Abstract
Building a three-dimensional model of the sucrose permease of Escherichia coli (CscB) with the X-ray crystal structure lactose permease (LacY) as template reveals a similar overall fold for CscB. Moreover, despite only 28% sequence identity and a marked difference in substrate specificity, the structural organization of the residues involved in sugar-binding and H+ translocation is conserved in CscB. Functional analyses of mutants in the homologous key residues provide strong evidence that they play a similar critical role in the mechanisms of CscB and LacY.
Keywords: membrane transporters, mutagenesis, structure, sugar binding, H+ translocation
Introduction
The major facilitator superfamily of membrane transport proteins is found ubiquitously in living organisms†. These transporters are involved in a multitude of functions that include uptake of essential nutrients and ions, excretion of metabolic end products and other toxic substances, and communication between the cell and the environment.1,2 The oligosaccharide/H+ symporter sub-family of the major facilitator superfamily2–4 contains sugar/H+ symporters that catalyse the coupled stoichiometric translocation of a sugar and an H+, thereby transducing free energy stored in the electrochemical H+ gradient (Δμ̄H+) into a sugar concentration gradient.
The lactose permease of Escherichia coli (LacY) is arguably the most extensively characterized member of the major facilitator superfamily,5 and only one of two for which X-ray crystal structures have been elucidated.6,7 Amino acid residues that play a critical role in the mechanism of galactose/H+ symport have been identified by extensive site-directed and Cys-scanning mutagenesis,8 and functional studies.5 The preceding paper9 focuses on similarities in the overall structure and the organization of amino acid residues involved in sugar binding and H+ translocation in prokaryotic, as well as eukaryotic transport proteins. The sucrose permease of E. coli (CscB),10,11 which transports sucrose but not lactose or other galactopyranosides, was chosen to test specific features described in the preceding paper.9 Alignment of CscB with the sequence of LacY reveals only 28% sequence identity with an overall homology of 51%. Nevertheless, most of the residues in LacY that are irreplaceable for transport activity are highly conserved in CscB.
As with LacY, previous mutational analysis of conserved Arg147 (Arg144 in LacY) and Asp129 (Glu126 in LacY) in CscB12 reveals a critical role for these residues, which are likely charge-paired and required for sugar binding.13–15 The charge pair Asp237/Lys358 is important for insertion of LacY into the membrane,16–19 and construction of an appropriate double mutant in CscB (N234D/S356K) results in increased insertion into the membrane.20 In addition, a cysteine-less CscB mutant has been engineered that exhibits a twofold increase in sucrose accumulation relative to wild-type CscB.21
Homology threading of the CscB protein sequence with the structure of LacY as template indicates that CscB, like LacY, has two symmetrically positioned six-helix bundles surrounding a large hydrophilic cavity open to the cytoplasm.9 Furthermore, the location and organization of the residues involved in sugar binding and H+ translocation in LacY appear to be similar in CscB. In this study, we target amino acid residues that should be important for sucrose/H+ symport based upon the predicted structure of CscB. Site-directed mutagenesis of the key residues indicates that they play a similar critical role in the mechanisms of action of CscB and LacY.
Results
Sugar-binding site residues
The major amino acyl residues that comprise the putative sugar-binding site of the CscB model exhibit striking conservation with respect to LacY X-ray structure (Figure 1). Thus, Arg147 and Asp129 in helices V and IV of CscB, respectively, which are probably charge-paired (Figure 1(b)), are in positions similar to Arg144 and Glu126 in LacY (Figure 1(a)). Glu270 in CscB (helix VIII), which is one helical turn closer to the cytoplasm than the irreplaceable Glu269 in LacY, aligns with Asn272 in the latter,9 but is in the vicinity of the sugar-binding site on the same face of helix VIII as in LacY. Tyr154, which is homologous to Trp151 in LacY, appears to be in virtually the same position. Moreover, Trp26 in CscB (homologous to Met23 in LacY) may be a component of the sugar-binding site, providing a larger hydrophobic surface. Highly conserved Phe23 in CscB is in a position homologous to Phe20 in LacY and may also play a role in sugar binding. The variations observed in some of the residues and their placement in the sugar-binding sites of CscB and LacY are likely to reflect differences in specificity and affinity for different sugar substrates.
Figure 1.
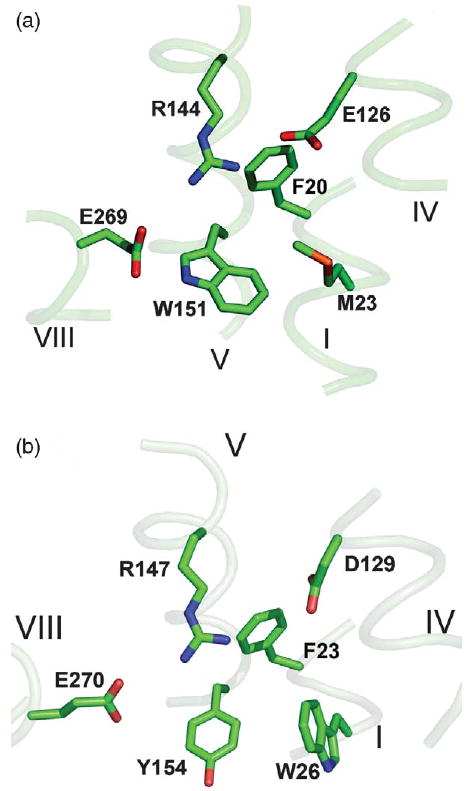
Substrate-binding site in (a) LacY (PDB ID: 1PV7) and (b) the CscB model. Transmembrane helices are numbered in roman numerals. Homologous amino acid residues are shown as sticks.
