Abstract
Stem-loop 2 (SL2) of the 5′-untranslated region of the mouse hepatitis virus (MHV) contains a highly conserved pentaloop (C47-U48-U49-G50-U51) stacked on a 5-bp stem. Solution nuclear magnetic resonance experiments are consistent with a 5′-uYNMG(U)a or uCUYG(U)a tetraloop conformation characterized by an anti-C47-syn-G50 base-pairing interaction, with U51 flipped out into solution and G50 stacked on A52. Previous studies showed that U48C and U48A substitutions in MHV SL2 were lethal, while a U48G substitution was viable. Here, we characterize viruses harboring all remaining single-nucleotide substitutions in the pentaloop of MHV SL2 and also investigate the degree to which the sequence context of key pentaloop point mutations influences the MHV replication phenotype. U49 or U51 substitution mutants all are viable; C47 substitution mutants also are viable but produce slightly smaller plaques than wild-type virus. In contrast, G50A and G50C viruses are severely crippled and form much smaller plaques. Virus could not be recovered from G50U-containing mutants; rather, only true wild-type revertants or a virus, G50U/C47A, containing a second site mutation were recovered. These functional data suggest that the Watson-Crick edges of C47 and G50 (or A47 and U50 in the G50U/C47A mutant) are in close enough proximity to a hydrogen bond with U51 flipped out of the hairpin. Remarkably, increasing the helical stem stability rescues the previously lethal mutants U48C and G50U. These studies suggest that SL2 functions as an important, but rather plastic, structural element in stimulating subgenomic RNA synthesis in coronaviruses.
Mouse hepatitis virus (MHV), a prototype group 2 coronavirus (CoV), is the most extensively studied CoV (26). CoVs are characterized by very large positive-sense RNA genomes of ≈30 kb, and during the course of replication they express seven to nine subgenomic RNAs (sgRNAs), each containing a common 3′-untranslated region (UTR) and 5′ leader sequence, the latter of which is identical to the 5′ end of the genomic RNA. The 3′ end of the leader sequence contains a short (6- to 8-nucleotide [nt]) sequence, the transcriptional regulatory sequence (TRS-L), which also is present in the genome just 5′ of the coding sequence for each subgenomic mRNA (TRS-B) (4). Among several models that have been proposed for CoV transcription and replication (2, 35, 38), the discontinuous transcription model during minus-strand synthesis is supported by significant accumulating genetic evidence (33, 45, 47, 48). In this model, a set of 5′ coterminal negative-strand sgRNAs are transcribed from positive-strand genomic RNA by the viral transcriptase/replicase complex (TRC), which then serve as templates for mRNAs synthesis. Genetic evidence is consistent with a process by which the complement to TRS-B on nascent or newly synthesized minus strands forms Watson-Crick base pairs with TRS-L within the 5′UTR of the genome to regulate mRNA synthesis (33, 45, 47, 48).
The biochemical details of sgRNA synthesis remain poorly understood. Since the same RNA template potentially can be used for both full-length genome replication and sgRNA transcription, the factors or physical features of the replication/transcription complex that stimulate one process relative to the other are not currently understood. It has been hypothesized that cellular factors bind to cis-acting RNA elements in the 5′- and 3′UTRs of the genomic RNA, which leads to genome circularization and facilitates the eventual base pairing between TRS-L and the nascent complementary TRS-B sequence that leads to template switching by the TRC at some frequency (48). Recent studies of the group 1a CoV porcine transmissible gastroenteritis virus reveal evidence for a transcriptional enhancer just 5′ of the TRS-B sequence upstream of the N gene that stimulates N sgRNA synthesis (28).
The characterization of cis-acting RNA elements in the 5′- and 3′UTRs and their functional roles in viral transcription and replication have been reviewed recently (24). We previously proposed a consensus RNA secondary structure model for the 5′ 140 nt of the CoV 5′UTR, characterized by three stem-loop structures, SL1, SL2, and SL4 (20). SL1 and SL2 are strongly supported by genetic and biophysical studies (23, 25, 27). Some CoVs contain a third stem-loop, SL3, coincident with the TRS-L (25). Note that the 3′ end of the leader RNA is coincident with the 3′ side of the body TRS and defines the fusion junction of the TRS-B and TRS-L sequences. With the exception of the TRS, SL2 is the most highly conserved structure in the CoV 5′UTR and typically contains a pentaloop sequence (C47-U48-U49-G50-U51 in MHV) stacked on a 5-bp stem, with some CoV sequences containing an additional U 3′ to U51 (25).
The consensus loop sequence that emerges from a comparison of all coronaviral group 1, group 2, and group 3 5′ leader RNAs is 5′ YUUGY(U)n, where n = 0 or 1, stacked on a closing Watson-Crick Y-R (U-A or C-G) base pair, i.e., yYUUGY(U)nr (Fig. 1A). SL2 therefore possesses sequence features consistent with at least two structurally distinct loop conformations. One is a U-turn-like structure (25) similar to that found in the hammerhead ribozyme (13), in which a UNR triloop is stacked on noncanonical C47-U51 base pairs (Fig. 1B). U-turn-like structures are characterized by a sharp turn in the phosphodiester backbone between U48 and U49 that is stabilized by Mg2+ binding near what would be the U48 and U49 phosphodiester groups in MHV SL2 (Fig. 1B). In this conformation, the imino proton of U51 will be protected from exchange with solvent by base pairing with C47, and U48 would donate two hydrogen bonds that stabilize the loop, one between its 2′ OH and the G50 N7 and the other its H3 imino proton to a nonbridging oxygen of the U51 phosphodiester linkage (3′ to G50) (6). These canonical U-turn hydrogen bonding interactions also can be found in a pentaloop with the 3′ U of the loop extruded into solvent; in this type of structure, the UNR motif will be formed by the 5′ three nucleotides in the loop and therefore will lack a noncanonical Y-Y base pair (3, 5). Previous nuclear magnetic resonance (NMR) studies failed to detect a C47-U51 base pair in the MHV pentaloop (25).
FIG. 1.
