Abstract
MicroRNAs (miRNAs) interfere with translation of specific target mRNAs and are thought to thereby regulate many cellular processes. Recent studies have suggested that miRNAs might play a role in osteoblast differentiation and bone formation. Here, we identify a new miRNA (miR-2861) in primary mouse osteoblasts that promotes osteoblast differentiation by repressing histone deacetylase 5 (HDAC5) expression at the post-transcriptional level. miR-2861 was found to be transcribed in ST2 stromal cells during bone morphogenetic protein 2–induced (BMP2-induced) osteogenesis, and overexpression of miR-2861 enhanced BMP2-induced osteoblastogenesis, whereas inhibition of miR-2861 expression attenuated it. HDAC5, an enhancer of runt-related transcription factor 2 (Runx2) degradation, was confirmed to be a target of miR-2861. In vivo silencing of miR-2861 in mice reduced Runx2 protein expression, inhibited bone formation, and decreased bone mass. Importantly, miR-2861 was found to be conserved in humans, and a homozygous mutation in pre–miR-2861 that blocked expression of miR-2861 was shown to cause primary osteoporosis in 2 related adolescents. Consistent with the mouse data, HDAC5 levels were increased and Runx2 levels decreased in bone samples from the 2 affected individuals. Thus, our studies show that miR-2861 plays an important physiological role in osteoblast differentiation and contributes to osteoporosis via its effect on osteoblasts.
Introduction
MicroRNAs (miRNAs) are a growing class of small (~22 nucleotides), single-stranded noncoding RNAs found in diverse organisms (1). They negatively regulate translation of specific mRNAs by base pairing with partially or fully complementary sequences in target mRNAs (2–4). Many miRNAs are evolutionarily conserved and have well-defined developmental and cell type–specific expression patterns (5).
Although the biological functions of most miRNAs are not yet fully understood, they may play a key role in the regulation of various biological processes, including developmental timing, cellular differentiation, proliferation, apoptosis, gene regulation, insulin secretion, cholesterol biosynthesis, and cancer development (4–7). As the function of miRNAs in bone metabolism has not been thoroughly explored, we have focused on the influence of miRNAs in osteoblastic differentiation, which is important for bone remodeling. As described above, involvement in various cell events suggests that miRNAs might also have a regulatory function in osteoblast differentiation and bone formation. Recently, results from several studies have supported this hypothesis (8–10), but the role of miRNAs in osteoblasts still remains unclear.
Osteoblastic differentiation can be strongly induced by bone morphogenetic protein (BMP) from stromal cells (11). Here, we cloned and identified what we believe is a novel miRNA from osteoblasts and investigated its role in BMP2-induced osteoblast differentiation. We then silenced the expression of this miRNA in mice to explore the effects on bone formation. In addition, we found a mutation in this miRNA gene that may be involved in the onset of osteoporosis.
Results
Identification of an miRNA from primary mouse osteoblasts.
In this study, we cloned short RNA molecules from primary mouse osteoblasts using a small RNA-cloning method developed by Ro et al. (12). A total of 162 clones were subsequently characterized by DNA sequencing and database searching. Results indicated that several kinds of cellular RNA fragments were present. More than 50% of the cloned RNAs represented breakdown products of abundant noncoding RNAs such as transfer RNA, ribosomal RNA, small nuclear RNA (snRNA), and small nucleolar RNA. Some were known miRNAs (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI39832DS1). The remaining novel RNA sequences were compared using BLAST on the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and flanking sequences were used to predict secondary structure by means of mfold (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) (13).
One of the fragments was classified as a putative novel miRNA (Supplemental Table 1) because it was located in a noncoding region and the predicted precursor sequences showed the characteristic miRNA precursor stem-loop secondary structure according to mfold (Figure 1A). The new miRNA was termed “miR-2861” and is located on chromosome 2 and conserved in the human sequence.
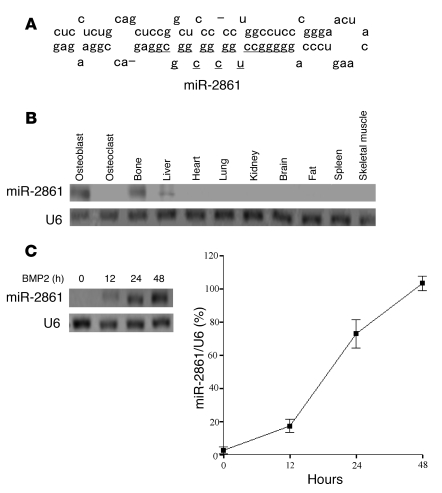
Figure 1. miR-2861 is primarily expressed in osteoblasts.
(A) Schematic diagram of the secondary structure of pre–miR-2861. The structures were predicted by mfold, and the mature miR-2861 sequence is underlined. (B) Northern blot analysis of miR-2861 expression in osteoblasts, osteoclasts, and various mouse tissues. (C) miR-2861 expression during BMP2-induced ST2 osteogenic differentiation. ST2 cells were treated with BMP2 (300 ng/ml) for the indicated durations, and miR-2861 expression was measured by Northern blotting. The blots were stripped and rehybridized with 32P-labeled oligonucleotide probes for U6. U6 snRNA was used as a loading control. Circles represent the percentage of U6 expression level at various time points.
miR-2861 is primarily expressed in osteoblasts.
The expression of miR-2861 was examined by Northern blotting using total RNA from primary mouse osteoblasts in addition to total RNA prepared from primary mouse osteoclasts, bone, liver, heart, lung, kidney, brain, fat, spleen, and skeletal muscle. This analysis confirmed that miR-2861 was primarily expressed in osteoblasts. In mouse tissue, miR-2861 was preferentially expressed in bone and detected at lower levels in liver, but was not found in other tissues (Figure 1B). In cultured bone cells, miR-2861 was detected in primary mouse osteoblasts, but not osteoclasts (Figure 1B).
To better understand the role of miR-2861 in osteogenic differentiation, we determined the miR-2861 expression profile of the stromal cell line ST2 after BMP2 treatment. Expression of miR-2861 was detected after treatment with BMP2 for 12 hours and increased progressively after 24–48 hours of treatment (Figure 1C).
Regulation of BMP2-induced ST2 osteogenic differentiation by miR-2861.
