Abstract
To determine the origin, phylogenetic relationships, and evolutionary dynamics of rabbit hemorrhagic disease virus (RHDV), we examined 210 partial and complete capsid gene nucleotide sequences. Using a Bayesian Markov chain Monte Carlo approach, we estimated that these sequences evolved at a rate of 3.9 × 10−4 to 11.9 × 10−4 nucleotide substitutions per site per year. This rate was consistent across subsets of data, was robust in response to recombination, and casts doubt on the provenance of viral strains isolated from the 1950s to the 1970s, which share strong sequence similarity to modern isolates. Using the same analysis, we inferred that the time to the most recent common ancestor for a joint group of RHDV and rabbit calicivirus sequences was <550 years ago and was <150 years ago for the RHDV isolates that have spread around the world since 1984. Importantly, multiple lineages of RHDV were clearly circulating before the major Chinese outbreak of 1984, a finding indicative of an early evolution of RHDV virulence. Four phylogenetic groups within RHDV were defined and analyzed separately. Each group shared a common ancestor in the mid-1960s or earlier, and each showed an expansion of populations starting before 1984. Notably, the group characterized by the antigenic variant RHDVa harbors the greatest genetic diversity, compatible with an elevated fitness. Overall, we contend that the high virulence of RHDV likely evolved once in the early part of the 20th century, well before the documented emergence of rabbit hemorrhagic disease in 1984.
Novel infectious diseases can emerge either by a species-jump into a new host or by mutation of an existing microorganism to a more virulent form. An example of each type of emerging disease has occurred in the European rabbit (Oryctolagus cuniculus): myxomatosis, where the poxvirus myxoma virus jumped to O. cuniculus from the tapeti (a lagomorph, Sylvilagus brasiliensis), in which it caused an innocuous cutaneous fibroma, and rabbit hemorrhagic disease (RHD), where a preexisting avirulent virus of European rabbits appears to have mutated to the lethal rabbit hemorrhagic disease virus (RHDV) that has spread around the world since 1984. Although the epidemiology and evolution of RHDV since 1984 have been studied in detail, the evolutionary history and geographic origins of the virus remain uncertain. In particular, neither the timing nor the location of the emergence of virulent RHDV is well understood, with one important idea being that virulence emerged multiple times independently around 1984 (13, 16, 24).
RHD is an acute or peracute hepatitis of European rabbits that is lethal in up to 90% of infections of rabbits older than 8 weeks of age and caused by a single-strand, positive-sense RNA virus of the family Caliciviridae (genus Lagovirus). Kittens younger than this become infected and shed virus but are not clinically ill. European rabbits are native to the Iberian peninsula and are found in the wild throughout Western Europe, Australia, New Zealand, Chile, Argentina, and various islands. Domestic breeds of European rabbits are farmed throughout the world for meat and fur. RHD emerged in China in 1984 in Angora rabbits imported from Germany. From China it spread around the world in either infected rabbits or rabbit products. RHD occurred in Italy in 1986 in farmed domestic rabbits and subsequently in farmed rabbits in other parts of Europe, Russia, India, the Americas, and the Middle East (reviewed in reference 6). In Europe, RHDV is believed to have spread into the wild rabbit population from domestic rabbits and is now established in wild rabbit populations throughout continental Europe, the United Kingdom, and Ireland, with substantial ecological consequences, particularly in the Iberian peninsula (5, 6). After its accidental release in 1995, the Czech v351 strain of RHDV was deliberately spread in Australia and subsequently New Zealand as a biological control for the wild European rabbit (5, 6).
Serological studies on wild and farmed rabbits and archival sera indicated that caliciviruses antigenically related to RHDV had been circulating, apparently harmlessly, in rabbit populations in Europe, Australia, and New Zealand prior to the emergence of RHDV. In some cases immunity to these viruses may provide a degree of cross-protection against virulent RHDV (3, 4, 7, 21, 27, 29, 31, 32). Sequence data has been published for three of these avirulent viruses: from commercial rabbits in Italy (3), from wild rabbits on Lambay Island in Ireland (15), and from wild rabbits in Australia (35). A fourth, similarly diverse, virus was identified from the United Kingdom; however, this Ashington strain was obtained from a dead rabbit (24). Phylogenetically, these four viruses are relatively distant from virulent RHDV and from each other (3, 16, 24, 35) and have been named rabbit caliciviruses (RCV) to distinguish them from RHDV.
Partial sequence data for the capsid gene of RHDV has also been obtained from archival serum samples in the United Kingdom stored since the 1950s and 1970s and originally sampled from domestic and wild rabbits in which no disease had been recorded (24). Strikingly, these sequences are very closely related to those sampled from virulent RHDV strains that were circulating in the late 1990s. On the basis of this evolutionary pattern it was suggested that virulent RHDV emerged due to mutation of an avirulent virus that was present in German rabbits exported to China and that had been circulating in Europe for many years (16, 24). Because a number of viral lineages clearly predate the first description of RHD in 1984, it was also proposed that high-virulence RHDV evolved a number of times independently close to 1984 (13, 16, 24). To test this hypothesis and to examine the evolutionary origins and dynamics of RHDV in more detail, we undertook an extensive Bayesian Markov chain Monte Carlo (MCMC) analysis of the available sequence data for RHDV. Our analysis provides new insights into the genesis of this important animal disease.
MATERIALS AND METHODS
Sequence data.
All available sequences (complete or partial) for the capsid gene of RHDV and RCV were downloaded from GenBank. Obvious duplicate sequences were removed, as were sequences for which no date of isolation could be obtained from either GenBank or from a literature search. Similarly, a set of 26 unpublished sequences from the United Kingdom (labeled as “PIT05”) were removed since we had no information about their provenance. This pruning resulted in an initial data set of 217 dated capsid gene sequences ranging from 316 to 1,740 nucleotides (nt) in length. Dates given for isolation ranged from 1955 to 2008 (see Table S1 in the supplemental material). Subsequently, an additional set of seven United Kingdom samples were removed, leaving a data set of 210 sequences (see Results).