Schematic representation of the sequence conservation of SL2 in the CoV 5′ leader region (A), as well as cartoon renderings of a canonical UNR U-turn loop (B) and a CUUG tetraloop (C) structures, each of which is compatible with the SL2 sequence conservation of panel A. Nucleotide sequence and secondary structural models of SL2 derived from the two group 2 CoVs MHV (D) and SARS-CoV (E), which were studied here. Note that the cartoons are highly schematized; the internucleotide linkages shown are standard 5′-3′ phosphodiester linkages. The 16-nt NMR target corresponds to the SARS-CoV SL2 sequence, except that U57 is replaced with a nonnative 3′ unpaired A. See the text for details.
Another structure compatible with the sequence conservation in the SL2 loop is the very stable gCUUGc (C47-U48-U49-G50 in MHV) tetraloop, the prototype member of a gCNGG(N)nc family of tetraloops (1, 31) (Fig. 1C). If CoV SL2 adopts this conformation, C47 would be Watson-Crick base paired to a syn-G50 nucleotide, with U48 lining the minor groove of the stem with its imino proton potentially within hydrogen bonding distance to a 2′ OH group of the stem-closing base pair (A52 in this case). U49 would be stacked on the ribose of C47 and U51 flipped out on the opposite site of the helix. Recent structural studies reveal that the gCAGG(U)c tetraloop, another member of the gCNGG(N)c family of tetraloops, is quite accommodating of sequence variations with the two middle bases (corresponding to U48 and U49 in MHV SL2) found on the same or opposite sides of the structure; the nature of the stem-closing base pair also is variable, although typically it is a G-C base pair (36). This gCNGG(N)nc tetraloop structure bears significant similarity to the members of the yYNMGg tetraloop family (36), the most stable of which is the paradigm yUUCGg tetraloop (34). The yYNMGg tetraloop positions the N nucleotide (U48 in MHV) in roughly the same place as in the gCNGGc tetraloop, with the M base stacked on the 5′ Y base on the opposite side of the loop. Structural and functional studies reveal that the 3′ insertion in the YNMG tetraloop, like the CNGG and GNRA tetraloops, is well tolerated, and an anti-Y-syn-G base pair can be replaced with an anti-U-syn-A base pair (31, 42). Other structural studies reveal that the general yYNMGg fold can be adopted by a wide range of sequences, including yGYYAg, with the first and fourth bases in the loop sometimes forming a pseudo- or noncanonical base pair with zero or one hydrogen bond (12, 18, 27, 32, 42).
The CoV SL2 sequence conservation including the closing base pair (Fig. 1), as well as functional studies (25), clearly are incompatible with the third major tetraloop class, GNRA, but also seemingly are incompatible with the YNMG family, since the N nucleotide (which corresponds to U48 in SL2) could not be replaced with any other nucleotide in MHV. In fact, U48C- and U48A-containing viruses are not viable, while a U48G-containing virus is viable (25). In this report, we extensively investigate the sequence requirements in the context of the solution structural properties of the SL2 stem-loop in an effort to distinguish between multiple structural models of the pentaloop. Here, we present evidence that SL2 adopts a conventional yYNMG(N)r tetraloop structure with novel functional features.
MATERIALS AND METHODS
RNA preparation and NMR spectroscopy.
Wild-type (WT), U48G, U48C, and U49A SL2 RNAs were prepared by runoff transcription using SP6 or T7 RNA polymerase, purified by denaturing polyacrylamide gel electrophoresis, and subjected to multiple cycles of ethanol precipitation as described previously (8, 25, 30). 13C,15N-[U]-labeled WT SL2 and 13C,15N-[G]-labeled U48G RNAs also were prepared using 13C,15N-[UTP] and 13C,15N-[GTP] in transcription reactions, respectively. The final NMR buffer was 10 mM potassium phosphate, pH 6.0; RNA concentrations were between 0.8 to 2.0 mM. All RNAs were checked for the presence of a monomer-dimer equilibrium by nondenaturing polyacrylamide gel electrophoresis prior to extensive NMR analysis. All NMR experiments were performed on a Varian Inova 500- or 600-MHz spectrometer in the Texas A&M University Biomolecular NMR Laboratory. The proton resonances were referenced to an internal standard (100 μM DSS). The jump-return echo one-dimensional (1D) and Watergate 1H-1H nuclear Overhauser enhancement spectroscopy (NOESY) spectra (τmix = 200 ms, where τmix is NOE mixing time) were acquired to obtain imino proton resonance assignments, while 1H-1H D2O NOESY (τmix = 250 ms) and total correlation spectroscopy (TOCSY) experiments were performed to obtain nonexchangeable proton resonance assignments using standard methodologies (15). Base and ribose assignments were confirmed and extended for 13C,15N-[U]-labeled WT and 13C,15N-[G]-labeled U48G RNAs by acquiring a 2D 1H-13C constant time-heteronuclear single quantam coherence spectrum coupled with through-bond 2D HNCCCH (40) and H(CCN)H-TOCSY experiments acquired in H2O to correlate imino protons with uridine H6 and guanosine H8 protons, respectively, (41). Sensitivity-optimized HCN-HMQC (ribose moiety, H1′-C1′-N1/9), HCN-TROSY (base moiety, H6/8-C6/8-N1/9), and HCNCH experiments acquired in D2O were used to provide through-bond sugar to base connectivities (14). Complete ribose proton connectivities were obtained with 1H-13C HCCH-TOCSY experiments in 13C,15N-[U]-labeled WT and 13C,15N-[G]-labeled U48G SL2 RNAs to provide unique starting points for sequential base-ribose connectivities in a D2O NOESY spectrum (τm = 250 ms). Additional NOE restraints were obtained from the analysis of a 13C-edited NOESY spectrum, a 2D 1H-15N CPMG-NOESY (60 and 250 ms) (29), and 2JHN-HSQC experiments (39). All of the NMR data were processed using nmrPipe and analyzed using Sparky (11, 16). Complete resonance assignments and solution structures of the WT and U48G SL2 RNAs will be reported elsewhere.
Cells and viruses.
