Abstract
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) ac96 is a core gene, but its role in virus replication is still unknown. To determine its role in the baculovirus life cycle, we used the AcMNPV bacmid system to generate an ac96-null virus (vAc96null). Our analyses showed that the absence of ac96 does not affect budded virus (BV) production or viral DNA replication in infected Sf9 cells. Western blotting and confocal immunofluorescence analysis showed that AC96 is expressed in both the cytoplasm and the nucleus throughout infection. In addition, AC96 was detected in the envelope fractions of both BV and occlusion-derived virus. Injection of vAc96null BV into the hemocoel killed Trichoplusia ni larvae as efficiently as repaired and control viruses; however, vAc96null was unable to infect the midgut tissue of Trichoplusia ni larvae when inoculated per os. Therefore, the results of this study show that ac96 encodes a new per os infectivity factor (PIF-4).
The Baculoviridae comprise a large and diverse group of viruses that are pathogens of insects, mainly from the Lepidoptera, Hymenoptera, and Diptera. During the typical biphasic infection cycle, two structurally and functionally distinct enveloped virion phenotypes are produced: occlusion-derived virus (ODV) and budded virus (BV) (35). The primary infection cycle in animals begins in the midgut cell after occlusion bodies (OBs) are ingested. Upon ingestion, the OBs dissolve in the alkaline environment of the midgut, and the ODVs are released into the lumen of midgut (15, 16, 20). Virions pass through a disrupted peritrophic membrane, a process often facilitated by enhancins, a group of virus-encoded metalloproteases (38). Subsequently, ODVs bind to and fuse directly with the microvilli of midgut columnar epithelial cells. A protein receptor is proposed to mediate the process, since binding is proteinase sensitive and saturable (15, 16, 20). After the nucleocapsids are transported to the nuclei of the midgut cells, viral DNA is released, followed by gene expression, DNA replication, and assembly of progeny nucleocapsids. In the late phase of infection, newly formed nucleocapsids are transported to the cell membrane, bud from the cell, and acquire a new envelope from the basal membrane. The BVs spread via the hemolymph (16) and the tracheal system (8) into the other tissues of the insect, causing the secondary infection.
Baculoviruses encode per os infectivity factors (PIFs) on the envelope surface of ODV to initiate the efficient primary infection in midgut. So far, four highly conserved core genes, p74 (pif-0), pif-1, pif-2, and pif-3, have been identified. The deletion of the Autographa californica multiple nucleopolyhedrovirus (AcMNPV) p74 gene results in the complete elimination of the per os infectivity of OBs, while virions purified from mutant OBs were infectious when injected into the hemocoels of Trichoplusia ni or Heliothis virescens larvae (13, 17, 22). P74 is proposed to function as an ODV attachment protein that binds to a specific 30-kDa receptor protein on the primary target cells within the midgut (17, 39). PIF-1 was originally identified in Spodoptera littoralis NPV, where the deletion of pif-1 (spli7) resulted in viruses that were unable to infect S. littoralis larvae per os (21). PIF-2 was first identified in Spodoptera exigua MNPV, and the disruption of pif-2 resulted in the complete loss of per os infectivity for the host (11, 31). PIF-1 and PIF-2 have also been shown to participate in the binding of ODV to target cells in the midgut (28). PIF-3 (ac115) is also an essential factor for oral infection of AcMNPV. Although PIF-3 is not required for ODV attachment and fusion, it may mediate a critical downstream event, such as the translocation of ODV along microvilli during primary infection (28).
AcMNPV, the archetype Alphabaculovirus of the Baculoviridae, has a double-stranded DNA genome of approximately 134 kbp that contains 154 predicted open reading frames (ORFs) (1). Comparative analysis of the 49 completely sequenced baculovirus genomes reveals 31 core genes that are conserved in all baculovirus genomes and are therefore likely to serve important roles in baculovirus life cycles (14, 26, 32, 37). Most core genes are related either to DNA replication, gene expression, packaging and assembly, or per os infection (37). Four core genes, ac68, p33 (ac92), ac96, and ac109, still have no known function or sequence similarities to proteins of known functions.
In this study, an ac96-null mutant was constructed utilizing an AcMNPV bacmid, and the results showed that in tissue culture, ac96 was nonessential and was not required for viral DNA replication, ODV production, or BV production. However, in vivo assays demonstrated that the ac96-null virus was unable to infect midgut tissue when T. ni larvae were inoculated per os. The core gene ac96 therefore encodes a new per os infectivity factor, PIF-4.
MATERIALS AND METHODS
Viruses and cells.
Spodoptera frugiperda Sf9 cells were maintained in 10% fetal bovine serum-supplemented TC100 medium at 27°C. AcMNPV recombinant bacmids were derived from bacmid bMON14272 (Invitrogen Life Technologies) and propagated in Escherichia coli strain DH10B.
5′-RACE.
To map the transcription start and stop sites for ac96, RNA was extracted from Sf9 cells infected with AcMNPV-E2 virus and collected at 4 h postinfection (4 hpi) and 24 hpi using a Qiagen RNeasy kit. Rapid amplification of 5′ cDNA ends (5′-RACE) was conducted by following the GeneRacer kit manufacturer's protocol (Invitrogen). Gene-specific primer 1 (GSP1) 1739 (5′-TTGAAATCTTGTATTAGCGTTTC-3′) and GSP2 1740 (5′-CCAACATGATTGACAACATT-3′) were paired with the GeneRacer 5′ nested primers to amplify the 5′ end of the ac96-specific transcript. The ac96-specific PCR fragments were cloned into the pDrive vector (Qiagen) and sequenced with the M13 forward primer.
Construction of bMON14272 ac96-null and repaired bacmids.
