Abstract
Human cytomegalovirus (HCMV) evades T-cell recognition by down-regulating expression of major histocompatibility complex (MHC) class I and II molecules on the surfaces of infected cells. Contrary to the “missing-self” hypothesis, HCMV-infected cells are refractory to lysis by natural killer (NK) cells. Inhibition of NK cell function is mediated by a number of HCMV immune evasion molecules, which operate by delivering inhibitory signals to NK cells and preventing engagement of activating ligands. One such molecule is UL142, which is an MHC class I-related glycoprotein encoded by clinical isolates and low-passage-number strains of HCMV. UL142 is known to down-modulate surface expression of MHC class I-related chain A (MICA), which is a ligand of the activating NK receptor NKG2D. However, the mechanism by which UL142 interferes with MICA is unknown. Here, we show that UL142 localizes predominantly to the endoplasmic reticulum (ER) and cis-Golgi apparatus. The transmembrane domain of UL142 mediates its ER localization, while we propose that the UL142 luminal domain is involved in its cis-Golgi localization. We also confirm that UL142 down-modulates surface expression of full-length MICA alleles while having no effect on the truncated allele MICA*008. However, we demonstrate for the first time that UL142 retains full-length MICA alleles in the cis-Golgi apparatus. In addition, we propose that UL142 interacts with nascent MICA en route to the cell surface but not mature MICA at the cell surface. Our data also demonstrate that the UL142 luminal and transmembrane domains are involved in recognition and intracellular sequestration of full-length MICA alleles.
Human cytomegalovirus (HCMV), a member of the Herpesviridae family, is a ubiquitous human pathogen with seroprevalence ranging between 50 to 100% worldwide (46). Severe morbidity and mortality are associated with HCMV infection in individuals that are immunocompromised or immunologically immature (25), and the severity of HCMV disease in immunocompromised individuals correlates with the level of immune suppression. However, HCMV infection in healthy immunocompetent individuals is usually asymptomatic/subclinical (25). Like all herpesviruses, HCMV establishes lifelong latent infection, HCMV residues, in hematopoietic cells of the myeloid lineage (45). Thus, following primary infection, HCMV persists in the host despite a robust humoral and cell-mediated immune response.
HCMV down-modulates surface expression of host major histocompatibility complex (MHC) class I molecules in order to evade T-cell recognition (5, 7, 24), but in doing so, the virus risks natural killer (NK) cell activation due to the lack of inhibitory receptor signaling (30). However, there is now ample evidence that HCMV evades NK cell-mediated lysis by a variety of different mechanisms (58). These include the expression of molecules that engage inhibitory NK receptors, as well as the down-modulation of ligands for activating NK receptors. The HCMV UL40 open reading frame encodes a nonameric peptide that enables cell surface expression of mature HLA-E molecules, which bind the inhibitory NK receptor CD94/NKG2A (10, 51, 54). Another inhibitory ligand expressed on HCMV-infected cells is UL18, which is a virus-encoded MHC class I homologue that binds the inhibitory receptor LILRB1 (14) with 1,000-fold-higher affinity than MHC class I (13). Importantly, UL18 has been shown to inhibit LILRB1+ NK cells while activating LILRB1− NK cells (40). In order to prevent NK cell activation via the NKp30 receptor, the HCMV tegument protein pp65 binds the activating receptor and dissociates the CD3ζ adaptor molecule (3). HCMV also encodes the glycoprotein UL141, which retains CD155 (or polio virus receptor or nectin-like molecule 5) in an immature form in the endoplasmic reticulum (ER), thereby preventing engagement by the activating receptors CD226 (or DNAM-1) and CD96 (or TACTILE) (52).
Cytomegalovirus infection induces expression of MHC class I-related molecules that are ligands for the potent activating receptor NKG2D. NKG2D is an activating C-type lectin receptor expressed on NK cells, γδ T cells, CD8+ αβ T cells, and CD4+ T cells (6, 19, 21, 44). Human NKG2D has multiple ligands including MHC class I-related chains (MICs), UL16 binding proteins (ULBPs), and retinoic acid early inducible 1-like transcripts (RAET1s). The best-characterized high-affinity ligands are ULBP1, ULBP2, ULBP3, MICA, and MICB (4). There is now evidence that HCMV can evade NKG2D-mediated activation of NK cells as well as costimulation of γδ T cells, CD8+ αβ T cells, and CD4+ T cells. It has been shown previously that transcription of the MICB gene is down-regulated by a virus-encoded microRNA, designated hcmv-miR-UL112 (48). In addition, the HCMV UL16 glycoprotein retains MICB, ULBP1, and ULBP2 (but not MICA or ULBP3) in the ER and cis-Golgi apparatus (15, 42, 55, 57, 60).
Murine cytomegalovirus (MCMV) also prevents surface expression of murine NKG2D ligands. The MCMV m145, m152, and m155 gene products down-modulate surface expression of murine NKG2D ligands by intracellular retention and degradation (28, 29, 31). Interestingly, HCMV-encoded UL142 (a glycoprotein encoded by clinical isolates and low-passage-number strains), which has been shown previously to inhibit NK cell-mediated cytotoxicity, is structurally related to these MCMV molecules (16, 36). Thus, we postulated that UL142 also down-modulated surface expression of the ligand(s) of human NKG2D (59). Subsequently, Chalupny et al. determined that UL142 down-modulates the surface expression of full-length MICA alleles but not the truncated allele MICA*008 (12). However, no mechanism was identified.
In this study, we show that UL142 is localized predominantly to the ER by virtue of its transmembrane domain and to the cis-Golgi apparatus by virtue of its luminal domain. We also show that UL142 interferes with MICA surface expression by retaining full-length alleles in the cis-Golgi apparatus. A delay in UL142-mediated down-modulation of MICA surface expression indicates that UL142 interacts with nascent MICA en route to the cell surface but not mature MICA at the cell surface. We also demonstrate that down-modulation of MICA cannot occur in the absence of the UL142 luminal domain. Altogether, our data reveal the mechanism by which UL142 interferes with surface expression of full-length MICA alleles.
MATERIALS AND METHODS
Cell lines.
Cells of the HeLa line (a human epithelial carcinoma cell line) and U373 line (a human astrocytoma cell line) were maintained in RPMI 1640 medium plus 10% fetal calf serum (FCS), 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C in humidified 5% CO2. MRC-5, COS-7, and human fetal foreskin fibroblast (HFFF) cells were maintained in Eagle's minimal essential medium plus 10% FCS, 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C in humidified 5% CO2.
Plasmids.
The UL142 gene was amplified by PCR from genomic DNA of the HCMV strain Toledo (GenBank accession number U33331) and then cloned into a variety of mammalian vectors. N-terminally FLAG-tagged UL142 (FLAG-UL142) was generated by cloning the UL142 gene into the p3xFLAG-CMV-9 vector (Sigma-Aldrich) between the EcoRI and BamHI sites, downstream of a preprotrypsin leader sequence and three FLAG tag sequences. As described previously (59), green fluorescent protein (GFP)-tagged UL142 was generated by cloning the UL142 gene into the PK1 vector (8) between the XhoI and XbaI sites, downstream of a generic leader sequence, a GFP cassette (BD Clontech), and a Myc tag sequence. In addition, GFP-tagged versions of the full-length allele MICA*018 (50) and the truncated allele MICA*008 allele and HLA-A2 (8) were generated by cloning the corresponding coding sequences into the PK1 vector. Untagged MICA*018 was also generated by cloning the MICA*018 allele into the pcDNA3 vector (Invitrogen).
