate the EBV LMP1 promoter with EBNA2, whereas EBNA3A and EBNA3B cannot (3, 24, 37, 64). An EBNA3C SUMO interaction motif (SIM) and a domain that interacts with PU.1 have been implicated in LMP1 promoter coactivation with EBNA2 (3, 24, 37, 56, 64). Moreover, decreased EBNA3C levels in an EBNA3B-deficient LCL correlate with decreased TCL1 and ITGA4 RNAs and increased Jagged 1, NCALD, and FLNA RNA levels, whereas decreased EBNA3B levels correlate with increased CXCR4 and ENTH RNA levels (10). Conditional EBNA3C inactivation results in induction of p16INKINK4A and growth inhibition (42). EBNA3C can interact with other transcription factors or modifiers of transcription, including c-myc, prothymosin alpha, histone deacetylases, CtBP, SMN, DP103, NM23-H1, SCFSkp2, and p300, and these interactions may contribute to EBNA3C's unique transcriptional effects (5, 12, 15, 18, 27, 29-31, 34, 36, 52, 58). Identification of EBNA3C residues essential for LCL growth is necessary for confirming the importance of RBP-Jκ and for evaluating the significance of these other protein interactions.
An initial analysis of the importance of EBNA3C domains for maintaining LCL growth used LCLs transformed with an EBV recombinant that expresses a conditional EBNA3C, which has the last codon fused in frame to the first codon of a 4-hydroxy-tamoxifen (4HT)-dependent mutant estrogen receptor hormone binding domain (EBNA3C-HT) (38, 42, 43). In medium without 4HT, EBNA3C-HT is gradually degraded and LCL growth slows over 7 to 14 days, stopping at days 15 to 20. Expression of other EBNAs, LMP1, CD21, CD23, or c-myc does not change over this period (10, 42). EBV EBNA3C-HT-transformed LCLs transfected with an oriP-based vector that expresses wild-type (wt) EBNA3C continue to grow after transfer to medium without 4HT, whereas EBNA3A, EBNA3B, and control expression vector-transfected LCLs are unable to maintain growth (42). Surprisingly, 11 of 17 EBNA3C mutants, with deletions as large as 129 amino acids (aa), fail to maintain LCL growth, consistent with the possibility that 68% of EBNA3C is essential for maintaining LCL growth (43). As the initial results of these experiments emerged, the current study of 19 EBNA3C mutants with deletions smaller than 77 aa was initiated to more precisely define EBNA3C residues important for LCL growth.
MATERIALS AND METHODS
Cells.
BJAB is an EBV-negative B-lymphoma cell line (45). EBNA3C-HT-infected LCL clones 15, 19, and 51 were studied and found to be dependent on 4HT for continued growth. Clone 51 was used in these experiments to verify that the results obtained are not clone dependent and grew almost as rapidly as clone 19, which was used in previous complementation analyses (42, 43). LCLs were maintained in RPMI 1640 medium (Gibco, Grand Island, NY) supplemented with 15% fetal bovine serum (Gemini Bio, West Sacramento, CA), l-glutamine, streptomycin, penicillin, and 400 nM 4HT (Sigma, St. Louis, MO). Other cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, l-glutamine, streptomycin, and penicillin. Viable cell numbers were determined using a hemocytometer and trypan blue exclusion. Cultures were mycoplasma free according to the MycoAlert luciferase assay (Lonza, Rockland, ME).
Plasmids.
pCEP-EBNA3C (E3C), -E3CΔ4-50, -E3CΔ51-100, -E3CΔ101-150, -E3CΔ151-200, -E3C209mJκ, -E3CΔ201-248, -E3CΔ249-311, -E3CΔ311-364, -E3CΔ367-400, -E3CΔ401-450, -E3CΔ447-500, -E3CΔ501-544, -E3C509mSIM, -E3CΔ550-625, -E3CΔ632-707, -E3CΔ701-750, -E3C728mCtBP, -E3CΔ751-800, -E3CΔ801-850, -E3CΔ851-900, -E3CΔ901-950, -E3CΔ951-992, and green fluorescence protein (GFP) are expression cassettes for GFP, EBNA3C wt, or EBNA3C mutant proteins in the pCEP4 (Invitrogen, Carlsbad, CA) oriP plasmid (14, 40). EBNA3C wt or EBNA3C mutant expression constructs were made by subcloning the SalI fragments containing simian virus 40 promoter-driven expression cassettes from pSG5 (Stratagene, La Jolla, CA) into SalI-digested pCEP4 vector. Constructions with point mutations disrupting specific interacting proteins have been previously described: EBNA3C 209mJκ (T209FGC212 mutated to A209AAA212 [62]), EBNA3C 509mSIM (D509VIEVID515 mutated to A509VIAVIA515 [56]), and EBNA3C 728mCtBP (P728LDLS732 mutated to A728LDAS732 [59]). All constructs were verified by sequencing.
Complementation assay.
EBNA3C-HT-infected LCLs (7.5 × 106) were transfected with oriP plasmid DNA expressing wt EBNA3C, EBNA3C mutants, or GFP as a control. Log-phase LCLs in 4HT medium were harvested, washed with complete medium, resuspended in 400 μl of complete medium with 15 μg of DNA in a cuvette (0.4-cm gap; Bio-Rad, Hercules, CA) for 10 min at 25°C, pulsed with 220 V at 960 μF, and cultured in 14 ml of LCL-conditioned medium with 4HT for 2 to 3 days. LCL transfection efficiencies were 20 to 40%, measured by GFP expression after transfection with the GFP-oriP plasmid. Cells were washed twice in warm medium, and 1 × 106 cells were cultured in 10 ml of complete medium with or without 4HT in a 25-cm2 culture flask. Every 3 to 7 days, cells were counted and split to 1 × 105 cells/ml.
Immunoprecipitations and Western blot analysis.