Replacement of Tyr154 with Trp or Ala abolishes the ability of CscB to catalyse active sucrose transport (Figure 2). However, replacement with Phe results in sucrose transport at a markedly reduced rate, but essentially the same steady-state level of accumulation as the wild-type (Figure 2). As shown previously,22 Trp151 in LacY can be replaced with Phe or Tyr with only a minor loss of transport activity due to a relatively small increase in Km. However, binding affinity decreases by a factor of 50 or 20, respectively.
Figure 2.
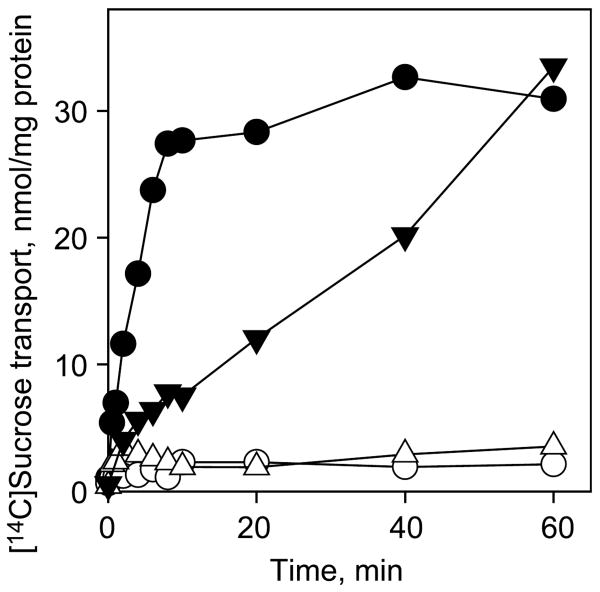
Effect of Y154 and E270 replacements on transport activity of CscB. Time-courses of sucrose accumulation by E. coli T184 expressing wild-type CscB (●), no permease (○) or CscB mutants Y154F (▼), and Y154W, Y154A, E270D or E270A (△) were measured in 50 μl aliquots of cell suspensions containing 70 μg of total protein and 4 mM [14C]sucrose as described in Materials and Methods.
Replacement of Glu270 with Ala or Asp abolishes sucrose transport activity (Figure 2) in a manner similar to that observed with Glu269 in LacY where removal of the carboxyl group eliminates binding and transport,23–25 and replacement with Asp dramatically decreases affinity.26
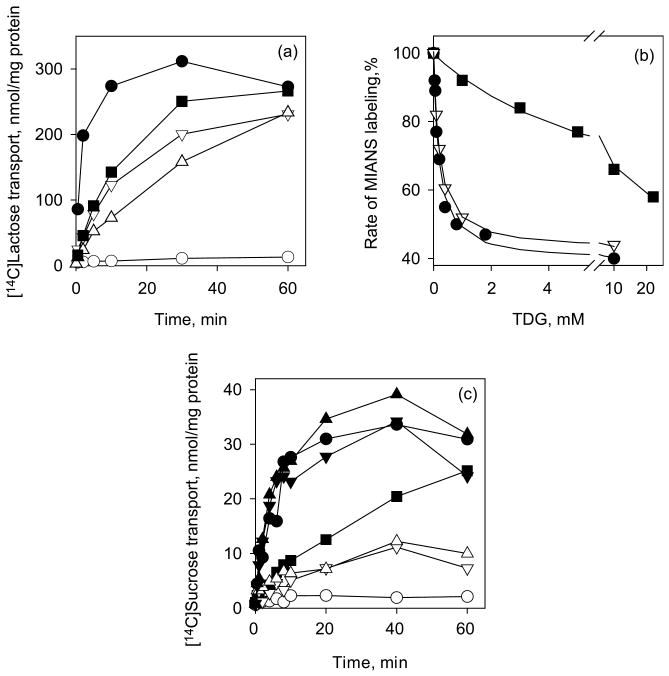
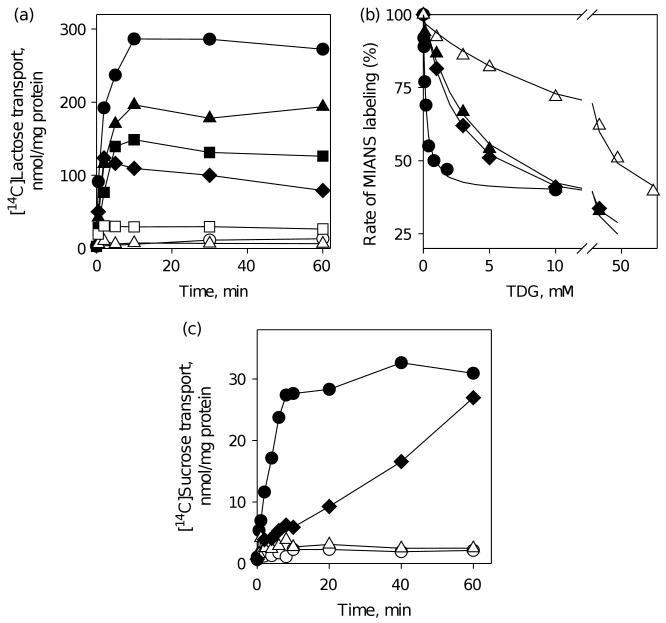
In LacY, Met23 lies within close proximity to the C6 atom of the galactopyranosyl ring of β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG), and Phe20 is close to Trp151.6 Replacement of Phe20 or Met23 with Ala decreases the initial rate of lactose accumulation without significant change in the steady state relative to the wild-type LacY (Figure 3(a)). The binding affinity of the F20A mutant for sugar in LacY, measured by TDG protection against alkylation,27–30 exhibits a 40-fold decrease, while the affinity of M23A is not changed compared to the wild-type LacY (Figure 3(b)). Cys replacements of Phe20 in LacY31 or Phe23 in CscB (Figure 3(c)) result in a significant decrease in the rate of sugar accumulation. Cys or Ala replacements for Trp26 in CscB (homologous to Met23 in LacY) decrease sucrose transport markedly; however, replacement with Phe or Met has little or no effect (Figure 3(c)). In contrast, corresponding replacement of Met23 in LacY with Trp significantly decreases the rate of lactose transport, relative to wild-type LacY (Figure 3(a)).