Chimeras of CD8 and UL142 were produced using the S5MY888 vector (a kind gift from Sean Munro, Laboratory of Molecular Biology, Cambridge, United Kingdom). N-terminally Myc-tagged CD8 (the CD8 wild-type [CD8-WT] construct) expressed from a cytomegalovirus promoter was generated by subcloning the CD8 gene from the S5MY888 vector into the pcDNA3.0 vector (Invitrogen) between the HindIII and XbaI sites. The cytoplasmic tail of CD8 was replaced with that of UL142 (yielding the CD8-UL142Tail construct) by cloning DNA encoding the UL142 tail into the CD8-WT plasmid between the AflII and XbaI sites. Chimeric CD8 with the UL142 transmembrane domain and cytoplasmic tail (CD8-UL142TMDTail) was generated by cloning DNA encoding the UL142 transmembrane and tail into the CD8-WT plasmid between the EcoRV and XbaI sites. In order to replace the CD8 transmembrane domain with that of UL142 (yielding CD8-UL142TMDonly), DNA encoding the CD8 extracellular domain and the UL142 transmembrane domain was amplified from the CD8-UL142TMDTail plasmid and then cloned into the CD8-WT plasmid between the XhoI and AflII sites. The luminal domain of CD8 was replaced with that of UL142 (yielding UL142Luminal-CD8TMDTail) by cloning DNA encoding the UL142 luminal domain into the CD8-WT plasmid between the XhoI and EcoRV sites. Chimeric CD8 with the UL142 luminal and transmembrane domains (UL142LuminalTMD-CD8Tail) was generated by cloning DNA encoding the UL142 luminal and transmembrane domains into the CD8-WT plasmid between the XhoI and AflII sites. Lastly, CD8 with the MICA*018 transmembrane domain and cytoplasmic tail (CD8-MICA*018TMDTail) was also generated.
Sequence analyses.
The analysis programs TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), PredictProtein (http://cubic.bioc.columbia.edu/predictprotein/) (43), and HMMTOP (http://www.enzim.hu/hmmtop/) (53) predicted that UL142 was a type I transmembrane protein. In addition, the transmembrane domain of UL142 was determined to comprise amino acids 271 to 290.
RAd.
Replication-deficient recombinant adenoviruses (RAd) 502 and 519 (23, 52) were obtained from Gavin Wilkinson and Peter Tomasec (University of Cardiff, United Kingdom). RAd 502 (designated RAd control GFP) encodes GFP, while RAd 519 (designated RAd UL142 GFP) expresses both GFP and UL142. HeLa or U373 cells were transduced with RAd 502 or 519 for 1 h at room temperature. Fresh medium was added, and then cells were incubated at 37°C in 5% CO2.
Anti-UL142 rabbit sera.
Sigma Genosys immunized two New Zealand White rabbits with the UL142 cytoplasmic tail peptide FTPVKFVYEVWRGQ conjugated at the N terminus to the carrier protein keyhole limpet hemocyanin.
Immunofluorescence microscopy.
HeLa cells were grown on glass coverslips and then transfected with plasmid DNA by using the TransIT-HeLaMONSTER transfection kit (Mirus/Cambridge Bioscience). Cells were analyzed 16 to 48 h posttransfection. In order to chase nascent proteins out of the ER and Golgi complex, transfected cells were incubated with 0.1 mg/ml cycloheximide (Sigma-Aldrich) in medium for 2 h at 37°C. Cells were fixed with 4% (para)formaldehyde, permeabilized with 0.5% Triton X-100-phosphate-buffered saline (PBS), and blocked in 10% FCS-0.5% Tween-PBS. Cells were stained with antibodies in blocking buffer for 1 h at room temperature. Antibodies to calreticulin (Calbiochem/Merck), GM130 (BD Biosciences), MHC class I (w6/32), UL142 (Sigma Genosys), Myc (9E10 [Santa Cruz] or ab9106 [Abcam]), FLAG (F2555 [Sigma-Aldrich]), and MICA (2C10 [Santa Cruz]) were detected with Alexa Fluor 488-, 568-, or 647-conjugated species-specific secondary antibodies (Molecular Probes/Invitrogen). The glass coverslips were mounted onto microscope slides. Stained cells were viewed using a Zeiss LSM510 META or Leica TCS SP confocal microscope, and images were analyzed using ImageJ.
For analysis of HCMV-infected cells, HFFF cells were infected with the HCMV strain TB40/e (a kind gift from Gerhart Jahn, University of Tubingen, Tubingen, Germany) at a multiplicity of infection of 3. HFFFs were infected for a total of 24, 48, and 72 h. Sixteen hours before the ends of these time periods, the cells were harvested and transfected with GFP-UL142 by using a Nucleofector kit (Amaxxa, Cologne, Germany). Transfected HFFFs were incubated for a further 16 h and then cytospun onto glass slides. Cells were fixed and permeabilized and then stained with antibodies to calreticulin (Calbiochem/Merck) and the HCMV immediate-early antigen (E13; Argene).
Flow cytometry.
Transduced cells were analyzed at 96 h posttransduction, while transfected cells were analyzed at 48 h posttransfection. Approximately 106 cells were incubated with antibodies against MICA (2C10 [Santa Cruz]), ULBP2 (R&D Systems), or Myc (ab9106 [Abcam]) for 1 h at 4°C. Mouse immunoglobulin G1 (IgG1; BD Biosciences) and IgG2a (R&D Systems) were isotype controls. Following washes in PBS, cells were incubated with Alexa Fluor 647-conjugated species-specific secondary antibodies (Molecular Probes/Invitrogen) for 1 h at 4°C. Stained cells were visualized using a Becton-Dickinson FACSCalibur four-color analyzer. The data were analyzed using Summit software.
Radiolabeling, immunoprecipitation, and endo H digestion.
HeLa cells transiently transfected with GFP-UL142 were pulsed with [35S]methionine and [35S]cysteine Pro-Mix (GE Healthcare) for 20 min; chased in medium containing excess nonradioactive methionine and cysteine for 0, 45, 180, or 360 min at 37°C; and then washed in ice-cold PBS. Cells from each chase time point were lysed in 1% digitonin lysis buffer (1% digitonin, 20 mM Tris-HCl [pH 7.4], 150 mM KCl, 1 mM EDTA, and 5 mM MgCl2) plus protease inhibitors (Roche) at 4°C. Cell nuclei and debris were removed by centrifugation at 17,000 × g for 10 min. Following preclearing with a mixture of Sepharose and protein G-Sepharose beads (GE Healthcare), GFP-UL142 was immunoprecipitated using rabbit anti-GFP antibody (ab290 [Abcam]) or rabbit anti-UL142 sera (Sigma Genosys) and protein G-Sepharose beads. The beads were washed in 0.1% digitonin lysis buffer, and bound proteins were eluted in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer plus reducing agent by being heated at 90°C for 5 min. Fractions of eluted proteins were digested with 1,000 U of endo Hf (a fusion of endoglycosidase H [endo H] and maltose binding protein; New England Biolabs) for 2 h at 37°C and then separated on an SDS-12% PAGE gel. The gel was fixed in 40% methanol-12% acetic acid and allowed to dry. Bands were visualized using a phosphor screen (Perkin-Elmer, Waltham, MA) and a storm scanner (Molecular Dynamics; Amersham Pharmacia) or BioMax MR film (Kodak).
Immunoblotting.
Whole-cell lysates of transfected HeLa cells in SDS-PAGE sample buffer plus 2.5% β-mercaptoethanol were prepared. Proteins were separated on an SDS-PAGE gel and wet transferred onto an Immobilon-P membrane (Millipore, Watford, United Kingdom). The membrane was blocked in 5% dried milk-0.05% Tween 20-PBS. Myc-tagged proteins were detected using rabbit anti-Myc antibody (ab9106 [Abcam]) followed by horseradish peroxidase-conjugated goat anti-rabbit Ig antibody (Dako Cytomation). Protein bands were visualized by chemiluminescence using ECL Western blotting detection reagents (GE Healthcare) and X-ray film (Kodak).
RESULTS
UL142 is localized predominantly to the ER and cis-Golgi apparatus.