The ac96 knockout was generated using the method described by Datsenko and Wanner (7). Briefly, a zeocin resistance gene was amplified using primers 1709 (5′-ATA TGCCACCGCATGCACGCCGGTCAGCAGCTTGACGCTAATTGAACAT TCGGATCTCTGCAGCAC-3′) and 1571 (5′-CACATCGAGAACGAGCGTGTGATCGGGCACGTTATTTTTTAATGTTGCAATCGAGGTCGACCCCCCTG-3′) with p2ZeoKS as the template. The primers contain 48 bp and 50 bp homologous to the C terminus of ac96, respectively. The PCR fragment of the zeocin resistance gene was gel purified and electroporated into Escherichia coli BW25113-pKD46 cells, which contained AcMNPV bacmid bMON14272 DNA. Electroporated cells were incubated at 37°C for 4 h in 3 ml of SOC medium (2% Bacto tryptone, 0.5% Bacto yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) and were placed on an agar medium containing zeocin (30 μg/ml) and kanamycin (50 μg/ml). Plates were incubated at 37°C overnight, and colonies resistant to zeocin and kanamycin were selected for further confirmation by PCR.
Two different pairs of primers from the ac96 locus of the AcMNPV bacmid genome were used to confirm that ac96 had been inactivated by the correct insertion of the zeocin cassette into the AcMNPV bacmid genome (see Fig. 2). Primers 1572 (5′-CTGTTCGCGTGTTTCT-3′) and 1014 (5′-CCGATATACTATGCCGATGAT T-3′) and primers 1573 (5′-ACAATGAAATAATACAAAAC-3′) and 1239 (5′-CTGACCGACGCCGACCAA-3′) were used to detect the correct insertion of the zeocin gene cassette at the two junctions of ac96 locus. Fragments of 536 bp and 407 bp, which were amplified with primers 1572 and 1014 and primers 1573 and 1239 from the ac96-null bacmid, confirmed the correct insertion of zeocin in the ac96 locus. One of the recombinant bacmids confirmed by PCR was selected and named bMON14272-ac96null. The bMON14272-ac96null bacmid was repaired with pFAcT-GFP (6), which contains polyhedrin and gfp as a marker, by Tn7-mediated transposition (25). This virus was named Ac96null.
FIG. 2.
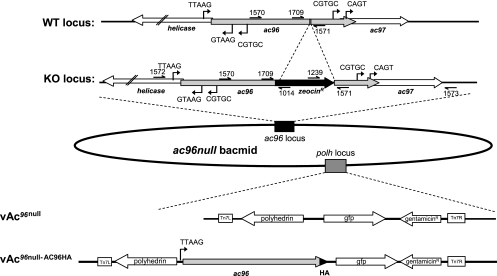
Schematic of the construction of the vAc96null and vAc96null-AC96HA bacmids. The ac96 ORF was inactivated by insertion of the zeocin resistance gene cassette (amplified with primers 1709 and 1571) between bp 84733 and 84734 of the AcMNPV genome (NC_001623.1) via homologous recombination in E. coli to generate the ac96-null bacmid. The correct insertion of the zeocin resistance cassette was confirmed using primers 1572 and 1014 and primers 1239 and 1573; their relative positions are indicated. The lower part of the figure shows the genes inserted into the polyhedrin (polh) locus of the ac96-null bacmid by Tn7-mediated transposition to generate vAc96null and vAc96null-AC96HA. WT, wild type; KO, knockout.
To tag AC96 with the influenza hemagglutinin (HA) epitope (CYPYDVPDYASL) at the C terminus, ac96 was amplified from bMON14272 with primers 1591 (5′-TGTTTCTCTGCAGGTTCTTC-3′) and 1592 (5′-GCGTCTA GATTAGGCGTAGTCGGGCACGTCGTAGGGGTATTCGGCCAACAAA TCCACGTA-3′). The ac96HA PCR fragments, which contain their native promoters and ORFs, were first digested with PstI and XbaI and then cloned into the same sites of pFAcT-GFP-Tnie1p(A) (27). The resulting plasmid, pFAct-ac96HA, contains ac96HA with the Trichoplusia ni SNPV ie1 poly(A) signal downstream, polyhedrin, and gfp as a marker. This plasmid was confirmed by restriction enzyme digestion and sequencing. Ac96HA, polyhedrin, and gfp were introduced back into bMON14272-ac96null as described above, and the resulting virus was named vAc96null-AC96HA. The control virus, AcBac, consisted of the bacmid bMON14272 repaired with polhedrin and gfp.
Time course analysis of BV production and DNA replication.
Sf9 cells (1.0 × 106/35-mm-diameter six-well plate) were transfected using liposomes (4) with 1.0 μg of each bacmid (vAc96null, vAc96null-AC96HA, and the control virus vAcBac). At various times posttransfection, the supernatant containing BV was harvested, and cell debris was removed by centrifugation (at 8,000 × g for 5 min). Infected cells were washed once with phosphate-buffered saline (PBS, consisting of 137 mM NaCl, 10 mM phosphate, and 2.7 mM KCl [pH 7.4]), scraped off with rubber policemen, and pelleted by centrifugation at 800 × g for 5 min; the supernatant was removed; and the pellets were stored at −80°C until analysis. BV production and viral DNA replication analyses were performed by quantitative PCR (qPCR) as described previously (27). BV titers at 6 and 72 h posttransfection (hpt) were also titrated in duplicate by endpoint dilution in Sf9 cells with 96-well microtiter plates.
Purification of BV and ODV.
Sf9 cells were infected with vAv96null-AC96HA at a multiplicity of infection (MOI) of 0.1 in two 50-ml spinner tissue culture flasks. Six days postinfection (p.i.), 80 ml of infected cells was harvested. BV and ODV were purified, and the envelope and nucleocapsids of BV and ODV were fractionated, as previously described (10).
Western blot assay.
Protein samples from Sf9 cell pellets and purified virions were subjected to Western blotting as described in a previous study (10). The antibodies used were (i) a mouse monoclonal anti-HA antibody (HA.11; Covance) (1:1,000), (ii) a mouse monoclonal antibody against Orgyia pseudotsugata MNPV (OpMNPV) VP39 (1:3,000) (29), (iii) mouse monoclonal antibody 4B7 against AcMNPV IE1 (1:5,000) (33), and (iv) a mouse monoclonal anti-AcV5 antibody recognizing GP64 (1:200) (18).
Immunofluorescence.