Log-phase BJAB cells (7.5 × 106) were electroporated with up to 15 μg oriP plasmid expressing wt EBNA3C, EBNA3C mutants, or a control GFP as described above for LCLs. After 48 h, cells were lysed at 4°C by vortexing in 150 mM NaCl, 1% NP-40, 50 mM Tris (pH 7.4), 2 mM EDTA with protease inhibitors (10 μg/ml aprotinin, 10 μg/ml leupeptin, and 0.5 μM phenylmethylsulfonylfluoride) and centrifuged to remove insoluble debris. The supernatant was incubated overnight with anti-RBP-Jκ rabbit antiserum (54) and protein A-Sepharose beads (Sigma) at 4°C. Beads were washed four times with lysis buffer. Total cell lysates or immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto a nitrocellulose membrane (Bio-Rad), and probed with anti-EBNA3C A10 monoclonal antibody (44), sheep polyclonal anti-EBNA3C antibody F125P (Exalpha Biologicals, Watertown, MA), rat RBP-Jκ-specific monoclonal antibody (T6709, Cosmo Bio, Tokyo, Japan), anti-TCL1 monoclonal antibody (Cell Signaling, Danvers, MA), or antitubulin monoclonal antibody (Sigma). After extensive washing, proteins were detected using species-specific horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and chemiluminescence reagent (Perkin Elmer, Waltham, MA).
Reporter assays.
BJAB cells (1 × 107) in log-phase growth were electroporated with 0.5 μg of pGK-βgal, 10 μg of pLuc-Cp reporter construct, and 1 μg of pSG5-EBNA2 alone or in combination with 10 μg of oriP plasmid expressing EBNA3C wt or mutant constructs (37). After 48 h, cells were lysed in reporter lysis buffer (luciferase assay system; Promega, Madison, WI). Clarified lysate luciferase and β-galactosidase (Galacto-Light Plus System; Applied Biosystems, Bedford, MA) activities were measured using an Optocomp I luminometer (MGM Instruments, Hamden, CT).
Flow cytometry analysis.
EBNA3C-HT LCLs transfected with control GFP, wt EBNA3C, or EBNA3C mutants were cultured in medium without 4HT. After 14 days, cells were fixed with 70% ethanol and stained with propidium iodide solution (50 μg/ml propidium iodide, 100 μg/ml RNase, and 3.8 mM sodium citrate in phosphate-buffered saline). Cell cycle distribution profiles were analyzed with a FACSCalibur system and CellQuest software (BD Biosciences, San Jose, CA) (44).
RESULTS
EBNA3C residues critical for LCL growth.
To identify EBNA3C residues important for maintaining LCL growth, wt EBNA3C and deletion or point mutants (Fig. 1) were expressed using oriP vectors and assessed for complementation of EBV EBNA3C-HT-transformed LCL growth under nonpermissive conditions for EBNA3C-HT (42, 43). EBNA3C-HT LCLs grew continuously in medium with 4HT, but growth slowed and stopped in medium without 4HT, whereas wt EBNA3C-transfected LCLs grew at similar rates in medium with or without 4HT (Fig. 1 and 2). Cells transfected with a null control oriP-based GFP expression vector continued to grow in the presence of 4HT but stopped growing and began dying at 14 to 20 days in the absence of 4HT (42, 43) (Fig. 2A). Western blotting showed that EBNA3C-HT was substantially decreased at day 7 and barely detectable by day 14, as previously described (42). Thus, EBNA3C-HT stability is 4HT dependent and EBNA3C expression in trans is required for continued LCL growth in medium without 4HT.
FIG. 1.
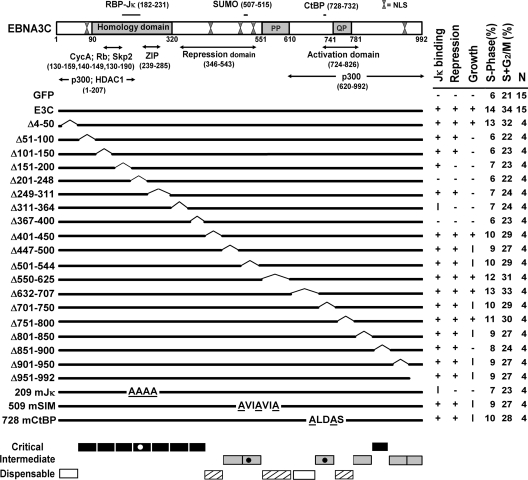
Schematic diagram of EBNA3C domains and mutants with summarized reverse genetic and biochemical data. Within the homology domain (aa 90 to 320), which has the highest level of identity (23 to 30%) among the EBNA3 proteins, aa T209FGC212 are critical for RBP-Jκ association. SUMO-1 and SUMO-3 interaction require D509VIEVID515, and P728LDLS732 mediates CtBP binding. Proline-rich (PP; aa 551 to 610) and glutamine-proline-rich (QP; aa 741 to 781) domains are indicated. Repression (aa 346 to 543) and activation (aa 724 to 826) domains were defined based on their effects when targeted to promoters by fusion to the Gal4 DNA binding domain. Putative leucine zipper (ZIP) and multiple functional nuclear localization signal (NLS) sequences are indicated. The results of wt EBNA3C (E3C) or mutant EBNA3C association with RBP-Jκ, repression of EBNA2 transcriptional activation of the Cp promoter, cell cycle distribution at day 14, and effects on LCL growth under EBNA3C-HT inactivation condition are shown in columns on the right side of the figure. +, wt; −, null; I, intermediate; N, number of complementation experiments. The regions of EBNA3C which are critical (black bars), intermediate (gray bars), and nearly (hashed bars) or completely (white bars) dispensable for LCL growth are indicated at the bottom.
FIG. 2.