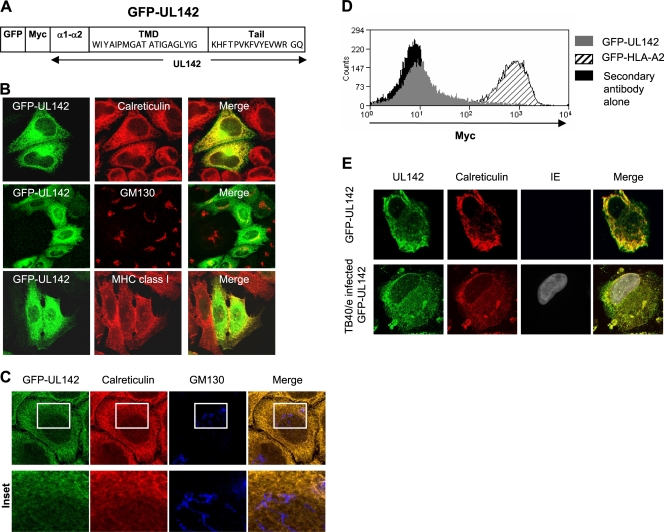
UL142 is an immune evasion molecule encoded by clinical isolates and low-passage-number strains of HCMV (59). In order to determine the mechanism by which UL142 mediates its function, it was necessary to characterize this viral glycoprotein. We reported previously that N- and C-terminally GFP-tagged UL142 proteins were completely sensitive to endo H digestion (59). This finding indicated that UL142 is unlikely to traffic beyond the cis-Golgi apparatus. However, GFP-UL142 (Fig. 1A) was also detected on the surfaces of transfected cells (59). In order to verify the subcellular localization of UL142, HeLa cells transfected with GFP-UL142 were stained with antibodies against markers for the ER (calreticulin), cis-Golgi apparatus (GM130), and cell surface (MHC class I) and then analyzed by immunofluorescence microscopy. At 48 h posttransfection, the majority of GFP-UL142 had colocalized with calreticulin in the ER and with GM130 in the cis-Golgi apparatus (Fig. 1B). A minor proportion of GFP-UL142 colocalized with MHC class I at the cell surface (Fig. 1B). Cells transfected with GFP-UL142 were treated with cycloheximide, which blocks further protein synthesis, in order to chase nascent proteins out of the ER and Golgi complex. Following treatment with cycloheximide, GFP-UL142 remained localized in the ER and cis-Golgi apparatus (data not shown); this result further supports the conclusion that GFP-UL142 was localized predominantly to the ER and cis-Golgi apparatus. A Z-projection of high-resolution scans of GFP-UL142-expressing HeLa cells stained for the ER marker calreticulin and the Golgi apparatus marker GM130 at 16 h posttransfection was generated. The three-dimensional reconstruction shows that GFP-UL142 is localized to the ER in transfected HeLa cells at 16 h posttransfection (Fig. 1C). Similar subcellular localization patterns were observed for GFP-UL142 expressed in cells of the lines U373 (a human astrocytoma cell line which expresses MICA*001) and MRC-5 (a human lung fibroblast cell line transformed with simian virus 40) (59) and COS-7 (an African green monkey kidney fibroblast cell line).
FIG. 1.
UL142 is localized predominantly to the ER and cis-Golgi apparatus. (A) Schematic of GFP-UL142, which is N-terminal GFP and Myc-tagged UL142. The amino acid sequences of the UL142 transmembrane domain (TMD) and cytoplasmic tail are shown. (B) HeLa cells transfected with GFP-UL142 (green) were fixed, permeabilized, and stained with antibodies against intracellular compartment markers (red) at 48 h posttransfection. Calreticulin served as a marker for the ER, GM130 was a cis-Golgi apparatus marker, and MHC class I was a marker for the cell surface. GFP-UL142 is localized predominantly to the ER and cis-Golgi apparatus. (C) Z-projection of high-resolution scans of GFP-UL142 (green)-expressing HeLa cells stained for the ER marker calreticulin (red) and cis-Golgi apparatus marker GM130 (blue). Magnified views of a small region showing the ER and cis-Golgi apparatus are also presented. The results show that GFP-UL142 is localized to the ER at 16 h posttransfection. (D) Mock-transfected HeLa cells and HeLa cells expressing GFP-UL142 or GFP-HLA-A2 were stained with rabbit anti-Myc antibody followed by Alexa Fluor 647-conjugated goat anti-rabbit IgG antibody. Surface expression of GFP-UL142 on a small proportion of transfected cells with very high levels of protein expression was observed. Control cell surface protein GFP-HLA-A2 was detected on all transfected cells regardless of the level of protein expression. As a control for the Myc staining, transfected cells were also stained with the Alexa Fluor 647-conjugated goat anti-rabbit IgG antibody alone. (E) HFFFs infected with HCMV for 72 h were transfected with GFP-UL142 (green) for the last 16 h of infection. Cells were stained for calreticulin as a marker of the ER (red) and immediate-early (IE) antigen as a marker of HCMV infection (gray). The results show that GFP-UL142 remains colocalized with the ER marker in virus-infected cells.
Analysis of HeLa cells transfected with GFP-UL142 by flow cytometry using anti-Myc tag antibody followed by Alexa Fluor 647-conjugated secondary antibody revealed that GFP-UL142 was expressed on the surfaces of transfected cells with very high levels of protein expression (Fig. 1D). The control surface protein GFP-HLA-A2 was detected on the surfaces of all transfected cells, regardless of the level of protein expression. This high level of GFP-UL142 expression is unlikely to be representative of endogenous UL142 protein expression during HCMV infection.
We were unable to detect endogenous UL142 expression in HCMV-infected cells. Consequently, transient expression of GFP-UL142 in HCMV-infected cells was set up. HFFFs were infected with HCMV strain TB40/e for 8, 32, and 56 h and then transfected with GFP-UL142 and incubated for a further 16 h. The cells were then stained for detection of calreticulin (an ER marker) and immediate-early antigen (a marker for virus infection). GFP-UL142 colocalized with calreticulin at 24, 48, and 72 h postinfection. Data for 72 h postinfection are shown in Fig. 1E. Thus, ER localization of GFP-UL142 was sustained in HCMV-infected cells.
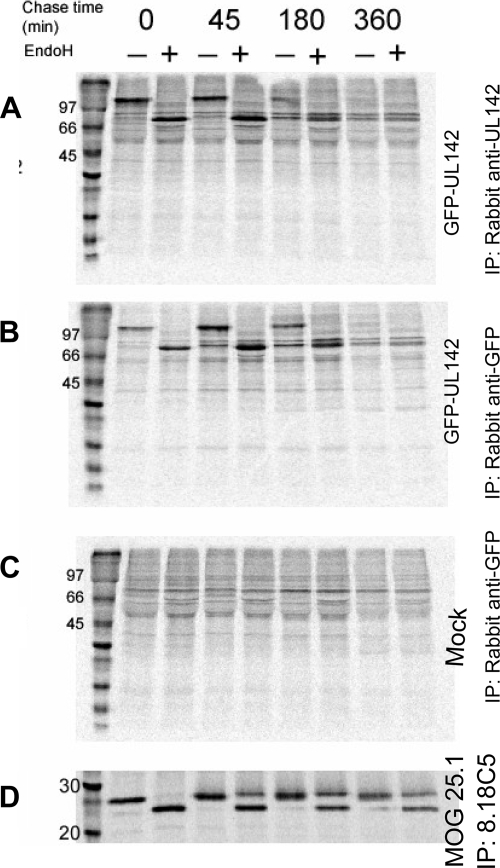
Immunofluorescence microscopy and flow cytometry detected GFP-UL142 at the surfaces of transfected HeLa cells. However, Western blotting indicated that GFP-UL142 was entirely endo H sensitive (59). It was possible that the proportion of cell surface-localized (and hence endo H-resistant) GFP-UL142 was too minor to detect by Western blotting. In order to resolve the apparent discrepancy in the cellular localization patterns of GFP-UL142, radiolabeling of HeLa cells transfected with GFP-UL142 was carried out. No endo H-resistant GFP-UL142 was visible following radiolabeling, immunoprecipitation, and endo H digestion after a 360-min chase (Fig. 2). Myelin oligodendrocyte glycoprotein 25.1 (MOG 25.1) (9), a control cell surface protein assayed in parallel, was endo H resistant at the 45-min chase time point. Thus, it is possible that GFP-UL142 migrates to the cell surface at a very slow rate. Alternatively, GFP-UL142 may be an endo H-sensitive cell surface protein, as has been reported previously (1, 41). Nonetheless, the immunofluorescence microscopy, Western blotting, and pulse-chase data together indicated that GFP-UL142 was predominantly intracellular (localized in the ER and cis-Golgi apparatus), compared with conventional cell surface molecules such as MHC class I.