Sequences were manually aligned by using SEAL (http://tree.bio.ed.ac.uk/software/seal/). The available capsid sequences were analyzed as either (i) the “total” sequence alignment containing sequences of various lengths (partial sequences ranged from 316 to 1,359 nt, while the complete capsid sequence was 1,740 nt in length); (ii) a “short” sequence alignment of 277 nt (spanning the region from nt 1111 to 1387), since this fragment was present in all available isolates; or (iii) the 63 sequences for which the “complete” 1,740-nt capsid sequence was available. The GenBank accession numbers of all sequences used in the present study are presented in Table S1 in the supplemental material.
Following our phylogenetic analysis (see Results), we also subdivided our RHDV data set into four distinct groups separated from each other by relatively long branch lengths and high posterior probability values on which we could undertake further analyses. These groups included group 1, comprising 25 sequences including the antigenic variant RHDVa sequences sampled in both Europe, America, and Asia (reference sequence, RHDVa_USA-Utah_UT-01_2001); group 2, comprising 36 sequences sampled mainly from Portugal and Spain (reference sequence, Spain_MC_89_1989); group 3, comprising 46 sequences sampled from diverse global locations including Australasia and the earliest Chinese isolate (reference sequence, WX_China_1984); and group 4, comprising 99 sequences of mainly European origin (reference sequence, Germany_Meiningen_1993).
Phylogenetic analysis.
To obtain an initial picture of the phylogenetic relationships of the RHDV and RV sequences, we inferred a maximum-likelihood (ML) tree for the total and short 217 sequence data sets using the PAUP* package (36) and using SPR branch swapping. For this analysis, the best-fit nucleotide substitution model was determined by MODELTEST (30) (parameter values are available from the authors on request). Support for key nodes on this phylogeny were determined by using bootstrap resampling based on 1,000 replicate neighbor-joining (NJ) trees inferred under the ML substitution model. Similarly, to obtain an initial measure of the degree of temporal structure in the “complete” capsid sequence data set, we performed a regression analysis of tree root-to-tip genetic distance against sampling time using the program Path-O-Gen (8; http://tree.bio.ed.ac.uk/software/pathogen/).
Bayesian MCMC analysis.
We estimated both the rate of nucleotide substitution per site and the time to the most recent common ancestor (TMRCA) for each data set by using the Bayesian MCMC approach available in the BEAST package (http://beast.bio.ed.ac.uk/) (9). In each case, we used both strict and relaxed (uncorrelated lognormal) molecular clocks, and a substitution model using the general-time-reversible substitution matrix with a different rate assigned to each codon position (although different substitution models produced similar results [data not shown, but available from the authors on request]). We also used the least constrained Bayesian skyline population coalescent prior in all cases. To reveal changes in relative genetic diversity through time (a measure of effective population size under neutral evolution), and particularly for the four subsets of RHDV, we inferred a Bayesian skyline plot for each data set (10). In each case, MCMC chains were run for sufficient time to achieve convergence (assessed by using the TRACER program [http://tree.bio.ed.ac.uk/software/tracer/]), with uncertainty in parameter estimates reflected in values of the 95% highest probability density (HPD). Because we could not achieve convergence for those data sets that included short (i.e., 277-nt) sequences, we based our final estimates of substitution rate on the “complete” capsid data set of 63 sequences from which we obtained very robust values. The mean rate and 95% HPD values obtained from this analysis (see Results) were then used as an empirical prior distribution on the substitution rate that is centered on the mean of the distribution; from this we were able to estimate all TMRCAs in the other data sets. Finally, for each data set we also used the BEAST program to compute the maximum clade credibility (MCC) tree from all of the plausible trees created during the BEAST run using the TreeAnnotator program, with the first 10% trees removed as burn-in.
Recombination analysis.
Recombination has been previously documented in RHDV (1, 14). To determine whether our estimates of substitution rate were adversely affected by recombination, we first screened for this process in the “complete” capsid sequence data set using the RPD, BOOTSCAN, and GENECOV programs available within the RDP3 package (22). This analysis revealed only the Germany_Hartmannsdorf_1996 sequence to be recombinant, as observed previously (see Results) (14). Therefore, we manually excluded the Germany_Hartmannsdorf_1996 sequence and repeated the BEAST analysis of substitution rates and TMRCAs as described above.
Analysis of ancient RHDV sequences.
We assessed the accuracy of reported sampling dates for seven RHDV capsid sequences sampled from the United Kingdom (Wellsbourne 1955, 1958, and 1959; Park Farm 1971, 1974, 1976, and 1981) by generating null distributions for their sampling dates. These null distributions represent the expected sampling time of these seven sequences given the observed sampling dates and genetic diversity of the 210 sequence “total” data set. The null distributions were estimated in BEAST by incorporating the sampling dates for each of the seven RHDV sequences in question as additional free parameters (with uniform priors of 0 to 100 years in the past [i.e., 1908 to 2008]). The remaining BEAST settings (i.e., relaxed uncorrelated lognormal molecular clock, nucleotide substitution model, and Bayesian skyline coalescent prior) were the same as those utilized above. The significance of the reported sampling dates for the seven RHDV capsid sequences was then calculated by comparison to their null distributions of sampling dates.
Spatial analysis.