Sf9 cells were infected with vAc96null-AC96HA at an MOI of 5. At 24, 36, and 48 hpi, the supernatant was removed, and the cells were washed once in PBS, followed by fixation in 3.5% paraformaldehyde in PBS for 10 min. The fixed cells were washed three times in PBS for 15 min, followed by permeabilization in 0.15% Triton X-100 in PBS for 15 min. The cells were then blocked for 60 min in blocking buffer (2% bovine serum albumin in PBS), followed by a 1-h incubation with the anti-HA antibody (1:200). Cells were washed three times, for 10 min each time, in blocking buffer, followed by a 1-h incubation with an Alexa 635-conjugated goat anti-mouse antibody (1:500; Molecular Probes). Cells were subsequently washed three times for 10 min in PBS, stained with 200 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) (Sigma), and examined using a Leica confocal microscope.
In vivo infectivity assays.
The infectivity of BV was examined by injecting 1,000, 30, or 3 TCID50 (50% tissue culture infective dosage) units of BV of vAc96null, vAc96null-AC96HA, or the control virus vAcBac into 4th-instar T. ni larvae. Grace's medium was used as a negative control. For the oral-infectivity assay, OBs of vAcBac, vAc96null, and vAc96null-AC96HA were purified from Sf9 cells and diseased larvae as described previously by Li et al. (23) and Erlandson et al. (9), respectively. Oral infection assays were conducted with 4th-instar T. ni using a leaf disc bioassay method (9) to administer doses of 5, 50, or 500 OBs per larva. Infected larvae were reared individually in 24-well plates and monitored daily until all larvae had either pupated or died. Tissue samples from all larval cadavers were examined by phase-contrast microscopy to determine viral infection status (the presence or absence of OBs). A separate experiment was conducted to assay for midgut infection; 4th-instar larvae were fed 50,000 OBs, and midgut epithelium preparations were isolated, using the technique of Braun and Keddie (2), at 48 hpi. The preparations were then observed with UV fluorescence microscopy to determine green fluorescent protein (GFP) expression.
RESULTS
Sequence analysis of ac96 and its homologues.
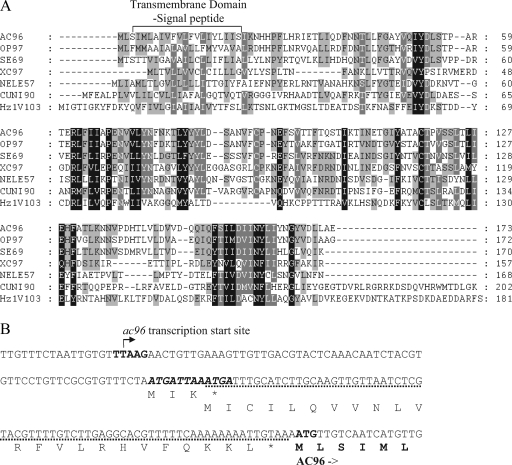
ac96 is a core gene, and its homologues are found in all baculoviruses whose genomic sequences have been published. In addition, a homologue of ac96 is also found in the distantly related Hz-1 virus (32). Figure 1A shows an alignment of a selection of AC96 homologues from each baculovirus genus, as well as a Nudivirus homologue. The proteins shown include OP97 (from OpMNPV, a group I alphabaculovirus), SE69 (from Spodoptera exigua MNPV, a group II alphabaculovirus), XC97 (from Xestia c-nigrum granulovirus, a betabaculovirus), NELE57 (from Neodiprion lecontei NPV, a gammabaculovirus), CUNI90 (from Culex nigripalpus NPV, a deltabaculovirus), and the unclassified Hz-1V103 protein of Hz-1 virus. It can be seen that AC96 has conserved features, including a highly hydrophobic transmembrane-signal peptide predicted at the N terminus, with a potential cleavage site between Ser21 and Ile22. In addition, a second transmembrane domain is weakly predicted between Thr119 and Leu133. These analyses therefore strongly suggested that AC96 is a membrane or envelope protein and potentially a structural protein.
FIG. 1.
Protein alignment of AC96 homologues and transcription start site of ac96. (A) Amino acid alignment of AC96 and selected baculovirus homologues. The predicted N-terminal transmembrane-signal peptide domain of AC96 (identified using the SignalP and TMPRED programs) and its homologues is labeled above the sequence. The alignment was generated using the AlignX program of Vector NTI (Invitrogen, Inc.). The GenBank accession numbers of the protein sequences used are as follows: NP_054126.1 for AC96, NP_046253.1 for OP97, NP_037829.1 for SE69, NP_059245.1 for XC97, YP_025257.1 for NELE57, NP_203393.1 for CUNI90, and NP_690522.1 for Hz1V103. Amino acids with a black background show identical or similar amino acids in all sequences, those with white letters and a gray background show identical or similar amino acids in six or seven sequences, and those with black letters and a gray background show identical or similar amino acids in five or fewer sequences. (B) 5′-RACE analysis of the ac96 transcriptional start site. The late promoter (TTAAG) and the AC96 codons are shown in boldface. The arrow indicates the location of the transcription initiation site for ac96 mRNA. Two minicistrons located in the 5′ UTR region are shown, one in bold italics and one with a dotted underline. The predicted amino acid sequence is given below the nucleotide sequence.
Mapping of the transcription start site of ac96.
The 5′ and 3′ ends of the ac96 ORF overlap with the 5′ end of helicase and the 5′ end of ac97, respectively (Fig. 2). Analysis of the promoter sequence of ac96 shows that a late gene transcriptional start site motif, TTAAG, is found 141 nucleotides upstream of the predicted start codon of ac96, within the helicase ORF. A polyadenylation signal motif, AATAAA, is found 159 nucleotides downstream of the ac96 stop codon. To map the transcription start, 5′-RACE was performed (Fig. 1B) and specifically amplified a product from RNA isolated from infected Sf9 cells at 24 hpi, but not from RNA isolated at 4 hpi and not from mock-infected cells. Sequencing of the 5′-RACE product showed that the ac96 transcript initiated at the predicted TTAAG motif, indicating that ac96 is a late gene.