Baby hamster kidney cells expressing MHV receptors (BHK-R) were maintained at 37°C and 5% CO2 in Dulbecco's modified essential medium (DMEM) supplemented with 10% calf serum, 10% tryptose phosphate broth, 800 μg/ml of Geneticin (G418 sulfate; Sigma), and 0.25 μg/ml of penicillin-streptomycin. DBT cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% calf serum. L2 cells were maintained at 37°C and 3% CO2 in DMEM supplemented with 10% calf serum. Wild-type MHV A59-1000 and all mutant viruses were propagated in DBT cells.
Construction and recovery of mutant viruses.
The construction and recovery of mutant viruses were carried out as described previously (25, 46). Briefly, the C47A, C47G, C47U, U49C, U49G, G50A, G50C, G50U, rG50U/C47A, U51A, U51C, U51G, ΔU51, MS-G50A, MS-G50C, MS-G50U, rMS-G50U/C47A, and MS-U48C mutations were constructed using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with WT MHV-A59-1000 plasmid A as the substrate for mutagenesis. Sequences of the mutagenic oligonucleotides are available on request. Mutagenized plasmids were sequenced, and fragments carrying the desired mutation were excised with MluI and SacII and religated into MluI-SacII-digested parental WT plasmid A to generate mutant plasmids that would be free of unintended mutations. DNA fragments from digested and gel-purified plasmids A to G were sequentially ligated to generate a cDNA corresponding to the whole MHV genome. This cDNA was transcribed in vitro using the Ambion T7 mMESSAGE mMACHINE kit (Applied Biosystems), and the transcription reaction subsequently was electroporated into BHK-R cells, which were overlaid onto DBT cells to recover mutant virus. Recovered mutant viruses were plaque purified on L2 cell monolayers. Plaque isolates were expanded in DBT cells, and their complete 5′ and 3′ UTRs were sequenced to confirm the presence or absence of additional mutations. Before a mutant was declared to be nonviable, at least three independent electroporations followed by three blind passages of each electroporation were done without recovering virus.
Plaque size determination and growth curve assays.
Plaque sizes of mutant and WT viruses were determined in L2 cells as described previously (19). Growth curves experiments in DBT cells were performed in triplicate wells of DBT cells in 96-well plates infected with mutant or WT viruses at a multiplicity of infection (MOI) of 3 and harvested at 0, 4, 8, 12, 16, and 24 h post infection (hpi). For some experiments an MOI of 0.01 was used, and cultures were harvested at 0, 8, 12, 16, 24, and 36 hpi. Virus titers were determined by plaque assays. Error bars represent the standard errors of the mean.
RESULTS
Previous functional studies established that a U48G substitution of the MHV SL2 stem-loop was compatible with virus replication, while other (A and C) substitutions were not (25). Here we compare key NMR spectral fingerprints of the WT and U48G RNAs that establish the basic architecture of SL2 in solution. Note that these spectra were acquired with the severe acute respiratory syndrome (SARS)-CoV SL2 sequence, which differs from the MHV RNA only in the nature of 2 of the 5 bp of the stem (Fig. 1D and E). The stem of SL2 in SARS and MHV is nearly identical. The SARS stem was chosen in order to take advantage of the more stable stem sequence. For NMR studies, the closing GC base pair significantly reduces fraying at the closing base pair as well as facilitating in vitro transcription. Both WT and U48G SL2s contained a nonnative 3′ A in an effort to stabilize the base of the stem (43).
The pentaloop is dynamic in solution.
Although the SL2 RNA adopts a single average conformation under low-salt conditions in solution, like other stem-loops, it is characterized by significant conformational dynamics or flexibility in the loop that is faster than overall macromolecular tumbling. This is readily apparent in the relative intensities of the aromatic C6-H6 cross-peaks of pentaloop residues versus helical stem residues in the [15N,13C-]U-WT SL2 RNA observed in a 1H-13C CT-HSQC spectrum (Fig. 2). U48, U49, and U51 are far more intense (more dynamic) than the corresponding stem residues, with U51 being the most dynamic of any loop residue (Fig. 2). Flexibility also manifests itself in ribose sugar conformation, with nucleotides in A-form helical stems confined to a C3′-endo (north) conformation (15, 37); all five loop nucleotides adopt a C2′-endo (south) conformation, or are in rapid exchange between C3′-endo and C2′-endo conformations, as evidenced by large 3J(H1′, H2′) vicinal couplings in a homonuclear TOCSY spectrum (data not shown). These data suggest that a folded loop conformation in SL2 is more dynamic than the helical stem on which it is stacked; this result is consistent with other canonical stem-loop sequences, except for the most stable tetraloops, e.g., cUUCGg (34).
FIG. 2.
Nucleobase-optimized 1H-13C HSQC spectra acquired for the [15N,13C-]U-WT SL2 RNA. The nucleotides labeled with stable isotopes are highlighted in boldface and italics in the structural model (inset). The conditions were 25.0°C and 10 mM potassium phosphate, pH 6.0.
G50 in both RNAs adopts a syn-glycosidic bond angle and is stacked on A52, with U51 flipped out of the helical stack.