Figure 3.
Functional significance of F20 and M23 residues in LacY and homologous residues F23 and W26 in CscB. (a) Time-courses of lactose accumulation by E. coli T184 expressing wild-type LacY (●), no permease (○) or LacY mutants: F20A (■), M23A (▽) and M23W (△) were measured in 50 μl aliquots of cell suspension containing 35 μg of total protein and 0.4 mM [14C]lactose as described in Materials and Methods. (b) Affinity of purified LacY mutants WT/C154G (●), F20A/C154G (■) or M23A/C154G (▽) detected by TDG protection against MIANS labeling at 0.5 μM protein and 1 μM MIANS as described in Materials and Methods. Apparent Kd values are 0.17 mM, 0.20 mM and 7 mM for WT/C154G, M23A/C154G and F20A/C154G, respectively. (c) Time-courses of sucrose accumulation by E. coli T184 expressing wild-type CscB (●), no permease (○) or CscB mutants: F23C (■), W26A (▽), W26C (△), W26F (▼) and W26M (▲) were obtained as described for Figure 2.
The C154G mutation in wild-type LacY results in a thermostable protein with high substrate affinity, but has little or no transport activity.29 Gly154 (helix V) is in close proximity to Gly24 (helix I), suggesting that the mutation may decrease conformational flexibility. Remarkably, activity and low thermostability are restored in the double mutant G24C/C154G.30 Since CscB has a Gly residue homologous to Cys154 in LacY, we introduced Gly replacements for Ser27 and/or Ser31 in attempts to engineer a conformationally stable mutant similar to C154G LacY.29 These two residues are in helix I facing Gly157 (helix V) in the CscB model and aligned with Cys154 in LacY. Transport activity and thermostability of either S27G or S27G/S31G CscB mutants are very similar to the wild-type (data not shown), which likely reflect no change in conformational flexibility.
Residues involved in H+ translocation
The spatial organization of the residues involved in H+ translocation in LacY (Figure 4(a)) appear to be very similar in CscB (Figure 4(b)). Clearly, Arg300, His320, Glu323 and Tyr233 in CscB (Figure 4(b)), respectively, are positioned almost exactly as Arg302, His322, Glu325 and Tyr236 in LacY (Figure 4(a)). The Lys319–Asp240 charge pair in LacY (Figure 4(a))6,18,32 also has a counterpart in CscB (Lys317–Asp237) (Figure 4(b)). The salt-bridge/H-bond network observed in LacY, which is comprised of Tyr236 (helix VII), Arg302 (helix IX), His322 and Glu325 (helix X) appears in the CscB model as well (Figure 4(b)).
Figure 4.
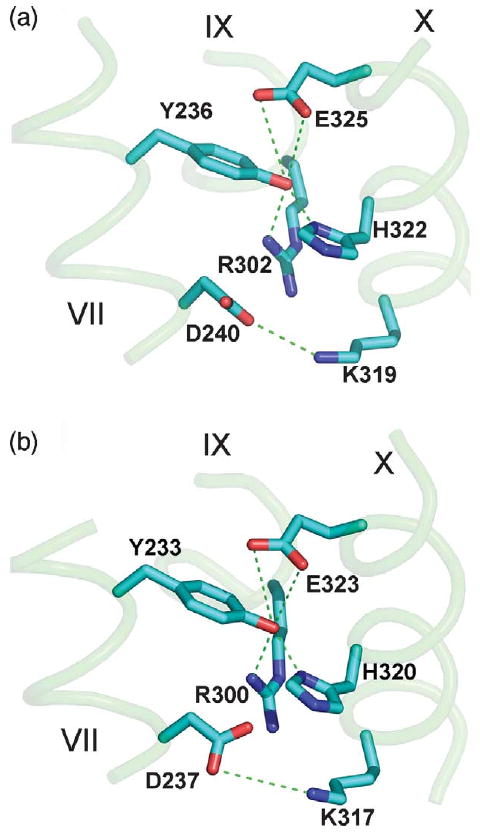
Amino acid residues involved in H+ translocation in (a) the LacY structure (1PV7.pdb) or (b) the CscB model. Transmembrane helices are numbered in roman numerals. Homologous amino acid residues are shown as sticks. Possible H-bonds and salt-bridges are shown as broken lines.