These results show that ac96 mRNA has a surprisingly long untranslated leader sequence. Analysis of the untranslated region (UTR) revealed the presence of two minicistrons of 12 bp and 72 bp (Fig. 1B). These minicistrons are conserved in Bombyx mori NPV but not in the more distantly related viruses, such as OpMNPV. Previous studies have shown that minicistrons in the 5′ UTR can impact the translation of the main ORF of a baculovirus mRNA (5, 36).
Construction of ac96-null and repaired viruses.
To determine the function of ac96, an ac96-null virus was constructed based on homologous recombination in E. coli using the AcMNPV bMON14272 bacmid (Fig. 2). The ac96 gene has an ORF of 522 nucleotides that, as indicated above, overlaps helicase and ac97 at the 5′ and 3′ ends by 14 and 29 nucleotides, respectively. The early promoter motif CGTGC and the late promoter motif TTAAG for helicase are located 163 and 83 nucleotides inside the ac96 ORF, respectively. At the 3′ end, there are no identifiable promoter elements, such as TATAA-CAGT or DTAAG, to identify either an early or a late promoter of ac97 within the ac96 ORF. To preserve the promoter of helicase and the potential promoter of ac97, primers 1570 and 1571 were initially used to construct an ac96-null virus (data not shown). This construct was nonviable in transfected cells, and attempts to rescue the primer 1570-primer 1571 knockout by reinsertion of ac96 at the polyhedrin locus were unsuccessful. This suggested that expression of helicase or ac97 was also interrupted and that the observed phenotype was not due to ac96 (data not shown). Therefore, a second ac96 knockout virus was designed using primers 1709 and 1571. In the second construct, the ac96 knockout bacmid was generated by insertion of the zeocin resistance cassette between the 386th and 387th nucleotides of the ac96 ORF. To confirm the correct insertion of the zeocin cassette, the ac96-null bacmid was examined by PCR (data not shown). Two pairs of primers, primers 1572 and 1014 and primers 1573 and 1239, were used to examine the recombination junctions of upstream and downstream flanking regions. The PCR results confirmed that the zeocin gene was inserted into ac96 as expected (data not shown). This construct could be rescued by ac96 and was used for further studies.
To monitor the effect of the ac96 mutation on virus infection and OB morphogenesis, the polyhedrin and gfp genes were inserted into the polyhedrin locus of the ac96-null virus by transposition, and this virus was named vAc96null (Fig. 2). To rescue the ac96-null virus, vAc96null-AC96HA was generated; in addition to polyhedrin and gfp, this virus contains the ac96 ORF driven by its native late promoter with sequences encoding an HA epitope at the 3′ terminus. vAcBac, which serves as the wild-type control virus, was constructed by transposing the polyhedrin and gfp genes into the polyhedrin locus of AcMNPV bacmid bMON14272 (Fig. 2).
Analysis of ac96-null and repaired viruses in transfected Sf9 cells.
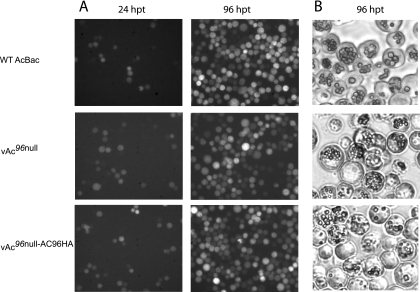
To determine the effect of ac96 inactivation on virus replication, vAc96null, vAc96null-AC96HA, and vAcBac bacmid DNAs were transfected into Sf9 cells. The expression of gfp in all the constructs driven by the OpMNPV ie1 promoter enables the monitoring of transfection by fluorescence microscopy (Fig. 3A). At 24 hpt, similar numbers of cells with GFP fluorescence were observed for all three viruses, indicating equal transfection efficiencies. By 96 hpt, every cell showed GFP fluorescence for all three viruses, indicating that infectious BV was produced from the initially transfected cells.
FIG. 3.
Analysis of viral replication in Sf9 cells. (A) Fluorescence microscopy showing the progression of infection in Sf9 cells transfected with vAc96null, vAc96null-AC96HA, or vAcBac. WT, wild type. (B) Light microscopy showing OB production in Sf9 cells transfected with vAc96null, vAc96null-AC96HA, or vAcBac at 96 hpt.
At 96 hpt, light microscopy analysis showed that vAc96null, vAc96null-AC96HA, and vAcBac produced OBs of normal appearance (Fig. 3B). In addition, equivalent numbers of OBs were observed in most cells. Western blot analysis of OBs to detect capsid protein confirmed the presence of ODV for all three viruses (data not shown). These results, therefore, suggest that AC96 does not affect OB formation.
Virus growth curves.
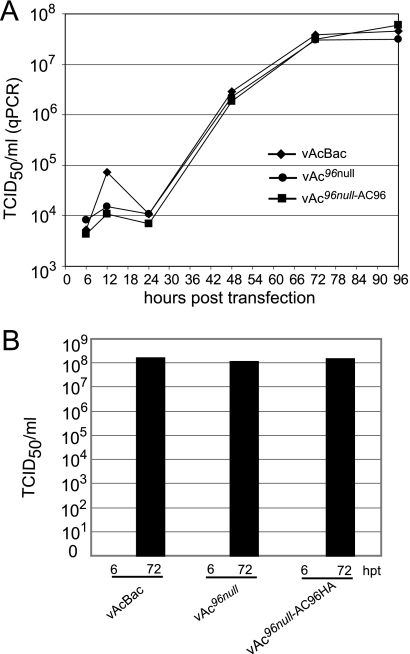
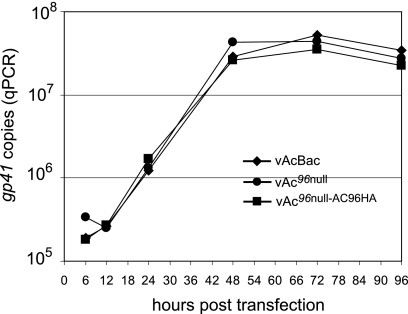
To determine if inactivation of ac96 quantitatively affects infectious BV production or replication kinetics, qPCR and TCID50 assays were used to estimate BV production at various times posttransfection. Figure 4A shows the replication kinetics of BV production from Sf9 cells transfected with vAc96null, vAc96null-AC96HA, or vAcBac as determined by qPCR, which detects viral genomes regardless of infectivity. No significant difference was observed in the BV titers assayed by qPCR from vAc96null-, vAc96null-AC96HA-, and vAcBac-transfected cells from 6 to 96 hpt.