To determine the extent and pattern of geographic structure in RHDV, we assigned each sequence in the “total” data set a single-letter character state reflecting the country of origin. The minimum number of changes in character state needed to produce the observed distribution of character states (excluding ambiguous changes) on the MCC tree for these data was then estimated by using the parsimony method in PAUP* (36). To determine the number expected under the null hypothesis of random mixing among countries, the character states of all isolates were randomized 1,000 times and the analysis repeated. Because MCC trees are automatically rooted, this analysis also allows us to infer the direction of migration events. As an additional assessment of the strength of geographical clustering, we computed the association index and parsimony score statistics of clustering strength by using the BaTS method (28) that examines all of the plausible trees produced by BEAST (1,000 replicates, with the first 10% of trees removed as burn-in) and therefore accounts for phylogenetic error. Both the association index and the parsimony score statistics measure the degree of association between a trait, in this case the sampling location, and the underlying phylogeny.
RESULTS
Phylogenetic relationships of RHDV.
Our provisional phylogenetic analysis generally corroborated those proposed previously (16, 20, 24, 26) (Fig. 1). In particular, we note that (i) there is a clear distinction between viruses assigned as RHDV and those described as RCV and (ii) that RHDV can be further subdivided into a number of phylogenetically distinct groups (see Fig. 2 for more details). In addition, our tip-to-root regression analysis of the “complete” capsid gene data set revealed that there is a clear temporal structure in these data, with viruses sampled earlier in time generally having shorter distances to the root of the tree (see Fig. S1 in the supplemental material).
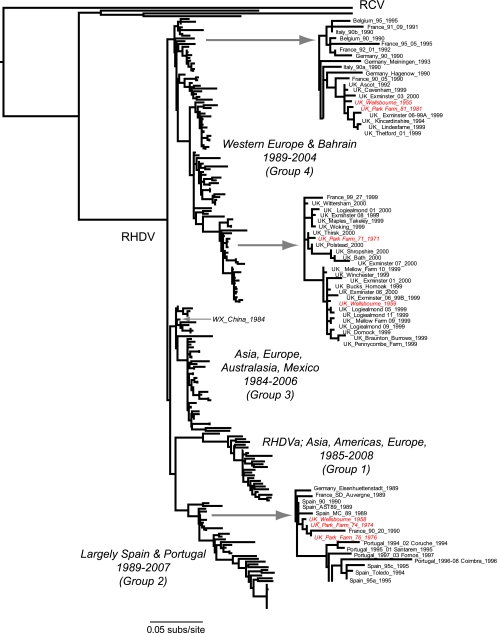
FIG. 1.
ML tree of 217 sequences of the capsid gene of RHDV and RCV. The tree is drawn to a scale of nucleotide substitutions per site and is midpoint rooted for purposes of clarity. The magnified portions of the tree show the positions of the seven United Kingdom isolates sampled before 1984. Note their close relationship to more contemporaneous isolates. A more comprehensive analysis of these data is presented in Fig. 2.
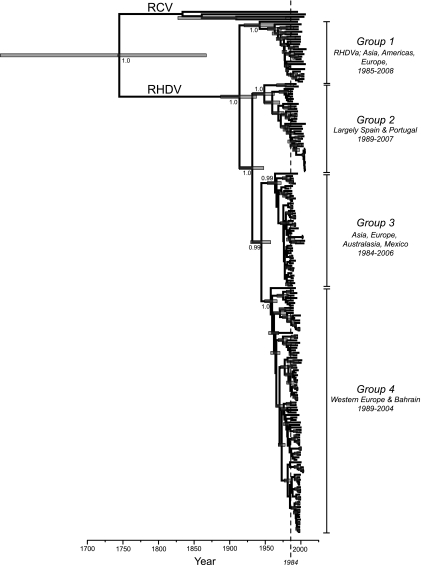
FIG. 2.
MCC tree of the “total” data set of 210 capsid gene sequences of RHDV and RCV. The time-scale (x axis) is given in years since the most recent sample. The four phylogenetic groups used in the subsequent analysis of RHDV population dynamics are indicated. Node numbers depict posterior probability values (a measure of node support), while node bars the depict the 95% HPD values on node height (age). The year 1984, indicating the year of the Chinese outbreak, is marked with a dotted line.
However, the most notable observation from the phylogenetic analysis, and one that clearly contradicts the notion of temporal structure, concerns seven of the United Kingdom sequences that were obtained by nested reverse transcription-PCR from archival samples of blood dating from as early as 1955: Wellsbourne 1955 (GenBank accession number, AF454040), Wellsbourne 1958 (AF454049), Wellsbourne 1959 (AF454007), Park Farm 1971 (AF454021), Park Farm 1974 (AF454047), Park Farm 1976 (AF454048), and Park Farm 1981 (AF454042). These sequences have been taken as evidence for the longstanding circulation of RHDV prior to the Chinese outbreak of 1984 (16, 24). However, our phylogenetic analysis reveals that although they fall into a variety of groups, these seven sequences are extremely closely related to the currently circulating virulent epidemic strains of RHDV, particularly those sampled from the United Kingdom, Spain, and France (Fig. 1). That these viruses show essentially no evolutionary change over periods of more than 20 years and cluster within groups of sequences sampled far more recently is highly anomalous and strongly suggests that they are modern contaminants (19). This possibility is examined in more detail below. Because of the uncertain provenance of these seven sequences, they were excluded from all subsequent analyses.
Rates of evolutionary change in RHDV.