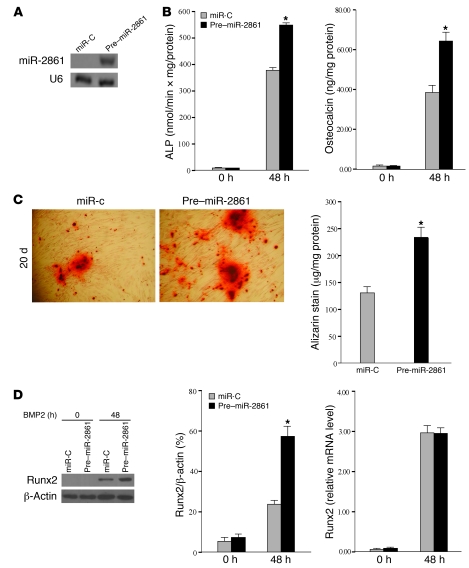
To investigate the role of miR-2861 during osteoblast differentiation, we stably transfected ST2 stromal cells with the miRNA precursor (pre–miR-2861) and assessed the level of mature miR-2861 by Northern blotting (Figure 2A). Osteoblastic differentiation was induced by the addition of 300 ng/ml BMP2. Alkaline phosphatase (ALP) activity, osteocalcin secretion, and expression of runt-related transcription factor 2 (Runx2) were measured after 48 hours. Compared with control cells, the levels of ALP activity and osteocalcin were increased by transfection of pre–miR-2861 (Figure 2B). As shown in Figure 2C, stable transfection of miR-2861 promoted mineralized nodule formation in 20-day cultures. Runx2 protein levels were also enhanced by transfection of miR-2861, whereas type II Runx2 (14) mRNA levels showed no considerable change (Figure 2D).
Figure 2. miR-2861 promotes BMP2-induced ST2 osteogenic differentiation.
(A) Northern blot analysis of miR-2861 levels in ST2 cells after transfection with pre–miR-2861 or miR-C. U6 was used as a loading control. (B) Overexpression of miR-2861 enhanced BMP2-induced ST2 osteogenic differentiation. ST2 cells were stably transfected with pre–miR-2861 or miR-C, then treated with BMP2 for 48 hours. ALP activity and osteocalcin secretion were determined. Data are shown as means ± SD. *P < 0.05 vs. miR-C, n = 5. (C) Microscopic view of the effects of miR-2861 on matrix mineralization in ST2 cells. Shown is a representative microscopic view at a magnification of ×200 of cells transfected with pre–miR-2861 or miR-C after 20 days of culture. Quantification of Alizarin Red S stain via extraction with cetyl-pyridinium chloride. Data are shown as means ± SD. *P < 0.05 vs. miR-C, n = 3. (D) Levels of Runx2 mRNA were determined using qRT-PCR and are shown as fold induction relative to control. Runx2 protein expression was determined by Western blot and expressed as densitometry of Runx2/β-actin. Data are shown as means ± SD. *P < 0.05 vs. miR-C, n = 3.
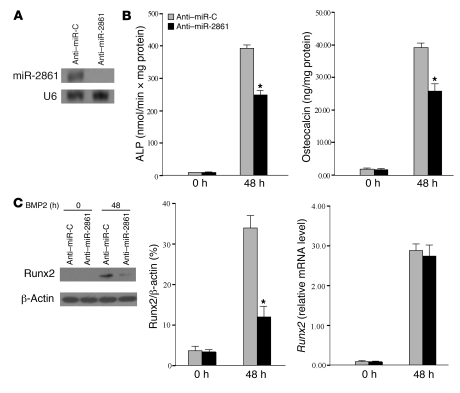
Next, we transfected BMP2-induced ST2 cells with 2′-O-methyl antisense inhibitory oligoribonucleotides (anti–miR-2861) (15). Anti–miR-2861 blocked miR-2861 expression in BMP2-induced ST2 cells (Figure 3A). Treatment with anti–miR-2861 attenuated osteogenic differentiation and reduced the increase in Runx2 protein induced by BMP2, but it did not influence the level of Runx2 mRNA (Figure 3, B and C). Decreased calcium accumulation was observed at day 5 of ST2 cell culture when miR-2861 expression was blocked by anti–miR-2861 (Supplemental Figure 1).
Figure 3. Inhibition of miR-2861 limits BMP2-induced osteogenic differentiation.
ST2 cells were treated with BMP2 and transiently transfected with anti–miR-2861 or anti–miR-C. (A) Northern blotting showed that anti–miR-2861 repressed the expression of miR-2861 in BMP2-induced ST2 cells. (B) ALP activity and osteocalcin secretion were measured at 48 hours as described in Figure 2B. (C) Runx2 mRNA and protein expression were detected as described in Figure 2D. *P < 0.05 vs. anti–miR-C.
All of these results suggest that miR-2861 acts to promote osteoblast differentiation.
miR-2861 directly targets histone deacetylase 5.
Recently Tay et al. (16) demonstrated that miRNAs exercise their effects on mRNAs through binding the coding regions of target genes. We used Rna22 (17) to predict the targets of miR-2861 because Rna22 does not need validated targets for training and neither requires nor relies on cross-species conservation. It is therefore an ideal tool for determining miRNA targets in amino acid coding sequences (CDSs) and 3′ untranslated regions (UTRs) of transcripts. Rna22 predicted 1 miR-2861 target in the CDS of histone deacetylase 5 (HDAC5) (Figure 4A).
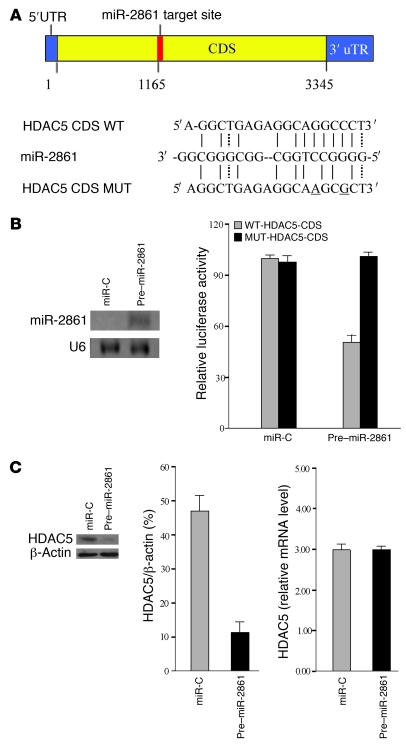
Figure 4. miR-2861 functional activity on target genes.
(A) Schematic of the miR-2861 putative target site in mouse HDAC5 CDS and alignment of miR-2861 with WT and MUT CDS regions of HDAC5 showing complementary pairing. The 2 mutated nucleotides are underlined. (B) miR-2861 targeted the HDAC5 CDS. ST2 cells were cotransfected with the luciferase reporter carrying WT-pGL3-HDAC5 or MUT-pGL3-HDAC5, phRL-null (Renilla plasmid), and miR-C or pre–miR-2861. Effects of miR-2861 and miR-C on the reporter constructs were determined 48 hours after transfection. Firefly luciferase values, normalized for Renilla luciferase, are presented. Data are shown as means ± SD. *P < 0.05 vs. MUT-pGL3-HDAC5, n = 3. (C) miR-2861 regulated HDAC5 expression at the post-transcriptional level. ST2 cells were transfected with pre–miR-2861 or miR-C. Northern blot analysis identified miR-2861 level in ST2 cells. Total cell lysates were collected and analyzed 48 hours after transfection. The level of HDAC5 mRNA was determined using qRT-PCR and normalized to β-actin. Data are shown as means ± SD. HDAC5 protein expression was determined by Western blot, using β-actin as a loading control. Results were expressed as densitometry of HDAC5/β-actin. Data are shown as means ± SD. *P < 0.05 vs. miR-C, n = 3.