FIG. 4.
BV production from Sf9 cells transfected with vAc96null, vAc96null-AC96HA, or vAcBac. (A) BV growth curves assayed by quantifying the number of viral genomes using qPCR. Titers were determined from supernatants of Sf9 cells transfected with vAc96null, vAc96null-AC96HA, or vAcBac at the designated time points. (B) Infectious virus titers at 6 and 72 hpt, determined by a TCID50 endpoint dilution assay. Each titer is derived from two independent TCID50 assays.
To confirm that equivalent amounts of infectious BV were produced, the virus titers at 6 and 72 hpt were analyzed by TCID50 endpoint dilution (Fig. 4B). No infectious BV was detected from any construct at 6 hpt. At 72 hpt, the average titers of vAc96null, vAc96null-AC96HA, and vAcBac were equivalent, results similar to those obtained using qPCR. The titration assays demonstrated that ac96 is not required for the production of infectious BV.
DNA replication assay of ac96-null and repaired viruses in Sf9 cells.
To quantitatively analyze the effect of ac96 on viral DNA replication, Sf9 cells transfected with bacmid vAc96null, vAc96null-AC96HA, or vAcBac were harvested at various times posttransfection, and total DNA was extracted and analyzed by qPCR (Fig. 5). The onset of DNA replication occurred between 12 and 24 hpt for all constructs, and DNA synthesis continued to increase to 48 hpt for vAc96null, vAc96null-AC96HA, and vAcBac. The DNA levels of all three viruses showed no difference throughout the infection process. These results show that the inactivation of ac96 does not affect the onset or level of DNA replication.
FIG. 5.
Quantitative real-time PCR analysis of viral DNA replication. Sf9 cells were transfected with 1.0 μg of vAc96null, vAc96null-AC96HA, or vAcBac DNA. At various times posttransfection, total DNA was extracted and analyzed by qPCR. Each time point represents the average for two independent replication assays.
Subcellular localization of AC96 during infection.
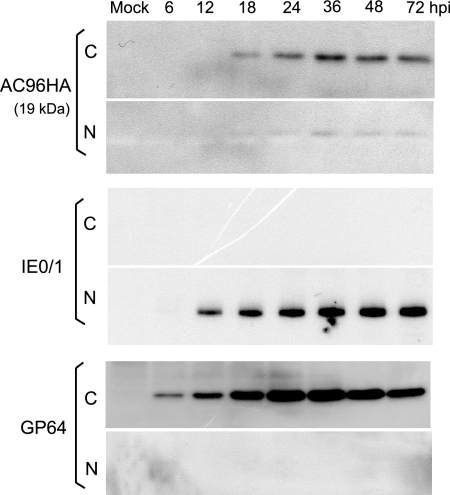
To further understand the function of AC96 in the viral life cycle, it is necessary to establish its temporal expression and cellular localization. An initial time course analysis of AC96 expression was performed using vAc96null-AC96HA. Infected Sf9 cells were collected at various time points p.i., and the nuclear and cytoplasmic fractions were separated. Western blot analysis showed that AC96 was translated from 12 to 72 hpi and was distributed in both the nucleus and the cytoplasm, but to a significantly greater extent in the cytoplasm (Fig. 6). The protein was detected at the predicted size of 19 kDa, suggesting that no major posttranslational modification occurs. As a control to test the efficiency of the fractionation process, the same samples were probed with an anti-IE1 antibody and an anti-GP64 antibody. The presence of IE1 and GP64 exclusively in the nuclear and cytoplasmic fractions, respectively, confirmed efficient cell fractionation.
FIG. 6.
Cellular localization of AC96HA as determined by Western blot analysis of cytoplasmic and nuclear fractions of vAc96null-AC96HA-infected cells. Sf9 cells were infected with vAc96null-AC96HA at an MOI of 2. Cells were harvested and separated into the nuclear (N) and cytoplasmic (C) fractions. Fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-HA, anti-IE1, and anti-GP64 monoclonal antibodies to detect HA-tagged AC96, IE1, or GP64, respectively.
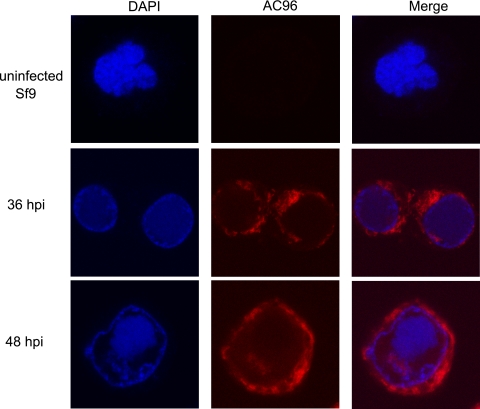
The cellular localization of AC96 was also analyzed by immunofluorescence and confocal microscopy. Uninfected Sf9 cells show no background fluorescence, demonstrating the specificity of the anti-HA antibody (Fig. 7). At 36 hpi, AC96 could be detected mainly in the cytoplasm and at low levels in the nucleus, where it was unevenly distributed at the periphery of the virogenic stroma. By 48 hpi, the AC96 fluorescence signal showed greater intensity in the cytoplasm, and the nuclear AC96 was more apparent as part of the virogenic stroma. The AC96 localization pattern from immunofluorescence was consistent with the results obtained from the fractionation and Western blot analyses.
FIG. 7.
Localization of AC96HA by immunofluorescence. Sf9 cells were infected with vAc96null-AC96HA at an MOI of 5. At different times p.i., cells were fixed, probed with a mouse monoclonal anti-HA antibody to detect AC96HA, and visualized by Alexa 635-conjugated goat anti-mouse immunoglobulin G (red). Additionally, cells were stained with DAPI to directly visualize nuclear DNA (blue).