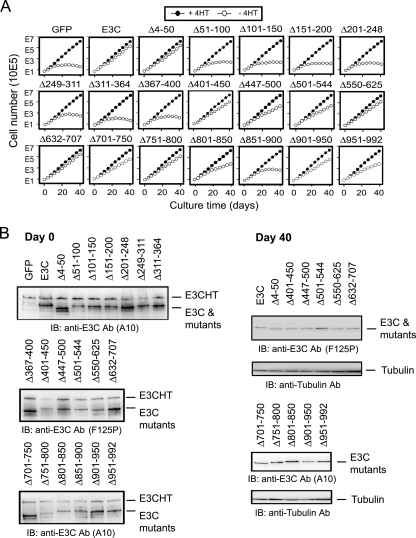
Growth complementation effects of wt or mutant EBNA3C in EBNA3C-HT LCLs under EBNA3C-HT inactivation condition. (A) EBNA3C-HT LCL cells were transfected with 15 μg of the oriP plasmid expressing control GFP, wt EBNA3C (E3C), or the indicated EBNA3C mutant. After 2 days, the cells were washed and resuspended at 1 × 106 cells/10 ml of complete medium with (+4HT) or without (−4HT) 4HT in 25-cm2 culture flasks (day 0). Every 3 or 4 days, cells were counted and pelleted and 1 × 106 cells were resuspended in 10 ml fresh medium. Total numbers of viable cells derived from the initial cultures were calculated and plotted at each time point. Data shown are representative of results from four experiments (Table 1). (B) Protein lysates made from each complementation assay on day 0 (2 days after transfection) and on day 40 for complementations displaying at least intermediate growth. Western blot analyses were performed with anti-EBNA3C antibodies (A10 or F125P) to detect wt or mutant EBNA3C. IB, immunoblotting; Ab, antibody.
EBNA3C-HT LCLs were transfected with oriP-based wt or mutant EBNA3C expression vectors and cultured in medium without 4HT for 40 days. For each mutant, four independent growth assays were performed (Table 1). Based on cumulative results, the growth phenotype was assigned as wt, intermediate, or null (Fig. 1). EBNA3C deletion mutants Δ4-50 and Δ632-707 enabled EBNA3C-HT LCLs to grow continuously in medium without 4HT at a rate indistinguishable from that of wt EBNA3C-transfected cells (Fig. 1 and 2A and Table 1). EBNA3C Δ401-450, Δ550-625, and Δ751-800 also supported robust growth of EBNA3C-HT LCLs in the absence of 4HT at a rate that was either equivalent to or just below that seen with wt EBNA3C-transfected cells (Fig. 1 and 2A and Table 1). EBNA3C mutants Δ447-500, Δ501-544, Δ701-750, Δ801-850, Δ901-950, and Δ951-992 also supported growth in medium without 4HT but at a noticeably reduced growth rate compared to wt EBNA3C (Fig. 1 and 2A and Table 1). In contrast, EBNA3C Δ51-100, Δ101-150, Δ151-200, Δ201-248, Δ249-311, Δ311-364, Δ367-400, and Δ851-900 were similar to the GFP vector control in not maintaining EBNA3C-HT LCL growth in medium without 4HT (Fig. 1 and 2A and Table 1). The growth curves shown in Fig. 2A are representative of results from four independent experiments with each EBNA3C mutant (Table 1). Overall, these data indicate that EBNA3C aa 51 to 400 and 851 to 900 are essential for LCL growth, while aa 447 to 544, 701 to 750, 801 to 850, and 901 to 992 are important for full wt growth rates, and aa 4 to 50, 401 to 450, 550 to 707, and 751 to 800 are dispensable for LCL growth (Fig. 1). Thus, these data reduce the EBNA3C residues essential for LCL growth from 68% to 40% of the EBNA3C open reading frame.
TABLE 1.
Summary of wt and mutant EBNA3C transcomplemenation experiments
| Mutant or EBNA construct | Growtha |
||
|---|---|---|---|
| wt | INT | None | |
| GFP | 15 | ||
| EBNA3C | 15 | ||
| Δ4-50 | 4 | ||
| Δ51-100 | 4 | ||
| Δ101-150 | 4 | ||
| Δ151-200 | 4 | ||
| Δ201-248 | 4 | ||
| Δ249-311 | 4 | ||
| Δ311-364 | 4 | ||
| Δ367-400 | 4 | ||
| Δ401-450 | 1 | 3 | |
| Δ447-500 | 4 | ||
| Δ501-544 | 4 | ||
| Δ550-625 | 1 | 3 | |
| Δ632-707 | 4 | ||
| Δ701-750 | 4 | ||
| Δ751-800 | 1 | 3 | |
| Δ801-850 | 4 | ||
| Δ851-900 | 4 | ||
| Δ901-950 | 4 | ||
| Δ951-992 | 3 | 1 | |
| 209mJK | 4 | ||
| 509mSIM | 4 | ||
| 728mCtBP | 4 | ||
For each mutant, the results of four independent complementation experiments were scored as wild-type (wt), intermediate (INT), or no growth (None). Based on these results, summary phenotypes were assigned as summarized in Fig. 1.
The inability of some EBNA3C deletion mutants to maintain LCL growth is not due to abnormal expression levels. In experiments similar to those shown in Fig. 2A, expression of EBNA3C Δ51-100, Δ101-150, Δ151-200, Δ201-248, Δ249-311, Δ311-364, Δ367-400, or Δ851-900 did not differ significantly from that of wt EBNA3C at 2 days after transfection (Fig. 2B, day 0; compare expression levels of EBNA3C mutants to the internal control of EBNA3C-HT expression in each lane). Expression of EBNA3C mutants that partially or completely maintained cell growth was also similar to wt EBNA3C expression assessed at day 0 and day 40 after complementation (Fig. 2B). The fact that EBNA3C-HT LCLs grown in medium without 4HT and complemented with mutants that supported cell growth intrinsically selected for similar levels of wt or mutant EBNA3C expression from oriP-based plasmids at day 40 is evidence that these mutants are similar to wt EBNA3C in efficiency in maintaining LCL growth.
Cell cycle distribution of EBNA3C-HT-complemented LCLs.
The cell cycle distribution of EBV EBNA3C-HT-transformed LCLs was evaluated at day 14 after complementation and shift to medium without 4HT to determine if specific EBNA3C residues had effects on cell cycle entry, cell cycle exit, or apoptotic cell death. Six percent of EBV EBNA3C-HT LCLs growing in medium without 4HT and complemented with wt EBNA3C were hypodiploid, 60% were in G0/G1, 14% were in S, and 20% were in G2/M, whereas 10% of cells complemented with GFP null control plasmid were hypodiploid, 69% were in G0/G1, 6% were in S, and 15% were in G2/M (Table 2 and Fig. 1). EBNA3C mutants that were wt in maintaining EBNA3C-HT LCL growth in medium without 4HT were similar to wt EBNA3C in cell cycle distribution, whereas EBNA3C mutants which were similar to GFP in not maintaining cell growth were similar to GFP in cell cycle distribution. EBNA3C mutants that were intermediate in maintaining cell growth were intermediate in cell cycle distribution (Table 2 and Fig. 1). Thus, these data are consistent with similar roles for EBNA3C essential domains in maintaining LCL growth and cell cycle distribution.