To test whether miR-2861 can directly regulate HDAC5, a luciferase reporter construct containing the CDS of HDAC5 was generated and mutations were introduced into the predicted miRNA binding site. We introduced the HDAC5 luciferase expression vector with pre–miR-2861 (WT-pGL3-HDAC5) into ST2 cells and measured the level of luciferase enzyme activity to determine the effects of miR-2861 on luciferase translation. Overexpression of miR-2861 suppressed the luciferase activity of the HDAC5 CDS reporter gene (Figure 4B). Mutation of 2 nucleotides within the putative target site in the HDAC5 CDS (MUT-pGL3-HDAC5) abolished this repression, confirming the specificity of the action (Figure 4B).
To directly test the validity of the putative target, ST2 cells were transfected with pre–miR-2861. The mRNA and protein levels of HDAC5 were measured by quantitative real-time PCR (qRT-PCR) and Western blotting. Relative to the control, overexpression of miR-2861 downregulated endogenous HDAC5 protein (Figure 4C). By contrast, no change in HDAC5 mRNA levels was noted (Figure 4C). The presence of endogenous HDAC5 mRNA may affect the reaction between pre–miR-2861 and the HDAC5 luciferase expression vector, thus resulting in the discrepancy between the effects of miR-2861 on endogenous HDAC5 protein and luciferase activity of the HDAC5 CDS reporter gene.
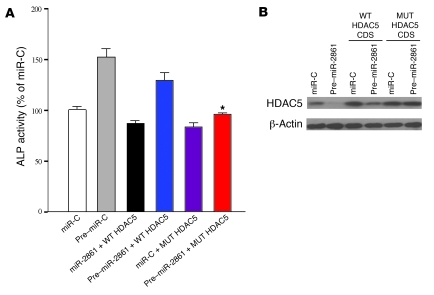
We next cotransfected the WT or mutant HDAC5 CDS construct with pre–miR-2861 into BMP2-induced ST2 cells to determine the effects on ALP activity and HDAC5 protein expression. Induction of ALP activity by pre–miR-2861 was rescued by the mutant HDAC5 CDS construct (Figure 5A). Western blotting showed that the mutant HDAC5 CDS construct was also able to rescue the pre–miR-2861–induced downregulation of HDAC5 protein (Figure 5B). These results suggested that HDAC5 is the most important target of miR-2861 in osteoblast differentiation.
Figure 5. The mutant HDAC5 CDS construct rescues miR-2861–modulated ST2 osteogenic differentiation.
BMP2-induced ST2 cells were cotransfected with the WT or mutant HDAC5 CDS construct and pre–miR-2861 or miR-C. (A) Transfection of mutant HDAC5 into BMP2-induced ST2 cells rescued the pre–miR-2861–induced increase of ALP activity. Data are shown as means ± SD. *P < 0.05 vs. pre–miR-2861 + WT HDAC5, n = 5. (B) Transfection of the mutant HDAC5 into BMP2-induced ST2 cells rescued the pre–miR-2861–induced downregulation of HDAC5 protein.
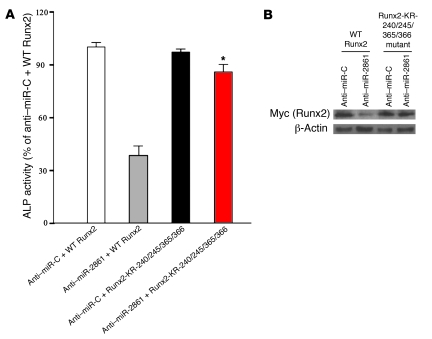
Jeon et al. (18) reported that HDAC5 deacetylates target residues lysine (K) 225, 230, 350, and 351 in type I Runx2. These positions correspond to K 240, 245, 365, and 366 in type II Runx2. We therefore constructed a type II Runx2 mutant with lysine-to-arginine (KR) substitutions (Runx2-KR-240/245/365/366) and cotransfected BMP2-induced ST2 cells with anti–miR-2861 and the Myc-tagged Runx2-KR-240/245/365/366 mutant. As shown in Figure 6, transfection of the Runx2-KR-240/245/365/366 mutant rescued the anti–miR-2861 inhibition of ALP activity. This showed that miR-2861 regulates Runx2 protein levels by targeting HDAC5 in osteoblast differentiation.
Figure 6. Runx2 mutant rescues anti–miR-2861–inhibited ALP activity.
BMP2-induced ST2 cells were cotransfected with anti–miR-C or anti–miR-2861 and Myc-tagged WT type II Runx2 or a Runx2 mutant containing KR substitutions in the HDAC5 deacetylation targets (Runx2-KR-240/245/365/366 mutant). (A) Transfection of the Runx2-KR-240/245/365/366 mutant into BMP2-induced ST2 cells rescued the anti–miR-2861 inhibition of ALP activity. Data are shown as means ± SD. *P < 0.05 vs. anti–miR-2861 + WT Runx2, n = 5. (B) Transfection of the Runx2 mutant into BMP2-induced ST2 cells rescued the anti–miR-2861–inhibited Runx2 protein levels. The abundance of Runx2 or Runx2 mutant was determined by Western blot using an anti-Myc antibody.
miR-2861 regulates BMP2-induced mouse bone marrow stromal cell osteogenic differentiation.
We also studied the function of miR-2861 in nontransformed mouse bone marrow stromal cells (BMSCs). Northern blotting showed that BMP2 induced the expression of miR-2861 in BMSCs (Supplemental Figure 2A). ALP activity and osteocalcin secretion were increased by transfection of pre–miR-2861 (Supplemental Figure 2B). The level of Runx2 protein was enhanced, whereas HDAC5 protein level was reduced after transfection of pre–miR-2861. The mRNA level of type II Runx2 and HDAC5 did not change (Supplemental Figure 2C). In contrast, treatment with anti–miR-2861 in BMSCs inhibited BMP2-induced osteogenic differentiation (Supplemental Figure 2D).
miR-2861 regulates bone formation in vivo.