Western blot analysis of AC96 in purified BV and ODV.
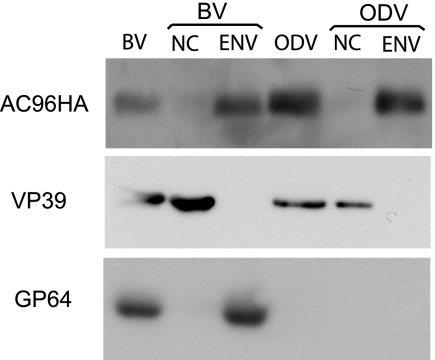
Previous reports have shown by mass spectrometry that CUNI69, an AC96 homologue, is associated with Culex nigripalpus NPV ODV. However, AC96 was not detected in AcMNPV ODV by similar analytical methods (3, 30). To determine if AC96 is a structural protein and to investigate the localization of AC96 in virions, BV and ODV were purified and analyzed by Western blotting for the presence of AC96. The results showed that AC96 could be detected in both BV and ODV (Fig. 8, lanes BV and ODV). The BV and ODV particles were also biochemically fractionated into nucleocapsid and envelope fractions (Fig. 8, lanes NC and ENV), and Western blot analysis indicated that AC96 was localized specifically in envelope fractions. As a control to confirm the efficiency of the fractionation, the known nucleocapsid protein VP39 and the BV envelope-specific protein GP64 were analyzed by Western blotting. Both VP39 and GP64 were observed in the expected fractions. These results showed that AC96 is associated with both BV and ODV virions and that it localizes in the envelopes of BV and ODV, consistent with the prediction that AC96 contains an N-terminal transmembrane-signal domain.
FIG. 8.
Analysis of AC96HA in purified and fractionated virions. BV and ODV were purified, fractionated, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. Duplicate blots were probed with an anti-HA antibody to detect AC96HA, with an anti-AcV5 monoclonal antibody to detect the BV envelope protein GP64, and with anti-VP39 to detect the nucleocapsid protein VP39. NC, nucleocapsid fraction; ENV, envelope fraction.
Bioassays.
To examine the effect of ac96 inactivation on viral infection in vivo, BV and OBs of vAc96null, vAc96null-AC96HA, and vAcBac from transfected Sf9 cells were assayed in 4th-instar T. ni larvae. Injection of the BV supernatant (1,000 TCID50 units/larva) of vAc96null into the hemocoels of larvae resulted in levels of mortality similar (100%) to those with vAc96null-AC96HA and vAcBac (Table 1). Similar results were obtained by injection of 30 or 3 TCID50 units/larva. These results, therefore, suggest that there was no difference in BV virulence when the midgut was bypassed by direct hemocoel injection. However, ingestion of OBs of vAc96null produced either no death or 2 deaths out of 24 larvae at the highest dose rate (500 OBs/larva). In contrast, vAc96null-AC96HA and vAcBac produced 24 or 20/23 deaths (Table 1). Similarly, at the lower doses, vAc96null was unable to infect larvae. These assays indicate that AC96 is required for per os infection of AcMNPV.
TABLE 1.
Infectivities of ac96 null and repaired viruses in 4th-instar T. ni larvae
| Dosage and virus | Inoculation method | Total no. of larvae | No. of larvae dead by day 8 p.i. |
|
|---|---|---|---|---|
| Expt 1 | Expt 2 | |||
| No virus | 24 | 0 | 0 | |
| 1,000 TCID50 | ||||
| vAcBac | Injection | 24 | 24 | 24 |
| vAc96null | Injection | 24 | 24 | 24 |
| vAc96null-AC96HA | Injection | 24 | 24 | 24 |
| 30 TCID50 | ||||
| vAcBac | Injection | 12 | 12 | NDa |
| vAc96null | Injection | 12 | 12 | ND |
| vAc96null-AC96HA | Injection | 12 | 12 | ND |
| 3 TCID50 | ||||
| vAcBac | Injection | 12 | 12 | ND |
| vAc96null | Injection | 12 | 11 | ND |
| vAc96null-AC96HA | Injection | 12 | 11 | ND |
| 5 OBs | ||||
| vAcBac | Per os | 24 | 3 | 1 |
| vAc96null | Per os | 24 | 1b | 0 |
| vAc96null-AC96HA | Per os | 24 | 6 | 4 |
| 50 OBs | Per os | |||
| vAcBac | Per os | 24 | 12 | 7 |
| vAc96null | Per os | 24 | 0 | 1 |
| vAc96null-AC96HA | Per os | 24 | 19 | 18 |
| 500 OBs | Per os | |||
| vAcBac | Per os | 24 | 24 | 20 |
| vAc96null | Per os | 24 | 0 | 2 (1b) |
| vAc96null-AC96HA | Per os | 24 | 24 | 23 |
ND, not done.
Larval death not due to virus infection (microscopy and PCR analysis).
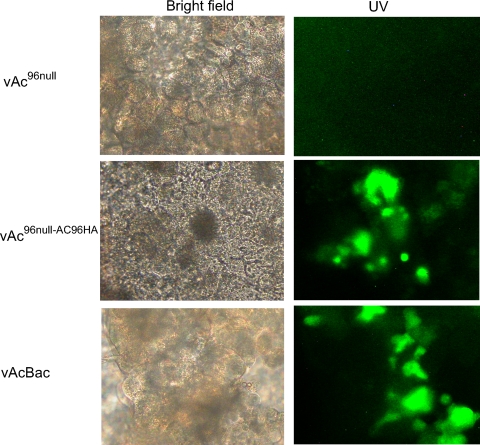
To test if the occlusions of vAc96null fail to initiate infection in vivo, 4th-instar T. ni larvae were fed 50,000 OBs of vAc96null, vAc96null-AC96HA, or vAcBac, and midgut epithelium preparations were examined at 48 hpi. Fluorescence microscopy analysis showed significant levels of infection (GFP expression) in midgut cells from vAc96null-AC96HA- and vAcBac-infected larvae but not in larvae infected with vAc96null (Fig. 9). These results demonstrate that vAc96null was unable to infect T. ni midgut epithelial cells, and AC96 appears to be required for the attachment and/or fusion of ODV to midgut epithelial cells.