TABLE 2.
Cell cycle profiles of EBNA3C-HT LCLs transfected with wt EBNA3C or EBNA3C mutants on day 14a
| Mutant or EBNA construct | % of cells at growth stage |
|||
|---|---|---|---|---|
| Sub-G1 | G0/G1 | S | G2/M | |
| GFP | 10 | 69 | 6 | 15 |
| EBNA3C | 6 | 60 | 14 | 20 |
| Δ4-50 | 6 | 62 | 13 | 19 |
| Δ51-100 | 9 | 69 | 6 | 16 |
| Δ101-150 | 9 | 68 | 6 | 17 |
| Δ151-200 | 9 | 68 | 7 | 16 |
| Δ201-248 | 9 | 69 | 6 | 16 |
| Δ249-311 | 8 | 68 | 7 | 17 |
| Δ311-364 | 7 | 69 | 7 | 17 |
| Δ367-400 | 7 | 70 | 6 | 17 |
| Δ401-450 | 6 | 65 | 10 | 19 |
| Δ447-500 | 6 | 67 | 9 | 18 |
| Δ501-544 | 6 | 65 | 10 | 19 |
| Δ550-625 | 6 | 63 | 12 | 19 |
| Δ632-707 | 6 | 61 | 13 | 20 |
| Δ701-750 | 6 | 65 | 10 | 19 |
| Δ751-800 | 6 | 64 | 11 | 19 |
| Δ801-850 | 7 | 66 | 9 | 18 |
| Δ851-900 | 7 | 69 | 8 | 16 |
| Δ901-950 | 6 | 67 | 9 | 18 |
| Δ951-992 | 7 | 66 | 9 | 18 |
EBNA3C-HT transfected with control GFP, wt EBNA3C, or the indicated EBNA3C mutants were cultured in medium without 4HT. Cells were fixed and stained with propidium iodide, and cell cycle profile distribution patterns were analyzed with a FACSCalibur on day 14. Results for control GFP, wt EBNA3C, and EBNA3C mutants are shown, together with the percentage of cells in each stage of the cell cycle.
EBNA3C interaction with RBP-Jκ is essential for LCL growth, whereas EBNA3C interaction with SUMO or CtBP is important for full wt growth.
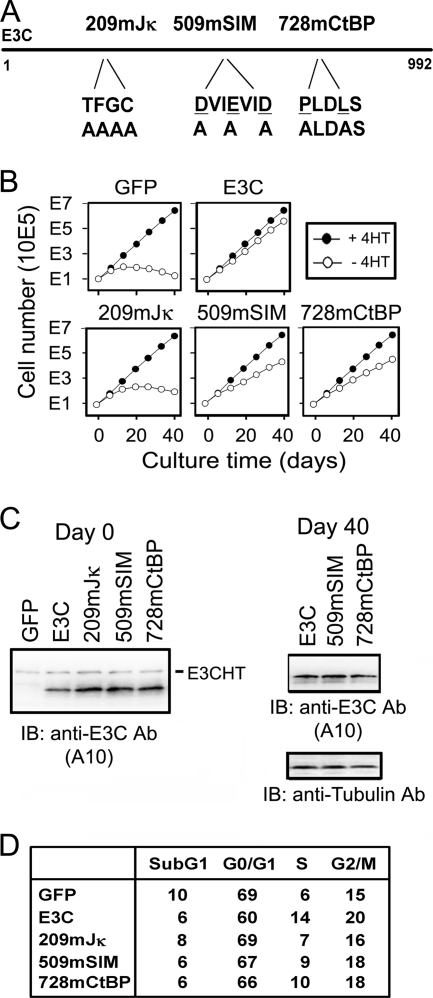
The importance of EBNA3C interactions with RBP-Jκ, SUMO, or CtBP in maintaining LCL growth was also tested in the EBNA3C-HT complementation assay. EBNA3C interaction site point mutants that are deficient in interactions with RBP-Jκ (T209FGC212 mutated to A209AAA212 [62]), SUMO (D509VIEVID515 mutated to A509VIAVIA515 [56]), or CtBP (P728LDLS732 mutated to A728LDAS732 [59]) (Fig. 3A) were tested for maintenance of LCL growth in medium without 4HT. The EBNA3C RBP-Jκ point mutant (EBNA3C 209mJκ) did not support EBNA3C-HT LCL growth in medium without 4HT. LCLs stopped growing by day 20 and thereafter decreased in number (Fig. 3B). In contrast, the EBNA3C SUMO (EBNA3C 509mSIM) and CtBP (EBNA3C 728mCtBP) interaction site mutants supported continued LCL growth at moderately reduced rates (Fig. 3B). In complementation tests, all three point mutants were expressed at levels similar to that of wt EBNA3C at day 0 (Fig. 3C), but only the SUMO and CtBP mutants grew and could be assayed at day 40. At day 40, EBNA3C 509mSIM and EBNA3C 728mCtBP were expressed at levels similar to that of wt EBNA3C (Fig. 3C).
FIG. 3.
Growth characteristics of EBNA3C-HT LCLs transfected with EBNA3C point mutants. (A) Diagram of EBNA3C point mutants, indicating the RBP-Jκ binding mutant, SUMO binding mutant, and CtBP binding mutant. (B) Growth curves of EBNA3C-HT LCLs transfected with 15 μg of the oriP plasmid expressing control GFP, wt EBNA3C (E3C), or the indicated EBNA3C point mutant. These data are representative of results from four experiments (Table 1). (C) Western blotting for EBNA3C from complemented EBNA3C-HT LCLs on day 0 (2 days after transfection) and day 40. (D) Percentage of EBNA3C-HT LCLs in each stage of cell cycle after complementation with the indicated construct cultured in the absence of 4HT for 14 days.