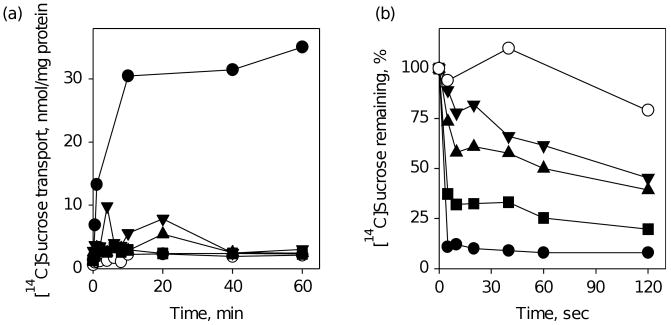
The residues in CscB (Arg300 and Glu323) homologous to Arg302 and Glu325 in LacY, which are critical for H+ translocation, were replaced individually with Ala. Both mutants are completely defective in active sucrose transport (Figure 5(a)), but catalyse equilibrium exchange, albeit at different rates (Figure 5(b)), which likely reflects different affinities for sucrose.33 The properties are similar to the Arg302 and Glu325 mutants in LacY,34,35 which bind sugar normally and catalyse exchange, but are specifically defective in all reactions that involve net H+ translocation.5 Moreover, as observed with His322 mutants in LacY,26,36,37 the H320A mutant in CscB is defective in sucrose transport (Figure 5(a)), and on the basis of the low rate of equilibrium exchange (Figure 5(b)), probably has much lower affinity for sucrose, unlike the Arg300 and Glu323 mutants.
Figure 5.
Functional significance of CscB residues R300, H320 or E323. (a) Time-courses of sucrose accumulation by E. coli T184 expressing wild-type CscB (●), no permease (○) or CscB mutants: R300A (■), H320A (▼) and E323A (▲) measured as described for Figure 2. (b) Time-courses of sucrose equilibrium exchange by RSO vesicles prepared from E. coli T184 cells expressing the same mutants as shown in (a), measured after 200-fold dilution of vesicle suspensions loaded with 100 mM [14C]sucrose into 100 mM unlabeled sucrose solution as described in Materials and Methods.
In LacY, Tyr236 participates in a charge-pair/H-bond network in the middle of a triangle formed by Arg302, His322 and Glu325,6 and replacement of Tyr236 with Phe, Lys, Glu, Gln, His or Ser inactivates lactose transport. In contrast, replacement with Cys, Thr or Ala allows lactose accumulation, although at a significantly lower level than wild-type LacY (Figure 6(a)). Substrate affinity is about 20–25 times lower for Y236A and Y236C mutants, and at least two orders of magnitude lower for Y236F mutant compared to wild-type LacY (Figure 6(b)). Replacements of Tyr233 in CscB (homologous to Tyr236 in LacY) result in complete loss of sucrose transport in the Y233F mutant and a markedly decreased rate of transport in Y233A mutant (Figure 6(c)).
Figure 6.
Functional significance of Y236 in LacY and homologous residue Y233 in CscB. (a) Time-courses of lactose accumulation by E. coli T184 expressing wild-type LacY (●), no permease (○) or LacY mutants: Y236C (▲), Y236T (■), Y236A (◆), Y236S (□), and Y236F, K, Q, E, H (△) measured as described for Figure 3(a). (b) Substrate affinity of purified LacY mutants: WT/C154G (●), Y236C/C154G (▲), Y236A/C154G (◆) and Y236F/C154G (△) detected by TDG protection against MIANS labeling as described for Figure 3(b). Apparent Kd values are 0.17 mM, 2.9 mM, 4.4 mM and 17.2 mM for WT/C154G, Y236A/C154G, Y236C/C154G, and Y236F/C154G, respectively. (c) Time-courses of sucrose accumulation by E. coli T184 expressing wild-type CscB (●), no permease (○) or CscB mutants: Y233A (◆) and Y233F (△) were obtained as described for Figure 2.
Discussion
The results presented here indicate that homology threading is a powerful approach for targeting important residues in membrane transport proteins. Comparative analysis reveals that all amino acid residues involved in H+ translocation in LacY are conserved in the amino acid sequence of CscB. Moreover, the threaded CscB model shows a practically identical spatial organization of the residues involved in H+ translocation in both permeases (Figure 4). Furthermore, CscB mutants R300A, H320A or E323A are unable to accumulate sucrose (Figure 5(a)), but catalyse equilibrium exchange (Figure 5(b)) in a manner similar to comparable mutants of LacY.34,35,37 The results indicate that the Arg300 and Glu323 mutants in CscB are also able to bind and translocate sugar in a process that does not involve net H+ translocation. The network of H-bonds and salt-bridges between Tyr236, Arg302, His322 and Glu325 observed in the LacY structure is conserved in CscB (Figure 4). Replacement of Tyr233 with Phe in CscB completely blocks active transport, as in LacY (Y236F mutant), while significant transport activity remains in both permeases when Tyr is replaced with Ala (Figure 6(a) and (c)).