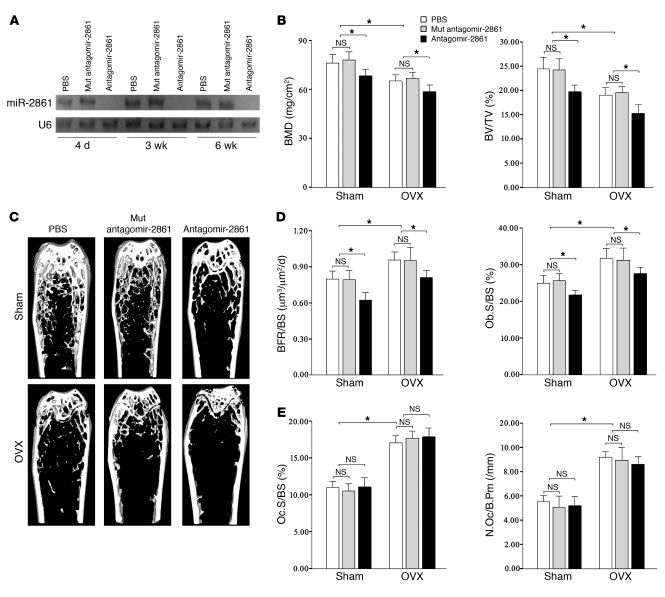
To investigate the function of miR-2861 in vivo, we performed an experiment in which a chemically modified antisense oligonucleotide (19) specific to miR-2861 (antagomir-2861) (80 mg/kg) was injected via a single tail vein injection into mice that had undergone sham operation or ovariectomy (OVX). Mutant antagomir-2861 (80 mg/kg) and PBS (0.2 ml) were used as controls. When mice were treated with antagomir-2861 for 3 consecutive days, Northern blotting showed that miR-2861 expression in bone was repressed for up to 3 weeks (Figure 7A). In contrast, mutant antagomir-2861 or PBS had no effect on miR-2861 expression (Figure 7A). These results indicated that antagomir-2861 efficiently downregulated miR-2861 expression in bone. To maintain the effects of miR-2861 on mice, the mice received another injection of antagomir-2861 on days 1–3 on the fourth week after the first injection. All mice were euthanized for analysis by the end of the sixth week after the first injection.
Figure 7. Antagomir-2861–treated mice have decreased bone mass and reduced osteoblast activity.
(A) No expression of miR-2861 was detected in bones of antagomir-2861–treated mice. Mice were injected intravenously with PBS, mutant (Mut) antagomir-2861, or antagomir-2861 (3 injections of 80 mg/kg/d in the first week, and another injection on days 1–3 of the fourth week) and bones were harvested at 4 days, 3 weeks, and 6 weeks after the first injection. Expression of miR-2861 was analyzed by Northern blot. (B) Silencing of miR-2861 resulted in decreased BMD. Structural parameters of femurs were measured by micro-CT. (C) micro-CT section of distal and midfemoral diaphyses. (D) Bone formation parameters of femurs measured by histological analysis. (E) Bone resorption parameters of mouse femur. Data are means ± SD of 8 mice per group. *P < 0.05.
The results showed that in SHAM groups, antagomir-2861–treated mice exhibited a significant reduction in femur bone mineral density (BMD) by Piximus in comparison with mice treated with mutant antagomir-2861 or PBS (Figure 7B). OVX mice treated with mutant antagomir-2861 or PBS treatment also presented decreased femur BMD (Figure 7B). Treatment with antagomir-2861 in OVX mice had minimum BMD (Figure 7B).
Quantification of the bone volume/tissue volume ratio (BV/TV), trabecular number, trabecular spacing, and trabecular thickness was conducted using micro-CT. The micro-CT images of distal and midfemoral diaphyses are shown in Figure 7C. The assay revealed that absence of miR-2861 significantly decreased bone parameters, including BV/TV and trabecular number, with a concomitant increase in trabecular spacing (Figure 7B and Supplemental Figure 3A). OVX mice treated with mutant antagomir-2861 or PBS also exhibited a significant decrease in BV/TV and trabecular number and increase in trabecular spacing (Figure 7B and Supplemental Figure 3A). After treatment with antagomir-2861 in OVX mice, these indices altered more obviously (Figure 7B and Supplemental Figure 3A). Trabecular thickness was only reduced in OVX mice with antagomir-2861 treatment (Supplemental Figure 3A). These results suggested that miR-2861 is involved in the regulation of bone metabolism.
We next measured several parameters of bone formation and resorption by histomorphometric analysis. Results showed that in SHAM groups, antagomir-2861–treated mice exhibited decreased bone formation rate per bone surface (BFR/BS), mineral appositional rate, osteoblast surface per bone surface (Ob.S/BS), and number of osteoblasts per bone perimeter (N.Ob/B.Pm) (Figure 7D and Supplemental Figure 3B). A significant increase in these bone formation parameters were detected in OVX mice treated with mutant antagomir-2861 or PBS (Figure 7D and Supplemental Figure 3B). However, antagomir-2861 decreased the levels of these bone formation parameters in OVX mice (Figure 7D and Supplemental Figure 3B). We also assessed bone resorption parameters, i.e., osteoclast surface per bone surface (Oc.S/BS) and the number of osteoclasts per bone perimeter (N.Oc/B.Pm), by histomorphometric analysis. OVX significantly augmented the levels of Oc.S/BS and N.Oc/B.Pm (Figure 7E). However, antagomir-2861 had no effect on Oc.S/BS or N.Oc/B.Pm (Figure 7E).
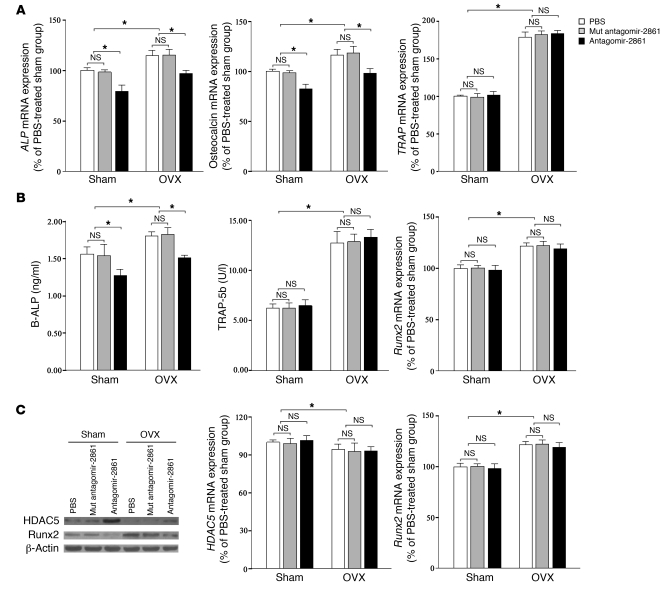
Our data showed that antagomir-2861 treatment reduced the levels of osteoblast activity markers such as ALP and osteocalcin in bone tissue and did not affect the level of an osteoclast activity marker (tartrate-resistant acid phosphatase [TRAP]) (Figure 8, A and B). Similarly, levels of serum bone-specific ALP were decreased in antagomir-2861–treated mice, whereas no change was observed in serum TRAP5b levels (Figure 8, A and B).
Figure 8. Osteoblast markers but not osteoclast markers are affected by the absence of miR-2861 in mice.
(A) qRT-PCR analysis of osteoblast and osteoclast markers in bone extracts from mice. (B) Serum concentrations of bone-specific ALP and TRAP5b. (C) Bone mRNA and protein expression levels of HDAC5 and Runx2. Level of HDAC5 and Runx2 mRNA were determined by qRT-PCR, and the results are expressed as a percentage of the PBS-treated SHAM group. HDAC5 and Runx2 protein expression was determined using Western blotting and normalized to total protein. β-Actin was used as a loading control. Data are means ± SD of 8 mice per group. *P < 0.05.