Fourteen days after transfection with EBNA3C 209mJκ, 23% of cells were in S+G2/M, similar to the 21% of S+G2/M cells in the GFP null control and substantially less than the 34% of cells in S+G2/M in the wt EBNA3C-transfected cells. EBNA3C 509mSIM- or EBNA3C 728mCtBP-transfected cells were intermediate, with 27 to 28% in S+G2/M at day 14 (Fig. 3D). Thus, EBNA3C interaction with RBP-Jκ is essential for continued LCL cell growth and cell cycle progression, whereas EBNA3C interactions with SUMO or CtBP are less significant determinants of cell growth and cell cycle progression.
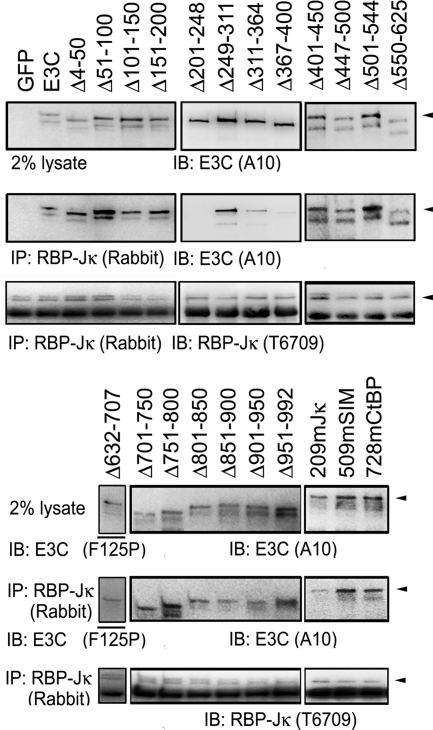
To further examine the effect of EBNA3C mutant interactions with RBP-Jκ on LCL growth, EBNA3C mutants were compared to wt EBNA3C in their association with RBP-Jκ, in their inhibition of EBNA2 activation of the EBV Cp promoter through RBP-Jκ, and in their maintenance of LCL growth. EBNA3C mutant association with RBP-Jκ was assessed in BJAB cells transfected, in parallel, with a vector expressing wt EBNA3C as a positive control and GFP as a negative control. After 48 h, cells were lysed and EBNA3C association with RBP-Jκ was evaluated by RBP-Jκ immunoprecipitation and Western blotting for RBP-Jκ and EBNA3C. Although wt EBNA3C and EBNA3C mutants differed slightly in expression in any one experiment, persistent differences were not noted (Fig. 4 and data not shown). Further, RBP-Jκ antibody immunoprecipitated similar amounts of RBP-Jκ and coimmunoprecipitated about 2% of wt EBNA3C. In contrast, EBNA3C Δ201-248, EBNA3C Δ311-364, EBNA3C Δ367-400, and EBNA3C 209mJκ were partially or substantially deficient in their immunoprecipitation with RBP-Jκ (Fig. 4). These data agree with previous results and indicate that EBNA3C aa 201 to 248, 311 to 364, 367 to 400, and 209 to 212 are important for EBNA3C association with RBP-Jκ. EBNA3C deleted for aa 366 to 400 was less deficient in RBP-Jκ coimmunoprecipitation in previous experiments (43). Most importantly, all EBNA3C mutants that were deficient in RBP-Jκ association were null for maintaining LCL growth (Fig. 1, 2, and 4).
FIG. 4.
EBNA3C residues required for RBP-Jκ association. BJAB cells were transfected with 15 μg of the oriP plasmid expressing wt EBNA3C (E3C), EBNA3C mutants, or control GFP. At 48 h after transfection, protein complexes were immunoprecipitated (IP) with polyclonal rabbit sera raised against RBP-Jκ and Western blotted (IB) with anti-EBNA3C antibodies (A10 or F125P) or RBP-Jκ-specific rat monoclonal antibody (T6709). Arrowheads indicate the full-sized protein identified by the immunoblot.
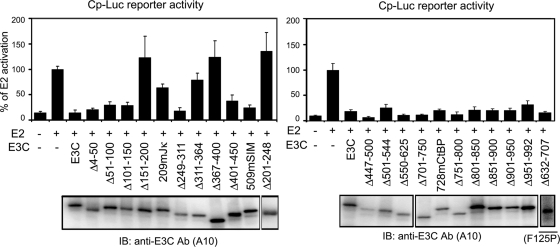
Analysis of EBNA3C residues critical for blocking EBNA2 activation of the EBNA Cp promoter in transient-transfection assays identified EBNA3C Δ151-200, Δ201-248, Δ311-364, Δ367-400, and 209mJκ as deficient in blocking EBNA2 activation (Fig. 5). All EBNA3C mutants deficient in RBP-Jκ association were defective in blocking EBNA2 activation of the Cp reporter and failed to maintain LCL growth (Fig. 1, 4, and 5). Mutants competent for RBP-Jκ association but defective in blocking EBNA2 activation of the Cp reporter assay (e.g., Δ151-200) also failed to maintain LCL growth.
FIG. 5.
EBNA3C Δ151-200, 209mJκ, Δ201-248, Δ311-364, and Δ367-400 are deficient in inhibition of EBNA2 activation of a multimerized C promoter. BJAB cells were transfected with the pLuc-Cp reporter construct containing eight copies of the RBP-Jκ binding site along with EBNA2 alone or with the indicated EBNA3C construct. Transfection efficiency was monitored by β-galactosidase activity using cotransfected pGK-βgal. The results are representative of results from three independent experiments.
In contrast, EBNA3C Δ51-100, Δ101-150, Δ249-311, and Δ851-900 were wt in association with RBP-Jκ and in repression of EBNA2 activation of Cp but were null for maintaining LCL growth. Thus, EBNA3C aa 51 to 100, 101 to 150, 249 to 311, and 851 to 900 are prime candidates to mediate essential interactions with proteins other than RBP-Jκ.
EBNA3C interaction with RBP-Jκ upregulates TCL1 protein expression.