The mean rate of nucleotide substitution estimated for the “complete” RHDV capsid sequence (63 sequences) using our Bayesian MCMC approach (relaxed molecular clock) was 7.7 × 10−4 nucleotide substitutions per site per year (subs/site/year), with 95% HPD values of 3.9 × 10−4 to 11.9 × 10−4 substitutions/site/year. Very similar mean rates (with strongly overlapping HPD values) were obtained using the strict molecular clock (mean = 6.1 × 10−4 substitutions/site/year) and with the divergent RCV sequences removed (mean = 7.0 × 10−4 substitutions/site/year). These rates also fall within the range previously observed for a diverse range of RNA viruses (17, 18). Similarly, our rate estimates do not seem to be adversely affected by recombination. Our screen for recombination revealed only the Germany_Hartmannsdorf_1996 sequence to be putatively recombinant (P = 2 × 10−3 under the BOOTSCAN method). Excluding this sequence had only a minor impact on evolutionary dynamics, producing in a mean substitution rate of 7.2 × 10−4 substitutions/site/year.
However, we were unable to obtain reliable rate estimates (i.e., nonconvergence) for the “total” or “short” 210 sequence RHDV data sets (results not shown, but available from the authors on request). This is most likely a function of the fact that the 277-nt region contains too few informative sites to undertake complex analyses of this kind. Therefore, we used the substitution rate estimated for the “complete” data set (i.e., mean of 7.7 × 10−4 subs/site/year) as a prior distribution to obtain estimates of the TMRCAs and MCC trees for these larger data sets, as well as those of the four groups of RHDV sequences.
Reliability of ancient RHDV sequences.
Given a mean substitution rate of 7.7 × 10−4 substitutions/site/year we would expect approximately one substitution per year in the 1,740-nt complete capsid sequence (equivalent to two lineages diverging for 0.5 years) and ∼1 substitution per 10 years in the 277-nt sequence (equivalent to two lineages diverging for 5 years). As such, the similarity of the seven United Kingdom sequences sampled prior to 1984 to the more recently sampled viruses is highly anomalous. To explore this issue in more detail, we extended our Bayesian MCMC approach. Our tests of the accuracy of reported sampling dates indicated that five of the seven United Kingdom sequences possess sampling dates that are inconsistent with the sampling dates and genetic diversity of the “total” data set of 210 RHDV capsid sequences analyzed here. Specifically, our tests reject (P < 0.05) the reported sampling dates of the Wellsbourne 1955 (mean = 1993.6, 95% HPD = 1981.3 to 2006.4), Wellsbourne 1958 (mean = 1986.0, HPD = 1969.7 to 2001.8), Wellsbourne 1959 (mean = 1999.9, HPD = 1993.7 to 2006.9), Park Farm 1971 (mean = 2001.5, HPD = 1994.5 to 2008), and Park Farm 1981 (mean 1993.9, HPD = 1982.3 to 2006.0) sequences; all of these sequences have reported sampling dates that fall outside of the estimated 95% HPD intervals for their null distributions. The reported sampling dates for the Park Farm 1974 (mean = 1985.4, HPD = 1968.7 to 2001.4) and Park Farm 1976 (mean = 1987.1, HPD = 1971.2 to 2003.1) sequences were both within the 95% HPDs of the null distributions and thus could not be rejected as inaccurate in this analysis, although it is notable that they fall toward the end of the credible distribution. Importantly, since substitution rates act as scalars upon time estimates, higher substitution rates than those used here would shift the null distributions toward the present and thus increase the significance (i.e., decrease the accuracy) of the reported sampling dates, whereas lower rates would produce the opposite effect.
Timing of RHDV evolution.
Our Bayesian MCMC analysis also allowed us to obtain robust estimates for times to common ancestry across the RHDV phylogeny. These are shown graphically as MCC trees for the “total” (Fig. 2), “short” 210 (see Fig. S2 in the supplemental material), and “complete” (see Fig. S3 in the supplemental material) capsid gene data sets.
In all data sets the TMRCAs across all of the viruses analyzed (i.e., both RHDV and RCV) was <550 years (mean values of 263, 318 and 215 years for the “total,” “short,” and “complete” data sets, respectively; 95% HPDs across all data sets, 66 to 515 years) and <150 years for the core group of RHDV isolates that represent the virulent form of virus that has spread around the world since 1984 (means of 94, 87, and 92 years for the “total,” “short,” and “complete” data sets, respectively; 95% HPDs across all data sets, 51 to 142 years). Hence, RHDV may have emerged early in the 20th century and there were clearly multiple lineages of RHDV circulating long before the Chinese outbreak of 1984 (represented by the dotted lines on the MCC trees).
The “RCV-like” viruses comprising RCV Italy 1996, RCV-A Australia 2008, Ashington UK 1998, and Lambay Island Ireland 2005 are characterized by long branch lengths, indicating considerable divergence both from each other and from the RHDV capsid sequences, with mean TMRCAs estimated to fall in 19th or early 20th centuries. Interestingly, the mean estimates for the TMRCA of the Ashington (i.e., United Kingdom) and Australian RCV strains fall in the 19th century (Fig. 2; see also Fig. S2 and S3 in the supplemental material) and are therefore compatible with the known history of the European rabbit; the earliest introduction of domestic European rabbits to Australia was in 1788, and wild European rabbits were introduced at least twice from 1859 (33).
Phylodynamic analysis of individual groups of RHDV.
Our phylogenetic analysis allowed us to identify four distinct groups of RHDV sequences (Fig. 2). To reveal the population dynamics of these groups, and hence key aspects of the epidemiological history of RHDV, we inferred both phylogenetic trees and Bayesian skyline plots for each group separately.
Group 1 is of particular interest because it contains the antigenic variants known as RHDVa which have caused four recent independent epidemics in the United States (23), as well as outbreaks in Cuba, Korea, China, Japan, Reunion Island, and Europe (2, 11, 20, 34, 37, 38) (see Fig. S4 in the supplemental material). RHDVa was first identified in Italy in 1997 (2), but related viruses were clearly present in China as early as 1985. Indeed, as with the other three groups identified here this group has relatively deep roots, with a mean estimated date for the MRCA of around 1966 (95% HPD = 1958 to 1974).