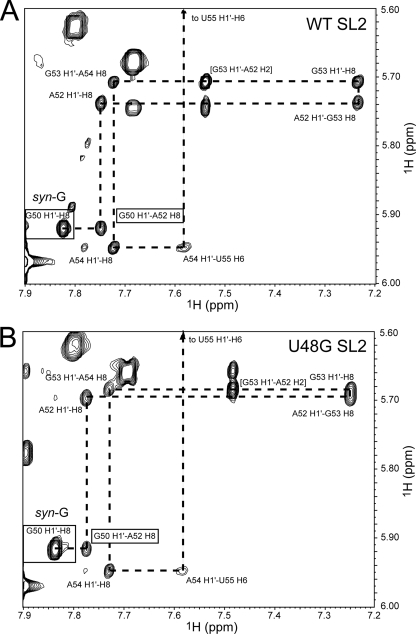
Homonuclear 1H-1H NOESY spectra were acquired across a range of mixing times in the both D2O and H2O solutions in the presence and absence of Mg2+. Preliminary experiments revealed that spectra acquired in the presence of Mg2+ or Co(NH3)63+ at a few millimolars were characterized by two U-U base pairs, which are possible only in an SL2 intermolecular duplex conformation, rather than stabilizing a new fold in the tetraloop (data not shown); this suggests that the structure that SL2 RNAs adopt here are not strongly stabilized by the addition of salt. NOESY experiments allowed us to establish the base pairing in each of the molecules as well as carry out a sequential intra- and interresidue aromatic base-ribose proton walk down each strand of the SL2 helix. The spectral region taken from a 1H-1H NOESY experiment acquired in D2O establishes that both the WT SL2 (Fig. 3A) and the U48G SL2 (Fig. 3B) RNAs exhibit continuous base stacking from G50 to A52 and sequentially down the remainder of the stem. No such stacking characterizes the U48-U49 and U49-G50 steps, with a sharp turn induced in the polynucleotide backbone between U48 and U49 as evidenced by the strongly upfield-shifted H4′ (3.85 ppm), H5′ (3.89 ppm), H5″ (3.85 ppm), and H2′ (3.96 ppm) protons of U49 (9, 10, 36). In addition, the intensity of the G50 intraresidue H8-H1′ correlation relative to those of others shown establishes that G50 adopts a syn- or high-anti-glycosidic bond angle (36). Since the syn-G50 nucleotide is stacked on A52, this requires that U51 be flipped out of the helix. Further support for this is a nearly complete lack of interresidue NOEs for U51, very sharp resonances (Fig. 2), and a strongly downfield-shifted H5 chemical shift for U51 (5.97 ppm).
FIG. 3.
Sequential intra- and interresidue NOEs (dashed line) obtained from 1H-1H NOESY spectra (τm = 200 ms) acquired in D2O that establish continuous base stacking from G50 to U55 on the 5′ strand of WT (A) and U48G (B) RNAs. The conditions were 25.0°C and 10 mM potassium phosphate, pH 6.0.
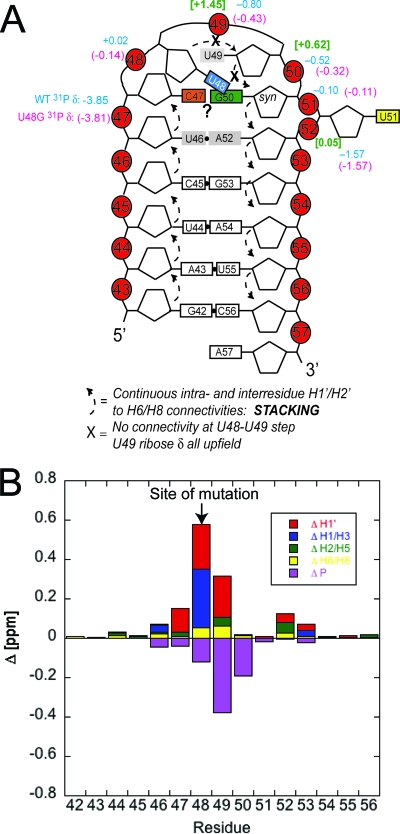
Figure 4A summarizes all of the NMR data in cartoon form, and it also provides a summary of the 31P chemical shifts of phosphodiester groups in the WT and U48G SL2s obtained from a 1H-31P HP-COSY experiment acquired on unlabeled RNAs. These chemical shifts are compared to those previously obtained for an RNA stem-loop from the VS ribozyme that is known to adopt a U-turn conformation in solution in the presence of Mg2+ (3). 1H-31P correlations are very sensitive reporters of the structures of individual phosphodiester linkages and thus loop conformation, with 1H chemical shift differences indicative of changes in the local environment. This summary, therefore, makes two important points. The high similarity of the WT and U48G RNA 31P resonances suggests that their loop structures are very similar and strikingly different from that measured for the U-turn-containing RNA. Figure 4B summarizes all of the 1H and 31P chemical shift differences between the WT and U48G RNAs and reveals that the differences are small and localized to the site of the substitution.
FIG. 4.
Summary of the NMR data obtained for WT and U48G SL2 RNAs. (A) Cartoon representation of SARS-CoV SL2 that summarizes all of the NMR data. 31P δ are indicated for the WT and U48G RNAs in blue and magenta, respectively, above the corresponding P atom; the 31P δ for the U-turn-containing RNA (3) is shaded green and is in brackets. (B) A plot of the absolute value chemical shift differences (designated Δ) between the WT and U48G SL2 RNAs; the Δ for 31P is multiplied by −1 for display purposes only.
The NMR data acquired for both WT and U48G RNAs (Fig. 4A) are incompatible with a canonical U48-U49-G50 U-turn loop structure, as we see no evidence for a C47-U51 base pair and Mg2+ is not required to fold this RNA (data not shown). A direct comparison of the 31P δ in the loop (Fig. 3A) provides further evidence against a U-turn-containing loop structure. The NMR data are, however, largely compatible with either a uCUYG(U)a or a uYNMG(U)a-like tetraloop, with U51 flipped out of the stack and a syn-G50 stacked on A52 with its Watson-Crick face oriented toward the Watson-Crick face of C47. The series of experiments summarized below address the degree to which these structural insights are supported by the functional data.
C47, U49, and U51 are remarkably tolerant of substitution in mutant viruses.