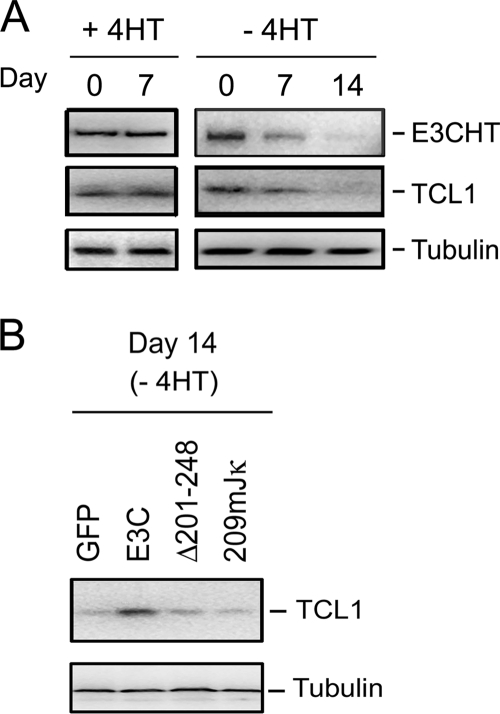
Since an LCL with low EBNA3C expression was deficient in T-cell leukemia 1 (TCL1) RNA (10) and TCL1 is implicated in the pathogenesis of aggressive B-cell chronic lymphocytic leukemia (49), TCL1 could be important for LCL growth. We therefore investigated whether TCL1 protein levels change following EBNA3C inactivation in EBNA3C-HT-transformed LCLs. Indeed, over 14 days in medium without 4HT, levels of TCL1 protein progressively decreased until TCL1 was barely detectable (Fig. 6). Comparison of RNA abundances in LCLs maintained in medium with or without 4HT or complemented with wt EBNA3C revealed that TCL1 levels correlate with EBNA3C expression (unpublished observation).
FIG. 6.
EBNA3C upregulation of TCL1 requires RBP-Jκ interaction. (A) Western blot for EBNA3C-HT (E3CHT), TCL1, and tubulin of EBNA3C-HT LCLs at days 0 and 7 after growth in the presence of 4HT and at days 0, 7, and 14 after growth in the absence of 4HT. (B) TCL1 and tubulin Western blots of EBNA3C-HT LCLs grown for 14 days in the absence of 4HT after transfection with GFP, EBNA3C, or the indicated EBNA3C mutants expressed from an oriP vector.
To investigate whether EBNA3C upregulated TCL1 expression through RBP-Jκ, TCL1 levels in EBV EBNA3C-HT-transformed LCLs growing in medium without 4HT for 14 days after complementation with wt EBNA3C, GFP null control, EBNA3C Δ201-248, or EBNA3C 209mJκ were determined. wt EBNA3C expression sustained TCL1 protein levels, whereas GFP expression failed to sustain TCL1 protein levels. Importantly, EBNA3C Δ201-248, which does not associate with RBP-Jκ, and EBNA3C 209mJκ, which is deficient in interactions with RBP-Jκ, were expressed at wt EBNA3C levels at day 0 but failed to sustain TCL1 expression (Fig. 6). These data indicate that EBNA3C upregulates TCL1 through interaction with RBP-Jκ.
DISCUSSION
The experiments presented here were undertaken in conjunction with recently reported experiments (43) and use smaller deletion mutants to more precisely identify those EBNA3C residues that are essential, less important, or dispensable for maintaining LCL growth. In finding that EBNA3C deleted for aa 4 to 50, 401 to 450, 447 to 500, 501 to 544, 550 to 625, 632 to 707, 701 to 750, 751 to 800, and 901 to 950 can support LCL growth at or near wt rates and that EBNA3C deleted for aa 801 to 850 and 951 to 992 can support growth at substantially reduced rates, we have narrowed down the essential EBNA3C residues to 40% of the EBNA3C open reading frame.
Our data support a trend evident in previous data (43) which link an extended domain that can affect RBP-Jκ interactions to a null growth phenotype. As was observed for EBNA3A (41) and for EBNA3C (43), EBNA3C deletion or point mutants were deficient in RBP-Jκ association or in inhibiting EBNA2 activation of the Cp promoter and failed to support LCL growth. Further, EBNA3C deleted for aa 151 to 200 was wt in RBP-Jκ association but deficient in inhibiting Cp activation and failed to maintain LCL growth, further supporting the notion that EBNA3C repression of EBNA2-mediated Cp activation is a more sensitive indicator of an EBNA3C activity through RBP-Jκ that is necessary for EBNA3C-mediated cell growth. Since RBP-Jκ is a transcription factor which interacts specifically with accessory factors, including repressors and activators (7, 9, 32, 33), the assay may be detecting a transcriptional effect relevant to cell growth, even though EBNA3C repression of EBNA2 activation of the Cp promoter itself may not be essential to EBNA3C's unique role in supporting LCL growth, since inhibition of EBNA2 activation of Cp is a shared function of EBNA3A, EBNA3B, and EBNA3C and EBNA3C loss cannot be overcome by EBNA3A or EBNA3B overexpression (42).
Nevertheless, these and previous EBNA3C deletion and point mutation data (42) indicate that EBNA3C interactions through RBP-Jκ are essential for LCL growth and that these effects are likely to be transcriptional. EBNA3C aa 280 to 525 have strong repressive effects, aa 580 to 992 have modest repressive effects, and aa 724 to 780 have activating effects when positioned next to a promoter by fusion to a Gal4 DNA binding domain; these effects are likely to be important for EBNA3C transcriptional regulation through RBP-Jκ (42). EBNA3C aa 580 to 992 include the CtBP binding site; however, an EBNA3C mutant deleted for aa 701 to 750 or null point mutant 728ALDAS in the CtBP repressor binding site (42) maintained similar levels of moderately impaired continued LCL growth, indicating that the interaction with the CtBP repressor is important, but not essential, for LCL growth. This contrasts with the striking CtBP binding site-dependent oncogene-like effects noted with EBNA3C overexpression in primary rodent fibroblasts, in which CtBP or CtIP may have an important role (11, 47, 48, 59). Similarly, an EBNA3C mutant deleted for aa 501 to 544, which includes part of the strong repressive domain, and a mutant with null point mutation 509AVIAVIA in the SUMO interaction motif in this domain (42) maintained similar levels of moderately impaired LCL growth, indicating that the SUMO interaction motif is also important, but not essential, for LCL growth (Fig. 1 and 3).