FIG. 2.
UL142 remains endo H sensitive following synthesis. (A, B, and C) HeLa cells transfected with GFP-UL142 or mock transfected were labeled with [35S]methionine and [35S]cysteine for 20 min and then chased in unlabeled methionine and cysteine for 0, 45, 180, or 360 min. GFP-UL142 was immunoprecipitated using either rabbit anti-UL142 sera (A) or monoclonal anti-GFP antibody (B). Fractions of the eluted proteins were digested with endo H and analyzed by gel electrophoresis. GFP-UL142 remained endo H sensitive and had disappeared by the 360-min chase time point. (D) The control cell surface protein MOG 25.1, which was analyzed in parallel, acquired endo H resistance after the 45-min chase. IP, immunoprecipitant; +, present; −, absent.
Radiolabeling also provided some insight into the kinetics of GFP-UL142 expression. Little or no GFP-UL142 was detected at the 180- and 360-min chase time points (Fig. 2). However, proportions of control protein MOG 25.1 remained approximately constant (Fig. 2). Similar kinetics were observed whether anti-UL142 sera or anti-GFP antibody was used to immunoprecipitate GFP-UL142. The disappearance of GFP-UL142 was indicative of secretion or degradation. However, since no GFP-UL142 was detected in the culture supernatants of transfected cells (data not shown), it is likely that the loss of GFP-UL142 was due to degradation. Interestingly, we were consistently unable to detect endogenous UL142 in HCMV-infected cells or untagged UL142 in transfected or transduced cells. Any cellular proteins that coprecipitated with GFP-UL142 were undetectable under these conditions (Fig. 2).
The transmembrane domain of UL142 mediates ER retention.
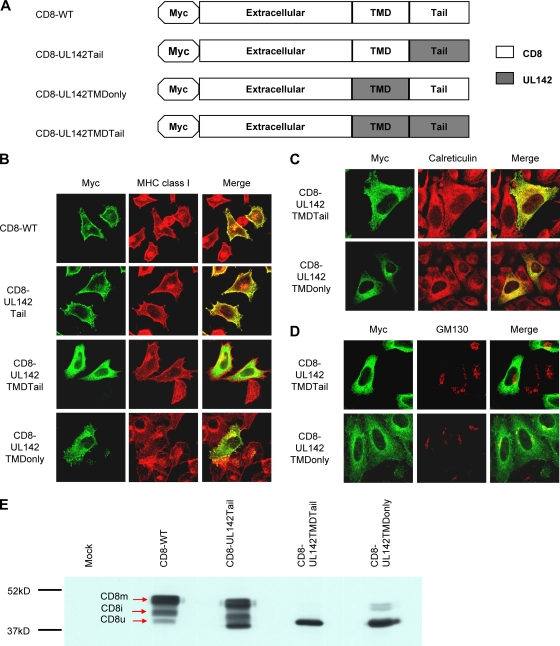
The cytoplasmic tails and transmembrane domains of type I transmembrane proteins are often responsible for their subcellular localization (49). In order to determine which domain of UL142 was responsible for its subcellular localization, chimeric CD8 reporter constructs with the cytoplasmic tail (CD8-UL142Tail), the transmembrane domain (CD8-UL142TMDonly), or both the transmembrane and tail (CD8-UL142TMDTail) of UL142 were generated (Fig. 3A). HeLa cells transfected with these CD8 constructs were stained with antibodies against the Myc tag and MHC class I (which served as a marker for the cell surface and Golgi apparatus) and then analyzed by immunofluorescence microscopy. Full-length CD8 (CD8-WT) and CD8-UL142Tail colocalized with MHC class I at the cell surface and Golgi apparatus (Fig. 3B). In contrast, CD8-UL142TMDTail was never detected at the cell surface (Fig. 3B). Instead, CD8-UL142TMDTail perfectly colocalized with calreticulin in the ER but not with GM130 in the Golgi apparatus (Fig. 3C and D). Interestingly, the majority of CD8-UL142TMDonly was retained intracellularly, while only a small proportion of this chimera was detected on the cell surface (Fig. 3B). Indeed, intracellular CD8-UL142TMDonly colocalized with calreticulin in the ER and with GM130 in the cis-Golgi apparatus (Fig. 3C and D). Control CD8 chimeras with the transmembrane domain and/or cytoplasmic tail of HLA-A2 were all expressed at the cell surface (data not shown).
FIG. 3.
The UL142 transmembrane domain (TMD) mediates ER localization. (A) Schematic of CD8-WT and CD8-UL142 chimeras, which have an N-terminal Myc tag. (B) HeLa cells transfected with the CD8-WT, CD8-UL142Tail, CD8-UL142TMDTail, or CD8-UL142TMDonly construct were fixed and permeabilized and then stained with antibodies against the Myc tag (green) and MHC class I (red). MHC class I served as a marker for both the cell surface and the Golgi apparatus. CD8-UL142TMDTail and CD8-UL142TMDonly were localized predominantly intracellularly. (C) HeLa cells transfected with the CD8-UL142TMDTail or CD8-UL142TMDonly construct were fixed and permeabilized and then stained with antibodies against the Myc tag (green) and the ER marker calreticulin (red). CD8-UL142TMDTail and CD8-UL142TMDonly colocalized with calreticulin. (D) HeLa cells transfected with CD8-UL142TMDTail or CD8-UL142TMDonly were fixed and permeabilized and then stained with antibodies against the Myc tag (green) and the cis-Golgi apparatus marker GM130 (red). CD8-UL142TMDonly, but not CD8-UL142TMDTail, colocalized with GM130. (E) Western blot of mock-transfected HeLa cells and transfected HeLa cells expressing CD8-WT or a CD8-UL142 chimera. CD8 progresses through three maturation states as it traffics through the cell. CD8u is located in the ER, CD8i is located in the cis-Golgi apparatus, and CD8m is located at the cell surface. The chimeras CD8-UL142TMDTail and CD8-UL142TMDonly were predominantly in the CD8u state. Molecular mass differences among the maturation states of CD8-WT and those of the CD8-UL142 chimeras are due to the compositions of their domains.
CD8 progresses through three maturation states, with different molecular weights, as it traffics through the cell (17, 37, 38). The unglycosylated precursor, CD8u, is synthesized in the ER and then traffics to the cis-Golgi apparatus. CD8i is an intermediate species that is palmitoylated and initially O glycosylated in the cis-Golgi apparatus. CD8m is the mature doublet form with elongated glycans that is generated in the late part of the Golgi complex and then traffics to the cell surface. CD8u has the lowest molecular weight, while CD8m has the highest. Thus, analysis of the maturation states of the CD8-UL142 chimeras would give further insight into posttranslational modifications, trafficking, and subcellular localization patterns of these molecules.
All three maturation species were observed for CD8-WT and CD8-UL142Tail by Western blotting (Fig. 3E). As expected, only the CD8u species was observed for CD8-UL142TMDTail (Fig. 3E), thereby confirming the ER localization observed for this chimera by immunofluorescence microscopy (Fig. 3B). Interestingly, the majority of CD8-UL142TMDonly was also in the CD8u form (Fig. 3E). Thus, both CD8-UL142TMDonly and CD8-UL142TMDTail were retained in the CD8u form in the ER by virtue of the UL142 transmembrane domain. It is likely that cell surface expression of CD8-UL142TMDonly was possible only because the CD8 cytoplasmic tail alleviated the ER retention effects of the UL142 transmembrane domain (27). Taken together, the data indicate that the UL142 transmembrane domain mediates ER retention. The absence of CD8-UL142TMDTail from the Golgi apparatus (Fig. 3C and D) and the absence of CD8i forms of this chimera (Fig. 3E) suggest that the UL142 luminal domain is involved in localization of UL142 to the cis-Golgi apparatus. It is possible that an interaction between the UL142 luminal domain and (i) MICA or (ii) other cellular proteins enables trafficking of UL142 beyond the ER.