By Western blotting, we noticed that HDAC5 and Runx2 protein levels were elevated and reduced, respectively, in bone extracts of antagomir-2861–treated mice (Figure 8C). Treatment with antagomir-2861 had no effect on HDAC5 or Runx2 mRNA levels in bone tissue (Figure 8C). These results collectively suggest that miR-2861 deletion could decrease bone mass by inhibiting bone formation.
We also performed biomechanical testing on mouse femurs. Antagomir-2861 treatment or OVX decreased biomechanical index value, such as ultimate load, ultimate stress, elastic modulus, and stiff index (Supplemental Figure 3C). These values were further reduced in antagomir-2861–treated OVX mice (Supplemental Figure 3C).
Clinical characteristics and miR-2861 expression in adolescents with primary osteoporosis.
Ten patients (6 males and 4 females) fulfilling the criteria for primary osteoporosis were included in the study (Table 1). All had normal 25-hydroxyl vitamin D serum concentrations.
Table 1 .
Clinical features and bone turnover biochemical markers in primary osteoporosis patients
Osteoporosis patient 7 (OP7) was a 15-year-old boy with a history of peripheral fractures. X-rays showed metaphyseal fractures and compression fractures, all of which resulted from low-impact trauma. By inquiring into family history, we learned that his 17-year-old sister also had a history of repeated fracture, including metaphyseal fractures and compression fractures. Her BMD Z score was –4.5, and she fulfilled the criteria for primary osteoporosis in adolescents. About 5 months after the last fracture of the proband, his sister fractured her left femur again in a fall.
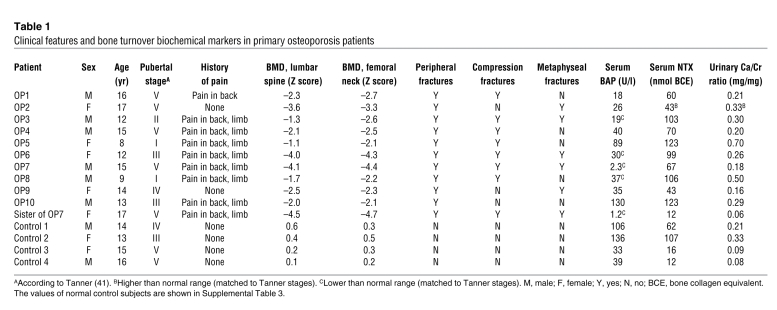
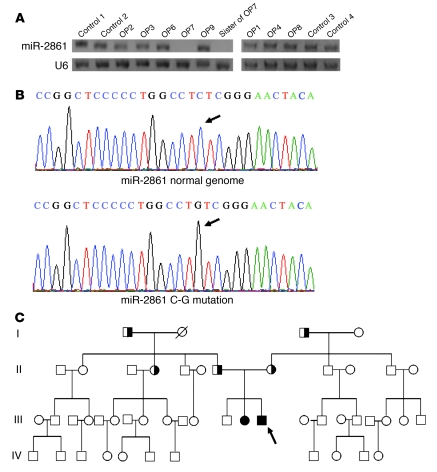
Northern blotting showed that bone miR-2861 expression was undetectable in OP7 and his sister. A representative picture is shown in Figure 9A. OP5 and OP10 did not provide bone samples.
Figure 9. Identification of a mutation in pre–miR-2861.
(A) miR-2861 expression in adolescent primary osteoporosis patients and controls. miR-2861 expression was analyzed by Northern blotting with total RNA isolated from bones of patients and controls. U6 was used as a loading control. Representative results are presented. (B) Chromatograms for the normal genome and the mutated miR-2861 samples. A fragment of an approximately 600-bp genomic sequence flanking the pre–miR-2861 at both the 5′ and 3′ ends was amplified, purified, and sequenced. Arrows indicate the location of the mutation. (C) Schematic representation of the miR-2861 genotypes of the studied kindred, consisting of 49 members from 4 generations. The proband is indicated by an arrow. Subjects with the homozygous C-G mutation are indicated by closed symbols; heterozygous carriers are indicated by half-shaded symbols.
We thus focused our attention on the kindred of OP7 and measured their BMD, except for the child less than 5 years old. The proband’s brother had a negative fracture history, and his BMD Z score was 0.1. In this kindred, the proband’s parents as well as I1, I3, and II4 (grandparents of the proband) had osteoporosis and presented BMD T scores less than –2.5 (Supplemental Table 2). The BMD T or Z scores for other family members ranged from –0.3 to 1.6. These members had no history of fracture.
Identification of a mutation in pre–miR-2861.
Since there was no expression of miR-2861 in the proband or his sister, both of whom had primary osteoporosis, we hypothesized that miR-2861 may be involved in the pathogenesis of primary osteoporosis. Recently, several studies have identified that mutations and SNPs in pre-miRNAs can affect miRNA expression and disease development by influencing miRNA processing (20–23). We therefore proposed that mutations or SNPs in pre–miR-2861 can decrease miR-2861 expression and induce osteoporosis. In OP7 and his sister, we found what we believe is a novel homozygous mutation residing at position +33 relative to the first nucleotide of pre–miR-2861 (Figure 9B). In addition, we discovered a heterozygous mutation in the same position in the proband’s parents and I1, I3, and II4 in the kindred (Figure 9C). These family members with the heterozygous mutation all suffered from osteoporosis (Supplemental Table 2). This result suggested that miR-2861 plays an important role in the development of osteoporosis.
We then extended this analysis to a larger cohort including 357 normal children (Supplemental Table 3), 396 healthy adults, and 369 adult patients with osteoporosis, but did not find the same mutation in pre–miR-2861. These results indicated that this mutation is a rare variant rather than a SNP in the general population.
The miR-2861 mutation represses miR-2861 expression and attenuates miRNA-mediated translational suppression.
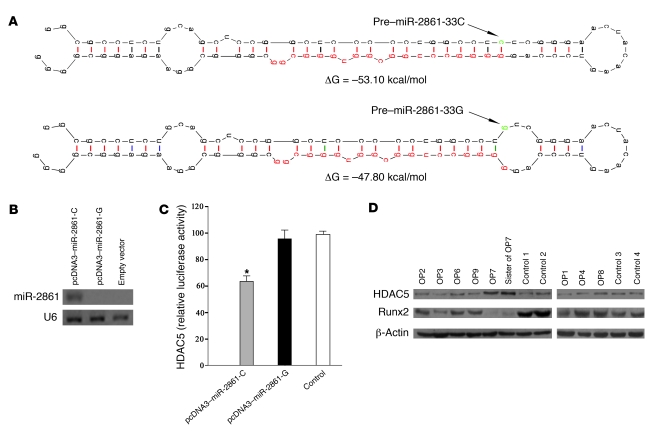
The mutation in pre–miR-2861 led to a base-pairing mismatch, decreased the free energy value from –53.1 kcal/mol to –47.8 kcal/mol, and created an enlarged RNA bulge in the predicted stem-loop structure (Figure 10A).
Figure 10. The miR-2861 mutation represses miRNA expression and attenuates miRNA-mediated translational suppression.