FIG. 9.
Infection of midgut epithelia of T. ni larvae at 48 hpi. The midguts of 4th-instar larvae ingesting 50,000 OBs of vAc96null, vAc96null-AC96HA, or vAcBac were isolated and observed under a bright-field (left) or a UV fluorescence (right) microscope. As indicated by GFP expression, infection with vAc96null-AC96HA or vAcBac, but not with vAc96null, was observed.
DISCUSSION
In this study, the baculovirus core gene ac96 is identified as a new per os infectivity factor, pif-4. The inactivation of ac96 in vAc96null abolished the infectivity of OBs (Table 1), and ODVs were unable to initiate an infection of the midgut (Fig. 9). However in tissue culture, loss of ac96 had no impact on virus replication or BV production; this is the same phenotype observed with other pif genes (21, 22, 28, 31). Three PIFs have previously been determined to play a role in the specific binding of ODV to midgut cells. P74 has been shown to function as an attachment protein and to bind to a 30-kDa host receptor protein on primary target cells within the midgut (17, 40). PIF-1 and PIF-2 also mediate the specific binding of ODV to midgut target cells (28). Although PIF-3 is not required for ODV binding, it is involved in another downstream event in primary infection (28). It is possible that AC96 (PIF-4), P74, PIF-1, and PIF-2 may form complexes to bind host receptors or may act sequentially to mediate ODV entry to initiate primary infection. This possibility is supported by a recent study by Song et al. (34), which showed that the PIF genes had to be present in the same virion in order to permit virus infection of the midgut.
A transmembrane signal peptide is predicted at the hydrophobic N terminus of AC96; it is conserved in the homologues from all baculoviruses and the distantly related insect virus Hz-1. This suggests that membrane association is a highly conserved function and is in agreement with our results using purified ODV, which showed that AC96 specifically localized to the ODV envelope (Fig. 8). Similarly, all the other known PIFs—P74, PIF-1, PIF-2, and PIF-3—also locate to the ODV envelope (12, 21, 34). The hydrophobic N terminus of PIF1, PIF-2, PIF-3, and the ODV envelope proteins ODV-E25 and ODV-E66 (11, 31) can act as a sorting motif, which is sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles, and ODV envelope within baculovirus-infected cells. The N-terminal transmembrane-signal sequence of AC96 has two features of a sorting motif, namely, a hydrophobic sequence and associated charged amino acids oriented on the cytoplasmic or nucleoplasmic face (3, 19). The vAc96null mutant in this study may express the N-terminal 128 amino acids of AC96 (Fig. 2), due to the location of the zeocin cassette, which inserts between Glu128 and His129. This effectively deletes the C-terminal half of AC96, indicating that the N-terminal region that contains the transmembrane domain is not sufficient for AC96 function. However, it is possible that the remaining amino acids, if expressed and stable, may maintain the infectivity of BV in tissue culture or in intrahemocoelically injected larvae.
AC96 was also detected in the envelope fraction of purified BV; to our knowledge, this is the first PIF that is also associated with BV. Immunofluorescence analysis using confocal microscopy showed that AC96 localized to a greater extent in cytoplasm but was also present to a lesser degree in the nucleus (Fig. 7). AC96 in the cytoplasm is most likely associated with the endoplasmic reticulum and is then transported into the inner nuclear membrane, ultimately to become part of the ODV envelope. It is also possible that AC96 could be delivered to the plasma membrane, like GP64 (24), and form part of the BV membrane. Analysis of vAc96null BV in tissue culture and by in vivo intrahemocoelic injections (Fig. 4; Table 1) did not reveal any phenotypic differences from the control virus with regard to replication or infectivity. This suggests that AC96 does not have a function in BV, but future studies may reveal that it plays a role or possibly is inadvertently packaged into BV. However, analysis of PIF-4 homologues in other baculovirus species may reveal additional or alternate functions.
All the PIF genes, including PIF-4, are core baculovirus proteins, indicating that the midgut infection process is an ancient and highly conserved process. Determining the function of the PIF genes and how they interact with each other and the insect midgut will be critical for the understanding of baculovirus virulence.
Acknowledgments
This study was supported in part by the Crops Genomics Initiative of Agriculture and Agri-Food Canada and by funding from the Natural Science and Engineering Research Council.