UL142 retains full-length, but not truncated, MICA forms in the cis-Golgi apparatus.
We originally proposed that UL142 might interfere with surface expression of NKG2D ligands (59). This proposal was based on the structural relatedness of UL142 to MCMV-encoded MHC class I-related proteins (34, 47), which interfere with surface expression of murine NKG2D ligands. Subsequently, Chalupny et al. reported that UL142 down-modulates surface expression of full-length MICA alleles but not the truncated allele MICA*008 (12). However, no mechanism was identified. Consequently, we sought to verify the initial observations by Chalupny et al., as well as to determine the mechanism by which UL142 down-modulates surface expression of full-length MICA alleles.
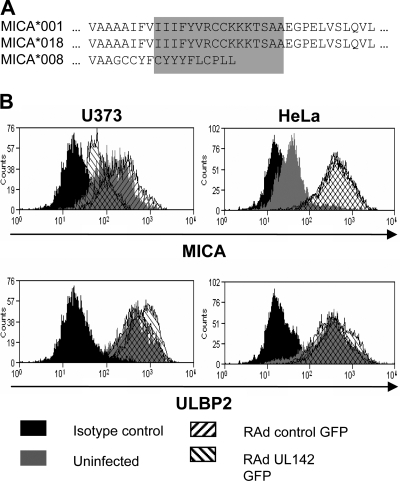
HeLa cells express the truncated allele MICA*008 (20), while U373 cells express the full-length allele MICA*001 (50, 62). In comparison with full-length MICA alleles, MICA*008 has an altered transmembrane domain and a truncated cytoplasmic tail (Fig. 4A). In order to investigate the effect of UL142 on surface expression of MICA, HeLa and U373 cells were transduced with RAd that expressed UL142 as well as GFP (designated RAd UL142 GFP). Transductions with Ad that expressed only GFP (designated RAd control GFP) were carried out as a control. In comparison with RAd control GFP, RAd UL142 GFP down-modulated surface expression of MICA*001 on U373 cells but not that of MICA*008 on HeLa cells (Fig. 4B). As a control, cell surface expression of ULBP2 (an NKG2D ligand sequestered by the HCMV protein UL16) in the presence of UL142 was evaluated. RAd UL142 GFP had no effect on surface expression of ULBP2 (Fig. 4B).
FIG. 4.
UL142 down-modulates surface expression of full-length MICA alleles. (A) Sequence alignment of portions of the transmembrane domains (highlighted in gray) and cytoplasmic tails of full-length and truncated MICA alleles. (B) U373 cells express the full-length allele MICA*001, while HeLa cells express the truncated allele MICA*008. HeLa and U373 cells were transduced with RAd UL142 GFP, which expresses both UL142 and GFP, or RAd control GFP, which expresses GFP alone. Transduction efficiency of ∼100% was achieved. Ninety-six hours posttransduction, the cells were stained with anti-MICA, anti-ULBP2, or an isotype control antibody, followed by Alexa Fluor 647-conjugated secondary antibody. UL142 down-modulated expression of MICA*001 on the surfaces of U373 cells but not that of MICA*008 on the surfaces of HeLa cells. Surface expression of ULBP2 on cells of both lines was unaffected by UL142. Overlapping of histograms is depicted by cross-hatching.
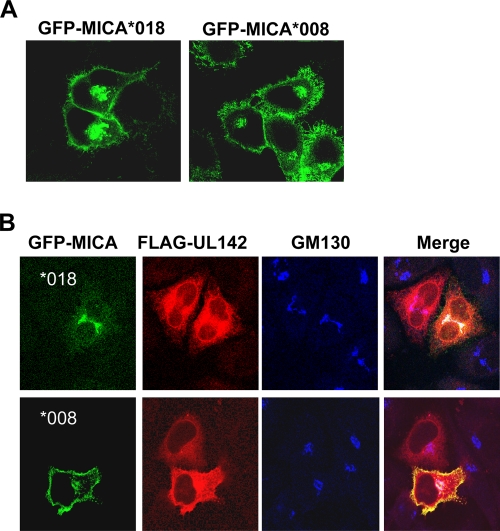
UL142-mediated down-modulation of MICA surface expression was observed from 96 h posttransduction. The delay in the removal of MICA from the cell surface suggested that UL142 interacted with nascent MICA en route to the cell surface but not mature MICA at the cell surface. This pattern is in agreement with the predominant ER and cis-Golgi apparatus localization of UL142 (Fig. 1B). MICA present on the cell surface prior to UL142 expression would eventually be degraded as a result of the cell's natural protein turnover but would not be replaced with newly synthesized molecules in the presence of UL142. In order to determine the cellular compartment in which UL142 and MICA interact, HeLa cells were transfected with GFP-tagged MICA alone or in conjunction with FLAG-tagged UL142 and then analyzed by immunofluorescence confocal microscopy. FLAG-UL142, like GFP-UL142, localized predominantly to the ER and cis-Golgi apparatus. When expressed in the absence of FLAG-UL142, GFP-tagged full-length MICA (GFP-MICA*018) and GFP-tagged truncated MICA (GFP-MICA*008) localized to the cell surface and Golgi apparatus (Fig. 5A). However, cotransfection with FLAG-UL142 resulted in the retention of GFP-MICA*018 in a perinuclear compartment at 16 h posttransfection but had no effect on surface expression of GFP-MICA*008 (Fig. 5B). Intracellular GFP-MICA*018 (and a proportion of FLAG-UL142) colocalized with the cis-Golgi apparatus marker GM130 (Fig. 5B); thus, UL142 retained the full-length MICA allele in the cis-Golgi apparatus.
FIG. 5.
UL142 retains full-length MICA alleles in the cis-Golgi apparatus. (A) HeLa cells transfected with either GFP-MICA*018 (GFP-tagged full-length MICA) or GFP-MICA*008 (GFP-tagged truncated MICA) were analyzed by confocal microscopy. Both alleles of MICA were localized to the cell surface and Golgi apparatus. (B) Immunofluorescence microscopy images of HeLa cells cotransfected with GFP-MICA*018 or GFP-MICA*008 (green) and FLAG-UL142 (red). Sixteen hours posttransfection, cells were fixed, permeabilized, and stained with antibodies against the FLAG tag and GM130 (blue), which served as a marker for the cis-Golgi apparatus. FLAG-UL142 prevents surface expression of GFP-MICA*018 by retaining the full-length MICA allele in the cis-Golgi apparatus. However, FLAG-UL142 has no effect on surface expression of the truncated allele GFP-MICA*008.
The UL142 luminal domain is required for intracellular retention of MICA.