The residues comprising the putative sugar-binding site in the model of CscB are very similar in character to those in LacY (Figure 1), although only two are strictly conserved (Arg and Phe). However, other amino acid residues with homologous properties in CscB (Asp129, Tyr154, Trp26 and Glu270) are spatially arranged in almost the same manner as in LacY (Glu126, Trp151, Met23 and Glu269). The importance of the charge pair Asp129/Arg147 for sucrose transport by CscB has been shown.12 Glu270 is aligned with Asn272 in LacY, but seems to play the same role functionally as Glu269. Thus, replacement of Glu270 with Ala or Asp abolishes sucrose transport (Figure 2). Trp26 is aligned with Met23 in LacY, and only Met or Phe replacements are fully functional in CscB (Figure 3(c)), suggesting the importance of a hydrophobic moiety in this position. Phe20 in LacY is in close proximity to Trp151 and is important for substrate binding, as mutant F20A binds sugar with much lower affinity (Figure 3(b)). The homologous residue, Phe23 in CscB, is positioned very similar to Phe20 in LacY and may play the same role, since replacement with Cys decreases the rate of sugar transport markedly in CscB (Figure 3(c)) as well as in LacY.31
Tyr154 in CscB also appearstoplayarole similar to that of homologous Trp151 in LacY, where Trp151 stacks hydrophobically with the galactoside moiety of disaccharide substrates.6,22,38 The nature of the aromatic residue at this position in both permeases is important, since mutant Y154F exhibits a markedly decreased rate of sucrose transport, and mutant Y154W is completely inactive (Figure 2). Notably, in several sugar-binding protein structures (PDB ID codes 1A78, 1LT5, 1A3K, 1DLL and 1ULC), binding of the galactosidic ring of TDG or lactose involves Trp in a manner similar to LacY (PDB ID 1PV7). However, binding of the glucopyranoside ring of sucrose in CscB appears to utilize a Tyr residue, as in several other proteins with bound sucrose or maltose (PDB ID codes 1AF6, 1BYH, and 1ANF).
There are striking differences in substrate specificity for sucrose and lactose permeases. LacY is absolutely specific for galactosidic sugars and does not bind or transport glucosides,39,40 while CscB is specific for sucrose and does not transport galactosides.21 The differences in the affinity and specificity of LacY and CscB are likely defined by differences in the structure of lactose and sucrose, respectively, that lead to alterations in the side-chains comprising the binding sites. Protein-bound lactose (PDB ID codes 1DLL, 1ULC) or TDG6 is in an extended conformation. In contrast, protein-bound sucrose (PDB ID codes 1AF6, 1PT2 and 1IW0) is in a bent conformation where glucosidic and fructosidic rings are positioned at approximately a right-angle. Moreover, in proteins with bound sucrose or maltose (PDB ID codes 1AF6, 1BYH, 1ANF), the C6 atom of the glucopyranoside ring is in the orientated opposite to that of the C6 atom of the galactopyranoside ring of bound lactose or TDG (PDB ID codes 1PV7, 1A78, 1LT5, 1ULC) with respect to the aromatic residue involved in sugar binding. When sucrose is docked into the sugar-binding site of the CscB homology model (Figure 7), the glucopyranosyl moiety of sucrose occupies a position homologous to that of the galactopyranosyl moiety of TDG in the structure of LacY (PDB ID 1PV7). In CscB, Arg147 likely interacts with the C3–OH group of glucopyranosyl ring, and Glu270 may interact with the hydroxyl groups of both sugar rings in sucrose, as this residue is one helical turn closer to the cytoplasmic face of the membrane than Glu269 in LacY.
Figure 7.
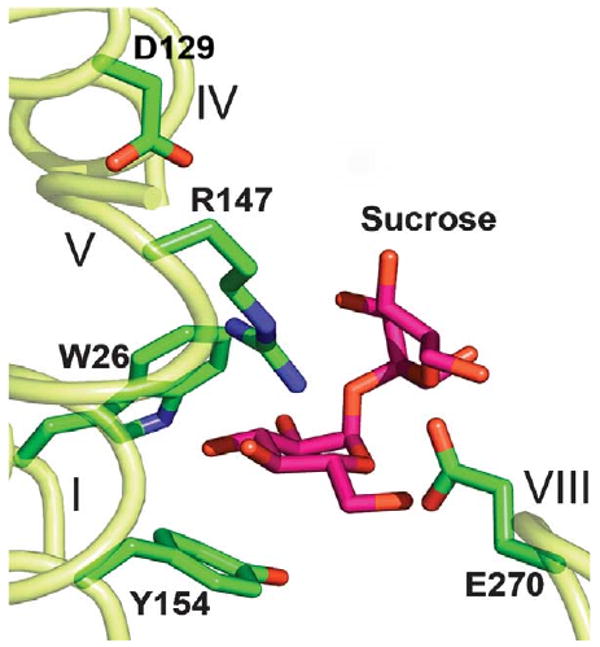
Sucrose molecule docked into the putative sugar-binding site of the CscB model. The sucrose molecule (coordinates from PDB, ID 1IW0), is modeled in the putative sugar-binding site of CscB using the program XtalView.45 Essential amino acid residues are shown as sticks. Transmembrane helices are numbered in roman numerals.