(A) The stem-loop structures of pre–miR-C and pre–miR-G, as predicted by mfold. Mature miRNAs are highlighted in red, and the mutated nucleotide is highlighted in green (and indicated by arrows). The free energy (ΔG) calculated by mfold is indicated. (B) The C-G mutation in the stem of pre–miR-2861 inhibited mature miR-2861 expression. Human 293 cells were transfected with pcDNA3-miR-2861-C, pcDNA3-miR-2861-G, or empty vector. The cells were harvested 48 hours after transfection, and miR-2861 expression was determined by Northern blotting. U6 was used as a loading control. (C) The miR-2861 mutation attenuated miRNA-mediated translational suppression. 293 cells were cotransfected with the luciferase reporter carrying HDAC5 CDS and pcDNA3-miR-2861-C, pcDNA3-miR-2861-G, or miR-C. Effects of the WT, MUT pre–miR-2861, or control on the reporter constructs were determined 48 hours after transfection. Firefly luciferase values, normalized for Renilla luciferase, are presented. Data are shown as means ± SD. *P < 0.05 vs. control, n = 3. (D) HDAC5 and Runx2 protein levels in human bone. HDAC5 and Runx2 protein expression levels were determined by Western blotting with proteins extracted from bone of patients and controls and normalized to total protein. β-Actin was used as a loading control. Representative results are presented.
To study a possible molecular effect of the C-G substitution, we prepared vectors containing either the WT pre–miR-2861 (pcDNA3-miR-2861-C) or the mutated pre–miR-2861 (pcDNA3-miR-2861-G). The 2 vectors were transfected into human 293 cells that lack endogenous miR-2861 expression. Northern blotting showed that miR-2861 was expressed in the WT transfectant. However, miR-2861 expression was not detected in the mutant transfectant (Figure 10B). These results indicated that the C-G substitution blocked the expression of mature miR-2861.
To explore the functional effects of the miR-2861 mutation on its target gene, we cotransfected the HDAC5 CDS luciferase expression vector with pcDNA3-miR-2861-C or pcDNA3-miR-2861-G into 293 cells. After 48 hours, we performed dual-luciferase assays. As expected, the luciferase activity of the HDAC5 expression vector was repressed by pcDNA3-miR-2861-C, but not by pcDNA3-miR-2861-G or the control (Figure 10C). This suggests that the pre–miR-2861 mutation attenuates miRNA-mediated translational suppression.
We further tested HDAC5 and Runx2 protein expression levels in bone samples of the proband, his sister, other primary osteoporosis patients, and controls. As expected, HDAC5 protein levels were elevated and Runx2 protein expression was decreased in the proband and his sister in comparison with controls (Figure 10D).
Serum bone alkaline phosphatase (BAP) levels (marker of osteoblast activity) were lower than the normal range, but serum cross-linked N-telopeptides of type I collagen (NTX) levels (a marker of osteoclast activity) and urinary Ca/Cr ratios were normal in the proband and his sister (Table 1). In the family members with heterozygous pre–miR-2861 mutations, serum BAP levels, but not NTX levels or urinary Ca/Cr ratios, were less than those of normal controls (Supplemental Table 4). This suggested that the pre–miR-2861 mutation can result in decreased osteoblastic activity, which causes osteoporosis.
Discussion
In the present study, we cloned and identified what is to our knowledge a novel miRNA, miR-2861, which promotes osteoblast differentiation by repressing HDAC5 expression at the post-transcriptional level.
To date, there are 988 human miRNAs and 754 mouse miRNAs in the miRBase miRNA registry (http://www.mirbase.org/) (24–26). However, thousands of miRNA genes in various genomes still need to be identified (27). Here, we identify what we believe is a novel miRNA from mouse osteoblasts that is conserved in the human sequence. Many miRNAs are reported to have unique tissue- or development-specific expression patterns, so we investigated the expression profile of miR-2861 in various tissues. miR-2861 was preferentially expressed in osteoblasts and bone, more weakly expressed in liver, but not detected in other tissues, osteoclasts, or ST2 cells. This pattern suggests that miR-2861 may play a role in osteoblast differentiation.
To determine whether miR-2861 is directly coupled to osteoblast differentiation, we investigated the action of miR-2861 in the process of BMP2-induced ST2 osteoblastogenesis. miR-2861 overexpression promoted BMP2-induced osteoblastogenesis, i.e., increased levels of ALP, osteocalcin, and mineralized nodule formation (28, 29). In contrast, inhibition of miR-2861 expression attenuated BMP2-induced osteoblastogenesis. miR-2861 overexpression also increased protein, but not mRNA, levels of the osteoblast differentiation transcription factor Runx2. The same effects were observed in mouse BMSCs by overexpressing or inhibiting miR-2861.
Generally, miRNAs repress gene expression through sequence-specific base pairing with a binding site in the 3′ UTRs of target transcripts (3, 7). However, recent studies have shown that miRNAs can exert their repression of mRNAs by combining with binding sites residing beyond the 3′ UTRs of target genes (16, 30, 31). Tay et al. (16) demonstrated that miR-134, miR-296, and miR-470 target the CDS region of transcription factors to regulate embryonic stem cell differentiation. Using Rna22, we predicted a potential binding site of miR-2861 in the CDS of HDAC5. Several lines of evidence indicated that HDAC5 is the most important target of miR-2861. First, miR-2861 bound to the CDS of HDAC5 mRNA with complementarity to the miR-2861 seed region, suggesting that it directly regulates HDAC5 expression. Second, miR-2861 overexpression inhibited the activity of WT-pGL3-HDAC5 but not MUT-pGL3-HDAC5. Third, HDAC5 protein levels decreased following miR-2861 overexpression, whereas mRNA levels did not change, suggesting a post-transcriptional control. Finally, transfection of mutant HDAC5 rescued the increased ALP activity and reduced HDAC5 protein expression induced by pre–miR-2861.
Studies have revealed that HDAC5 is involved in Runx2 degradation. BMP2 signaling stimulates Runx2 acetylation and inhibits Smurf1-mediated degradation of Runx2 (18, 32–35). In contrast, HDAC5 deacetylates Runx2, allowing the protein to undergo Smurf-mediated degradation (18, 32). Inhibition of HDAC5 increases Runx2 acetylation and potentiates BMP2-stimulated osteoblast differentiation. This is likely why Runx2 mRNA levels did not change after overexpression of miR-2861. Our study showed that inhibition of miR-2861 had no effect on cells expressing a form of Runx2 containing mutated HDAC5 deacetylation target sites. Therefore, we presumed that miR-2861 induces osteoblast differentiation by repressing HDAC5, an enhancer of Runx2 degradation.
Since miR-2861 induces osteoblast differentiation in vitro, it may play an important role in bone formation in vivo. Antagomirs, a novel class of chemically engineered oligonucleotides, can silence endogenous miRNAs with high efficiency and specificity in mice (19). Hence, we generated mice lacking miR-2861 through intravenous administration of antagomir-2861. Our results showed that silencing of miR-2861 led to a decrease in bone mass in SHAM mice and enhanced bone mass loss in OVX mice. Bone formation indices and markers of osteoblast activity in bone were reduced by antagomir-2861 treatment. However, markers of osteoclast activity displayed no obvious changes. Concomitant with the decreased bone formation in antagomir-2861–treated mice, bone HDAC5 protein expression was increased and Runx2 protein expression was reduced. These data suggest that miR-2861 influences bone mass by regulating bone formation, predominantly through its effect on HDAC5 and Runx2.