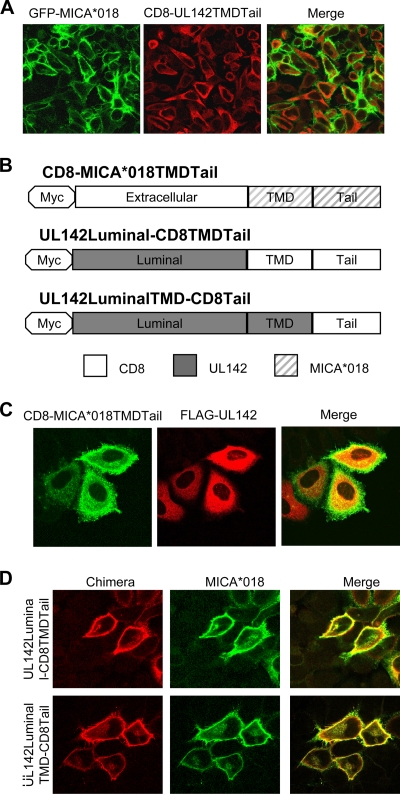
It is important that while FLAG-UL142 sequestered GFP-MICA*018 in the cis-Golgi apparatus (Fig. 5B), GFP-UL142 had no effect on surface expression of FLAG-MICA*018 (data not shown). This finding suggested that the large GFP tag on the UL142 luminal (i.e., α1-α2) domain might have prevented binding to MICA*018 and, thus, that the UL142 luminal (i.e., α1-α2) domain might be involved in intracellular retention of full-length MICA alleles. In support of this possibility, we observed that the CD8-UL142TMDTail chimera (Fig. 3A) was unable to intracellularly retain GFP-MICA*018 (Fig. 6A). Thus, the transmembrane domain and cytoplasmic tail of UL142 were insufficient for intracellular sequestration of full-length MICA alleles. With the aid of several sequence analysis programs (see Materials and Methods), UL142 was predicted to be a type I transmembrane protein. Accordingly, the involvement of the UL142 luminal domain in intracellular sequestration of full-length MICA alleles would suggest an interaction with the MICA extracellular domain. In support of this hypothesis, we observed that FLAG-UL142 was unable to intracellularly retain a CD8 chimera with the transmembrane domain and cytoplasmic tail of the full-length allele MICA*018 (Fig. 6B and C). Thus, the data suggest that an interaction between the UL142 luminal domain and the MICA extracellular domain is involved in UL142 intracellular retention of full-length MICA alleles.
FIG. 6.
The UL142 luminal domain is required for intracellular retention of full-length MICA alleles. (A) HeLa cells cotransfected with GFP-MICA*018 (green) and CD8-UL142TMDTail (red) were fixed, permeabilized, and incubated with anti-UL142 rabbit sera. CD8-UL142TMDTail had no effect on surface expression of GFP-MICA*018. (B) Schematic of CD8-MICA*018TMDTail (a CD8 chimera with the transmembrane domain [TMD] and cytoplasmic tail of the full-length allele MICA*018), UL142Luminal-CD8TMDTail (a CD8 chimera with the luminal domain of UL142), and UL142LuminalTMD-CD8Tail (a CD8 chimera with the luminal and transmembrane domains of UL142). (C) HeLa cells cotransfected with CD8-MICA*018TMDTail and FLAG-UL142 were fixed and permeabilized and then stained with antibodies against the Myc (green) and FLAG (red) tags. FLAG-UL142 had no effect on the cell surface expression of CD8-MICA*018TMDTail. (D) HeLa cells cotransfected with UL142Luminal-CD8TMDTail or UL142LuminalTMD-CD8Tail (red) and untagged MICA*018 (green) were fixed and permeabilized and then stained with antibodies against the Myc tag (red) and MICA (green). Both chimeras were unable to intracellularly sequester MICA*018.
CD8 chimeras containing the UL142 luminal domain (with or without the UL142 transmembrane domain) (Fig. 6B) were localized predominantly to the cell surface and had no effect on the surface expression of the full-length MICA*018 allele (Fig. 6D) due to the presence of the CD8 cytoplasmic tail (27). The inability of these UL142-CD8 chimeras to intracellularly retain MICA*018 suggests that UL142 must be localized to the ER/cis-Golgi apparatus in order to intracellularly sequester full-length MICA alleles.
DISCUSSION
The HCMV glycoprotein UL142 inhibits NK cell-mediated lysis by down-modulating surface expression of full-length, but not truncated, MICA alleles (12, 59). UL142 is structurally related to MCMV-encoded MHC class I-related proteins, which down-modulate ligands of murine NKG2D. The m145 protein interferes with MULT-1 (a murine NKG2D ligand) in a compartment beyond the ER-Golgi apparatus intermediate compartment or cis-Golgi apparatus (29). Another MCMV MHC class I-related protein, m155, interferes with the NKG2D ligand H60 following exit from the ER-Golgi apparatus intermediate compartment or cis-Golgi apparatus (22) and/or targets H60 for proteasomal degradation (31). Our data suggest that UL142 functions intracellularly like these MCMV proteins because it is localized predominantly to the ER and cis-Golgi apparatus. Furthermore, we demonstrate that UL142 sequesters full-length, but not truncated, MICA alleles in the cis-Golgi apparatus.
The transmembrane domain of UL142 mediates localization to the ER. ER localization signals such as KDEL or K(X)KXX (49), which are normally found in the cytoplasmic tail of ER resident proteins, are absent from UL142. Thus, ER localization of UL142 may be mediated by (i) unknown ER retention signals within the transmembrane domain, (ii) intramembrane interactions with ER resident cellular proteins, or (iii) the length of the transmembrane domain (11, 56). Cholesterol, whose concentration in lipid bilayers increases along the secretory pathway, is proposed to regulate protein sorting in the absence of dominant luminal or cytoplasmic associations (11, 35). Consequently, plasma membrane proteins usually possess transmembrane domains that are 23 amino acids or more in length, while the transmembrane domains of Golgi proteins are 17 amino acids or less. Thus, the 20-amino-acid transmembrane domain of UL142 may be compatible only with the cholesterol content of ER membranes. Despite the observed ER/cis-Golgi apparatus localization of tagged UL142, it was possible that the subcellular localization of UL142 might be altered in the presence of other proteins expressed during HCMV infection. This phenomenon was recently described for UL18, which localizes predominantly to the ER and cis-Golgi apparatus when expressed in isolation (32). However, we observed that GFP-UL142 transiently expressed during HCMV infection remained localized to the ER.
UL142 cannot intracellularly sequester the truncated allele MICA*008. There are high degrees of sequence identity among known MICA alleles (2). However, MICA*008 has a truncated cytoplasmic tail due to a premature stop codon resulting from a frameshift mutation (18, 33). In addition, the frameshift mutation alters the sequence of the MICA transmembrane domain. Therefore, any interaction between full-length MICA alleles and UL142 is likely to involve the transmembrane domain sequence AAAIFVIIIFYV and/or the cytoplasmic tail sequence absent in MICA*008. Furthermore, intracellular sequestration of MICA is likely to involve the UL142 transmembrane domain and/or cytoplasmic tail since UL142 is a type I transmembrane protein. In support of this hypothesis, it is interesting that Chalupny et al. observed that UL142 down-modulates surface expression of a full-length MICA allele (GenBank accession number AAD52060) that is identical to MICA*008 in the extracellular domain (12).
We observed that intracellular sequestration of full-length MICA alleles cannot occur in the absence of the UL142 luminal domain or the MICA extracellular domain. One possible interpretation of the data is that the UL142 luminal domain binds the MICA extracellular domain, thereby distinguishing MICA from other NKG2D ligands. In this case, the UL142 luminal domain would recognize both full-length and truncated MICA alleles while the UL142 transmembrane and/or cytoplasmic tail would interact only with full-length MICA alleles. Furthermore, in the absence of the initial recognition of MICA by the UL142 luminal domain, intracellular retention of full-length MICA alleles by the UL142 transmembrane and/or cytoplasmic tail might be impossible. Another possibility is that UL142 cannot intracellularly sequester full-length MICA alleles if the viral protein cannot traffic to the cis-Golgi apparatus. We propose that trafficking of UL142 from the ER to the cis-Golgi apparatus is mediated by an interaction between the UL142 luminal domain and (i) MICA or (ii) other cellular proteins. If this is the case, then it further highlights the importance of the ER/cis-Golgi apparatus localization of UL142 to its immune evasion function.