Sequence alignments reveal two major variations in the amino acid ensembles comprising the sugar-binding sites.9 LacY and several other putative sugar transporters have Glu126 and Glu269 (LacY numbering), while CscB and many other putative transporters have an Asp in the position homologous to Glu126 and a Glu that aligns with Asn272. Possibly, this interchange plays an important role in sugar specificity. Indeed, in CscB both carboxyl groups at homologous positions (129 or 270) are essential for sucrose transport. Also, LacY mutant E126D exhibits a low rate of lactose transport as a result of decreased affinity for sugar,41 while mutant D129E in CscB does not significantly alter sucrose transport by CscB.12 In LacY, the galactosidic ring of TDG forms multiple contacts with residues in the N-terminal six-helix bundle and the sugar at the anomeric position extends toward the C-terminal six-helix bundle, making additional contacts with Lys358 and possibly Asp237 to increase affinity.6 However, it is unlikely that binding of sucrose in the bent conformation makes significant contacts with the C-terminal six-helix bundle of CscB, which may explain why Ser and Asn, respectively, in CscB are in homologous positions to Lys358 and Asp237 of LacY. To the contrary, the fructose moiety may make additional contacts with residues in the N-terminal six-helix bundle.
In summary, the results presented here provide functional evidence for the argument that the amino acid residues involved in both sugar binding and H+ translocation, as well as their organization are conserved in LacY and CscB.
Materials and Methods
Materials
All oligonucleotides were synthesized by Integrated DNA Technologies, Inc (Coralville, IA). [14C]lactose was purchased from Amersham Biosciences (Piscataway, NJ) and [14C]sucrose was purchased from Perkin Elmer (Boston, MA). Penta-His antibody-horseradish peroxidase (HRP) conjugate was obtained from Qiagen (Valencia, CA). All restriction enzymes were purchased from New England Biolabs (Beverly, MA). The Quick Change II kit was from Stratagene (La Jolla, CA). Talon superflow resin was obtained from BD Clontech (Palo Alto, CA). All other materials were of reagent grade and were obtained from commercial sources.
Construction of mutants
All mutants were constructed by oligonucleotide, site-specific mutagenesis using a Quikchange® II kit. Generally, 30–40 bp direct and reverse primers bearing the mutated triplet in the middle of the primer were designed by using the Vector NTI 9.1 Suit program (Invitrogen, Carlsbad, CA). All mutagenesis procedures were carried out according to the Quickchange II manual, except that the temperature of the extension reaction was lowered to 68–70 °C during PCR-directed mutagenesis to reduce possible primer duplication. Mutagenic constructs were sequenced over the entire gene to confirm mutations introduced and to discard unwanted mutations.
Expression analysis
The pSP72/CscB and pT7-5/LacY constructs were engineered with a C-terminal His-tag to enable identification by Western blot analysis using penta-His HRP conjugated antibody (Qiagen, Valencia, CA). All mutants described were expressed at levels similar to those of the appropriate wild-type.
Transport assays
E. coli T184 [lacI+O+Z−Y−(A), rspL, met−, thr−, recA, hsdM, hsdR/F′ lacIqO+ZD118(Y+A+)] was transformed with the appropriate expression vector and grown aerobically overnight at 37 °C in Luria-Bertani culture medium containing 100 μg/ml of ampicillin. A tenfold dilution of the culture was grown for 2 h before induction with 1 mM IPTG. Following induction, growth was continued for a further 2 h, after which the cells were harvested and washed in 100 mM potassium phosphate (KPi; pH 7.2), 10 mM MgSO4, and adjusted to an absorbance at 420 nm (A420) of 20 (approximately 1.4 mg/ml of protein) for sucrose, or to A420=10 (approximately 0.7 mg/ml of protein) for lactose transport measurements. Transport of [14C]sucrose or [14C]lactose (5–10 mCi/mmol) at a final concentration of 4 mM and 0.4 mM, respectively, was assayed by rapid filtration.42
Right-side-out (RSO) membrane vesicles were used for equilibrium exchange.35 RSO vesicles were prepared as described,43,44 and resuspended in 100 mM KPi (pH 7.5), 10 mM MgSO4 at a protein concentration of 35 mg/ml, frozen in liquid nitrogen and stored at −80 °C. [14C]sucrose (10 mCi/mmol specific activity; 100 mM final concentration) was added to RSO vesicles and equilibration was carried out overnight on ice in the presence of 50 μM valinomycin and 1 μM nigericin. Equilibrium exchange was initiated by addition of 2 μl aliquots of RSO vesicles to 0.4 ml (200-fold dilution) of 100 mM KPi (pH 7.5) containing 100 mM sucrose. Reactions were quenched at given times with 100 mM KPi (pH 7.5), 100 mM LiCl, and assayed by rapid filtration.
LacY purification
LacY mutants were purified from E. coli XL1-Blue cells transformed with given plasmids essentially as described by using Co(II) affinity chromatography (BD Clontech, Palo Alto, CA).30 The C154G mutation was introduced in order to increase protein stability in dodecyl-β,d-maltopyranoside (DDM).29 All protein preparations were at least 95% pure, as judged by staining with silver after SDS-PAGE. A micro BCA method (Pierce, Rockford, IL) was used for measurements of protein concentration.