We examined miR-2861 expression in 2 adolescents (from the same family) with primary osteoporosis and found that miR-2861 expression was undetectable in bone. These patients also displayed elevated HDAC5 protein levels and reduced Runx2 expression in bone. These results suggested that decreased miR-2861 production contributed to osteoporosis in these patients and prompted us to identify the causes of the reduction.
Recently, several studies reported that sequence variations within miRNA genes, including primary miRNAs (pri-miRNAs), precursor miRNAs (pre-miRNAs), and mature miRNAs, could modify the biological activities of miRNAs (20–23, 36). Calin et al. identified several sequence mutations in pre-miRNA and pri-miRNA regions in patients with chronic lymphocytic leukemia; these mutations might influence miRNA production and lead to cell transformation (21). In this study, we found a homozygous C-G mutation in the stem of pre–miR-2861 in the 2 primary osteoporosis patients who lacked miR-2861 expression in bone. We confirmed that the C-G mutation in the stem of pre–miR-2861 blocked the expression of mature miR-2861 in vitro. Heterozygous mutations in the same position were detected in the proband’s parents and 3 other family members. These family members all suffered from osteoporosis. In the subjects with homozygous or heterozygous pre–miR-2861 mutations, values of a serum marker of osteoblast activity were lower than in normal controls, but the marker of osteoclast activity was unchanged. This finding indicated that the mutation in pre–miR-2861 can result in decreased osteoblast activity, likely resulting in osteoporosis. However, this mutation was not found in healthy people and other osteoporosis patients, suggesting that the mutation contributes to primary osteoporosis but is a rare variant. Therefore, our results imply that the miR-2861 mutation plays a crucial role in the pathogenesis of primary osteoporosis by influencing the expression of miR-2861.
In conclusion, the present study provides evidence that an miRNA primarily expressed in osteoblasts functions in osteoblast differentiation by repressing its target gene HDAC5 and promotes bone formation in vivo. Furthermore, our study shows that a mutation in pre–miR-2861 may result in primary osteoporosis in adolescents by influencing the expression of miR-2861. Together, our studies show that miR-2861 serves an important physiological function in osteoblast differentiation and contributes to osteoporosis.
Methods
Cell culture.
See Supplemental Methods.
Small RNA isolation and cloning.
Small RNAs were isolated from mouse osteoblasts using a mirVana miRNA isolation kit (Ambion) according to the manufacturer’s instructions. Small RNA isolation and cloning were performed as previously described (12). Briefly, small RNAs were polyadenylated and a 5′ adapter was ligated to poly(A)-tailed RNA using T4 RNA ligase (Invitrogen). The ligation products were reverse transcribed to make small RNA cDNAs, which were then amplified using PCR. The PCR products were directly subcloned into pcDNA3.1 TOPO vector (Invitrogen) for sequencing analysis. Oligo sequences are listed in Supplemental Table 5.
Bioinformatic analysis.
See Supplemental Methods.
Northern blotting.
See Supplemental Methods.
Plasmid constructs.
See Supplemental Methods and Supplemental Tables 7–9.
Transfection assay.
The miRNA inhibitors 2′-O-methyl antisense oligonucleotide targeted toward miR-2861 (anti–miR-2861) and miRNA inhibitor negative control (anti–miR-C) were purchased from GenePharma Co.
For stable transfection of pre–miR-2861, ST2 cells were transfected with the plasmid construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Stably transfected cells were selected using puromycin (1 μg/ml).
For transient transfection, a complex of Lipofectamine 2000 (Invitrogen) and plasmids or RNA oligonucleotides was prepared according to the manufacturer’s instructions and directly mixed with cells in 24-well cell culture plates at a density of 3 × 104 cells per well.
ALP activity and osteocalcin secretion assay.
See Supplemental Methods.
Measurement of mineralized matrix formation.
See Supplemental Methods.
qRT-PCR analysis.
See Supplemental Methods and Supplemental Table 6.
Western blotting.
See Supplemental Methods.
Luciferase reporter assay.
ST2 cells were transfected using Lipofectamine 2000 (Invitrogen) with either WT or mutant pGL3-HDAC5 constructs (200 ng) and pre–miR-2861 or pre–miR-C for 48 hours. As a positive control, the modified pGL3 control vector was used without a CDS insert. Cells treated solely with Lipofectamine served as negative controls. 293 cells were cotransfected with human WT HDAC5 CDS luciferase expression vector and pcDNA3-miR-2861-C or pcDNA3-miR-2861-G.
The Dual Luciferase Reporter Assay System (Promega) was used to quantify luminescent signal using a luminometer (Glomax; Promega). Each value from the firefly luciferase assay was normalized to the Renilla luciferase value from the cotransfected phRL-null vector (Promega).
Mice.
All procedures involving mice were approved by the Central South University Animal Management Committee. Six-week-old female WT C57BL/6 mice were used in all experiments and were housed under specific pathogen–free conditions (22°C, 12-hour light/12-hour dark cycles, and 50%–55% humidity) with free access to food pellets and tap water.
Six-week-old female mice were randomly divided into 6 groups and underwent either sham operation or bilateral OVX under general anesthesia by the dorsal approach. These mice received, on days 1 to 3 for 3 consecutive weeks, either antagomir-2861 or mutant antagomir-2861 at a dose of 80 mg/kg body weight or a comparable volume of PBS (0.2 ml) through tail vein injection. Measurement of miR-2861 levels in bone was performed 4 days, 3 weeks, and 6 weeks after the last injection. Six weeks after the first injection, mice were euthanized and blood was taken by dorsal aortic puncture. Serum was collected and stored at –70°C until analysis. Bone samples were removed for further analysis. Antagomir-2861 (CCGCCCGCCGCCAGGCCCC) and mutant antagomir-2861 (CCGCCCGCCGCCTCGGGGC) were synthesized by RiboBio Co.
BMD measurement.
The left femur of each mouse was fixed onto the scanning table along the longitudinal axis, and the whole femur was scanned by dual-energy X-ray absorptiometry (DXA) using a PIXImus densitometer (GE Lunar) to determine the BMD (37).
Human BMD was measured using a DXA fan-beam bone densitometer (Hologic QDR 4500A; Hologic) at lumbar spine (L1–L4) and the left femoral neck as previously described (38, 39). All BMD results are expressed in g/cm2. The control spine phantom scan performed each day had a long-term (more than 10 years) coefficient of variation of less than 0.43%.
Histomorphometric analysis.
See Supplemental Methods.
Micro-CT analysis.
See Supplemental Methods.
Study population.