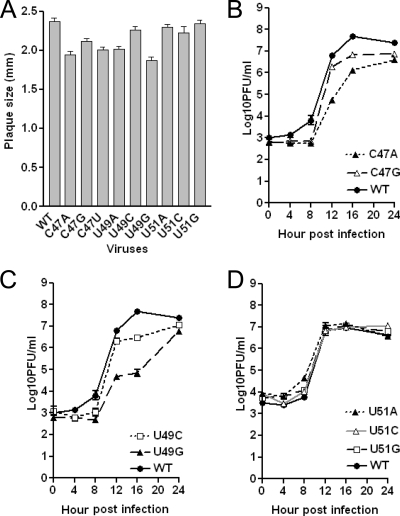
Using a reverse genetic system, we constructed MHV genomic RNAs harboring one of all possible single-nucleotide substitutions of C47, U49, and U51 and recovered viable viruses from cells into which these RNAs were introduced. Viruses were plaque purified, and the complete 5′- and 3′UTRs were sequenced to verify the integrity of the recovered viruses. The virus plaque size and growth phenotypes were determined essentially as described previously (23, 25). The replacement of C47 with any other nucleotide induces a very small but measurable effect on virus plaque size (Fig. 5A) and a modest effect on growth kinetics in a one-step growth curve experiment (Fig. 5B). Likewise, the replacement of U49 has a similarly small effect on the plaque size and growth phenotype, with the U49G virus exhibiting properties comparable to those of C47 substitution mutants (Fig. 5A and C). Finally, substitutions at U51 have no measurable effect on mutant virus plaque size and growth phenotype (Fig. 5A and D).
FIG. 5.
Phenotypes of C47, U49, and U51 substitution mutant viruses. (A) Average plaque size (± standard errors of the means) of indicated mutant and WT viruses. (B) Growth curves of C47A and C47G substitution mutants at an MOI of 3. (C) Growth curves of U49C and U49G substitution mutants at an MOI of 3. (D) Growth curves for the U51A, U51C, and U51G viruses performed at an MOI of 3.
Since the structural data reveal that U51 is flipped out of the hairpin loop, we also characterized a ΔU51 deletion mutant. Although we recovered virus from cells electroporated with RNA genomes carrying this deletion, all five plaque-purified viral isolates recovered from two separate electroporations contained the WT sequence, with U51 added back in. Since all of our U51 substitution mutants are viable and had phenotypes in cell culture similar to that of the WT virus, the results with the ΔU51 mutation suggest that U51 is a spacer element that is required to organize the stem-loop architecture. Interestingly, despite the distinct structural context, analogous findings characterize substitution and deletion mutants of a loop 3′ U (U700) in the Mg2+-stabilized conformation of U-turn-containing stem-loop V of the VS ribozyme, where U700 is extruded to solvent (3, 5, 6). In striking contrast, the thermodynamic stabilities of a YNMG-like tetraloop from the mutant human telomerase RNA with and without a 3′ U within a 5′-YNMG(U) pentaloop sequence are essentially identical, and the examination of the pentaloop solution structure reveals that the 3′ U is flipped out into the major groove of the loop (42). Thus, the CoV SL2 may adopt something of a hybrid structure, with the sequence conservation and mutagenesis data (see below) being largely consistent with a YNMG(U) or CUYG(U) tetraloop, but one that seems to functionally require a 3′ U51 spacer residue. Indeed, RNA stem-loops that adopt this general structure show considerable variability in sequence. The most common structural feature is that nucleotides in the first (C47 in SL2) and fourth (G50 in SL2) positions form Watson-Crick (C-G or U-A) or noncanonical (U-G or G-A) base pairs, with the middle second (U48) and third (U49) nucleotides being capable of adopting a range of orientations in the loop (36).
G50 is important for viral function.
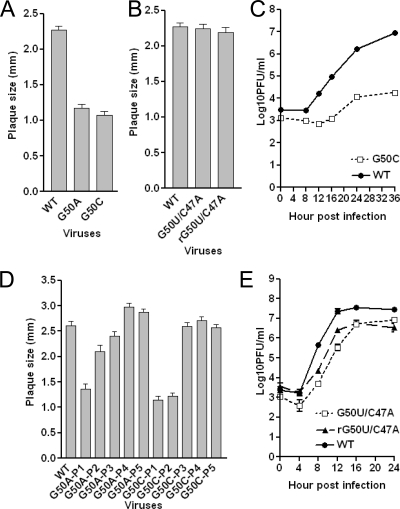
Virtually every common tetraloop motif, namely GNRA, YNMG, and CUYG, incorporates a purine in the fourth position of the loop that adopts a syn-glycosidic bond angle, in this case G50. We therefore examined the functional importance of G50, again by the characterization of single-nucleotide substitution mutant-containing viruses. In striking contrast to the other nucleotides in the pentaloop, the replacement of G50 with either adenosine (G50A) or cytidine (G50C) leads to severely crippled mutant viruses with significantly smaller plaque sizes (Fig. 6A). G50A- and G50C-containing viruses revert to WT virus (sequence data not shown) by passage three and regain the WT plaque size and morphologies (Fig. 6D). We could not perform one-step or multistep growth curves for the G50A mutant virus because of low virus titer at passage one. We did obtain a sufficient titer of passage one virus to perform a multistep growth curve experiment (MOI = 0.01) with the G50C mutant virus; the kinetics of growth were strongly delayed relative to those of the WT virus, and the final yield was approximately 1,000-fold less than that of the WT (Fig. 6C). Although the plaque sizes of the output virus in this experiment were a somewhat more heterogeneous than those of the input G50C virus used in this experiment, few WT-sized plaques were observed, making it unlikely that WT revertants contributed significantly to the output virus. The replacement of G50 with uridine results in either WT plaque isolates (8 out of 10 plaques sequenced) or in a specific second-site mutation (G50U/C47A) (2 out of 10 plaques sequenced) from three separate electroporations. Plaque isolates of the G50U/C47A virus and its reengineered recombinant counterpart, rG50U/C47A, show almost identical plaques and only a slightly impaired growth phenotype compared to that of the WT virus (Fig. 6B and E). Thus, the G50U mutant is nonviable, rapidly reverts to a WT virus, or is stable only with a compensating second-site C47A mutation. Taken together, these data demonstrate that G50 substitutions are strongly selected against and suggest a range of Watson-Crick or non-Watson-Crick base-pairing interactions between G50 and C47 that are important for MHV replication.
FIG. 6.