Group 2 is the most geographically distinct of the four groups, consisting predominantly of isolates sampled from the Iberian peninsula (Spain and Portugal), the south of France and one German isolate (see Fig. S5 in the supplemental material). This group also has a relatively old TMRCA, with a mean date of 1948 (95% HPD = 1931 to 1963). In addition, there are two reasonably well supported subgroups within this group, each with a mean TMRCA around 1962 but with clearly defined temporal structure; viruses in one of the subgroups were all isolated from 2000 to 2007, whereas viruses in the other group were isolated between 1989 and 1997.
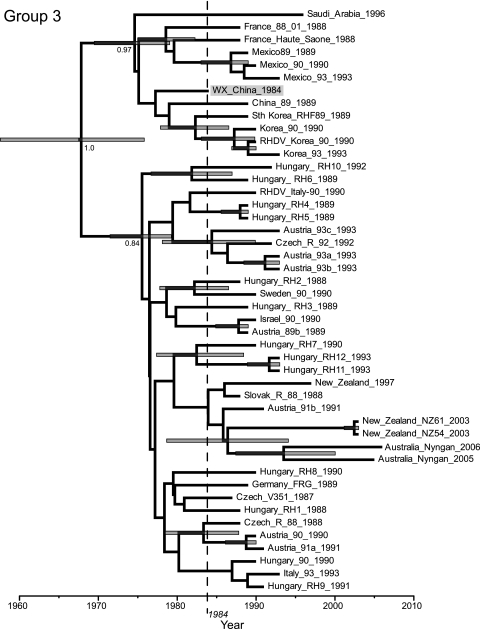
Group 3 contains the oldest RHDV isolate, dating to China in 1984, and hence representing the documented beginning of the RHDV epidemic (Fig. 3). However, this group also includes viruses isolated in central Europe (the Czech Republic, Slovakia, Germany, Austria, and Hungary), Italy, France, and Sweden isolated from 1987 to 1993, as well as viruses sampled from epidemics in Korea and Mexico. The mean estimate for the TMRCA of this group was 1967 (95% HPD = 1958 to 1976). Interestingly, the viruses isolated in France in 1988 and Mexico in 1989 form a strongly defined subcluster with the earliest Chinese virus (along with viruses from Korea and Saudi Arabia) that we estimate to have existed in the 1970s (95% HPD = 1970 to 1979). Group 3 also includes the small number of sequences from Australia and New Zealand, where virus derived from the Czech v351 strain spread from 1995 and 1997, respectively (5), although this part of the tree is relatively poorly supported.
FIG. 3.
MCC tree of RHDV group 3 which contains the earliest Chinese isolate from 1984 (shaded). The time-scale (x axis) is given in years since the most recent sample. Node numbers depict posterior probability values, while node bars the depict the 95% HPD values on node height. The year 1984, indicating the year of the Chinese outbreak, is marked with a dotted line.
The final phylogenetic group, group 4, contained the largest group (99 sequences) and contains RHDV isolates from central Europe, France, Belgium, Holland, the United Kingdom, and Ireland isolated between 1989 to 2004, as well as a single isolate from Bahrain (see Fig. S6 in the supplemental material). Although the mean TMRCA dates to 1959 (95% HPD = 1949 to 1968), many of the lineages in this group seem to have originated in the 1970s to early 1980s.
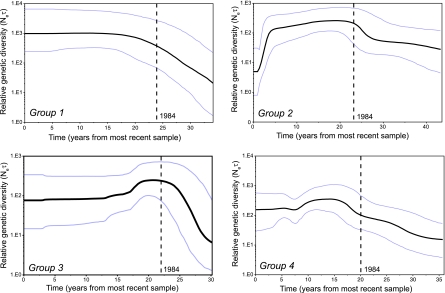
These four phylogenetic groups have contrasting phylodynamic patterns (Fig. 4). Specifically, groups 1, 3, and 4 show fairly flat dynamics; that is, after a period of increasing genetic diversity 25 to 30 years ago, that was trending upward before the 1984 outbreak, there has been no obvious increase in diversity in recent years. In contrast, group 2, which mostly contains sequences from the Iberian peninsula, is apparently decreasing in relative genetic diversity (and hence likely in population size). This pattern is compatible with the control of RHD in domestic rabbits by vaccination and the reported decline in the wild rabbit population of the Iberian peninsula due to RHD (5). This group also shows a clear geographic barrier to entry of RHDV from the rest of Europe (25); it is notable that the emerging RHDVa strains have seemingly not contributed to viral populations in this region or at least have not been sampled. Finally, it is striking that group 1, which contains the sequences related to the antigenic variant RHDVa, possesses the greatest genetic diversity (y axis) and hence likely possess the largest effective population size. This is compatible with elevated fitness of this group compared to the others defined in the analysis.
FIG. 4.
Bayesian skyline plots depicting the changing levels of relative genetic diversity through time (Neτ, where Ne is the effective population size and τ the generation time between subsequent transmission events). Note that because the date of the most recent sample differs between groups, the skyline plots for each group are on slightly different absolute time-scales. The year 1984, indicating the year of the Chinese outbreak, is marked with a dotted line.
Phylogeography of RHDV.
Both the parsimony and Bayesian analyses revealed a strong geographical clustering by country in the RHDV and RCV sequences analyzed here (P < 0.001 in all cases). Hence, although there was clearly a widespread geographical dispersal of RHDV (and presumably of infected rabbits) during the 20th century, there is still stronger clustering within specific geographical regions than expected under random movement (panmixis). Hence, within-country evolution is the most important phylodynamic pattern in RHDV. A more detailed examination of the potential movements between specific geographic localities reveals that France is the most important source population for RHDV among the available data, including some clear migration to Germany (see Table S2 in the supplemental material). The only other viral migration of note is between Portugal and Spain. However, the small and biased nature of sampling dictates that all conclusions pertaining to viral phylogeography should be drawn with caution.