AD169, an HCMV strain that lacks UL142, down-modulates surface expression of full-length MICA alleles, but not MICA*008, at 24 h postinfection (62). The AD169-encoded immunoevasin(s) is reported to mediate degradation, rather than sequestration, of full-length MICA alleles (62). Thus, UL142 is one of two (or more) HCMV proteins involved in down-modulating MICA surface expression. UL142 is expressed at 72 h postinfection (59). It is not apparent why HCMV has evolved to encode proteins that down-modulate MICA surface expression at both 24 and 72 h postinfection. Nonetheless, by down-modulating surface expression of full-length MICA alleles, HCMV has evolved to inhibit activation of NK cells as well as costimulation of γδ T cells, CD8+ αβ T cells, and CD4+ T cells, all of which express the potent activating NK receptor NKG2D (6, 19, 21, 44). In addition, down-modulation of MICA surface expression may have an impact on factors in acquired immunity, such as primed T cells. Specifically, robust interference with NKG2D ligand surface expression would provide HCMV with a long window of opportunity to replicate and spread to further individuals following reactivation from latency.
The polymorphism and redundancy of NKG2D ligands, coupled with the redundancy within HCMV mechanisms for evasion of NKG2D-mediated activation, may reflect selective pressures promoting the evolution of both the human immune system and HCMV. Indeed, the emergence of the MICA*008 allele may be a host defensive adaptation selected to evade HCMV. The inability of AD169 to prevent surface expression of MICA*008 results in NK cell-mediated lysis (62)—thus, MICA*008 is able to activate NK cells via NKG2D despite its truncation. It is possible that individuals with the MICA*008 allele are less susceptible to HCMV infection than those with full-length MICA alleles; indeed, there is some evidence in support of this idea (26). Interestingly, this truncated variant of MICA is the most prevalent allele in most populations studied (39, 61). Understanding the mechanisms underlying the regulation of NK cell-mediated lysis, as well as the mechanisms that HCMV has evolved to defend the infected cell, will be beneficial in the development of prophylactic drugs, vaccines, and antiviral therapies.
Acknowledgments
We thank Hugh Reyburn for his helpful advice and Fiona Morgan for her technical assistance.
O.A. was funded by a Wellcome Trust 4-year Ph.D. studentship. This work was also supported by the Wellcome Trust project grant 079591/Z/06/Z.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Ackerman, A. L., C. Kyritsis, R. Tampe, and P. Cresswell. 2003. Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc. Natl. Acad. Sci. USA 100:12889-12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony Nolan Trust. 2008. MICA nucleotide sequence alignments. Anthony Nolan Research Institute, London, United Kingdom. http://hla.alleles.org/data/mica.html.
- 3.Arnon, T. I., H. Achdout, O. Levi, G. Markel, N. Saleh, G. Katz, R. Gazit, T. Gonen-Gross, J. Hanna, E. Nahari, A. Porgador, A. Honigman, B. Plachter, D. Mevorach, D. G. Wolf, and O. Mandelboim. 2005. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 6:515-523. [DOI] [PubMed] [Google Scholar]
- 4.Bahram, S., H. Inoko, T. Shiina, and M. Radosavljevic. 2005. MIC and other NKG2D ligands: from none to too many. Curr. Opin. Immunol. 17:505-509. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, P. D., and J. E. Grundy. 1992. Down-regulation of the class I HLA heterodimer and beta 2-microglobulin on the surface of cells infected with cytomegalovirus. J. Gen. Virol. 73(Pt. 9):2395-2403. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, S., V. Groh, J. Wu, A. Steinle, J. H. Phillips, L. L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727-729. [DOI] [PubMed] [Google Scholar]
- 7.Beersma, M. F., M. J. Bijlmakers, and H. L. Ploegh. 1993. Human cytomegalovirus down-regulates HLA class I expression by reducing the stability of class I H chains. J. Immunol. 151:4455-4464. [PubMed] [Google Scholar]
- 8.Boyle, L. H., A. K. Gillingham, S. Munro, and J. Trowsdale. 2006. Selective export of HLA-F by its cytoplasmic tail. J. Immunol. 176:6464-6472. [DOI] [PubMed] [Google Scholar]
- 9.Boyle, L. H., J. A. Traherne, G. Plotnek, R. Ward, and J. Trowsdale. 2007. Splice variation in the cytoplasmic domains of myelin oligodendrocyte glycoprotein affects its cellular localisation and transport. J. Neurochem. 102:1853-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braud, V. M., D. S. Allan, D. Wilson, and A. J. McMichael. 1998. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Bretscher, M. S., and S. Munro. 1993. Cholesterol and the Golgi apparatus. Science 261:1280-1281. [DOI] [PubMed] [Google Scholar]
- 12.Chalupny, N. J., A. Rein-Weston, S. Dosch, and D. Cosman. 2006. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 346:175-181. [DOI] [PubMed] [Google Scholar]
- 13.Chapman, T. L., A. P. Heikeman, and P. J. Bjorkman. 1999. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11:603-613. [DOI] [PubMed] [Google Scholar]
- 14.Cosman, D., N. Fanger, L. Borges, M. Kubin, W. Chin, L. Peterson, and M. L. Hsu. 1997. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 7:273-282. [DOI] [PubMed] [Google Scholar]
- 15.Cosman, D., J. Mullberg, C. L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N. J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14:123-133. [DOI] [PubMed] [Google Scholar]
- 16.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J. Gen. Virol. 73(Pt. 10):2709-2713. [DOI] [PubMed] [Google Scholar]
- 17.Erra, M. C., L. Iodice, L. V. Lotti, and S. Bonatti. 1999. Cell fractionation analysis of human CD8 glycoprotein transport between endoplasmic reticulum, intermediate compartment and Golgi complex in tissue cultured cells. Cell Biol. Int. 23:571-577. [DOI] [PubMed] [Google Scholar]
- 18.Gaudieri, S., C. Leelayuwat, D. C. Townend, J. Mullberg, D. Cosman, and R. L. Dawkins. 1997. Allelic and interlocus comparison of the PERB11 multigene family in the MHC. Immunogenetics 45:209-216. [DOI] [PubMed] [Google Scholar]
- 19.Groh, V., A. Bruhl, H. El-Gabalawy, J. L. Nelson, and T. Spies. 2003. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 100:9452-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groh, V., R. Rhinehart, H. Secrist, S. Bauer, K. H. Grabstein, and T. Spies. 1999. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc. Natl. Acad. Sci. USA 96:6879-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groh, V., K. Smythe, Z. Dai, and T. Spies. 2006. Fas-ligand-mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat. Immunol. 7:755-762. [DOI] [PubMed] [Google Scholar]
- 22.Hasan, M., A. Krmpotic, Z. Ruzsics, I. Bubic, T. Lenac, A. Halenius, A. Loewendorf, M. Messerle, H. Hengel, S. Jonjic, and U. H. Koszinowski. 2005. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J. Virol. 79:2920-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengel, H., T. Flohr, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1996. Human cytomegalovirus inhibits peptide translocation into the endoplasmic reticulum for MHC class I assembly. J. Gen. Virol. 77(Pt. 9):2287-2296. [DOI] [PubMed] [Google Scholar]
- 25.Ho, M. 2008. The history of cytomegalovirus and its diseases. Med. Microbiol. Immunol. 197:65-73. [DOI] [PubMed] [Google Scholar]
- 26.Huang, B. S., Q. Z. Luo, B. Mei, and P. Yu. 2007. Correlation between MHC class I-related chain A gene *008 allele and human cytomegalovirus infection. Nan Fang Yi Ke Da Xue Xue Bao 27:509-511. (In Chinese.) [PubMed] [Google Scholar]
- 27.Iodice, L., S. Sarnataro, and S. Bonatti. 2001. The carboxyl-terminal valine is required for transport of glycoprotein CD8 alpha from the endoplasmic reticulum to the intermediate compartment. J. Biol. Chem. 276:28920-28926. [DOI] [PubMed] [Google Scholar]