Phenotypes of G50 substitution mutant viruses. (A) Average plaque size (± standard errors of the means) of G50A and G50C mutant viruses. (B) Average plaque size (± standard errors of the means) of G50U/C47A and rG50U/C47A mutant viruses. (C) Growth curves were obtained for the G50C substitution mutant at an MOI of 0.01. (D) Average plaque size (± standard errors of the means) of G50A and G50C mutant passages 1 through 5. (E) Growth curves of recovered G50U/C47A and rG50U/C47A mutant viruses at an MOI of 3.
A careful analysis of the exchangeable 1H NMR data of both WT and U48G RNAs provides no direct evidence for or against a C47-G50 base pair, since the G50 imino proton exchanges too rapidly with solvent to observe; alternatively, these two nucleotides may not directly base pair or may form a noncanonical pair with a single hydrogen bond (27, 31, 42). On the other hand, since C47 is stacked on U46 and extends the helical geometry of the 5′ strand in SL2 and G50 is stacked on A52 on the opposite strand (Fig. 3), their Watson-Crick edges likely are in close proximity.
Increasing the stability of the SL2 stem partially rescues otherwise lethal mutants.
The predicted thermodynamic stability of the 5-bp stem of MHV SL2 is lower than that of all other CoV sequences and contains just one G-C base pair and three consecutive A-U pairs at the base of the stem (Fig. 1); in contrast, all other SL2s contain two G-C pairs (25). As a result of this, SL2 is not base paired in the minimum free energy model of the 5′UTR of MHV, a unique feature that distinguishes MHV from other CoV sequences. The fact that the replacement of the MHV SL2 with the SARS-CoV SL2 (Fig. 1) in an otherwise all-MHV genome results in a viable WT-like virus, however, argues for an independently folded SL2 in each case (20). Nonetheless, this suggests the possibility that a more stable SL2 stem suppresses the previously observed phenotypes observed in mutant SL2 all-MHV viruses in the event that loop stability and stem stability cooperate to maintain replication function.
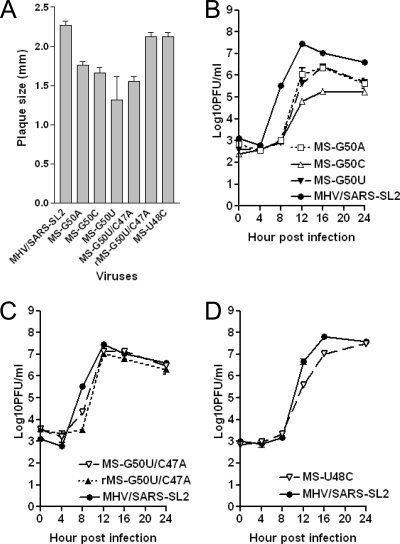
To test this idea, we reconstructed the lethal mutants U48C (25), G50U (as described above), and ΔU51, as well as the severely impaired mutants G50A and G50C, in the context of the SARS-CoV SL2 stem. As discussed above, MHV and SARS-CoV SL2 possess identical pentaloop sequences and differ only in their stem sequence and composition (Fig. 1). The recovered chimeric mutant viruses, designated MS-G50A and MS-G50C, show significantly improved plaque sizes relative to those of the corresponding all-MHV SL2 mutants (Fig. 7A); however, each remains detectably adversely affected by these substitutions. The MS-G50A and MS-G50C viruses grew to considerably higher titers than the corresponding all-MHV SL2 mutants, enabling us to perform a one-step growth curve experiment. As shown in Fig. 7B, the growth kinetics and maximal titers were moderately impaired relative to those of their MHV/SARS-SL2 parent virus. The MS-G50U mutant also is impaired and is characterized by small plaques and a delayed growth phenotype compared to that of its parental WT chimeric mutant MHV/SARS-SL2 (Fig. 7A and B). Consistently with this impairment of function, we again were able to recover a second-site chimeric mutant, denoted MS-G50U/C47A. In this context however, it is characterized by a much smaller plaque size but a similar growth phenotype relative to that of the parental virus, MHV/SARS-SL2 (Fig. 7A and C). To further examine the phenotype of this second-site mutant virus, we reconstructed this second site mutation using reverse genetics to produce a new recombinant virus, rMS-G50U/C47A, that contained both the G50U and C47A mutations. Unlike the MS-G50U/C47A second-site mutant virus, this virus produced plaques virtually identical in size to that of the parental MHV/SARS-SL2 virus (Fig. 7A). However, it grew with WT (MHV/SARS-SL2) kinetics, as did the MS-G50U/C47A second-site mutant virus (Fig. 7C). These results suggest that the MS-G50U/C47A virus contains an additional unidentified mutation that adversely affects plaque size that is not found in the reconstructed rG50U/C47A virus. In contrast to the G50 substitutions, the chimeric MS-U48C mutant is characterized by a nearly WT plaque size and a growth phenotype essentially as robust as that of the MHV/SARS-SL2 virus (Fig. 7A and D). We also were able to recover an rMS-ΔU51 virus, although that required two blind passages. Like the MS-U48C mutant, the rMS-ΔU51 virus was fairly robust and produced plaques that were 89% the size of those produced by the parental MHV/SARS-SL2 virus.
FIG. 7.
Phenotypes of MHV-SARS-CoV (MS) chimeric viruses (see the text for details). r, recombinant. (A) Average plaque size (± standard errors of the means) of MHV-SARS-CoV chimeric viruses. (B) Growth curve of MS-G50A, MS-G50C, and MS-G50U mutant viruses at an MOI of 3. (C) Growth curve of MS-G50U/C47A and rMS-G50U/C47A mutants at an MOI of 3. (D) Growth curve of the MS-U48C mutant virus at an MOI of 3.
To determine if the substitution mutants affect viral RNA synthesis, DBT cells were infected with representative mutant viruses and the virus-specific RNAs were metabolically labeled in the presence of actinomycin D with [32P]orthophosphate from 6 to 12 hpi. The labeled RNAs then were extracted and analyzed by formaldehyde agarose gel electrophoresis (Fig. 8). The amount of RNA synthesized in cells infected with each mutant largely reflects the viral growth phenotype. The ratios of sgRNAs to genomic RNAs generally are similar to that of the WT virus. To summarize, the originally lethal mutants in an all-MHV context, G50U and U48C, are viable in the context of a pentaloop supported by the SARS-CoV SL2 stem, although it gives rise to viruses with distinct phenotypes. We do not know yet whether these mutations give rise to other second-site substitutions outside of the 5′- and 3′UTRs; in any case, it is interesting that these stem mutations were not recovered from an all-MHV U48C virus (25).