Although EBNA2 upregulation of c-myc through RBP-Jκ is critical for LCL growth (1, 14, 26, 57, 63) and EBNA3C can associate with c-myc (5), c-myc levels are unaffected by EBNA3C inactivation in EBV EBNA3C-HT-transformed LCLs (43), indicating that c-myc is not the critical target of EBNA3C effects through RBP-Jκ.
Instead, our data indicate that EBNA3C regulation of transcription through RBP-Jκ increases TCL1 protein expression in LCLs. EBNA3C deletion and point mutants deficient in RBP-Jκ interaction were unable to restore TCL1 levels or maintain LCL growth. Indeed, following the switch of EBNA3C-HT LCLs to medium without 4HT, TCL1 protein levels fell in parallel with slowed cell growth. LCL growth is fundamentally dependent on EBNA2 upregulation of c-myc, and c-myc-driven lymphocyte proliferation requires antiapoptotic support. Given the prominent prosurvival role of TCL1 interactions with AKT in B-cell chronic lymphocytic leukemia and T-cell leukemia and lymphoma (6, 35, 49), TCL1 may indeed be important in the effects of EBNA3C on LCL growth, possibly by also increasing AKT-mediated downmodulation of c-myc transcriptional effects (65). The role of TCL1 can be tested by assessing the effects of a short hairpin RNA knockdown of TCL1 in LCLs or by assessing the extent to which forced TCL1 expression rescues growth of EBV EBNA3C-HT-transformed LCLs grown in medium without 4HT.
Together with the previous EBNA3C genetic analyses (65), the current analyses substantially limit the EBNA3C residues essential for cell growth and thereby also identify candidate residues for interactions with cell proteins other than RBP-Jκ that are likely to mediate EBNA3C growth effects. EBNA3C aa 51 to 100, 100 to 150, 249 to 311, and 851 to 900 were identified in this study and EBNA3C aa 130 to 159, 251 to 300, and 827 to 909 were identified in a previous study (43) as residues required for LCL growth but not for RBP-Jκ interaction. These EBNA3C residues are likely to interact with proteins, other than RBP-Jκ, which are essential for the EBNA3C effects on cell growth and merit further genetic and biochemical investigation to identify their binding partners. EBNA3C aa 51 to 150 include residues implicated in cullin and cyclin interactions (30, 31), which may well be important for cell growth. Thus, transcriptional regulation of virus and cell genes through RBP-Jκ is a key, but likely nonexclusive, mediator of the EBNA3C effects in maintaining LCL growth. Until such genetic and biochemical investigations are complete, we cannot exclude the possibility that a candidate residue for interaction with other cell proteins is affecting intra- or intermolecular interactions through RBP-Jκ.
Surprisingly, EBNA3C mutations that were null for maintaining LCL growth through RBP-Jκ interaction as well as through aa 51 to 100, 100 to 150, 249 to 311, and 851 to 900 result in the same fluorescence-activated cell sorter analysis cell cycle phenotype, with more cells in G0/G1 and fewer in S+G2/M, as with EBNA3C inactivation. Since previous investigations of the time course of growth arrest following EBNA3C inactivation indicate that p16INK4A RNA and protein are progressively induced, pRb becomes progressively hypophosphorylated, cell cycle entry is inhibited, and cyclin A protein levels decrease (42), we are now investigating whether these null complementation mutants have similar effects on p16INK4A induction. In some instances, this may be due to intra- rather than intermolecular interactions of these candidate residues with other EBNA3C residues essential for LCL growth. Although EBV LMP1 expression can repress p16INK4A (46, 61) and EBNA3C can be required for wt LMP1 levels (2, 39), LMP1 levels did not decrease following EBNA3C inactivation in the EBV EBNA3C-HT-transformed LCLs (42). Further experiments using the EBNA3C-HT EBV genome to transform peripheral blood B cells from genetically deficient p16INK4A individuals would clarify the potentially central role of p16INK4A in causing growth arrest upon EBNA3C inactivation (19).
Analogous p16INK4A-repressive effects may be mediated by EBNA3A, since p16INK4A is also induced in cells infected with an EBV that has a null mutation in EBNA3A expression or conditional EBNA3A inactivation in EBV EBNA3A-HT LCLs has similar effects on cell cycle distribution (22, 40). Since p16INK4A transcription is frequently repressed by H3K27 trimethylation or dimethylation and bmi-1 or other Polycomb proteins (16, 23), we speculate that EBNA3C and EBNA3A may both be necessary for effective p16INK4A repression.
Acknowledgments
This research was supported by grants from the National Cancer Institute of the USPHS (CA47006 and CA87661).
Footnotes
Published ahead of print on 23 September 2009.