DISCUSSION
Virulent RHDV most likely evolved from an avirulent RCV (6). Indeed, sequences of RCV-like viruses that are antigenically cross-reactive with RHDV have been characterized from Italy, the United Kingdom, Ireland, and Australia (3, 15, 24, 35). With the possible exception of the Ashington virus (United Kingdom), which was isolated from a dead rabbit (24), these viruses appear to circulate “harmlessly” in wild and domestic O. cuniculus. Our molecular clock analysis of capsid nucleotide sequences reveals that RCV-like viruses are clearly divergent from each other and from RHDV, with a common ancestor for all viruses over 200 years ago. Similarly, our mean estimates for the TMRCA of the main cluster of RHDV sequences place this in the early 20th century. In other words, these RCV-like viruses have not recently given rise to RHDV. However, the circulation of these RCV-like viruses, probably in an oral-fecal cycle with a gut tropism (3, 35), does explain the detection of antibodies in sera stored prior to 1984 that react with RHDV and in rabbits that had never been exposed to RHDV (3, 4, 21, 26, 27, 31, 32). It can be assumed that these viruses spread to Australia and New Zealand in wild and/or domestic rabbits introduced from the United Kingdom. Presumably, similar viruses were introduced with European rabbits to other countries, but these have not been identified due to a lack of surveillance.
All four of the phylogenetic groups defined in our analysis contain virulent RHDV. A common feature of all of these groups is that many lineages likely originated during the 1970s, suggesting that there was a period of viral radiation at this time. This is shown clearly on the Bayesian skyline plots, which all depict an increase in genetic diversity (and hence population size) prior to 1984. Crucially, this also means that there were already multiple separate lineages of RHDV before the documented emergence of RHD in China in 1984. The central puzzle of RHDV evolution is that each of these lineages contains virulent viruses, and each predates the documented emergence of RHD in 1984. This implies either that high virulence evolved multiple times in multiple viral lineages close to 1984 or (more plausibly) that virulence emerged earlier in the 20th century but the disease was not documented until 1984 when the trade in rabbits provided the opportunity for RHDV to spread from an established, but apparently cryptic, transmission cycle.
Prior phylogenetic work led to suggestions that RHDV with sequences closely related to those that emerged from China in 1984 were circulating harmlessly in the United Kingdom and other European localities during the 1950s; hence, it was suggested that virulence emerged at least twice during the late 20th century: once in Europe and once in China (13, 16, 24). However, there are at least two problems with this conceptual model. First, we show here that the sequences from the 1950s and 1970s from the United Kingdom appear to be modern contaminants: given the rate of RHDV evolution documented here and that of RNA viruses more generally, these early RHDV sequences are expected to be far more divergent from their modern counterparts. If we remove these sequences from the analysis, there is now no evidence for RHDV-like viruses, as distinct from RCV-like viruses, in Europe prior to 1986. Second, this model requires that high virulence emerged independently multiple times in all of the lineages containing virulent virus that date prior to 1984 (and not just two independent introductions as previously proposed). This model seems untenable given that our molecular clock studies reveal that a multitude of viral lineages date back this far in epidemiological time (i.e., fall to the left of the dotted line on the tree figures).
Another scenario is that there is so much recombination in RHDV that it is impossible to infer evolutionary history and hence the evolution of virulence. Although recombination has clearly played some role in the evolution of RHDV (1, 14), all capsid phylogenies presented to date show generally congruent groups, and our observation of clear geographic structure argues against large-scale recombination. In addition, recombination seemed to have little effect on our estimates of times to common ancestry. We therefore do not regard rampant recombination as a viable explanation.
Similarly, the possibility that RHDV is not a virus of European rabbits but has recently jumped from another lagomorph species can be discounted since the RCV-like and RHDV isolates are clearly closely related; any virus that had recently jumped species boundaries is likely to have much deeper roots. For example, European brown hare syndrome virus is a relative of RHDV that causes a similar disease to RHD in European hare species. However, European brown hare syndrome virus capsid sequences are much more divergent from RHDV than are the RCV-like viruses (26), and the virus does not infect European rabbits. The only scenario that cannot be totally excluded in this context is that a virus of European rabbits jumped species and then diverged in its new host before jumping back into European rabbits but with a newly evolved virulent phenotype. However, the shift back into European rabbits either had to occur 80 or more years ago or the virus must have diverged in the alternative host and then crossed into European rabbits multiple times in recent years to explain the phylogenetic patterns observed. In addition, there is no evidence for such a reservoir host, and all testing to date has indicated that RHDV is unable to replicate in species other than O. cuniculus (reviewed in reference 6).
Therefore, we propose that the most likely scenario is that virulent RHDV strains evolved once, early in the 20th century, but were not detected until 1984. Although this seems to be the most parsimonious explanation on the currently available data, it does require an explanation of why virulent disease was not observed before 1984. One scenario is that evolution of virulent RHDV did not occur in Europe, but rather took place in Asia in farmed European rabbits. This model is supported by several lines of evidence. First, RHD clearly emerged in China. This emergence was also coincident with a large expansion of rabbit production; rabbit meat exported from China increased from 308 tonnes in 1975 to 53,200 tonnes in 1983 (12; cited in reference 6). Given the difficult sociopolitical conditions in China and neighboring countries in the first half of the 20th century, it is plausible that a virulent disease in rabbits was able to evolve in this region without leaving a clear record. The peculiar epidemiology of RHDV may have aided this emergence since kittens less than 8 weeks of age can be infected and shed virus but do not exhibit clinical disease; moreover, the virus is relatively robust and persists in the environment (6). It has been noted that diagnosis of RHD in young rabbits in China is difficult since it does not have the same clinical presentation as RHD in older animals (38). In addition, avirulent strains of virus may have provided similar cross-protection to that observed or inferred with RCV (3, 31, 35). This may have allowed the coexistence of virulent and avirulent viral strains so that large-scale disease outbreaks may have been unusual until the intensification of production. Indeed, it is interesting that RHD was first described in imported rabbits that may have been immunologically naive. Alternatively, it is possible (although less likely) that viral lineages somehow circulated undetected in Europe until 1986 but spread to China in or prior to 1984, and then around the world from China, either spreading back to Europe or emerging from cryptic foci in Europe.