FIG. 8.
Analysis of total genomic RNA (RNA 1) and sgRNA (RNAs 2 to 7) in mutant virus-infected cells. DBT cells infected with WT and mutant viruses at an MOI of 1 and RNAs were metabolically labeled from 6 to 12 hpi in the presence of actinomycin D. Total RNAs were extracted and resolved by formaldehyde agarose gel electrophoresis.
DISCUSSION
In this study, we combine NMR data with an exhaustive mutagenesis analysis to infer the structural determinants and functional requirements of SL2 for the replication of MHV in cultured cells. All of the data taken collectively are consistent with the proposal that the SL2 loop adopts a tetraloop structure that exhibits key hallmarks of a canonical CUYG(N) and YNMG(N)-like tetraloop previously investigated by NMR methods, with the 3′ U51 (N) flipped out of the stack. The functional data are consistent with an as-yet structurally undefined anti-C47 and a syn-G50 base-pairing interaction, which is a hallmark of essentially all previously characterized tetraloop folds. The isolation of G50U/C47A second-site revertants with a near-WT phenotype suggests that an A-U base-pairing interaction substitutes for the C47-G50 interaction. Further, the structural and functional data reveal that the identity of the base at position 51 is immaterial. Although this nucleotide is required to obtain infectious virus in the context of the less stable MHV SL2, viable virus was obtained with the ΔU51 mutation in the context of the somewhat more stable SARS SL2. This suggests that the presence of a nucleotide at position 51 facilitates the folding of SL2 rather than the bulged U51 mediating a long-range RNA-RNA or RNA-protein interaction required for replication. This is consistent with previous structural studies of RNA pentaloops that collectively show that the bulged nucleotide is dispensable, with its deletion imparting little change in structure and stability (31, 36, 42).
We previously showed that U48C MHV supports the synthesis of both positive- and negative-sense genomic RNAs but fails to direct the synthesis of positive- and negative-sense sgRNAs in infected cells in culture (25). Surprisingly, when chimeric viruses are engineered such that the SARS-CoV SL2 stem replaces the MHV SL2 stem (Fig. 1), both originally lethal U48C and G50U mutations in an all-MHV context now are viable. Furthermore, the originally feeble G50A and G50C mutant viruses are more robust in this context. Although previous results reveal that SL2 plays a critical role in sgRNA transcription and adopts a defined structure in solution (25), the current findings reveal that this role is not precisely dependent on any one particular interaction within the loop. This may be consistent with the recognized structural plasticity in tetraloops in general, which are known to be quite tolerant of multiple base substitutions (36). The fact that what are formally second-site substitutions introduced into the immediately adjacent SL2 stem rescues the effect of destabilizing mutations in the pentaloop suggests that SL2 plays a generic structural role in stabilizing a higher-order structure within the 5′UTR or a 5′UTR-′UTR complex that is important specifically for sgRNA synthesis. Although we cannot completely rule out a structure-dependent interaction of SL2 with RNA or protein binding partners, the functional plasticity of the nucleotide sequence of SL2, at least as it relates to the requirements for RNA replication/transcription in infected cells in culture, argues against a specific role of SL2 in engaging in a highly specific RNA-RNA or RNA-protein complex formation that might be essential for template switching during discontinuous sgRNA synthesis (7, 17, 21, 22, 44). We hypothesize that a minimally stable SL2 structure serves as a scaffold that directs the folding of the immediately adjacent elements that ultimately mediate genome circularization and thus facilitate base scanning by the viral transcriptase/replicase complex to regulate sgRNA synthesis. Current efforts are in progress in our laboratories to further define these molecular determinants of SL2 function.
Acknowledgments
We gratefully acknowledge support from National Institutes of Health grants AI040187, AI067416, and AI051493.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Aviv, T., A. N. Amborski, X. S. Zhao, J. J. Kwan, P. E. Johnson, F. Sicheri, and L. W. Donaldson. 2006. The NMR and X-ray structures of the Saccharomyces cerevisiae Vts1 SAM domain define a surface for the recognition of RNA hairpins. J. Mol. Biol. 356:274-279. [DOI] [PubMed] [Google Scholar]
- 2.Baric, R. S., S. A. Stohlman, and M. M. Lai. 1983. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J. Virol. 48:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchard, P., J. Lacroix-Labonte, G. Desjardins, P. Lampron, V. Lisi, S. Lemieux, F. Major, and P. Legault. 2008. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA 14:736-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budzilowicz, C. J., S. P. Wilczynski, and S. R. Weiss. 1985. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3′ end of the viral mRNA leader sequence. J. Virol. 53:834-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, D. O., P. Bouchard, G. Desjardins, and P. Legault. 2006. NMR structure of varkud satellite ribozyme stem-loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry 45:10591-10605. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, D. O., and P. Legault. 2005. Nuclear magnetic resonance structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry 44:4157-4170. [DOI] [PubMed] [Google Scholar]
- 7.Choi, K. S., P. Huang, and M. M. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. [DOI] [PubMed] [Google Scholar]
- 8.Cornish, P. V., M. Hennig, and D. P. Giedroc. 2005. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 102:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cromsigt, J., B. van Buuren, J. Schleucher, and S. Wijmenga. 2001. Resonance assignment and structure determination for RNA. Methods Enzymol. 338:371-399. [DOI] [PubMed] [Google Scholar]
- 10.Cromsigt, J. A., C. W. Hilbers, and S. S. Wijmenga. 2001. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR 21:11-29. [DOI] [PubMed] [Google Scholar]
- 11.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 12.Du, Z., J. Yu, N. B. Ulyanov, R. Andino, and T. L. James. 2004. Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry 43:11959-11972. [DOI] [PubMed] [Google Scholar]
- 13.Feig, A. L., W. G. Scott, and O. C. Uhlenbeck. 1998. Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb. Science 279:81-84. [DOI] [PubMed] [Google Scholar]
- 14.Fiala, R., J. Czernek, and V. Sklenar. 2000. Transverse relaxation optimized triple-resonance NMR experiments for nucleic acids. J. Biomol. NMR 16:291-302. [DOI] [PubMed] [Google Scholar]
- 15.Fürtig, B., C. Richter, J. Wohnert, and H. Schwalbe. 2003. NMR spectroscopy of RNA. Chembiochem 4:936-962. [DOI] [PubMed] [Google Scholar]
- 16.Goddard, T. D., and D. G. Kneller. Sparky 3. University of California, San Francisco, CA.