- 28.Krmpotic, A., D. H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A. A. Scalzo, U. H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529-535. [DOI] [PubMed] [Google Scholar]
- 29.Krmpotic, A., M. Hasan, A. Loewendorf, T. Saulig, A. Halenius, T. Lenac, B. Polic, I. Bubic, A. Kriegeskorte, E. Pernjak-Pugel, M. Messerle, H. Hengel, D. H. Busch, U. H. Koszinowski, and S. Jonjic. 2005. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J. Exp. Med. 201:211-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ljunggren, H. G., and K. Karre. 1990. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 11:237-244. [DOI] [PubMed] [Google Scholar]
- 31.Lodoen, M. B., G. Abenes, S. Umamoto, J. P. Houchins, F. Liu, and L. L. Lanier. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J. Exp. Med. 200:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maffei, M., F. Ghiotto, M. Occhino, M. Bono, A. De Santanna, L. Battini, G. L. Gusella, F. Fais, S. Bruno, and E. Ciccone. 2008. Human cytomegalovirus regulates surface expression of the viral protein UL18 by means of two motifs present in the cytoplasmic tail. J. Immunol. 180:969-979. [DOI] [PubMed] [Google Scholar]
- 33.Mizuki, N., M. Ota, M. Kimura, S. Ohno, H. Ando, Y. Katsuyama, M. Yamazaki, K. Watanabe, K. Goto, S. Nakamura, S. Bahram, and H. Inoko. 1997. Triplet repeat polymorphism in the transmembrane region of the MICA gene: a strong association of six GCT repetitions with Behcet disease. Proc. Natl. Acad. Sci. USA 94:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., Jr. 2004. Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major histocompatibility system. Cell. Microbiol. 6:707-717. [DOI] [PubMed] [Google Scholar]
- 35.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14:4695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novotny, J., I. Rigoutsos, D. Coleman, and T. Shenk. 2001. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J. Mol. Biol. 310:1151-1166. [DOI] [PubMed] [Google Scholar]
- 37.Pascale, M. C., M. C. Erra, N. Malagolini, F. Serafini-Cessi, A. Leone, and S. Bonatti. 1992. Post-translational processing of an O-glycosylated protein, the human CD8 glycoprotein, during the intracellular transport to the plasma membrane. J. Biol. Chem. 267:25196-25201. [PubMed] [Google Scholar]
- 38.Pascale, M. C., N. Malagolini, F. Serafini-Cessi, G. Migliaccio, A. Leone, and S. Bonatti. 1992. Biosynthesis and oligosaccharide structure of human CD8 glycoprotein expressed in a rat epithelial cell line. J. Biol. Chem. 267:9940-9947. [PubMed] [Google Scholar]
- 39.Petersdorf, E. W., K. B. Shuler, G. M. Longton, T. Spies, and J. A. Hansen. 1999. Population study of allelic diversity in the human MHC class I-related MIC-A gene. Immunogenetics 49:605-612. [DOI] [PubMed] [Google Scholar]
- 40.Prod'homme, V., C. Griffin, R. J. Aicheler, E. C. Wang, B. P. McSharry, C. R. Rickards, R. J. Stanton, L. K. Borysiewicz, M. Lopez-Botet, G. W. Wilkinson, and P. Tomasec. 2007. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1− NK cells. J. Immunol. 178:4473-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riteau, B., N. Rouas-Freiss, C. Menier, P. Paul, J. Dausset, and E. D. Carosella. 2001. HLA-G2, -G3, and -G4 isoforms expressed as nonmature cell surface glycoproteins inhibit NK and antigen-specific CTL cytolysis. J. Immunol. 166:5018-5026. [DOI] [PubMed] [Google Scholar]
- 42.Rolle, A., M. Mousavi-Jazi, M. Eriksson, J. Odeberg, C. Soderberg-Naucler, D. Cosman, K. Karre, and C. Cerboni. 2003. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J. Immunol. 171:902-908. [DOI] [PubMed] [Google Scholar]
- 43.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 44.Saez-Borderias, A., M. Guma, A. Angulo, B. Bellosillo, D. Pende, and M. Lopez-Botet. 2006. Expression and function of NKG2D in CD4+ T cells specific for human cytomegalovirus. Eur. J. Immunol. 36:3198-3206. [DOI] [PubMed] [Google Scholar]
- 45.Sinclair, J., and P. Sissons. 2006. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87:1763-1779. [DOI] [PubMed] [Google Scholar]
- 46.Sissons, J. G., A. J. Carmichael, N. McKinney, J. H. Sinclair, and M. R. Wills. 2002. Human cytomegalovirus and immunopathology. Springer Semin. Immunopathol. 24:169-185. [DOI] [PubMed] [Google Scholar]
- 47.Smith, H. R., J. W. Heusel, I. K. Mehta, S. Kim, B. G. Dorner, O. V. Naidenko, K. Iizuka, H. Furukawa, D. L. Beckman, J. T. Pingel, A. A. Scalzo, D. H. Fremont, and W. M. Yokoyama. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA 99:8826-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stern-Ginossar, N., N. Elefant, A. Zimmermann, D. G. Wolf, N. Saleh, M. Biton, E. Horwitz, Z. Prokocimer, M. Prichard, G. Hahn, D. Goldman-Wohl, C. Greenfield, S. Yagel, H. Hengel, Y. Altuvia, H. Margalit, and O. Mandelboim. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27-54. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, M., J. M. Boname, S. Field, S. Nejentsev, M. Salio, V. Cerundolo, M. Wills, and P. J. Lehner. 2008. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 105:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomasec, P., V. M. Braud, C. Rickards, M. B. Powell, B. P. McSharry, S. Gadola, V. Cerundolo, L. K. Borysiewicz, A. J. McMichael, and G. W. Wilkinson. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. [DOI] [PubMed] [Google Scholar]
- 52.Tomasec, P., E. C. Wang, A. J. Davison, B. Vojtesek, M. Armstrong, C. Griffin, B. P. McSharry, R. J. Morris, S. Llewellyn-Lacey, C. Rickards, A. Nomoto, C. Sinzger, and G. W. Wilkinson. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 54.Ulbrecht, M., S. Martinozzi, M. Grzeschik, H. Hengel, J. W. Ellwart, M. Pla, and E. H. Weiss. 2000. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164:5019-5022. [DOI] [PubMed] [Google Scholar]
- 55.Vales-Gomez, M., H. Browne, and H. T. Reyburn. 2003. Expression of the UL16 glycoprotein of human cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol. 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson, R. T., and J. E. Pessin. 2001. Transmembrane domain length determines intracellular membrane compartment localization of syntaxins 3, 4, and 5. Am. J. Physiol. Cell Physiol. 281:C215-C223. [DOI] [PubMed] [Google Scholar]
- 57.Welte, S. A., C. Sinzger, S. Z. Lutz, H. Singh-Jasuja, K. L. Sampaio, U. Eknigk, H. G. Rammensee, and A. Steinle. 2003. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 33:194-203. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson, G. W., P. Tomasec, R. J. Stanton, M. Armstrong, V. Prod'homme, R. Aicheler, B. P. McSharry, C. R. Rickards, D. Cochrane, S. Llewellyn-Lacey, E. C. Wang, C. A. Griffin, and A. J. Davison. 2008. Modulation of natural killer cells by human cytomegalovirus. J. Clin. Virol. 41:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wills, M. R., O. Ashiru, M. B. Reeves, G. Okecha, J. Trowsdale, P. Tomasec, G. W. Wilkinson, J. Sinclair, and J. G. Sissons. 2005. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 175:7457-7465. [DOI] [PubMed] [Google Scholar]
- 60.Wu, J., N. J. Chalupny, T. J. Manley, S. R. Riddell, D. Cosman, and T. Spies. 2003. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J. Immunol. 170:4196-4200. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y., A. M. Lazaro, B. Lavingia, and P. Stastny. 2001. Typing for all known MICA alleles by group-specific PCR and SSOP. Hum. Immunol. 62:620-631. [DOI] [PubMed] [Google Scholar]
- 62.Zou, Y., W. Bresnahan, R. T. Taylor, and P. Stastny. 2005. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J. Immunol. 174:3098-3104. [DOI] [PubMed] [Google Scholar]