Finally, it is notable that an antigenic variant of RHDV, termed RHDVa, was first described in 1997 (2) and has spread around the world causing epidemics in China, Korea, the United States, Mexico, Japan, Reunion Island, and Europe (11, 20, 23, 34, 37). It has been suggested that this strain is more competitive than the previous RHDV strains and is replacing these in the wild in parts of Europe (23). In support of this hypothesis we note that all RHDVa viruses form a distinct phylogenetic group and that this group has the highest relative genetic diversity within RHDV, indicative of the highest effective population size. One of the most intriguing aspects of RHDV evolution is that this virus appears to have maintained its very high virulence during the 25 years since it emerged. At face value this suggests that virulence is adaptive for transmission. If more competitive strains of RHDV are emerging, it will be of interest to understand both the molecular and epidemiological bases to this enhanced competitiveness. From a biological control perspective it would also be important to evaluate whether these strains could increase the impact of RHDV on Australian wild rabbit populations.
Supplementary Material
Footnotes
Published ahead of print on 16 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abrantes, J., P. J. Esteves, and W. van der Loo. 2008. Evidence for recombination in the major capsid gene VP60 of the rabbit haemorrhagic disease virus (RHDV). Arch. Virol. 159:329-335. [DOI] [PubMed] [Google Scholar]
- 2.Capucci, L., F. Fallacara, S. Grazioli, A. Lavazza, M. L. Pacciarini, and E. Brocchi. 1998. A further step in the evolution of rabbit haemorrhagic disease virus: the appearance of the first consistent antigenic variant. Vet. Res. 58:115-126. [DOI] [PubMed] [Google Scholar]
- 3.Capucci, L., P. Fusi, A. Lavazza, M. L. Pacciarini, and C. Rossi. 1996. Detection and preliminary characterization of a new calicivirus related to rabbit haemorrhagic disease virus but nonpathogenic. J. Virol. 70:8614-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capucci, L., A. Nardin, and A. Lavazza. 1997. Seroconversion in an industrial unit of rabbits infected with a non-pathogenic rabbit haemorrhagic disease-like virus. Vet. Rec. 140:647-650. [DOI] [PubMed] [Google Scholar]
- 5.Cooke, B. D. 2002. Rabbit haemorrhagic disease: field epidemiology and the management of wild rabbit populations. Rev. Sci. Tech. 21:347-358. [DOI] [PubMed] [Google Scholar]
- 6.Cooke, B. D., and F. Fenner. 2002. Rabbit haemorrhagic disease and the biological control of wild rabbits, Oryctolagus cuniculus, in Australia and New Zealand. Wildl. Res. 29:689-706. [Google Scholar]
- 7.Cooke, B. D., S. McPhee, A. J. Robinson, and L. Capucci. 2002. Rabbit haemorrhagic disease: does a pre-existing RHDV-like virus reduce the effectiveness of RHD as a biological control in Australia. Wildl. Res. 29:673-682. [Google Scholar]
- 8.Drummond, A., O. G. Pybus, and A. Rambaut. 2003. Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol. 54:331-358. [DOI] [PubMed] [Google Scholar]
- 9.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond, A. J., A. Rambaut, B. Shapiro, and O. G. Pybus. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185-1192. [DOI] [PubMed] [Google Scholar]
- 11.Farnos, O., D. Rodrıguez, O. Valdes, M. Chiong, F. Parra, J. R. Toledo, E. Fernandez, R. Lleonart, and M. Suarez. 2007. Molecular and antigenic characterization of rabbit hemorrhagic disease virus isolated in Cuba indicates a distinct antigenic subtype. Arch. Virol. 152:1215-1221. [DOI] [PubMed] [Google Scholar]
- 12.Feng-Yi, Z. 1990. The rabbit industry in China. J. Appl. Rabbit Res. 12:278-279. [Google Scholar]
- 13.Forrester, N. L., M. I. Abubakr, E. M. E. Abu Elzein, A. I. al-Afaleq, F. M. T. Housawi, S. R. Moss, S. L. Turner, and E. A. Gould. 2006. Phylogenetic analysis of Rabbit haemorrhagic disease virus strains from the Arabian peninsula: did RHDV emerge simultaneously in Europe and Asia? Virology 344:277-282. [DOI] [PubMed] [Google Scholar]
- 14.Forrester, N. L., S. R. Moss, S. L. Turner, H. Schirrmeier, and E. A. Gould. 2008. Recombination in rabbit haemorrhagic disease virus: possible impact on evolution and epidemiology. Virology 376:390-396. [DOI] [PubMed] [Google Scholar]
- 15.Forrester, N. L., R. C. Trout, and E. A. Gould. 2007. Benign circulation of rabbit haemorrhagic disease virus on Lambay Island, Eire. Virology 358:18-22. [DOI] [PubMed] [Google Scholar]
- 16.Forrester, N. L., R. C. Trout, S. L. Turner, D. Kelly, B. Boag, S. Moss, and E. A. Gould. 2006. Unravelling the paradox of rabbit haemorrhagic disease virus emergence, using phylogenetic analysis; possible implications for rabbit conservation strategies. Biol. Conserv. 131:296-306. [Google Scholar]