Measurement of binding affinity
Substrate protection of Cys148 against alkylation by 2-(4′-maleimidylanilino)naphthalene-6-sulfonic acid (sodium salt) (MIANS) in purified LacY mutants was measured in 50 mM NaPi (pH 7.5), 0.02% (w/v) DDM.27,28 MIANS (1 μM) was added to protein (0.5 μM) equilibrated with given concentrations of TDG and time-courses of fluorescence change were recorded using excitation and emission wavelengths of 330 nm and 415 nm, respectively, as described.29 The rates of the reactions were calculated from the slopes of the fluorescence increase. The reaction rates were plotted against concentration of TDG and fit with a hyperbolic equation using Sigmaplot 8.0 (SPSS Inc., Chicago, IL) in order to estimate apparent binding affinity.30
Molecular modeling
Homology threading was done as described in the preceding paper.9 The sucrose molecule (coordinates from PDB ID 1IW0, α-d-glucopyranosyl β-d-fructofuranoside), was manually docked into the putative sugar-binding site of the CscB model using the program XtalView.45
Acknowledgments
We thank Dr Jun-Yong Choe for help with docking the sucrose molecule into the putative sugar-binding site of the CscB model. This work was supported by NIH grants DK069463 and DK051131 to H.R.K and by the NSF grant 0450970.
Abbreviations used
- CscB
sucrose/H+ symporter from Escherichia coli
- LacY
lactose/H+ symporter from Escherichia coli
- TDG
β-d-galactopyranosyl-1-thio-β-d-galactopyranoside
- DDM
dodecyl-β-d-maltopyranoside
- HRP
horseradish peroxidase
- RSO
right side out membrane vesicles
- MIANS
2-(4′-maleimidylanilino)-naphthalene-6-sulfonic acid (sodium salt)
Footnotes
References
- 1.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analysis and information. Nucl Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 3.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–32. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaback HR, Sahin-Toth M, Weinglass AB. The kamikaze approach to membrane transport. Nature Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 6.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 8.Frillingos S, Sahin-Toth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 9.Kasho VN, Smirnova IN, Kaback HR. Sequence alignment and homology threading reveals prokaryotic and eukaryotic proteins similar to lactose permease. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.02.049. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bockmann J, Heuel H, Lengeler JW. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in E. coli EC3132. Mol Gen Genet. 1992;235:22–32. doi: 10.1007/BF00286177. [DOI] [PubMed] [Google Scholar]
- 11.Sahin-Tóth M, Frillingos S, Lengeler JW, Kaback HR. Active transport by the CscB permease in Escherichia coli K-12. Biochem Biophys Res Commun. 1995;208:1116–1123. doi: 10.1006/bbrc.1995.1449. [DOI] [PubMed] [Google Scholar]
- 12.Sahin-Tóth M, Kaback HR. Functional conservation in the putative substrate binding site of the sucrose permease from Escherichia coli. Biochemistry. 2000;39:6170–6175. doi: 10.1021/bi000125g. [DOI] [PubMed] [Google Scholar]
- 13.Venkatesan P, Kaback HR. The substrate-binding site in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1998;95:9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao M, Zen KC, Hubbell WL, Kaback HR. Proximity between Glu126 and Arg144 in the lactose permease of Escherichia coli. Biochemistry. 1999;38:7407–7412. doi: 10.1021/bi9906524. [DOI] [PubMed] [Google Scholar]
- 15.Wolin CD, Kaback HR. Thiol cross-linking of transmembrane domains IV and V in the lactose permease of Escherichia coli. Biochemistry. 2000;39:6130–6135. doi: 10.1021/bi0001269. [DOI] [PubMed] [Google Scholar]
- 16.King SC, Hansen CL, Wilson TH. The interaction between aspartic acid 237 and lysine 358 in the lactose carrier of Escherichia coli. Biochim Biophys Acta. 1991;1062:177–186. doi: 10.1016/0005-2736(91)90390-t. [DOI] [PubMed] [Google Scholar]
- 17.Dunten RL, Sahin-Tóth M, Kaback HR. Role of the charge pair formed by aspartic acid 237 and lysine 358 in the lactose permease of Escherichia coli. Biochemistry. 1993;32:3139–3145. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 18.Sahin-Tóth M, Kaback HR. Properties of interacting aspartic acid and lysine residues in the lactose permease of Escherichia coli. Biochemistry. 1993;32:10027–10035. doi: 10.1021/bi00089a019. [DOI] [PubMed] [Google Scholar]
- 19.He MM, Voss J, Hubbell WL, Kaback HR. Use of designed metal-binding sites to study helix proximity in the lactose permease of Escherichia coli: 1. Proximity of helix VII (Asp237 and Asp240) with helices X (Lys319) and XI (Lys358) Biochemistry. 1995;34:15661–15666. doi: 10.1021/bi00048a009. [DOI] [PubMed] [Google Scholar]
- 20.Frillingos S, Sahin-Tóth M, Lengeler JW, Kaback HR. Helix packing in the sucrose permease of Escherichia coli: properties of engineered charge pairs between helices VII and XI. Biochemistry. 1995;34:9368–9373. doi: 10.1021/bi00029a012. [DOI] [PubMed] [Google Scholar]
- 21.Sahin-Tóth M, Frillingos S, Lawrence MC, Kaback HR. The sucrose permease of Escherichia coli: functional significance of cysteine residues and properties of a cysteine-less transporter. Biochemistry. 2000;39:6164–6169. doi: 10.1021/bi000124o. [DOI] [PubMed] [Google Scholar]