The clinical study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University, and written informed consent was obtained from all participants. The diagnosis of primary osteoporosis in adolescents was based on the criteria described by Hartikka et al. (40): (a) low BMD, defined as a femoral neck and/or lumbar spine Z score less than –2.0; (b) history of increased bone fragility, defined as fractures caused by low-impact trauma; and/or (c) compression fractures. Patients with a known diagnosis or clinical features suggestive of osteogenesis imperfecta (blue sclerae, dentinogenesis imperfecta, joint hypermobility, hearing impairment, abnormal collagen biosynthesis established by skin biopsy) or of osteoporosis-pseudoglioma syndrome (impaired vision, ligamental laxity, mental retardation, or seizure disorder), as well as patients with an underlying chronic illness or systemic medication, were excluded from the study. Patients were clinically assessed for phenotypic features, height, weight, and pubertal status. Pubertal development was assessed according to Tanner stage (41). History of fractures was collected from hospital records and by interview at the clinical visit.
The normal control group for genomic DNA mutation analysis consisted of 357 normal children (176 female, age range 5–18; 181 male, age range 5–18) (Supplemental Table 3) and 396 healthy adults (200 female, age range 19–76; 196 male, age range 19–79) from the same geographic area as the patients.
In addition, we enrolled 369 adult osteoporosis patients (196 female, age range 40–75; 173 male, age range 45–80) for genomic DNA mutation analysis in our study. In adults, primary osteoporosis was diagnosed if the femoral neck and/or lumbar spine T score was less than –2.5 SD (38, 39).
Some data used in our study were derived from the osteoporosis epidemiology study conducted in our institute, which has been ongoing since 1996 (38, 39).
Tissue samples.
Bone samples were obtained from the osteoporosis patients who had undergone fracture surgery from the Second Xiangya Hospital. Bone samples obtained during surgery from victims of road traffic accidents (20 children, aged 9–18) were used as controls (control for bone sample analysis). The controls were clinically assessed for pubertal status and BMD value. Every sample was divided into 2 parts; one part was used for protein extraction, the other was snap-frozen in liquid nitrogen and stored at –80°C for RNA isolation. All subjects provided informed consent before surgical operation. OP5 and OP10 refused to offer bone samples, so we were not able to measure miR-2861 expression in their bone.
Biochemistry.
See Supplemental Methods.
Genetic studies.
Genomic DNA was isolated from peripheral blood using the QIAamp DNA extraction kit (Qiagen) according to the manufacturer’s protocol. An approximately 600-bp fragment of genomic sequence flanking pre–miR-2861 at both the 5′ and 3′ end was amplified. The product was purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced with an ABI 3730 automated DNA sequencer (PerkinElmer Applied Biosystems). Conformation-sensitive gel electrophoresis analysis of the PCR products was also performed as previously described (42). The PCR primers used for sequencing were: forward 5′-GAGGAGCAGGGCACTGCG-3′ and reverse 5′-CGAGCTTCTCGTATTTGGAAACTT-3′.
Statistics.
Data are presented as mean ± SD. Comparisons were made using a 1-way ANOVA. All experiments were repeated at least 3 times, and representative experiments are shown. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants 30870925, 30672197, and 30700891 from the China National Natural Scientific Foundation; grant 101035 from the Fok Ying Tung Education Foundation; and grant 20060533031 from the Specialized Research Fund for the Doctoral Program of Higher Education.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3666–3677 (2009). doi:10.1172/JCI39832.
References
- 1.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schramke V., Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301:1069–1074. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- 3.Noma K., et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 4.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Hagen J.W., Lai E.C. microRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–2332. doi: 10.4161/cc.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Li Z., et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oskowitz A.Z., et al. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuno Y., et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 2008;368:267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi A., Komori T., Suda T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr. Rev. . 2000;21:393–411. doi: 10.1210/er.21.4.393. [DOI] [PubMed] [Google Scholar]
- 12.Ro S., et al. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA. 2007;13:1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. . 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee C., et al. Differential regulation of the two principal Runx2/Cbfa1 n-terminal isoforms in response to bone morphogenetic protein-2 during development of the osteoblast phenotype. Endocrinology. 2001;142:4026–4039. doi: 10.1210/en.142.9.4026. [DOI] [PubMed] [Google Scholar]
- 15.Meister G., Landthaler M., Dorsett Y., Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 17.Miranda K.C., et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Jeon E.J., et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 19.Krützfeldt J., et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y., Cullen B.R. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin G.A., et al. A Micro-RNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. . 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 22.Duan R., Pak C., Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 23.Jazdzewski K., et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. U. S. A. . 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambros V., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S., et al. Rfam: annotating non-coding RNAs in complete genomes, Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S., et al. In vitro and in vivo synergistic interactions between the Runx2/Cbfa1 transcription factor and bone morphogenetic protein-2 in stimulating osteoblast differentiation. J. Bone Miner. Res. 2003;18:705–715. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi A., et al. Effects of BMP-2, BMP-4, and BMP-6 on osteoblastic differentiation of bone marrow-derived stromal cell lines, ST2 and MC3T3-G2/PA6. Biochem. Biophys. Res. Commun. 1996;220:366–371. doi: 10.1006/bbrc.1996.0411. [DOI] [PubMed] [Google Scholar]
- 30.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee I., et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang J.S., Alliston T., Delston R., Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao M., et al. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J. Biol. Chem. 2004;279:12854–12859. doi: 10.1074/jbc.M313294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao M., Qiao M., Oyajobi B.O., Mundy G.R., Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J. Biol. Chem. 2003;278:27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita M., et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory R. I., et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 37.Mazess R.B., Nord R., Hanson J.A., Barden H.S. Bilateral measurement of femoral bone mineral density. J. Clin. Densitom. 2000;3:133–140. doi: 10.1385/JCD:3:2:133. [DOI] [PubMed] [Google Scholar]
- 38.Liao E.Y., et al. Establishment and evaluation of bone mineral density reference databases appropriate for diagnosis and evaluation of osteoporosis in Chinese women. J. Bone Miner. Metab. 2003;21:184–192. doi: 10.1007/s007740300029. [DOI] [PubMed] [Google Scholar]
- 39.Wu X.P., et al. Establishment of BMD reference plots and determination of peak BMD at multiple skeletal regions in mainland Chinese women and the diagnosis of osteoporosis. Osteoporos. Int. 2004;15:71–79. doi: 10.1007/s00198-003-1517-x. [DOI] [PubMed] [Google Scholar]
- 40.Hartikka H., et al. Heterozygous mutations in the LDL receptor–related protein 5 (LRP5) gene are associated with primary osteoporosis in children. J. Bone Miner. Res. 2005;20:783–789. doi: 10.1359/JBMR.050101. [DOI] [PubMed] [Google Scholar]
- 41. Tanner, J.M. 1978.Growth at adolescence. Blackwell. Oxford, United Kingdom. 28–39. [Google Scholar]
- 42.Körkkö J., Annunen S., Pihlajamaa T., Prockop D.J., Ala-Kokko L. Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: Comparison with denaturing gradient gel electrophoresis and nucleotide sequencing. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1681–1685. doi: 10.1073/pnas.95.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.