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., D. H. Crawford, and J. A. Thomas. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361-369. [DOI] [PubMed] [Google Scholar]
- 3.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajaj, B. G., M. Murakami, Q. Cai, S. C. Verma, K. Lan, and E. S. Robertson. 2008. Epstein-Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J. Virol. 82:4082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boccellato, F., E. Anastasiadou, P. Rosato, B. Kempkes, L. Frati, A. Faggioni, and P. Trivedi. 2007. EBNA2 interferes with the germinal center phenotype by downregulating BCL6 and TCL1 in non-Hodgkin's lymphoma cells. J. Virol. 81:2274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borggrefe, T., and F. Oswald. 2009. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 66:1631-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourillot, P. Y., L. Waltzer, A. Sergeant, and E. Manet. 1998. Transcriptional repression by the Epstein-Barr virus EBNA3A protein tethered to DNA does not require RBP-Jkappa. J. Gen. Virol. 79:363-370. [DOI] [PubMed] [Google Scholar]
- 9.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 10.Chen, A., B. Zhao, E. Kieff, J. C. Aster, and F. Wang. 2006. EBNA-3B- and EBNA-3C-regulated cellular genes in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 80:10139-10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P.-L., F. Liu, S. Cai, X. Lin, A. Li, Y. Chen, B. Gu, E. Y.-H. P. Lee, and W.-H. Lee. 2005. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhuri, T., S. C. Verma, K. Lan, M. Murakami, and E. S. Robertson. 2007. The ATM/ATR signaling effector Chk2 is targeted by Epstein-Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J. Virol. 81:6718-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cludts, I., and P. J. Farrell. 1998. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J. Virol. 72:1862-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper, A., E. Johannsen, S. Maruo, E. Cahir-McFarland, D. Illanes, D. Davidson, and E. Kieff. 2003. EBNA3A association with RBP-Jκ down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J. Virol. 77:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil, J., and G. Peters. 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell Biol. 7:667-677. [DOI] [PubMed] [Google Scholar]
- 17.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 19.Hayes, M. J., A. Koundouris, N. Gruis, W. Bergman, G. G. Peters, and A. J. Sinclair. 2004. p16(INK4A)-independence of Epstein-Barr virus-induced cell proliferation and virus latency. J. Gen. Virol. 85:1381-1386. [DOI] [PubMed] [Google Scholar]
- 20.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 21.Henle, W., V. Diehl, G. Kohn, H. Zur Hausen, and G. Henle. 1967. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science 157:1064-1065. [DOI] [PubMed] [Google Scholar]
- 22.Hertle, M. L., C. Popp, S. Petermann, S. Maier, E. Kremmer, R. Lang, J. Mages, and B. Kempkes. 2009. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathog. 5:e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 24.Jiménez-Ramírez, C., A. J. Brooks, L. P. Forshell, K. Yakimchuk, B. Zhao, T. Z. Fulgham, and C. E. Sample. 2006. Epstein-Barr virus EBNA-3C is targeted to and regulates expression from the bidirectional LMP-1/2B promoter. J. Virol. 80:11200-11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannsen, E., C. L. Miller, S. R. Grossman, and E. Kieff. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J. Virol. 70:4179-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaul, R., S. C. Verma, M. Murakami, K. Lan, T. Choudhuri, and E. S. Robertson. 2006. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J. Virol. 80:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieff, E. D., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 29.Knight, J. S., K. Lan, C. Subramanian, and E. S. Robertson. 2003. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 77:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight, J. S., N. Sharma, D. E. Kalman, and E. S. Robertson. 2004. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells. J. Virol. 78:12857-12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. USA 102:18562-18566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopan, R., and M. X. Ilagan. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovall, R. A. 2008. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27:5099-5109. [DOI] [PubMed] [Google Scholar]
- 34.Krauer, K. G., M. Buck, D. K. Belzer, J. Flanagan, G. M. Chojnowski, and T. B. Sculley. 2004. The Epstein-Barr virus nuclear antigen-6 protein co-localizes with EBNA-3 and survival of motor neurons protein. Virology 318:280-294. [DOI] [PubMed] [Google Scholar]
- 35.Künstle, G., J. Laine, G. Pierron, S.-I. Kagami, H. Nakajima, F. Hoh, C. Roumestand, M.-H. Stern, and M. Noguchi. 2002. Identification of Akt association and oligomerization domains of the Akt kinase coactivator TCL1. Mol. Cell Biol. 22:1513-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuppers, D. A., K. Lan, J. S. Knight, and E. S. Robertson. 2005. Regulation of matrix metalloproteinase 9 expression by Epstein-Barr virus nuclear antigen 3C and the suppressor of metastasis Nm23-H1. J. Virol. 79:9714-9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruo, S., E. Johannsen, D. Illanes, A. Cooper, and E. Kieff. 2003. Epstein-Barr Virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J. Virol. 77:10437-10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruo, S., E. Johannsen, D. Illanes, A. Cooper, B. Zhao, and E. Kieff. 2005. Epstein-Barr virus nuclear protein 3A domains essential for growth of lymphoblasts: transcriptional regulation through RBP-Jκ/CBF1 is critical. J. Virol. 79:10171-10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruo, S., Y. Wu, S. Ishikawa, T. Kanda, D. Iwakiri, and K. Takada. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 103:19500-19505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruo, S., Y. Wu, T. Ito, T. Kanda, E. D. Kieff, and K. Takada. 2009. Epstein-Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc. Natl. Acad. Sci. USA 106:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maunders, M. J., L. Petti, and M. Rowe. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J. Gen. Virol. 75:769-778. [DOI] [PubMed] [Google Scholar]
- 45.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 46.Ohtani, N., P. Brennan, S. Gaubatz, E. Sanij, P. Hertzog, E. Wolvetang, J. Ghysdael, M. Rowe, and E. Hara. 2003. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J. Cell Biol. 162:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541-2549. [PubMed] [Google Scholar]
- 48.Parker, G. A., R. Touitou, and M. J. Allday. 2000. Epstein-Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene 19:700-709. [DOI] [PubMed] [Google Scholar]
- 49.Pekarsky, Y., A. Palamarchuk, V. Maximov, A. Efanov, N. Nazaryan, U. Santanam, L. Rassenti, T. Kipps, and C. M. Croce. 2008. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc. Natl. Acad. Sci. USA 105:19643-19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pope, J. H. 1967. Establishment of cell lines from peripheral leucocytes in infectious mononucleosis. Nature 216:810-811. [DOI] [PubMed] [Google Scholar]
- 51.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 54.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosendorff, A., D. Illanes, G. David, J. Lin, E. Kieff, and E. Johannsen. 2004. EBNA3C coactivation with EBNA2 requires a SUMO homology domain. J. Virol. 78:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spender, L. C., G. H. Cornish, B. Rowland, B. Kempkes, and P. J. Farrell. 2001. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75:3537-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Touitou, R., M. Hickabottom, G. Parker, T. Crook, and M. J. Allday. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 75:7749-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J. Virol. 70:5909-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, X., Z. He, B. Xin, and L. Cao. 2000. LMP1 of Epstein-Barr virus suppresses cellular senescence associated with the inhibition of p16INK4a expression. Oncogene 19:2002-2013. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, B., S. Maruo, A. Cooper, R. C. M., E. Johannsen, E. Kieff, and E. Cahir-McFarland. 2006. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. USA 103:1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, J., J. Blenis, and J. Yuan. 2008. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. USA 105:6584-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]