- 22.Guan L, Hu Y, Kaback HR. Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry. 2003;42:1377–1382. doi: 10.1021/bi027152m. [DOI] [PubMed] [Google Scholar]
- 23.Ujwal ML, Sahin-Tóth M, Persson B, Kaback HR. Role of glutamate-269 in the lactose permease of Escherichia coli. Mol Membr Biol. 1994;11:9–16. doi: 10.3109/09687689409161024. [DOI] [PubMed] [Google Scholar]
- 24.Frillingos S, Ujwal ML, Sun J, Kaback HR. The role of helix VIII in the lactose permease of Escherichia coli. I. Cys-scanning mutagenesis. Protein Sci. 1997;6:431–437. doi: 10.1002/pro.5560060220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinglass AB, Sondej M, Kaback HR. Manipulating conformational equilibria in the lactose permease of Escherichia coli. J Mol Biol. 2002;315:561–571. doi: 10.1006/jmbi.2001.5289. [DOI] [PubMed] [Google Scholar]
- 26.Sahin-Tóth M, Karlin A, Kaback HR. Unraveling the mechanism of lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:10729–10732. doi: 10.1073/pnas.200351797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Kaback HR. Cysteine 148 in the lactose permease of Escherichia coli is a component of a substrate binding site. 2. Site-directed fluorescence studies. Biochemistry. 1994;33:12166–12171. doi: 10.1021/bi00206a020. [DOI] [PubMed] [Google Scholar]
- 28.le Coutre J, Whitelegge JP, Gross A, Turk E, Wright EM, Kaback HR, Faull KF. Proteomics on full-length membrane proteins using mass spectrometry. Biochemistry. 2000;39:4237–4242. doi: 10.1021/bi000150m. [DOI] [PubMed] [Google Scholar]
- 29.Smirnova IN, Kaback HR. A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry. 2003;42:3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 30.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Interhelical packing modulates conformational flexibility in the lactose permease of Escherichia coli. Biochemistry. 2005;44:7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 31.Sahin-Tóth M, Persson B, Schwieger J, Cohan M, Kaback HR. Cysteine scanning mutagenesis of the N-terminal 32 amino acid residues in the lactose permease of Escherichia coli. Protein Sci. 1994;3:240–247. doi: 10.1002/pro.5560030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahin-Tóth M, Dunten RL, Gonzalez A, Kaback HR. Functional interactions between putative intramembrane charged residues in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10547–10551. doi: 10.1073/pnas.89.21.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lolkema JS, Carrasco N, Kaback HR. Kinetic analysis of lactose exchange in proteoliposomes reconstituted with purified lac permease. Biochemistry. 1991;30:1284–1290. doi: 10.1021/bi00219a018. [DOI] [PubMed] [Google Scholar]
- 34.Carrasco N, Antes LM, Poonian MS, Kaback HR. Lac permease of Escherichia coli: histidine-322 and glutamic acid-325 may be components of a charge-relay system. Biochemistry. 1986;25:4486–4488. doi: 10.1021/bi00364a004. [DOI] [PubMed] [Google Scholar]
- 35.Sahin-Tóth M, Kaback HR. Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichia coli. Proc Natl Acad Sci USA. 2001;98:6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Püttner IB, Sarkar HK, Poonian MS, Kaback HR. Lac permease of Escherichia coli: His-205 and His-322 play different roles in lactose/H+ symport. Biochemistry. 1986;25:4483–4485. doi: 10.1021/bi00364a003. [DOI] [PubMed] [Google Scholar]
- 37.Püttner IB, Sarkar HB, Padan E, Lolkema JS, Kaback HR. Characterization of site-directed mutants in the lac permease of Escherichia coli. I. Replacement of histidine residues. Biochemistry. 1989;28:2525–2533. doi: 10.1021/bi00432a027. [DOI] [PubMed] [Google Scholar]
- 38.Vazquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahin-Tóth M, Akhoon KM, Runner J, Kaback HR. Ligand recognition by the lactose permease of Escherichia coli: specificity and affinity are defined by distinct structural elements of galactopyranosides. Biochemistry. 2000;39:5097–5103. doi: 10.1021/bi0000263. [DOI] [PubMed] [Google Scholar]
- 40.Sahin-Tóth M, Lawrence MC, Nishio T, Kaback HR. The C-4 hydroxyl group of galactopyranosides is the major determinant for ligand recognition by the lactose permease of Escherichia coli. Biochemistry. 2001;43:13015–13019. doi: 10.1021/bi011233l. [DOI] [PubMed] [Google Scholar]
- 41.Sahin-Tóth M, le Coutre J, Kharabi D, le Maire G, Lee JC, Kaback HR. Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry. 1999;38:813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 42.Consler TG, Tsolas O, Kaback HR. Role of proline residues in the structure and function of a membrane transport protein. Biochemistry. 1991;30:1291–1298. doi: 10.1021/bi00219a019. [DOI] [PubMed] [Google Scholar]
- 43.Kaback HR. Transport in isolated bacterial membrane vesicles. Methods Enzymol. 1974;31:698–709. doi: 10.1016/0076-6879(74)31075-0. [DOI] [PubMed] [Google Scholar]
- 44.Short SA, Kaback HR, Kohn LD. Localization of D-lactate dehydrogenase in native and reconstituted Escherichia coli membrane vesicles. J Biol Chem. 1975;250:4291–4296. [PubMed] [Google Scholar]
- 45.McRee DE. A visual protein crystallographic software system for X11/XView. J Mol Graph. 1992;10:44–46. [Google Scholar]