- 17.Hanada, K., Y. Suzuki, and T. Gojobori. 2004. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 21:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:152-161. [DOI] [PubMed] [Google Scholar]
- 19.Krasnitz, M., A. J. Levine, and P. Rabadan. 2008. Anomalies in the influenza virus genome database: new biology or laboratory errors? J. Virol. 82:8947-8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Gall-Recule, G., F. Zwingelstein, S. Laurent, C. de Boisseson, Y. Portejoie, and D. Rasschaert. 2003. Phylogenetic analysis of rabbit haemorrhagic disease virus in France between 1993 and 2000, and the characterisation of RHDV antigenic variants. Arch. Virol. 148:65-81. [DOI] [PubMed] [Google Scholar]
- 21.Marchandeau, S., G. Le Gall-Recule, S. Bertagnoli, J. Aubineau, G. Botti, and A. Lavazza. 2005. Serological evidence for a non-protective RHDV-like virus. Vet. Res. 36:53-62. [DOI] [PubMed] [Google Scholar]
- 22.Martin, D. P. 2009. Recombination detection and analysis using RDP3. Methods Mol. Biol. 537:185-205. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh, M. T., S. C. Behan, F. W. Mohamed, Z. Lu, K. E. Moran, T. G. Burrage, J. G. Neilan, G. B. Ward, G. Botti, L. Capucci, and S. A. Metwally. 2007. A pandemic strain of calicivirus threatens rabbit industries in the Americas. Virol. J. 4:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss, S. R., S. L. Turner, R. C. Trout, P. J. White, P. J. Hudson, A. Desai, M. Armesto, N. L. Forrester, and E. A. Gould. 2002. Molecular epidemiology of rabbit haemorrhagic disease virus. J. Gen. Virol. 83:2461-2467. [DOI] [PubMed] [Google Scholar]
- 25.Muller, A., J. Freitas, E. Silva, G. Le Gall-Recule, F. Zwingelstein, J. Abrantes, P. J. Esteves, P. C. Alves, W. van der Loo, J. Kolodziejek, N. Nowotny, and G. Thompson. 2009. Evolution of rabbit haemorrhagic disease virus (RHDV) in the European rabbit (Oryctolagus cuniculus) from the Iberian Peninsula. Vet. Microbiol. 135:368-373. [DOI] [PubMed] [Google Scholar]
- 26.Nowotny, N., C. Ros Bascuñana, A. Ballagi-Pordany, D. Gavier-Widen, M. Uhlen, and S. Belak. 1997. Phylogenetic analysis of rabbit haemorrhagic disease and European brown hare syndrome viruses by comparison of sequences from the capsid protein gene. Arch. Virol. 142:657-673. [DOI] [PubMed] [Google Scholar]
- 27.O'Keefe, J. S., J. E. Tempero, M. X. J. Motha, M. F. Hansen, and P. H. Atkinson. 1999. Serology of rabbit haemorrhagic disease virus in wild rabbits before and after the release of the virus in New Zealand. Vet. Microbiol. 66:29-40. [DOI] [PubMed] [Google Scholar]
- 28.Parker, J., A. Rambaut, and O. G. Pybus. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239-246. [DOI] [PubMed] [Google Scholar]
- 29.Parkes, J. P., G. L. Norbury, R. P. Heyward, and G. Sullivan. 2002. Epidemiology of rabbit haemorrhagic disease (RHD) in the South Island, New Zealand, 1997-2001. Wildl. Res. 29:543-555. [Google Scholar]
- 30.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, A. J., P. D. Kirkland, R. I. Forrester, L. Capucci, B. D. Cooke, and A. W. Philbey. 2002. Serological evidence for the presence of a calicivirus in Australian wild rabbits, Oryctolagus cuniculus, before the introduction of rabbit haemorrhagic disease virus (RHDV): its potential influence on the specificity of a competitive ELISA for RHDV. Wildl. Res. 29:652-655. [Google Scholar]
- 32.Rodak, L., B. Smid, L. Valicek, T. Vesely, J. Stepanek, J. Hampl, and E. Jurak. 1990. Enzyme linked immunosorbent assays to rabbit haemorrhagic disease virus and determination of its major structural proteins. J. Gen. Virol. 71:1075-1080. [DOI] [PubMed] [Google Scholar]
- 33.Rolls, E. C. 1969. They all ran wild. Angus and Robertson, Melbourne, Australia.
- 34.Schirrmeier, H., I. Reimann, B. Köllner, and H. Granzow. 1999. Pathogenic, antigenic and molecular properties of rabbit haemorrhagic disease virus (RHDV) isolated from vaccinated rabbits: detection and characterization of antigenic variants. Arch. Virol. 144:719-735. [DOI] [PubMed] [Google Scholar]
- 35.Strive, T., J. D. Wright, and A. J. Robinson. 2009. Identification and partial characterisation of a new lagovirus in Australian wild rabbits. Virology 384:97-105. [DOI] [PubMed] [Google Scholar]
- 36.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 37.Tian, L., J. Liao, J.-W. Li, W.-R. Zhou, X.-L. Zhang, and H.-N. Wang. 2007. Isolation and identification of a non-haemagglutinating strain of rabbit haemorrhagic disease virus from China and sequence analysis for the VP60 gene. Virus Genes 35:745-752. [DOI] [PubMed] [Google Scholar]
- 38.Yang, L., F. Wang, B. Hu, J. Xue, Y. Hua, B. Zhou, D. Wang, and W. Xu. 2008. Development of an RT-PCR for rabbit hemorrhagic disease virus (RHDV) and the epidemiology of RHDV in three eastern provinces of China. J. Virol. Methods 151:24-29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.