- 17.Huang, P., and M. M. C. Lai. 2001. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihle, Y., O. Ohlenschlager, S. Hafner, E. Duchardt, M. Zacharias, S. Seitz, R. Zell, R. Ramachandran, and M. Gorlach. 2005. A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA. Nucleic Acids Res. 33:2003-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. F., M. Feng, P. Liu, J. J. Millership, B. Yount, R. S. Baric, and J. L. Leibowitz. 2005. The effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570-14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, H., M. Feng, M. E. Schroeder, D. P. Giedroc, and J. L. Leibowitz. 2006. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 80:10600-10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H. P., P. Huang, S. Park, and M. M. Lai. 1999. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 73:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H. P., X. Zhang, R. Duncan, L. Comai, and M. M. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. USA 94:9544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, L., H. Kang, P. Liu, N. Makkinje, S. T. Williams, J. L. Leibowitz, and D. P. Giedroc. 2008. Structural lability in stem-loop 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J. Mol. Biol. 377:790-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, P., and J. L. Leibowitz. RNA higher-order structures within the coronavirus 5′ and 3′ untranslated regions and their roles in viral replication, in press. In S. Lal (ed.), Molecular biology of the SARS coronavirus. Springer-Verlag, Berlin, Germany.
- 25.Liu, P., L. Li, J. J. Millership, H. Kang, J. L. Leibowitz, and D. P. Giedroc. 2007. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA 13:763-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchers, W. J., J. Zoll, M. Tessari, D. V. Bakhmutov, A. P. Gmyl, V. I. Agol, and H. A. Heus. 2006. A GCUA tetranucleotide loop found in the poliovirus orilL by in vivo SELEX (un)expectedly forms a YNMG-like structures: extending the YNMG family with GYYA. RNA 12:1671-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno, J. L., S. Zuniga, L. Enjuanes, and I. Sola. 2008. Identification of a coronavirus transcription enhancer. J. Virol. 82:3882-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller, L., P. Legault, and A. Pardi. 1995. Improved RNA structure determination by detection of NOE contacts to exchange-broadened amino protons. J. Am. Chem. Soc. 117:11043-11048. [Google Scholar]
- 30.Nixon, P. L., A. Rangan, Y. G. Kim, A. Rich, D. W. Hoffman, M. Hennig, and D. P. Giedroc. 2002. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 322:621-633. [DOI] [PubMed] [Google Scholar]
- 31.Oberstrass, F. C., A. Lee, R. Stefl, M. Janis, G. Chanfreau, and F. H. Allain. 2006. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat. Struct. Mol. Biol. 13:160-167. [DOI] [PubMed] [Google Scholar]
- 32.Ohlenschläger, O., J. Wohnert, E. Bucci, S. Seitz, S. Hafner, R. Ramachandran, R. Zell, and M. Gorlach. 2004. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237-248. [DOI] [PubMed] [Google Scholar]
- 33.Pasternak, A. O., E. van der Born, W. J. M. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proctor, D. J., J. E. Schaak, J. M. Bevilacqua, C. J. Falzone, and P. C. Bevilacqua. 2002. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry 41:12062-12075. [DOI] [PubMed] [Google Scholar]
- 35.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthensis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 36.Schwalbe, M., O. Ohlenschlager, A. Marchanka, R. Ramachandran, S. Hafner, T. Heise, and M. Gorlach. 2008. Solution structure of stem-loop alpha of the hepatitis B virus post-transcriptional regulatory element. Nucleic Acids Res. 36:1681-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott, L. G., and M. Hennig. 2008. RNA structure determination by NMR. Methods Mol. Biol. 452:29-61. [DOI] [PubMed] [Google Scholar]
- 38.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simorre, J. P., G. R. Zimmermann, L. Mueller, and A. Pardi. 1996. Triple-resonance experiments for assignment of adenine base resonances in C-13/N-15-labeled RNA. J. Am. Chem. Soc. 118:5316-5317. [Google Scholar]
- 40.Simorre, J. P., G. R. Zimmermann, A. Pardi, B. T. N. Farmer, and L. Mueller. 1995. Triple resonance HNCCCH experiments for correlating exchangeable and nonexchangeable cytidine and uridine base protons in RNA. J. Biomol. NMR 6:427-432. [DOI] [PubMed] [Google Scholar]
- 41.Sklenár, V., T. Dieckmann, S. E. Butcher, and J. Feigon. 1996. Through-bond correlation of imino and aromatic resonances in C-13-,N-15-labeled RNA via heteronuclear TOCSY. J. Biomol. NMR 7:83-87. [DOI] [PubMed] [Google Scholar]
- 42.Theimer, C. A., L. D. Finger, and J. Feigon. 2003. YNMG tetraloop formation by a dyskeratosis congenita mutation in human telomerase RNA. RNA 9:1446-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theimer, C. A., Y. Wang, D. W. Hoffman, H. M. Krisch, and D. P. Giedroc. 1998. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J. Mol. Biol. 279:545-564. [DOI] [PubMed] [Google Scholar]
- 44.van Hemert, M. J., S. H. van den Worm, K. Knoops, A. M. Mommaas, A. E. Gorbalenya, and E. J. Snijder. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yount, B., R. S. Roberts, L. Lindesmith, and R. S. Baric. 2006. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl. Acad. Sci. USA 103:12546-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zúñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus sgRNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]