We thank Leslie Willis for outstanding laboratory support and Keith Moore for excellent technical assistance with insect bioassays.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 2.Braun, L., and B. A. Keddie. 1997. A new tissue technique for evaluating effects of Bacillus thuringiensis toxins on insect midgut epithelium. J. Invertebr. Pathol. 69:92-104. [DOI] [PubMed] [Google Scholar]
- 3.Braunagel, S. C., W. K. Russell, G. Rosas-Acosta, D. H. Russell, and M. D. Summers. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. USA 100:9797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, M. J. 1995. Lipofection reagents prepared by a simple ethanol injection technique. BioTechniques 18:1027-1032. [PubMed] [Google Scholar]
- 5.Chang, M. J., and G. W. Blissard. 1997. Baculovirus gp64 gene expression: negative regulation by a minicistron. J. Virol. 71:7448-7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai, X., T. M. Stewart, J. A. Pathakamuri, Q. Li, and D. A. Theilmann. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 78:9633-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelhard, E. K., L. N. Kam-Morgan, J. O. Washburn, and L. E. Volkman. 1994. The insect tracheal system: a conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc. Natl. Acad. Sci. USA 91:3224-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlandson, M. A., S. Newhouse, K. Moore, A. Janmatt, J. Myers, and D. A. Theilmann. 2007. Characterization of naturally occurring baculovirus isolates from Trichoplusia ni populations from vegetable greenhouses. Biol. Control 41:256-263. [Google Scholar]
- 10.Fang, M., X. Dai, and D. A. Theilmann. 2007. Autographa californica multiple nucleopolyhedrovirus EXON0 (ORF141) is required for efficient egress of nucleocapsids from the nucleus. J. Virol. 81:9859-9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, M., Y. Nie, Q. Wang, F. Deng, R. Wang, H. Wang, J. M. Vlak, X. Chen, and Z. Hu. 2006. Open reading frame 132 of Helicoverpa armigera nucleopolyhedrovirus encodes a functional per os infectivity factor (PIF-2). J. Gen. Virol. 87:2563-2569. [DOI] [PubMed] [Google Scholar]
- 12.Fang, M., H. Wang, L. Yuan, X. Chen, J. M. Vlak, and Z. Hu. 2003. Open reading frame 94 of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus encodes a novel conserved occlusion-derived virion protein, ODV-EC43. J. Gen. Virol. 84:3021-3027. [DOI] [PubMed] [Google Scholar]
- 13.Faulkner, P., J. Kuzio, G. V. Williams, and J. A. Wilson. 1997. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 78:3091-3100. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Maruniak, A., J. E. Maruniak, P. M. Zanotto, A. E. Doumbouya, J. C. Liu, T. M. Merritt, and J. S. Lanoie. 2004. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 78:7036-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granados, R. R. 1978. Early events in the infection of Heliothis zea midgut cells by a baculovirus. Virology 90:170-174. [DOI] [PubMed] [Google Scholar]
- 16.Granados, R. R., and K. A. Lawler. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297-308. [DOI] [PubMed] [Google Scholar]
- 17.Haas-Stapleton, E. J., J. O. Washburn, and L. E. Volkman. 2004. P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. J. Virol. 78:6786-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 19.Hong, T., M. D. Summers, and S. C. Braunagel. 1997. N-terminal sequences from Autographa californica nuclear polyhedrosis virus envelope proteins ODV-E66 and ODV-E25 are sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles and the envelope of occlusion derived virus. Proc. Natl. Acad. Sci. USA 94:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, H. M., and J. P. Burand. 1993. Saturable attachment sites for polyhedron-derived baculovirus on insect cells and evidence for entry via direct membrane fusion. J. Virol. 67:1860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikhno, I., S. Gutierrez, L. Croizier, G. Croizier, and M. L. Ferber. 2002. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J. Gen. Virol. 83:3013-3022. [DOI] [PubMed] [Google Scholar]
- 22.Kuzio, J., R. Jaques, and P. Faulkner. 1989. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology 173:759-763. [DOI] [PubMed] [Google Scholar]
- 23.Li, Q., L. Li, K. Moore, C. Donly, D. A. Theilmann, and M. Erlandson. 2003. Characterization of Mamestra configurata nucleopolyhedrovirus enhancin and its functional analysis via expression in an Autographa californica M nucleopolyhedrovirus recombinant. J. Gen. Virol. 84:123-132. [DOI] [PubMed] [Google Scholar]
- 24.Li, Z., and G. W. Blissard. 2009. The Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: analysis of transmembrane domain length and sequence requirements. J. Virol. 83:4447-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy, C. B., and D. A. Theilmann. 2008. AcMNPV ac143 (odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology 375:277-291. [DOI] [PubMed] [Google Scholar]
- 27.Nie, Y., M. Fang, and D. A. Theilmann. 2009. AcMNPV AC16 (DA26, BV/ODV-E26) regulates the levels of IE0 and IE1 and binds to both proteins via a domain located within the acidic transcriptional activation domain. Virology 385:484-495. [DOI] [PubMed] [Google Scholar]
- 28.Ohkawa, T., J. O. Washburn, R. Sitapara, E. Sid, and L. E. Volkman. 2005. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 79:15258-15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, M. N., R. L. Quant-Russell, G. F. Rohrmann, and G. S. Beaudreau. 1988. P39, a major baculovirus structural protein: immunocytochemical characterization and genetic location. Virology 167:407-413. [PubMed] [Google Scholar]
- 30.Perera, O., T. B. Green, S. M. Stevens, Jr., S. White, and J. J. Becnel. 2007. Proteins associated with Culex nigripalpus nucleopolyhedrovirus occluded virions. J. Virol. 81:4585-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pijlman, G. P., A. J. Pruijssers, and J. M. Vlak. 2003. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. J. Gen. Virol. 84:2041-2049. [DOI] [PubMed] [Google Scholar]
- 32.Rohrmann, G. F. 2008. Baculovirus molecular biology. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD. http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=bacvir.
- 33.Ross, L., and L. A. Guarino. 1997. Cycloheximide inhibition of delayed early gene expression in baculovirus-infected cells. Virology 232:105-113. [DOI] [PubMed] [Google Scholar]
- 34.Song, J., R. Wang, F. Deng, H. Wang, and Z. Hu. 2008. Functional studies of per os infectivity factors of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 89:2331-2338. [DOI] [PubMed] [Google Scholar]
- 35.Theilmann, D. A., G. W. Blissard, B. Bonning, J. A. Jehle, D. R. O'Reilly, G. F. Rohrmann, S. Thiem, and J. M. Vlak. 2005. Baculoviridae, p. 177-185. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom.
- 36.Theilmann, D. A., L. G. Willis, B. J. Bosch, I. J. Forsythe, and Q. Li. 2001. The baculovirus transcriptional transactivator ie0 produces multiple products by internal initiation of translation. Virology 290:211-223. [DOI] [PubMed] [Google Scholar]
- 37.van Oers, M. M., and J. M. Vlak. 2007. Baculovirus genomics. Curr. Drug Targets 8:1051-1068. [DOI] [PubMed] [Google Scholar]
- 38.Wang, P., and R. R. Granados. 1997. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. USA 94:6977-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao, L., W. Zhou, H. Xu, Y. Zheng, and Y. Qi. 2004. The Heliothis armigera single nucleocapsid nucleopolyhedrovirus envelope protein P74 is required for infection of the host midgut. Virus Res. 104:111-121. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, R., Z. H. Yu, X. Q. Li, F. Jia, J. H. Wu, and X. Chen. 2004. Heliocoverpa armigera single nucleocapsid nucleopolyhedrovirus induces Hz-AM1 cell cycle arrest at the G2 phase with accumulation of cyclin B1. Virus Res. 105:113-120. [DOI] [PubMed] [Google Scholar]